Abstract
Diterpenoid biosynthesis has been extensively studied in plants and fungi, yet cloning and engineering diterpenoid pathways in these organisms remain challenging. Bacteria are emerging as prolific producers of diterpenoid natural products, and bacterial diterpene synthases are poised to make significant contributions to our understanding of terpenoid biosynthesis. Here we will first survey diterpenoid natural products of bacterial origin and briefly review their biosynthesis with emphasis on diterpene synthases (DTSs) that channel geranylgeranyl diphosphate to various diterpenoid scaffolds. We will then highlight differences of DTSs of bacterial and higher organism origins and discuss the challenges in discovering novel bacterial DTSs. We will conclude by discussing new opportunities for DTS mechanistic enzymology and applications of bacterial DTS in biocatalysis and metabolic pathway engineering.
Introduction
Terpenoids comprise the largest, structurally most diverse family of natural products and play important roles in all living organisms. Among the ~60,000 members known to date, ~12,000 are diterpenoids, most of which are produced in plants and fungi. Diterpenoids of bacterial origin are known but rare, however recent advances in genomics have revealed that the biosynthetic potential for terpenoids in bacteria, particularly in the actinimycetes, may be significantly underestimated [1,2].
Diterpenoid biosynthesis has been extensively studied in plants and fungi [3–6], yet cloning the respective genes and characterizing and engineering diterpenoid pathways in these higher organisms remain challenging [7,8]. Scattering of the biosynthetic genes on the genomic DNA of these higher organisms substantially increases the effort to clone all the genes encoding the complete biosynthetic machinery for a given diterpenoid natural product. In contrast, genes encoding secondary metabolite biosynthesis in actinomycetes are nearly always arranged on the bacterial chromosome as a cluster. Recent characterization of terpene synthases (TSs) from several actinomycete species demonstrated that these enzymes are not membrane-bound and can be overproduced with relative ease as soluble, functional recombinant proteins in heterologous hosts such as E. coli [9]. Diterpenoid biosynthesis in bacteria therefore may provide new opportunities to characterize these enzymes and to engineer their biosynthetic machinery for diterpenoid natural product structural diversity.
Diterpenoids are all derived from (E,E,E)-geranylgeranyl diphosphate (GGDP). Diterpene synthases (DTSs), also known as diterpene cyclases, catalyze the critical step in diterpenoid biosynthesis by morphing GGDP into one of the many diterpenoid scaffolds, further transformations of which by the downstream enzymes afford the enormous structural diversity known for diterpenoid natural products. TSs in general, DTSs included, can display incredible fidelity, catalyzing multi-step cyclization reactions with exquisite regiochemical and stereochemical control [10] or display marked product promiscuity, with a single enzyme generating over fifty unique products from a single substrate [11]. It is the fidelity and promiscuity in this chemistry that has inspired a great interest in exploiting TSs for engineered biosynthesis of novel terpenoid natural products [7,8,12].
Here we will first survey diterpenoid natural products of bacterial origin and briefly review their biosynthesis with emphasis on DTSs that channel GGDP to various diterpenoid scaffolds. We will then highlight differences of DTSs of bacterial and higher organism origins and discuss the challenges in discovering novel bacterial DTSs. We will conclude by discussing new opportunities for DTS mechanistic enzymology and applications of bacterial DTS in biocatalysis and metabolic pathway engineering.
Bacterial diterpenoids
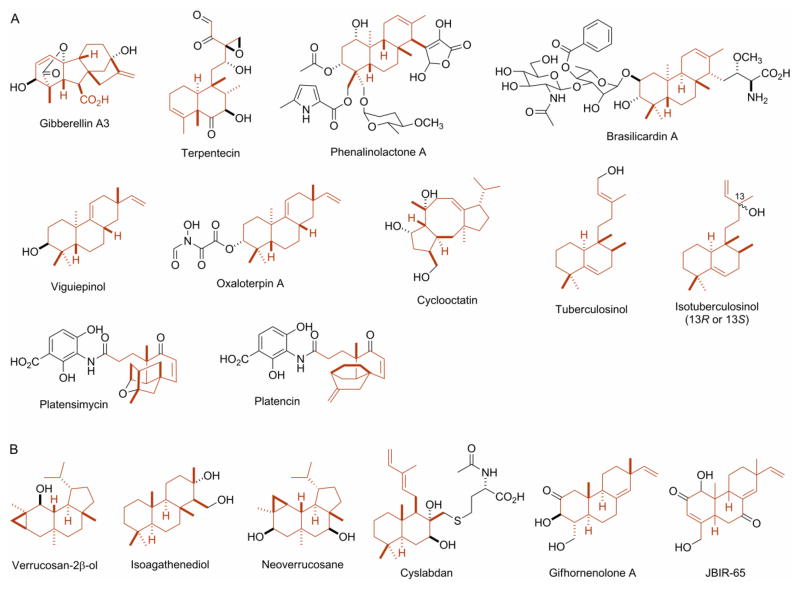
The discovery of gibberellins (GAs) from Rhizobium phaseoli in 1988, originally only known to fungi and plants, may represent the first report of bacterial diterpenoids [13,14]. It was followed by the discovery of verrucosan-2β-ol from Chloroflexus aurantiacus in 1993 [15] and isoagathenediol from Rhodospirillum rubrum in 1995 [16]. Since then, the list of bacterial diterpenoids has grown steadily, and Figure 1 summarizes the bacterial diterpenoids known to date. These include terpentecin from Streptomyces griseolosporeus MF730-N6 [17,18,19], the phenalinolactones from Streptomyces sp. Tu6071 [20], the brasilicardins from Nocardia brasiliensis IFM 0406 [21], viguiepinol and the oxaloterpins from Streptomyces sp. KO-3988 [22,23], cyclooctatin from Streptomyces melanosporofaciens MI614-43F2 [24], tuberculosinol and the isotuberculosinols from Mycobacterium tuberculosis H37Rv [25–29], platensimycin from Streptomyces platensis MA7327 [30–32], platencin from Streptomyces platensis MA7339 [33,34,35], the neoverrucosanes from Saprospira grandis [36,37], cyslabdan from Streptomyces sp. K04-0144 [38], the gifhornenolones from Verrucosispora gifhornesis YM28-088 [39], and JBIR-65 from Actinomadura sp. SpB081030SC-15 [40]. The actinomycetes have emerged as prolific producers of bacterial diterpenoids [1,2]. Bacterial producers of paclitaxel have also been reported, many of which were actinomycetes, however definitive evidence supporting their paclitaxel production remains elusive [41].
Figure 1.
Bacterial diterpenoid natural products with their diterpenoid carbon scaffolds highlighted in red: (A) the biosynthetic gene clusters for these natural products have been cloned and partially characterized and (B) biosynthesis for these natural products has not been studied.
Bacterial DTSs
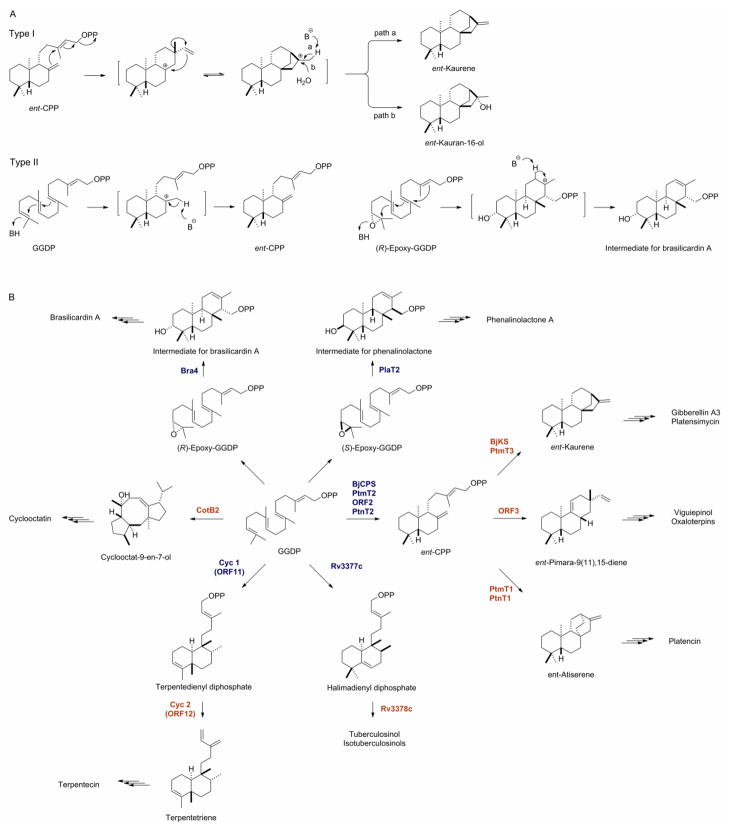
DTS classification follows other TSs. Type I TSs initiate a cyclization reaction via a heterolytic cleavage of the polyprenyl diphosphate, while type II TSs initiate the cyclization reaction via protonation of a double bond or an epoxide ring. In both cases, the resulting carbocation undergoes a cascade of cyclization, the fate of which is determined by a combination of steric and electrostatic forces within the active site cavity. The cyclization cascade is ultimately terminated by abstraction of a proton or electrophilic attack by water (Figure 2A) [4,5]. Because type II TSs leave the diphosphate group intact, their products can serve as substrates for further cyclization by type I TSs. The high frequency with which such two-step cyclizations are employed differentiates diterpenoid biosynthesis from that of smaller terpenoids, which rarely implement a type II mechanism.
Figure 2.
Bacterial diterpene synthases (DTSs): (A) Mechanisms of type I and type II DTSs and (B) pathways for bacterial diterpenoid natural product biosynthesis, highlighting known bacterial type I (blue) and type II DTSs (red) that convert GGDP to diverse diterpenoid scaffolds en route to the final natural products. See Figure 1 legend for structures of the diterpenoid natural products. See Figure 3 legend for accession numbers of the type I and type II DTSs. DTSs, diterpene synthases; ent-CPP, ent-copalyl diphosphate; GGDP, geranylgeranyl diphosphate.
Terpentedienyl diphosphate synthase and terpentetriene synthase were the first two characterized bacterial DTSs, reported in 2001 for terpentecin biosynthesis from S. griseolosporeus MF730-N6 [18,19]. Since then, a total of 16 bacterial DTSs have been identified from various organisms, with genome sequencing efforts unveiling many more candidates whose functions as DTSs require experimental confirmation. Figure 2B summarizes the individual transformations catalyzed by these DTSs en route to their respective diterpenoid natural products, highlighting the remarkable catalytic landscape covered by bacterial DTSs.
Bacterial type II DTSs
Terpentedienyl diphosphate synthase (Cyc1) from S. griseolosporeus MF730-N6, the first bacterial type II DTS reported, converts GGDP to terpentedienyl diphosphate en route to terpentecin (Figure 2B). Although it displays only a moderate sequence similarity (<30%) to the N-terminal halves of characterized eukaryotic DTSs, the presence of a DXDD motif solidified its bioinformatics-based functional assignment [17,18], which was subsequently confirmed experimentally in vitro [17,19]. Other bacterial type II DTSs identified since include halimadienyl diphosphate synthase (Rv3377c) from M. tuberculosis [25,26,29] and ent-copalyl diphosphate (ent-CPP) synthases from S. sp. KO-3988 (ORF2) [22], B. japonicum (BjCPS) [42], S. platensis MA7327 (PtmT2) [43], and S. platensis MA7339 (PtnT2) [43], respectively (Figure 2B), all of which share the characteristic DXDD motif. The type II DTSs, reported for brasilicardin A (Bra4) and phenalinolactone A (Plat2) from N. brasiliensis IFM 0406 [21] and S. sp. Tu6071 [20], respectively, are atypical, with their signature DXDD motif replaced with a (E/D)SA(E/N) motif. Intriguingly, both clusters contain a separate gene homologous to eukaryotic squalene epoxidase, which is thought to convert GGDP to epoxy-GGDP. The latter would support a sterol-like cyclization reaction, where the non-canonical type II DTS Bra4 or PlaT2 would initiate the cyclization reaction by protonating an epoxy group rather than a double bond [20] (Figure 2B).
Bacterial type I DTSs
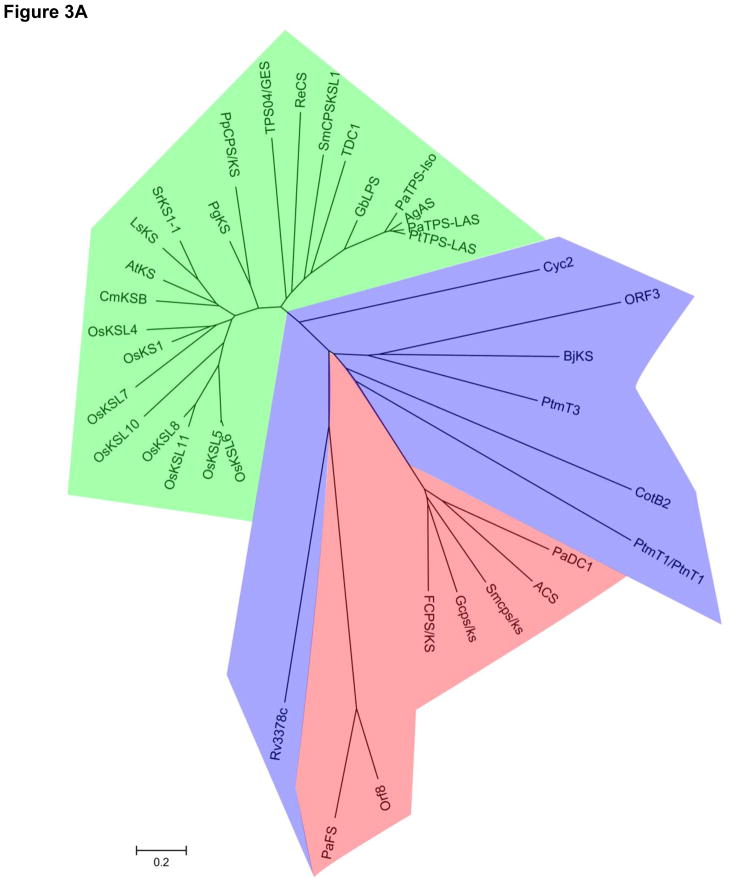
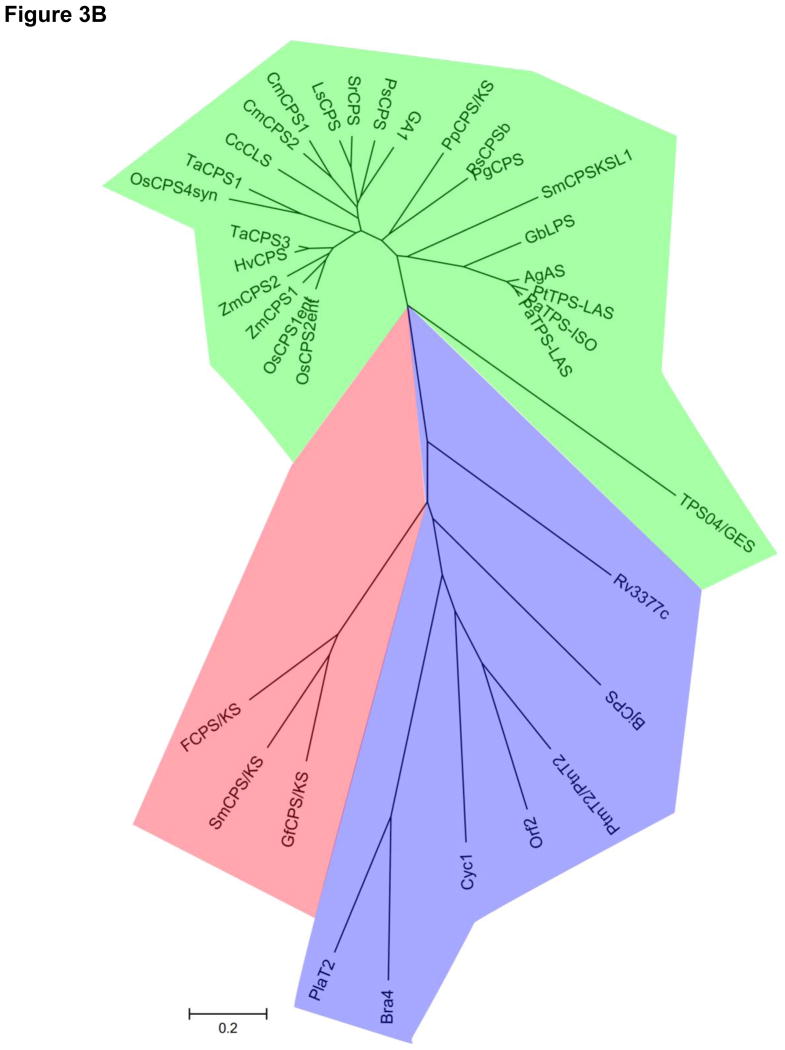
The staggering sequence diversity present in bacterial type I DTSs hinders sequence-gazing efforts but heightens our understanding of the minimal requirements for catalysis. Terpentetriene synthase (Cyc2) from S. griseolosporeus MF730-N6, the first bacterial type I DTS reported, was readily identified on the basis of its sequence homology to known bacterial type I TSs, and presence of the characteristic DDXXD and NSE/DTE motifs [18,19]. The other bacterial type I DTSs characterized since - including the tuberculosinol/isotuberculosinol synthase (Rv3378c) from M. tuberculosis [26,27,29], the cyclooctatenol synthase (CotB2) from S. melanosporofaciens [24], the pimaradiene synthase (ORF3) from S. sp. KO-3988 [22], and two ent-kaurene synthases (BjKS and PtmT3) from B. japonicum [42] and S. platensis MA7327 [43], respectively - however, have an average of just 13% sequence identity. No pairwise alignment displays greater than 20% sequence identity, despite the fact that the latter four enzymes act on a same substrate ent-CPP, and this is in contrast to plant and fungal type I DTSs that display an average of 28% and 25% pairwise identity, respectively. This sequence diversity is evident by the deep branching of bacterial type I DTSs, compared to plant and fungal enzymes, on a minimum-evolution phylogenetic tree (Figure 3A). Moreover, the canonical active site motifs have even been called into question with recent studies of bacterial type I DTSs. For example, the tuberculosinol/isotuberculosinol synthase Rv3378c retains the DDXXD motif but lacks the conserved NSE/DTE motif [26,29]. The two ent-atiserene synthases (PtmT1 and PtnT1) from S. platensis MA7327 and MA7339, respectively, lack both the DDXXD and NSE/DTE motifs, and contain instead two atypical DXXXD motifs [43]. These findings expand our understanding of the structural elements required for catalysis and will guide future mechanistic investigations and DTS discovery efforts.
Figure 3.
Minimum evolution trees of primary amino acid sequences from plant (green), fungal (red), and bacterial (blue) type I (A) and type II DTSs (B). The branch lengths illustrate the extent of sequence diversity found in bacterial DTSs compared with plant or fungal enzymes. The trees were constructed in Mega5.0 with a ClustalW-generated primary sequence alignment. Shown in parentheses are accession numbers. Bacterial type I DTSs: Cyc2 (BAB39207), ORF3 (BAD86798), BjKS (BAC47415), PtmT3 (ACO31279), CotB2 (BAI44338), PtmT1 (ACO31274), PtnT1 (ADD83014), Rv3378 (P_217895). Fungal type I DTSs: PaDC1 (BAG30961), ACS (BAB62102), Smcps/ks (CAP07655), Gfcps/ks (Q9UVY5), FCPS/KS (BAA22426), Orf8 (bsc8) (BAI44849), PaFS (BAF45925). Plant type I DTSs: OsKSL6 (ABH10733), OsKSL5 (ABH10732), OsKSL11 (AAZ76733), OsKSL8 (BAD34478), OsKSL10 (ABH10735), OsKSL7 (ABH10734), OsKS1 (AAQ72559), OsKSL4 (AAU05906), CmKSB (AAB39482), AtKS (AAC39443), LsKS (BAB12441), SrKS1-1 (AF097310_1), PgKS (ADB55708), PpCPS/KS (BAF61135), TPS04/GES (NP_564772), RcCS (XP_002513340), SmCPSKSL1 (AEK75338), TDC1 (AAC49310), GbLPS (AAL09965), PaTPS-Iso (AAS47690), AgAS (Q38710), PaTPS-LAS (AAS47691), PtTPS (AAX07435). Bacterial type II DTSs: Rv3377c (NP_217894), BjCPS (BAC47414), PtmT2 (ACO31276), PtnT2 (ADD83015), Orf2 (BAD86797), Cyc1 (BAB39206), Bra4 (BAG16278), PlaT2 (ABB69743). Fungal type II DTSs: FCPS/KS (BAA22426), SmCPS/KS (CAP07655), GfCPS/KS (Q9UVY5). Plant type II DTSs: OsCPS1ent (BAD42449), OsCPS2ent (Q6ET36), ZmCPS1 (AAT49065), ZmCPS2 (ADB55709), HvCPS (BAH56560), TaCPS3 (AAT70083), OsCPS4syn (NP_0010521), TaCPS1 (BAH56558), CcCLS (ADJ93862), CmCPS1 (AAD04292), CmCPS2 (AAD04923), LsCPS (BAB12440), SrCPS (AAB87091), PsCPS (AAB58822), GA1 (AAA53632), PpCPS/KS (BAF61135), PsCPSb (ADB55709), PgCPS (ADB55707), GbLPS (AAL09965), AgAS (Q38710), PtTPS-LAS (AAX07435), PaTPS-ISO (AAS47690), PaTPS-LAS (AAS47691), SmCPSKSL1 (AEK75338), TPS04/GES (NP_564772). DTSs, diterpene synthases.
Evolutionary relationship of bacterial and eukaryotic DTSs
Structures for a number of type I and type II TSs of bacterial, fungal, or plant origin are available [4,5], but no structure of a bacterial DTS is currently known. Nonetheless, structures of various known TSs coupled with detailed bioinformatic analyses of the bacterial variants now provide a clearer picture of DTS evolution. Bacterial type II DTSs are homologous to bacterial triterpene synthases for which the three-dimensional structure is known, including squalene hopene cyclase [44], and primary sequence alignments suggest a conservation of overall topology and active site location [45]. Thus, bacterial type II DTSs are hypothesized to contain the same βγ-didomain structure deriving from an ancient duplication of two (α/α)6 barrels. On the contrary, bacterial type I DTSs are predicted to have a single α-domain “isoprenoid fold” based on their homology and overall primary sequence alignments to the bacterial type I sesquiterpene synthases, pentalenene synthase [9,46] and epi-isozizaene synthase [47]. Plant DTSs likely evolved from an early fusion of the bacterial type I and II enzymes to form bifunctional αβγ-tridomain DTSs that can catalyze both types of cyclization but in separate active sites. Such tridomain DTSs are still seen today, for example, in the bifunctional ent-kaurene synthase from the moss Physcomitrella patens [48] and the bifunctional abietadiene synthase from Abies grandis [49]. Monofunctional type I or type II DTSs found in plants commonly retain vestigial domains from ancestral enzymes that, while lacking active site motifs, likely remain for structural support [50–52]. The relevance of these evolutionary roots to current studies in DTS biochemistry and enzymology is many-fold. For example, bacterial DTSs, in particular type I DTSs, tend to be significantly smaller than their eukaryotic counterparts, perhaps to a degree that makes the bacterial enzymes inherently easier to manipulate for biochemical studies. The ancient roots of DTSs in prokaryotes also explain the greater sequence diversity observed in these enzymes (Figure 3). Further examination and utilization of bacterial DTSs in future studies could facilitate interrogating how sequence divergence in core catalytic motifs affects enzyme mechanism and product diversity in TSs.
Discovery of new bacterial DTSs
Challenges in discovering bacterial DTSs
The lack of sequence conservation in bacterial DTSs makes sequence-based approaches for their discovery difficult, albeit still possible [53]. Many of the bacterial DTSs characterized to date were identified by their clustering with more readily identifiable terpene biosynthetic genes. For example, screening genomic DNA of S. griseolosporeus MF730-N6 for mevalonate pathway genes led to the identification of terpentedienyl diphosphate synthase (Cyc1) and terpentetriene synthase (Cyc2) for terpentecin biosynthesis [18]. A similar strategy yielded the pimaradiene synthase (ORF3) for viguiepinol biosynthesis in S. sp. KO-3988 [54]. Other bacterial diterpene gene clusters have been identified only by screening for genes required for the biosynthesis of separate chemical moieties [20,43] or those involved in tailoring reactions [55]. The requirement for the common substrate GGDP can be exploited to identify bacterial DTSs. Previous studies have attempted to determine the chain-length determining factors that distinguish GGDP synthase from shorter- and longer-chain polyprenyl diphosphate synthases [56–59] and several key amino acid residues have been identified and verified in vitro. From these data, predictive algorithms could be designed to selectively scan genomes for GGDP synthases, and such a strategy was used recently to clone the brasilicardin A gene cluster from N. brasiliensis [60].
Atypical DTSs in the platensimycin and platencin biosynthetic gene clusters
Platensimycin (PTM) and platencin (PTN) are composed of a substituted aminobenzoic acid and a diterpenoid-derived carboxylic acid, linked by an amide bond (Figure 1) [30–35]. The terpenoid moieties of PTM and PTN bear ent-kaurene and ent-atiserene scaffolds, respectively [61,62]. While ent-kaurene production has been studied in great detail in plants [63], fungi [64], and, more recently, bacteria [42], little is known about ent-atiserene production. Following the cloning of a PTM-PTN dual producing gene cluster from S. platensis MA7327, putative ent-CPP (Ptm2) and ent-kaurene synthases (Ptm3) were identified by sequence comparison with known DTSs. Comparison of the PTM-PTN gene cluster to a PTN-only producing gene cluster from S. platensis MA7339 ruled out the possibility that a promiscuous ent-kaurene synthase could provide both scaffolds. This led to the search and eventual discovery of PtmT1 and PtnT1 as the ent-atiserene synthases, novel type I DTSs that lack canonical active site motifs, for PTN biosynthesis [43]. A preliminary analysis of sequence databases has revealed other misannotated homologues, suggesting that the ent-atiserene synthase belongs to a previously unrecognized family of bacterial type I DTSs [43].
New opportunities for mechanistic enzymology and engineered biosynthesis
Mechanistic enzymology
Much work has been done in higher plants and fungi to attempt to fully understand the catalytic landscape of DTSs. Structural data point to the importance of precisely positioned amino acid side chains in the active site cavity that guide the folding of intermediates and stabilize carbocation intermediates. Mechanistic enzymology and structural biology of known bacterial DTSs, and of those yet to be discovered, will complement previous studies by providing a wealth of new and diverse sequences to populate the catalytic landscape (Figures 2 and 3). These studies will hopefully answer important questions such as how ent-kaurene synthases from B. japonicum and S. platensis can produce the same diterpene scaffold with such divergent primary sequences. Additionally, continued discovery and full characterization of non-canonical bacterial type I and type II DTSs will surely broaden our understanding of the mechanistic requirements for DTS reaction initiation.
Exploiting bacterial DTSs to generate structural diversity
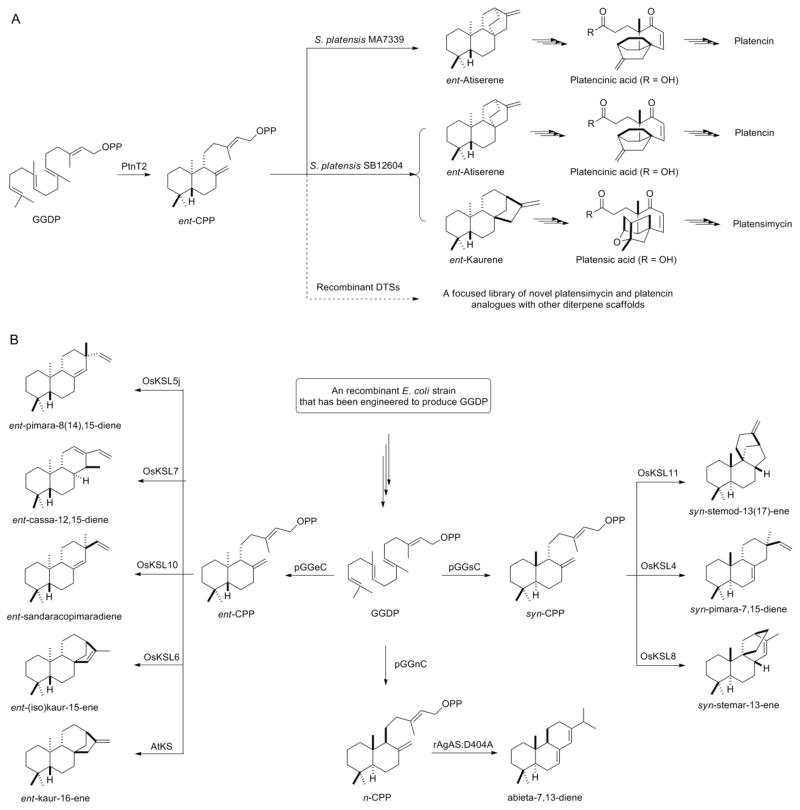
Since all diterpenoids derive from a common origin, GGDP, a degree of modularity exists among DTSs from different biosynthetic pathways. There is little evidence for protein-protein interactions between enzymes in a terpenoid biosynthetic pathway. This enables various type I and type II DTS to be mixed and matched to produce a variety of carbon scaffolds. For example, the PTN-producing strain, S. platensis MA7339, can be converted into a PTM-PTN dual producer by heterologously expressing the ent-kaurene synthase and other genes involved in PTM ether-ring formation, and this suggests that alternative ent-copalyl-derived diterpene scaffolds could be accepted into the PTM and PTN pathways (Figure 4A) [43]. The extent to which diverse diterpene scaffolds can replace native scaffolds in extant biosynthetic pathways depends on the ability of the downstream tailoring enzymes to accept the new scaffold. Structural similarities among different diterpene scaffolds may therefore dictate compatibility, as exemplified in the PTM and PTN biosynthesis. In order to fully explore the potential of diterpenoid biosynthetic pathways, we must increase the number of characterized DTSs to begin to approximate the number of diterpenoid scaffolds found in Nature.
Figure 4.
(A) Modularity of DTS biochemistry demonstrated by conversion of the platencin- producing strain, S. platensis MA7339, into a platensimycin and platencin dual-producing strain, S. platensis SB12604, by genetic engineering [43], and a proposal of producing new analogues by heterologous expression of additional type I DTSs in S. platensis MA7339. (B) Utility of DTS modularity demonstrated through expression of various type I and type II DTSs in a recombinant E. coli strain that has been engineered to produce GGDP to yield eight different diterpene scaffolds, which provide an entry point to engineered production of thousands of diterpenoid natural products. DTSs, diterpene synthases; ent-CPP, ent-copalyl diphosphate; n-CPP, normal-copalyl diphosphate; syn-CPP, syn-copalyl diphosphate; GGDP, geranylgeranyl diphosphate; pGGeC, pGGnC, and pGGsC, three engineered type II DTSs that convert GGDP to ent-CPP, n-CPP, and syn-CPP, respectively; AtKS, rAgAS:D404A, OsKSL4, OsKSL5j, OsKSL6, OsKSL7, OsKSL8, OsKSL10, and OsKSL11, 11 engineered type II DTSs that convert ent-CPP, n-CPP, or syn-CPP to the nine diterpenoid scaffolds, respectively [66].
Engineered production of diterpenoid natural products in bacteria
Bacteria provide convenient hosts for engineered production of terpenoid natural products with important commercial value. This strategy has received considerable attention [7,8] and led to recent success when principles from the emerging field of synthetic biology were applied to produce precursors of the sesquiterpene antimalarial drug, artemisinin, in a bacterial host [65]. All indications suggest that diterpenoid pathways are equally amenable to such metabolic engineering efforts. This was recently demonstrated through construction of a modular DTS expression system in E. coli. Three different type II DTSs, affording ent-, syn-, and normal-CPPs, were first introduced into a recombinant E. coli strain that was engineered to produce GGDP. Several product-specific type I DTS were then added to afford recombinant strains that produce nine diterpenoids representing eight distinct scaffolds (Figure 4B) [66]. These diterpenoid scaffolds, alone, provide an entry point to the engineered production of over 2,000 known diterpenoid natural products.
Conclusions and prospective
Bacteria are emerging as prolific producers of diterpenoids, and bacterial DTSs are poised to make significant contributions to our current understanding of terpenoid biosynthesis. Advantages of studying diterpenoid biosynthesis in bacteria include (i) access to mechanistic and structural studies as facilitated by the technical feasibility of working with bacterial enzymes, (ii) expansion of mechanistic understanding through the characterization of novel enzymes with non-canonical catalytic motifs, and (iii) opportunities for whole pathway engineering to produce complex diterpenoid natural products. Recent findings have already challenged the paradigm of TS biochemistry and mechanistic enzymology from studies in higher organisms and promise to expand the boundaries of DTS catalytic landscape. Each new DTS characterized will either extend these boundaries further or fill in the gaps between existing sequences. The number of possible natural or unnatural diterpenoid scaffolds, accessible from the common substrate GGDP by DTSs, is staggering, and gaining access to these structures by engineered biosynthesis will greatly aid both drug discovery efforts and development of biotechnology applications.
Highlights.
Cloning and engineering diterpenoid pathways in plants and fungi remain challenging.
Bacteria are emerging as prolific producers of diterpenoid natural products.
Bacterial diterpene synthases are poised to make significant contributions to our understanding of terpenoid biosynthesis.
Diterpenoid biosynthesis in bacteria provides new opportunities for pathway engineering to produce complex diterpenoid natural products.
Acknowledgments
Research on discovery, biosynthesis, and metabolic pathway engineering of terpenoid natural products in the Shen lab is supported in part by NIH grants AI079070 and GM086184. M.J.S was supported in part by NIH Predoctoral Training grant GM08505.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
• of special interests
•• of outstanding interests
- 1.Dairi T. Studies on biosynthetic genes and enzymes of isoprenoids produced by actinomycetes. J Antibiot. 2005;58:227–243. doi: 10.1038/ja.2005.27. [DOI] [PubMed] [Google Scholar]
- 2.Daum M, Herrmann S, Wilkinson B, Bechthold A. Genes and enzymes involved in bacterial isoprenoid biosynthesis. Curr Opinion Chem Biol. 2009;13:180–188. doi: 10.1016/j.cbpa.2009.02.029. [DOI] [PubMed] [Google Scholar]
- 3.Tudznski B. Gibberellin biosynthesis in fungi: genes, enzymes, evolution, and impact on biotechnology. Appl Microbiol Biotechnol. 2005;66:597–611. doi: 10.1007/s00253-004-1805-1. [DOI] [PubMed] [Google Scholar]
- 4.Christianson DW. Structural biology and chemistry of the terpenoid cyclases. Chem Rev. 2006;106:3412–3442. doi: 10.1021/cr050286w. [DOI] [PubMed] [Google Scholar]
- 5.Christianson DW. Unearthing the roots of the terpenome. Curr Opinion Chem Biol. 2008;12:141–150. doi: 10.1016/j.cbpa.2007.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peters RJ. Two rings in them all: the labdane-related diterpenoids. Nat Prod Rep. 2010;27:1521–1530. doi: 10.1039/c0np00019a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ajikumar PK, Tyo K, Carlsen S, Mucha O, Phon TH, Stephanopoulos G. Terpenoids: opportunities for biosynthesis of natural product drugs using engineered microorganisms. Mol Pharmaceutics. 2008;5:167–190. doi: 10.1021/mp700151b. [DOI] [PubMed] [Google Scholar]
- 8.Kirby J, Keasling JD. Biosynthesis of plant isoprenoids: perspectives for microbial engineering. Annu Rev Plant Biol. 2009;60:335–355. doi: 10.1146/annurev.arplant.043008.091955. [DOI] [PubMed] [Google Scholar]
- 9.Lesburg CA, Zhai G, Cane DE, Christianson DW. Crystal structure of pentalenene synthase: mechanistic insights on terpenoid cyclization reactions in biology. Science. 1997;277:1820–1824. doi: 10.1126/science.277.5333.1820. [DOI] [PubMed] [Google Scholar]
- 10.Felicetti B, Cane DE. Aristolochene synthase: mechanistic analysis of active site residues by site-directed mutagenesis. J Am Chem Soc. 2004;126:7212–7221. doi: 10.1021/ja0499593. [DOI] [PubMed] [Google Scholar]
- 11•.Steele CL, Crock J, Bohlmann J, Croteau R. Sesquiterpene synthases from Grand Fir (Abies grandis): comparison of constitutive and wound-induced activities and cDNA isolation, characterization, and bacterial expression of δ-selinene synthase and γ-humulene synthase. J Biol Chem. 1998;273:2078–2089. doi: 10.1074/jbc.273.4.2078. This paper describes the characterization of two sesquiterpene synthases from Grand Fir trees and provides a stunning example of the chemical diversity attainable by terpene synthases. One enzyme, δ-selinene synthase produces 34 products in vitro, while the other, γ-humulene, yields an incredible 52 unique products. [DOI] [PubMed] [Google Scholar]
- 12••.Yoshikuni Y, Ferrin TE, Keasling JD. Designed divergent evolution of enzyme function. Nature. 2006;440:1078–1082. doi: 10.1038/nature04607. The authors demonstrate the power of applying protein engineering to terpene synthases by rationally engineering a promiscuous γ-humulene synthase. Seven specific terpene synthases were generated, each with unique major products. [DOI] [PubMed] [Google Scholar]
- 13.Atzorn R, Crozier A, Wheeler C, Sandberg G. Production of gibberellins and indole 3-acetic acid by Rhizobium phaseoli in relation to nodulation of Phaseolus vulgaris roots. Planta. 1988;175:532–538. doi: 10.1007/BF00393076. [DOI] [PubMed] [Google Scholar]
- 14.Bottini R, Cassan F, Piccoli P. Gibberellin production by bacteria and its involvement in plant growth promotion and yield increase. Appl Microbiol Biotechnol. 2004;65:497–503. doi: 10.1007/s00253-004-1696-1. [DOI] [PubMed] [Google Scholar]
- 15.Hefter J, Richnow HH, Fischer U, Trendel JM, Michaelis W. (-)-Verrucosan-2β-ol from the phototrophic bacterium Chloroflexus aurantiacus: first report of a verrucosane-type diterpenoid from a prokaryote. J Gen Microbiol. 1993;139:2757–2761. [Google Scholar]
- 16.Chuck J, Barrow KD. The isolation of isoagathenediol: a new tricyclic diterpene from the lipids of Rhodospirillum rubrum. Microbiology. 1995;141:2659–2663. [Google Scholar]
- 17.Hamano Y, Dairi T, Yamamoto M, Kawasaki T, Keneda K, Kuzuyama T, Itoh N, Seto H. Cloning of a gene cluster encoding enzymes responsible for the mevalonate pathway from a terpenoid-antibiotic-producing Streptomyces strain. Biosci Biotechnol Biochem. 2001;65:1627–1635. doi: 10.1271/bbb.65.1627. [DOI] [PubMed] [Google Scholar]
- 18••.Dairi T, Hamano Y, Kuzuyama T, Itoh N, Furihata K, Seto H. Eubacterial diterpene cyclase genes essential for production of the isoprenoid antibiotic terpentecin. J Bacteriol. 2001;183:6085–6094. doi: 10.1128/JB.183.20.6085-6094.2001. An elegant account of the first in vivo and in vitro characterization of bacterial diterpene synthases. Orf11 and Orf12, together, were shown to convert GGDP into terpentetriene en route to the diterpene antibiotic, terpentecin. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hamano Y, Kuzuyama T, Itoh N, Furihata K, Seto H, Dairi T. Functional analysis of eubacterial diterpene cyclase responsible for biosynthesis of a diterpene antibiotic, terpentecin. J Biol Chem. 2002;277:37098–37104. doi: 10.1074/jbc.M206382200. [DOI] [PubMed] [Google Scholar]
- 20.Durr C, Schnell HJ, Luzhetskyy A, Murillo R, Weber M, Welzel K, Vente A, Bechtold A. Biosynthesis of the terpene phenalinolactone in Streptomyces sp. Tu6071: analysis of the gene cluster and generation of derivatives. Chem Biol. 2006;13:365–377. doi: 10.1016/j.chembiol.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 21.Hayashi Y, Matsuura N, Toshima H, Itoh N, Ishikawa J, Mikami Y, Dairi T. Cloning of the gene cluster responsible for the biosynthesis of brasilicardin A, a unique diterpenoid. J Antibiot. 2008;61:164–174. doi: 10.1038/ja.2008.126. [DOI] [PubMed] [Google Scholar]
- 22.Ikeda C, Hayashi Y, Itoh N, Seto H, Dairi T. Functional analysis of eubacterial ent-copalyl diphosphate synthase and pimara-9(11),15-diene synthase with unique primary sequences. J Biochem. 2007;141:37–45. doi: 10.1093/jb/mvm004. [DOI] [PubMed] [Google Scholar]
- 23.Motohashi K, Ueno R, Sue M, Furihata K, Matsumoto T, Dairi T, Omura S, Seto H. Studies on terpenoids produced by actinomycetes: oxaloterpins A, B, C, D, and E, diterpenes from Streptomyces sp. KO-3988. J Nat Prod. 2007;70:1712–1717. doi: 10.1021/np070326m. [DOI] [PubMed] [Google Scholar]
- 24.Kim SY, Zhao P, Igarashi M, Sawa R, Tomita T, Nishiyama M, Kuzuyama T. Cloning and heterologous expression of the cyclooctatin biosynthetic gene cluster afford a diterpene cyclase and two P450 hydroxylases. Chem Bio. 2009;16:736–743. doi: 10.1016/j.chembiol.2009.06.007. [DOI] [PubMed] [Google Scholar]
- 25.Nakano C, Hoshino T. Characterization of the Rv3377c gene product, a type-B diterpene cyclase, from the Mycobacterium tuberculosis H37 genome. ChemBioChem. 2009;10:2060–2071. doi: 10.1002/cbic.200900248. [DOI] [PubMed] [Google Scholar]
- 26•.Mann FM, Xu M, Chen X, Fulton DB, Russell DG, Peters RJ. Edaxadiene: a new bioactive diterpene from Mycobacterium tuberculosis. J Am Chem Soc. 2009;131:17526–17527. doi: 10.1021/ja9019287. Although the structure of edaxadiene was later revised to that of isotuberculosinol, the biological activity of the diterpene produced by Mycobacterium tuberculosis reflects a key role during pathogenesis. That diterpene natural products function in nature as virulence factors adds to the importance of a detailed mechanistic understanding of these diterpene synthases. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maugel N, Mann FM, Hillwig ML, Peters RJ, Snider BB. Synthesis of (±)-Nosyberkol (Isotuberculosinol, Revised Structure of Edaxadiene) and (±)-Tuberculosinol. Org Lett. 2010;12:2626–2629. doi: 10.1021/ol100832h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prch L, Kirby J, Keasking JD, Alber T. Diterpene production in Mycobacterium tuberculosis. FEBS J. 2010;277:3588–3595. doi: 10.1111/j.1742-4658.2010.07767.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakano C, Ootsuka T, Takayama K, Mitsui T, Sato T, Hoshino T. Characterization of the Rv3378c gene product, a new diterpene synthase for producing tuberculosinol and (13R, S)-isotuberculosinol (nosyberkol) from the Mycobacterium tuberculosis H37Rv genome. Biosci Biotechnol Biochem. 2011;75:75–81. doi: 10.1271/bbb.100570. [DOI] [PubMed] [Google Scholar]
- 30••.Wang J, Soisson SM, Young K, Shoop W, Kodali S, Galgoci A, Painter R, Parthasarathy G, Tang YS, Cummings R, Ha S, Dorso K, Motyl M, Jayasuriya H, Ondeyka J, Herath K, Zhang C, Hernanez L, Allocco J, Basilio A, Tormo JR, Genilloud O, Vicente F, Pelaez F, Colwell L, Lee SH, Michael B, Felcetto T, Gill C, Silver LL, Hermes JD, Bartizal K, Barrett J, Schmatz D, Becker JW, Cully D, Singh SB. Platensimycin is a selective FabF inhibitor with potent antibiotic properties. Nature. 2006;441:358–361. doi: 10.1038/nature04784. This discovery of a novel class of antibiotic reinforces that natural products remain an important source of new pharmacophores for drug discovery; however, exploiting this resource will require creative new approaches such as the antisense RNA technique described here. The diterpene-derived platensimycin is currently a lead candidate for the development of both antibacterial and antidiabetic agents. [DOI] [PubMed] [Google Scholar]
- 31.Singh SB, Jayasuriya H, Ondeyka JG, Herath KB, Zhang C, Zink DL, Tsou NN, Ball RG, Basilio A, Genilloud O, Diez MT, Vicente F, Pelaez F, Young K, Wang J. Isolation, structure, and absolute stereochemistry of platensimycin, a broad spectrum antibiotic discovered using an antisense differential sensitivity strategy. J Am Chem Soc. 2006;128:11916–11920. doi: 10.1021/ja062232p. [DOI] [PubMed] [Google Scholar]
- 32.Smanski MJ, Peterson RM, Rajski SR, Shen B. Engineered Streptomyces platensis strains that overproduce antibiotics platensimycin and platencin. Antimicrob Agents Chermother. 2009;53:1299–1304. doi: 10.1128/AAC.01358-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang J, Kodali S, Lee SH, Galgoci A, Painter R, Dorso K, Racine F, Motyl M, Hernandez L, Tinney E, Colletti SL, Herath K, Cummings R, Salazar O, Gonzalez I, Basilio A, Vicente F, Genilloud O, Pelaez F, Jayasuriya H, Young K, Cully DF, Singh SB. Discovery of platencin, a dual FabF and FabH inhibitor with in vivo antibiotic properties. Proc Natl Acad Sci USA. 2007;104:7612–7616. doi: 10.1073/pnas.0700746104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jayasuriya H, Herath KB, Zhang C, Zink DL, Basilio A, Genilloud O, Diez MT, Vicente F, Gonzalez I, Salazar O, Pelaez F, Cummings R, Ha S, Wang J, Singh SB. Isolation and structure of platencin: a FabH and FabF dual inhibitor with potent broad-spectrum antibiotic activity. Angew Chem Int Ed. 2007;46:4684–4688. doi: 10.1002/anie.200701058. [DOI] [PubMed] [Google Scholar]
- 35.Yu Z, Smanski MJ, Peterson RM, Marchillo K, Andes D, Rajski SR, Shen B. Engineering of Streptomyces platensis MA7339 for overproduction of platencin and congeners. Org Lett. 2010;12:1744–1747. doi: 10.1021/ol100342m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spyere A, Rowley DC, Jensen PR, Fenical W. New neoverrucosane diterpenoids produced by the marine gliding bacterium Saprospira grandis. J Nat Prod. 2003;66:818–822. doi: 10.1021/np0205351. [DOI] [PubMed] [Google Scholar]
- 37.Mincer TJ, Spyere A, Jensen PR, Fenical W. Phylogentic analysis and diterpenoid production by marine bacteria of the genus Saprospira. Curr Microbiol. 2004;49:300–307. doi: 10.1007/s00284-004-4358-8. [DOI] [PubMed] [Google Scholar]
- 38.Fukumoto A, Kim Y, Hanaki H, Shiomi K, Tomoda H, Omura S. Cyslabdan, a new potentiator of imipenem activity against methicillin-resistant Staphylococcus aureus, produced by Streptomyces sp K04-0144. J Antibiot. 2008;61:7–10. doi: 10.1038/ja.2008.102. [DOI] [PubMed] [Google Scholar]
- 39.Shirai M, Okuda M, Motohashi K, Imoto M, Furihata K, Matsuo Y, Katsuta A, Shizuri Y, Seto H. Terpenoids produced by actinomycetes: isolation, structrural elucidation and biosynthesis of new diterpenes, gifhornenolones A and B from Verrucosispora gifhornesis YM28-088. J Antibiot. 2010;63:245–250. doi: 10.1038/ja.2010.30. [DOI] [PubMed] [Google Scholar]
- 40.Takagi M, Motohashi K, Khan ST, Hashimoto J, Shin-ya K. JBIR-65, a new diterpene, isolated from a sponge-derived Actinomadura sp SpB081030SC-15. J Antibiot. 2010;63:401–403. doi: 10.1038/ja.2010.61. [DOI] [PubMed] [Google Scholar]
- 41.Flores-Bustamante ZR, Rivera-Orduna FN, Martinez-Cardenas A, Flores-Cotera LB. Microbial paclitaxel: advances and perspectives. J Antibiot. 2010;63:460–467. doi: 10.1038/ja.2010.83. [DOI] [PubMed] [Google Scholar]
- 42.Morrone D, Chambers J, Lowry L, Kim G, Anterola A, Bender K, Peters RJ. Gibberellin biosynthesis in bacteria: separate ent-copalyl diphosphate and ent-kaurene synthases in Bradyrhizobium japonicum. FEBS Lett. 2009;583:475–480. doi: 10.1016/j.febslet.2008.12.052. [DOI] [PubMed] [Google Scholar]
- 43••.Smanski MJ, Yu Z, Casper J, Lin S, Peterson RM, Chen Y, Wendt-Pienkowski E, Rajski SR, Shen B. Dedicated ent-kaurene and ent-atiserene synthases for platensimycin and platencin biosynthesis. Proc Natl Acad Sci USA. 2011;108:13498–13503. doi: 10.1073/pnas.1106919108. The natural product gene clusters encoding platensimycin and platencin biosynthesis in various strains of S. platensis showcase Nature s ability to exploit the modularity of terpene synthase biochemistry for structural diversity. The authors discover ent-atiserene synthase as a new family of diterpene synthases and demonstrate the conversion of a platencin producer to a platensimycin and platencin dual-producer through genetic engineering. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wendt KU, Poralla K, Schulz GE. Structure and function of a squalene cyclase. Science. 1997;277:1811–1815. doi: 10.1126/science.277.5333.1811. [DOI] [PubMed] [Google Scholar]
- 45••.Cao R, Zhang Y, Mann FM, Huang C, Mukkamala D, Hudock MP, Mead ME, Prisic S, Wang, Lin FY, Chang TK, Peters RJ, Oldfield E. Diterpene cyclases and the nature of the isoprenoid fold. Proteins. 2010;78:2417–2432. doi: 10.1002/prot.22751. The authors draw from many lines of evidence, including bioinformatics, site-directed mutagenesis, and domain-swapping to make structural predictions for diterpene synthases and describe likely routes by which they may have evolved. Many of these predictions were later validated when the structure of taxadiene synthase was solved. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seemann M, Zhai G, de Kraker JW, Paschall CM, Christianson DW, Cane DE. Pentalenene synthase: analysis of active site residues by site-directed mutagenesis. J Am Chem Soc. 2002;124:7681–7689. doi: 10.1021/ja026058q. [DOI] [PubMed] [Google Scholar]
- 47.Aaron JA, Lin X, Cane DE, Christianson DW. Structure of epi-isozizaene synthase form Streptomyces coelicolor A3(2), a platform for new terpenoid cyclization templates. Biochemistry. 2010;49:1787–1797. doi: 10.1021/bi902088z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hayashi KI, Kawaide H, Notomi M, Sakigi Y, Matsuo A, Nozaki H. Identification and functional analysis of bifunctional ent-kaurene synthase from the moss Physcomitrella patens. FEBS Lett. 2006;580:6175–6181. doi: 10.1016/j.febslet.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 49.Vogel BS, Wildung MR, Vogel G, Croteau R. Abietadiene synthase from grand fir (Abies grandis): cDNA isolation, characterization, and bacterial expression of a bifunctional diterpene cyclase involved in resin acid biosynthesis. J Biol Chem. 1996;271:23262–23268. doi: 10.1074/jbc.271.38.23262. [DOI] [PubMed] [Google Scholar]
- 50.Peters RJ, Carter OA, Zhang Y, Matthews BW, Croteau RB. Bifunctional abietadiene synthase: mutual structural dependence of the active sites for protonation-initiated and ionization-initiated cyclizations. Biochemistry. 2003;42:2700–2707. doi: 10.1021/bi020492n. [DOI] [PubMed] [Google Scholar]
- 51••.Koksal M, Jin Y, Coates RM, Croteau R, Christianson DW. Taxadiene synthase structure and evolution of modular architecture in terpene biosynthesis. Nature. 2011;469:116–120. doi: 10.1038/nature09628. The first example of a three-dimensional structure of a diterpene synthase is described. The taxadiene synthase structure provides clear connection between type I and type II terpene synthase enzymology in the evolution of DTSs in plants. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52•.Koksal M, Hu H, Coates RM, Peters RJ, Christianson DW. Structure and mechanism of the diterpene cyclase ent-copalyl diphosphate synthase. Nat Chem Biol. 2011;7:431–433. doi: 10.1038/nchembio.578. Comparing the structure of an ent-CPP synthase from Arabidopsis thaliana to the taxadiene synthase illustrates how the tridomain plant DTS can utilize distinct active sites to catalyze either the type I or type II initiation chemistry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nakano C, Okamura T, Sato T, Dairi T, Hoshino T. Mycobacterium tuberculosis H37Rv3377c encodes the diterpene cyclase for producing the halimane skeleton. Chem Comm. 2005:1016–1018. doi: 10.1039/b415346d. [DOI] [PubMed] [Google Scholar]
- 54.Kawasaki T, Hayashi Y, Kuzuyama T, Furihata K, Itoh N, Seto H, Dairi T. Biosynthesis of a natural polyketide-isoprenoid hybrid compound, furaquinocin A: identification and heterologous expression of the gene cluster. J Bacteriol. 2006;188:1236–1244. doi: 10.1128/JB.188.4.1236-1244.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tully RE, van Berkum P, Lovins KW, Keister DL. Identification and sequencing of a cytochrome P450 gene cluster from Bradyrhizobium japonicum. Biochim Biophys Acta. 1998;1398:243–255. doi: 10.1016/s0167-4781(98)00069-4. [DOI] [PubMed] [Google Scholar]
- 56•.Tarshis LC, Proteau PJ, Kellogg BA, Sacchettini JC, Poulter CD. Regulation of product chain length by isoprenyl diphosphate synthases. Proc Natl Acad Sci USA. 1996;93:15018–15023. doi: 10.1073/pnas.93.26.15018. A thorough application of site-directed mutagenesis, X-ray structural analysis of farnesyl diphosphate synthase, and bioinformatic analyses allowed the authors to develop a model describing how chain length is controlled in polyprenyltransferases. This work has led to more accurate predictions of enzyme function based on primary sequence information. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ohnuma SI, Hemmi H, Ohto C, Nakane H, Nishino T. Effects of random mutagenesis in a putative substrate-binding domain of geranylgeranyl diphosphate synthase upon intermediate formation and substrate specificity. J Biochem. 1997;121:696–704. doi: 10.1093/oxfordjournals.jbchem.a021642. [DOI] [PubMed] [Google Scholar]
- 58.Hemmi H, Noike M, Nakayama T, Nishino T. An alternative mechanism of product chain-length determination in type III geranylgeranyl diphosphate synthase. Eur J Biochem. 2003;270:2186–2194. doi: 10.1046/j.1432-1033.2003.03583.x. [DOI] [PubMed] [Google Scholar]
- 59.Ogura K, Koyama T. Enzymatic aspects of isoprenoid chain elongation. Chem Rev. 1998;98:1263–1276. doi: 10.1021/cr9600464. [DOI] [PubMed] [Google Scholar]
- 60.Toyomasu T, Niida R, Kenmoku H, Kanno Y, Miura S, Nakano C, Shiono Y, Mitsuhashi W, Toshima H, Oikawa H, Hoshino T, Dairi T, Kato N, Sassa T. Identification of diterpene biosynthetic gene clusters and functional analysis of labdane-related diterpene cyclases in Phomopsis amygdali. Biosci Biotech Biochem. 2008;72:1038–1047. doi: 10.1271/bbb.70790. [DOI] [PubMed] [Google Scholar]
- 61.Herath KB, Attygalle AB, Singh SB. Biosynthetic studies of platensimycin. J Am Chem Soc. 2007;129:15422–15423. doi: 10.1021/ja0758943. [DOI] [PubMed] [Google Scholar]
- 62.Herath K, Attygalle AB, Singh SB. Biosynthetic studies of platencin. Tetrahedron Lett. 2008;49:5755–5758. [Google Scholar]
- 63.Yamaguchi S, Saito T, Abe H, Yamane H, Murofushi N, Kamiya Y. Molecular cloning and characterization of a cDNA encoding the gibberellin biosynthetic enzyme ent-kaurene synthase B from pumpkin (Cucurbita maxima L) Plant J. 1996;10:203–213. doi: 10.1046/j.1365-313x.1996.10020203.x. [DOI] [PubMed] [Google Scholar]
- 64.Kawaide H, Imai R, Sassa T, Kamiya Y. ent-Kaurene synthase from the fungus Phaeosphaeria sp. L487: cDNA isolation, characterization, and bacterial expression of a bifunctional diterpene cyclase in fungal gibberellin biosynthesis. J Biol Chem. 1997;272:21706–21712. doi: 10.1074/jbc.272.35.21706. [DOI] [PubMed] [Google Scholar]
- 65.Keasling JD. Synthetic biology for synthetic chemistry. ACS Chem Biol. 2008;3:64–76. doi: 10.1021/cb7002434. [DOI] [PubMed] [Google Scholar]
- 66••.Cyr A, Wilderman PR, Determan M, Peters RJ. A modular approach for facile biosynthesis of labdane-related diterpenes. J Am Chem Soc. 2007;129:6684–6685. doi: 10.1021/ja071158n. The modularity of diterpene synthases is on display in this paper as the authors combine different type II and type I DTSs in E. coli to individually produce eight different diterpene scaffolds. This work serves as the foundation for engineered biosynthesis of many compounds of economic interest as the scaffolds produced are found in thousands of described natural products. [DOI] [PMC free article] [PubMed] [Google Scholar]