Abstract
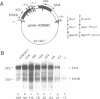
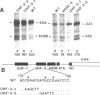
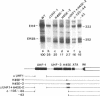
The early H4 (EH4) histone gene of the sea urchin, Strongylocentrotus purpuratus, is shown to contain at least five positive-responding sequence elements and one negative-responding site which control the level of in vitro transcription in an embryonic nuclear extract. The positive acting elements are: 1) the UHF-1 region, located between -133 and -102 (the site of a strong footprint, due at least in part to the binding of an 85 kD protein factor termed UHF-1); 2) the H4 specific element (H4SE), situated between -62 and -39; 3) a sequence corresponding to a TATA box between -33 and -26 (TAACAATA); 4) the transcriptional initiation site; and 5) an internal sequence element found between +19 and +50. Deletion of, or base changes in, the UHF-1, H4SE, initiation, or internal sequence sites resulted in significant decreases in transcription. Base substitutions in the TATA-like sequence had much less effect, resulting in no more than a 2-fold decrease in transcription, and there was no evidence that alternative initiation sites are utilized in the mutant templates. The negative element (termed the UHF-3 site) is contained within a footprinted region between nucleotides -75 and -56. Base substitutions in this area result in templates that were transcribed at a level 1.2-2.0-fold higher than the wild-type gene. Transcription levels of double UHF-1/H4SE and UHF-1/INR mutants were those expected from additive effects of the individual mutations and there was no suggestion of synergism.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Biggin M. D., Tjian R. Transcription factors that activate the Ultrabithorax promoter in developmentally staged extracts. Cell. 1988 Jun 3;53(5):699–711. doi: 10.1016/0092-8674(88)90088-8. [DOI] [PubMed] [Google Scholar]
- Busslinger M., Barberis A. Synthesis of sperm and late histone cDNAs of the sea urchin with a primer complementary to the conserved 3' terminal palindrome: evidence for tissue-specific and more general histone gene variants. Proc Natl Acad Sci U S A. 1985 Sep;82(17):5676–5680. doi: 10.1073/pnas.82.17.5676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busslinger M., Portmann R., Irminger J. C., Birnstiel M. L. Ubiquitous and gene-specific regulatory 5' sequences in a sea urchin histone DNA clone coding for histone protein variants. Nucleic Acids Res. 1980 Mar 11;8(5):957–977. doi: 10.1093/nar/8.5.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carcamo J., Maldonado E., Cortes P., Ahn M. H., Ha I., Kasai Y., Flint J., Reinberg D. A TATA-like sequence located downstream of the transcription initiation site is required for expression of an RNA polymerase II transcribed gene. Genes Dev. 1990 Sep;4(9):1611–1622. doi: 10.1101/gad.4.9.1611. [DOI] [PubMed] [Google Scholar]
- Chen W., Struhl K. Yeast mRNA initiation sites are determined primarily by specific sequences, not by the distance from the TATA element. EMBO J. 1985 Dec 1;4(12):3273–3280. doi: 10.1002/j.1460-2075.1985.tb04077.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs G., Maxson R., Kedes L. H. Histone gene expression during sea urchin embryogenesis: isolation and characterization of early and late messenger RNAs of Strongylocentrotus purpuratus by gene-specific hybridization and template activity. Dev Biol. 1979 Nov;73(1):153–173. doi: 10.1016/0012-1606(79)90144-1. [DOI] [PubMed] [Google Scholar]
- Childs G., Nocente-McGrath C., Lieber T., Holt C., Knowles J. A. Sea urchin (lytechinus pictus) late-stage histone H3 and H4 genes: characterization and mapping of a clustered but nontandemly linked multigene family. Cell. 1982 Dec;31(2 Pt 1):383–393. doi: 10.1016/0092-8674(82)90132-5. [DOI] [PubMed] [Google Scholar]
- Clerc R. G., Bucher P., Strub K., Birnstiel M. L. Transcription of a cloned Xenopus laevis H4 histone gene in the homologous frog oocyte system depends on an evolutionary conserved sequence motif in the -50 region. Nucleic Acids Res. 1983 Dec 20;11(24):8641–8657. doi: 10.1093/nar/11.24.8641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Concino M. F., Lee R. F., Merryweather J. P., Weinmann R. The adenovirus major late promoter TATA box and initiation site are both necessary for transcription in vitro. Nucleic Acids Res. 1984 Oct 11;12(19):7423–7433. doi: 10.1093/nar/12.19.7423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dailey L., Hanly S. M., Roeder R. G., Heintz N. Distinct transcription factors bind specifically to two regions of the human histone H4 promoter. Proc Natl Acad Sci U S A. 1986 Oct;83(19):7241–7245. doi: 10.1073/pnas.83.19.7241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dailey L., Roberts S. B., Heintz N. Purification of the human histone H4 gene-specific transcription factors H4TF-1 and H4TF-2. Genes Dev. 1988 Dec;2(12B):1700–1712. doi: 10.1101/gad.2.12b.1700. [DOI] [PubMed] [Google Scholar]
- Grosschedl R., Birnstiel M. L. Identification of regulatory sequences in the prelude sequences of an H2A histone gene by the study of specific deletion mutants in vivo. Proc Natl Acad Sci U S A. 1980 Mar;77(3):1432–1436. doi: 10.1073/pnas.77.3.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunstein M., Diamond K. E., Knoppel E., Grunstein J. E. Comparison of the early histone H4 gene sequence of Strongylocentrotus purpuratus with maternal, early, and late histone H4 mRNA sequences. Biochemistry. 1981 Mar 3;20(5):1216–1223. doi: 10.1021/bi00508a025. [DOI] [PubMed] [Google Scholar]
- Grunstein M. Hatching in the sea urchin Lytechinus pictus is accompanied by a shift in histone H4 gene activity. Proc Natl Acad Sci U S A. 1978 Sep;75(9):4135–4139. doi: 10.1073/pnas.75.9.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanly S. M., Bleecker G. C., Heintz N. Identification of promoter elements necessary for transcriptional regulation of a human histone H4 gene in vitro. Mol Cell Biol. 1985 Feb;5(2):380–389. doi: 10.1128/mcb.5.2.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hariharan N., Kelley D. E., Perry R. P. Equipotent mouse ribosomal protein promoters have a similar architecture that includes internal sequence elements. Genes Dev. 1989 Nov;3(11):1789–1800. doi: 10.1101/gad.3.11.1789. [DOI] [PubMed] [Google Scholar]
- Hariharan N., Perry R. P. Functional dissection of a mouse ribosomal protein promoter: significance of the polypyrimidine initiator and an element in the TATA-box region. Proc Natl Acad Sci U S A. 1990 Feb;87(4):1526–1530. doi: 10.1073/pnas.87.4.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentschel C. C., Birnstiel M. L. The organization and expression of histone gene families. Cell. 1981 Aug;25(2):301–313. doi: 10.1016/0092-8674(81)90048-9. [DOI] [PubMed] [Google Scholar]
- Hentschel C., Irminger J. C., Bucher P., Birnstiel M. L. Sea urchin histone mRNA termini are located in gene regions downstream from putative regulatory sequences. Nature. 1980 May 15;285(5761):147–151. doi: 10.1038/285147a0. [DOI] [PubMed] [Google Scholar]
- Hieter P. A., Hendricks M. B., Hemminki K., Weinberg E. S. Histone gene switch in the sea urchin embryo. Identification of late embryonic histone messenger ribonucleic acids and the control of their synthesis. Biochemistry. 1979 Jun 26;18(13):2707–2716. doi: 10.1021/bi00580a004. [DOI] [PubMed] [Google Scholar]
- Hultmark D., Klemenz R., Gehring W. J. Translational and transcriptional control elements in the untranslated leader of the heat-shock gene hsp22. Cell. 1986 Feb 14;44(3):429–438. doi: 10.1016/0092-8674(86)90464-2. [DOI] [PubMed] [Google Scholar]
- Hurt M. M., Pandey N. B., Marzluff W. F. A region in the coding sequence is required for high-level expression of murine histone H3 gene. Proc Natl Acad Sci U S A. 1989 Jun;86(12):4450–4454. doi: 10.1073/pnas.86.12.4450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones K. A., Luciw P. A., Duchange N. Structural arrangements of transcription control domains within the 5'-untranslated leader regions of the HIV-1 and HIV-2 promoters. Genes Dev. 1988 Sep;2(9):1101–1114. doi: 10.1101/gad.2.9.1101. [DOI] [PubMed] [Google Scholar]
- Kaumeyer J. F., Weinberg E. S. Sequence, organization and expression of late embryonic H3 and H4 histone genes from the sea urchin, Strongylocentrotus purpuratus. Nucleic Acids Res. 1986 Jun 11;14(11):4557–4576. doi: 10.1093/nar/14.11.4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemler I., Busslinger M. Characterization of two nonallelic pairs of late histone H2A and H2B genes of the sea urchin: differential regulation in the embryo and tissue-specific expression in the adult. Mol Cell Biol. 1986 Nov;6(11):3746–3754. doi: 10.1128/mcb.6.11.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles J. A., Childs G. J. Comparison of the late H1 histone genes of the sea urchins Lytechinus pictus and Strongelocentrotus purpuratus. Nucleic Acids Res. 1986 Oct 24;14(20):8121–8133. doi: 10.1093/nar/14.20.8121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles J. A., Childs G. J. Temporal expression of late histone messenger RNA in the sea urchin Lytechinus pictus. Proc Natl Acad Sci U S A. 1984 Apr;81(8):2411–2415. doi: 10.1073/pnas.81.8.2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles J. A., Lai Z. C., Childs G. J. Isolation, characterization, and expression of the gene encoding the late histone subtype H1-gamma of the sea urchin Strongylocentrotus purpuratus. Mol Cell Biol. 1987 Jan;7(1):478–485. doi: 10.1128/mcb.7.1.478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroeger P., Stewart C., Schaap T., van Wijnen A., Hirshman J., Helms S., Stein G., Stein J. Proximal and distal regulatory elements that influence in vivo expression of a cell cycle-dependent human H4 histone gene. Proc Natl Acad Sci U S A. 1987 Jun;84(12):3982–3986. doi: 10.1073/pnas.84.12.3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Lai Z. C., Childs G. Characterization of the structure and transcriptional patterns of the gene encoding the late histone subtype H1-beta of the sea urchin Strongylocentrotus purpuratus. Mol Cell Biol. 1988 Apr;8(4):1842–1844. doi: 10.1128/mcb.8.4.1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Z. C., Maxson R., Childs G. Both basal and ontogenic promoter elements affect the timing and level of expression of a sea urchin H1 gene during early embryogenesis. Genes Dev. 1988 Feb;2(2):173–183. doi: 10.1101/gad.2.2.173. [DOI] [PubMed] [Google Scholar]
- Mauron A., Kedes L., Hough-Evans B. R., Davidson E. H. Accumulation of individual histone mRNAs during embryogenesis of the sea urchin Strongylocentrotus purpuratus. Dev Biol. 1982 Dec;94(2):425–434. doi: 10.1016/0012-1606(82)90359-1. [DOI] [PubMed] [Google Scholar]
- Maxson R. E., Jr, Wilt F. H. Accumulation of the early histone messenger RNAs during the development of Strongylocentrotus purpuratus. Dev Biol. 1982 Dec;94(2):435–440. doi: 10.1016/0012-1606(82)90360-8. [DOI] [PubMed] [Google Scholar]
- Maxson R., Cohn R., Kedes L., Mohun T. Expression and organization of histone genes. Annu Rev Genet. 1983;17:239–277. doi: 10.1146/annurev.ge.17.120183.001323. [DOI] [PubMed] [Google Scholar]
- Maxson R., Mohun T., Gormezano G., Childs G., Kedes L. Distinct organizations and patterns of expression of early and late histone gene sets in the sea urchin. Nature. 1983 Jan 13;301(5896):120–125. doi: 10.1038/301120a0. [DOI] [PubMed] [Google Scholar]
- Maxson R., Mohun T., Gormezano G., Kedes L. Evolution of late H2A, H2B, and H4 histone genes of the sea urchin, Strongylocentrotus purpuratus. Nucleic Acids Res. 1987 Dec 23;15(24):10569–10582. doi: 10.1093/nar/15.24.10569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris G. F., Marzluff W. F. A factor in sea urchin eggs inhibits transcription in isolated nuclei by sea urchin RNA polymerase III. Biochemistry. 1983 Feb 1;22(3):645–653. doi: 10.1021/bi00272a019. [DOI] [PubMed] [Google Scholar]
- Morris G. F., Price D. H., Marzluff W. F. Synthesis of U1 RNA in a DNA-dependent system from sea urchin embryos. Proc Natl Acad Sci U S A. 1986 Jun;83(11):3674–3678. doi: 10.1073/pnas.83.11.3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mous J., Stunnenberg H., Georgiev O., Birnstiel M. L. Stimulation of sea urchin H2B histone gene transcription by a chromatin-associated protein fraction depends on gene sequences downstream of the transcription start site. Mol Cell Biol. 1985 Oct;5(10):2764–2769. doi: 10.1128/mcb.5.10.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatani Y., Brenner M., Freese E. An RNA polymerase II promoter containing sequences upstream and downstream from the RNA startpoint that direct initiation of transcription from the same site. Proc Natl Acad Sci U S A. 1990 Jun;87(11):4289–4293. doi: 10.1073/pnas.87.11.4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overton G. C., Weinberg E. S. Length and sequence heterogeneity of the histone gene repeat unit of the sea urchin, S. purpuratus. Cell. 1978 Jun;14(2):247–257. doi: 10.1016/0092-8674(78)90111-3. [DOI] [PubMed] [Google Scholar]
- Perkins K. K., Dailey G. M., Tjian R. In vitro analysis of the Antennapedia P2 promoter: identification of a new Drosophila transcription factor. Genes Dev. 1988 Dec;2(12A):1615–1626. doi: 10.1101/gad.2.12a.1615. [DOI] [PubMed] [Google Scholar]
- Perry M., Thomsen G. H., Roeder R. G. Genomic organization and nucleotide sequence of two distinct histone gene clusters from Xenopus laevis. Identification of novel conserved upstream sequence elements. J Mol Biol. 1985 Oct 5;185(3):479–499. doi: 10.1016/0022-2836(85)90065-8. [DOI] [PubMed] [Google Scholar]
- Roberts S. B., Weisser K. E., Childs G. Sequence comparisons of non-allelic late histone genes and their early stage counterparts. Evidence for gene conversion within the sea urchin late stage gene family. J Mol Biol. 1984 Apr 25;174(4):647–662. doi: 10.1016/0022-2836(84)90088-3. [DOI] [PubMed] [Google Scholar]
- Schaffner W., Kunz G., Daetwyler H., Telford J., Smith H. O., Birnstiel M. L. Genes and spacers of cloned sea urchin histone DNA analyzed by sequencing. Cell. 1978 Jul;14(3):655–671. doi: 10.1016/0092-8674(78)90249-0. [DOI] [PubMed] [Google Scholar]
- Sierra F., Stein G., Stein J. Structure and in vitro transcription of a human H4 histone gene. Nucleic Acids Res. 1983 Oct 25;11(20):7069–7086. doi: 10.1093/nar/11.20.7069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smale S. T., Baltimore D. The "initiator" as a transcription control element. Cell. 1989 Apr 7;57(1):103–113. doi: 10.1016/0092-8674(89)90176-1. [DOI] [PubMed] [Google Scholar]
- Smale S. T., Schmidt M. C., Berk A. J., Baltimore D. Transcriptional activation by Sp1 as directed through TATA or initiator: specific requirement for mammalian transcription factor IID. Proc Natl Acad Sci U S A. 1990 Jun;87(12):4509–4513. doi: 10.1073/pnas.87.12.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soeller W. C., Poole S. J., Kornberg T. In vitro transcription of the Drosophila engrailed gene. Genes Dev. 1988 Jan;2(1):68–81. doi: 10.1101/gad.2.1.68. [DOI] [PubMed] [Google Scholar]
- Sugarman B. J., Dodgson J. B., Engel J. D. Genomic organization, DNA sequence, and expression of chicken embryonic histone genes. J Biol Chem. 1983 Jul 25;258(14):9005–9016. [PubMed] [Google Scholar]
- Sures I., Levy S., Kedes L. H. Leader sequences of Strongylocentrotus purpuratus histone mRNAs start at a unique heptanucleotide common to all five histone genes. Proc Natl Acad Sci U S A. 1980 Mar;77(3):1265–1269. doi: 10.1073/pnas.77.3.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talkington C. A., Leder P. Rescuing the in vitro function of a globin pseudogene promoter. Nature. 1982 Jul 8;298(5870):192–195. doi: 10.1038/298192a0. [DOI] [PubMed] [Google Scholar]
- Tebb G., Mattaj I. W. Positionally exact initiation is required for the formation of a stable RNA polymerase II transcription complex in vivo. EMBO J. 1988 Dec 1;7(12):3785–3792. doi: 10.1002/j.1460-2075.1988.tb03263.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokunaga K., Hirose S., Suzuki Y. In monkey COS cells only the TATA box and the cap site region are required for faithful and efficient initiation of the fibroin gene transcription. Nucleic Acids Res. 1984 Feb 10;12(3):1543–1558. doi: 10.1093/nar/12.3.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung L., Morris G. F., Yager L. N., Weinberg E. S. Sea urchin early and late H4 histone genes bind a specific transcription factor in a stable preinitiation complex. Mol Cell Biol. 1989 Apr;9(4):1476–1487. doi: 10.1128/mcb.9.4.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzman J. A., Wilt F. H. The role of RNA polymerase initiation and elongation in control of total RNA and histone mRNA synthesis in sea urchin embryos. Dev Biol. 1984 Nov;106(1):174–180. doi: 10.1016/0012-1606(84)90073-3. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- Wang S. W., Robins A. J., d'Andrea R., Wells J. R. Inverted duplication of histone genes in chicken and disposition of regulatory sequences. Nucleic Acids Res. 1985 Feb 25;13(4):1369–1387. doi: 10.1093/nar/13.4.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg E. S., Hendricks M. B., Hemminki K., Kuwabara P. E., Farrelly L. A. Timing and rates of synthesis of early histone mRNA in the embryo of Strongylocentrotus purpuratus. Dev Biol. 1983 Jul;98(1):117–129. doi: 10.1016/0012-1606(83)90340-8. [DOI] [PubMed] [Google Scholar]
- Wells D. E. Compilation analysis of histones and histone genes. Nucleic Acids Res. 1986;14 (Suppl):r119–r149. doi: 10.1093/nar/14.suppl.r119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkfein R. J., Connor W., Mezquita J., Dixon G. H. Histone H4 and H2B genes in rainbow trout (Salmo gairdnerii). J Mol Evol. 1985;22(1):1–19. doi: 10.1007/BF02105800. [DOI] [PubMed] [Google Scholar]
- Yager L. N., Kaumeyer J. F., Weinberg E. S. Evolving sea urchin histone genes--nucleotide polymorphisms in the H4 gene and spacers of Strongylocentrotus purpuratus. J Mol Evol. 1984;20(3-4):215–226. doi: 10.1007/BF02104728. [DOI] [PubMed] [Google Scholar]
- Zhong R., Roeder R. G., Heintz N. The primary structure and expression of four cloned human histone genes. Nucleic Acids Res. 1983 Nov 11;11(21):7409–7425. doi: 10.1093/nar/11.21.7409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wijnen A. J., Stein J. L., Stein G. S. A nuclear protein with affinity for the 5' flanking region of a cell cycle dependent human H4 histone gene in vitro. Nucleic Acids Res. 1987 Feb 25;15(4):1679–1698. doi: 10.1093/nar/15.4.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wijnen A. J., Wright K. L., Lian J. B., Stein J. L., Stein G. S. Human H4 histone gene transcription requires the proliferation-specific nuclear factor HiNF-D. Auxiliary roles for HiNF-C (Sp1-like) and HiNF-A (high mobility group-like). J Biol Chem. 1989 Sep 5;264(25):15034–15042. [PubMed] [Google Scholar]