Abstract
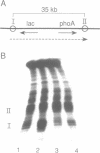
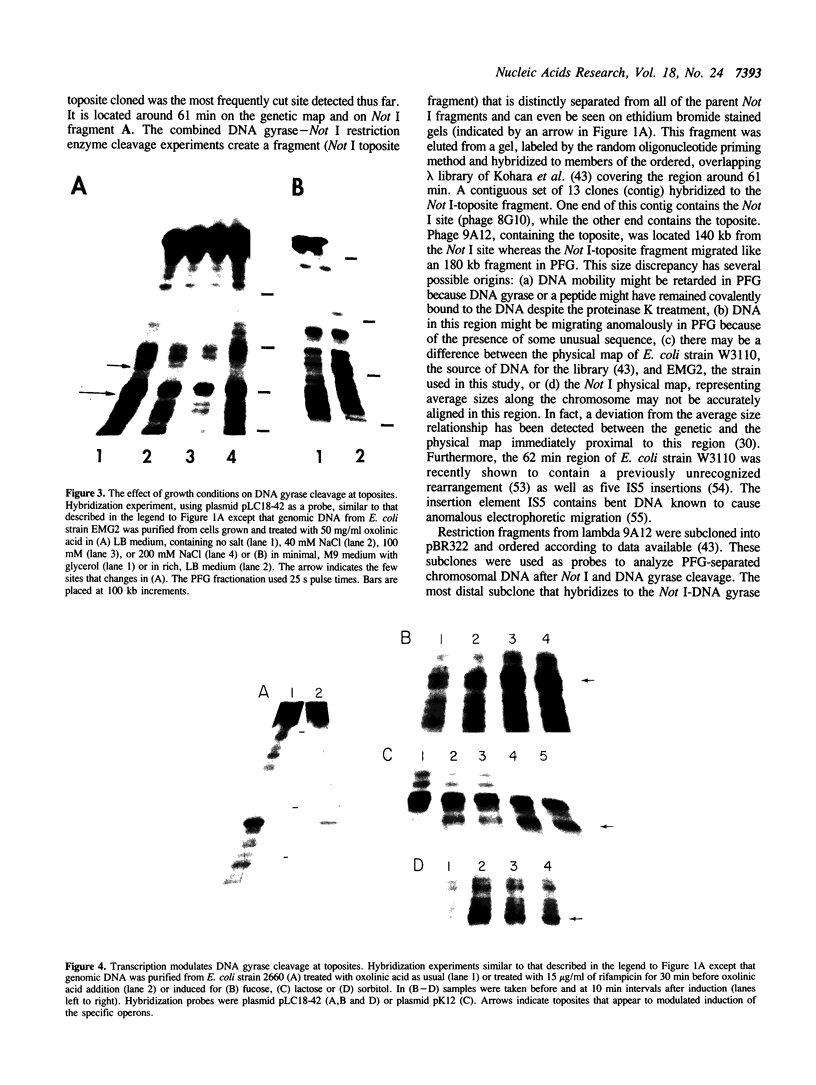
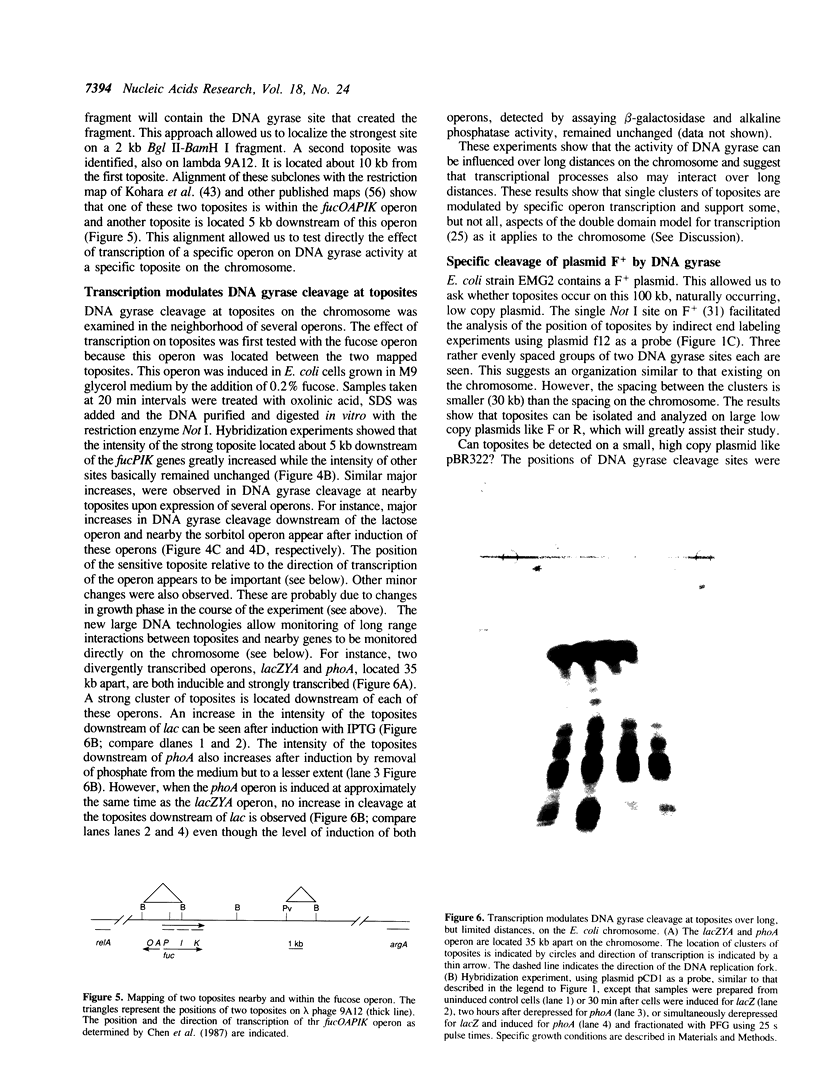
Prominent DNA gyrase-mediated cleavage sites, induced by oxolinic acid, occur at specific, but infrequent, locations on the Escherichia coli chromosome. These sites, which we call toposites, may represent high affinity DNA gyrase binding sites or may mark chromosomal regions that accumulate superhelical stress. Toposites are usually grouped in 5 to 10 kb clusters that are mostly 50 to 100 kb apart. The total number of clusters on the chromosome is between 50 and 100. The location of sites depends on the local sequence. The extent of DNA gyrase cleavage at toposites can be strongly modulated by transcription occurring at as far as 35 kb away.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Atherly A. G. Escherichia coli mutant containing a large deletion from relA to argA. J Bacteriol. 1979 May;138(2):530–534. doi: 10.1128/jb.138.2.530-534.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann B. J. Pedigrees of some mutant strains of Escherichia coli K-12. Bacteriol Rev. 1972 Dec;36(4):525–557. doi: 10.1128/br.36.4.525-557.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balke V. L., Gralla J. D. Changes in the linking number of supercoiled DNA accompany growth transitions in Escherichia coli. J Bacteriol. 1987 Oct;169(10):4499–4506. doi: 10.1128/jb.169.10.4499-4506.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borowiec J. A., Gralla J. D. Supercoiling response of the lac ps promoter in vitro. J Mol Biol. 1985 Aug 20;184(4):587–598. doi: 10.1016/0022-2836(85)90305-5. [DOI] [PubMed] [Google Scholar]
- Béjar S., Bouché J. P. The spacing of Escherichia coli DNA gyrase sites cleaved in vivo by treatment with oxolinic acid and sodium dodecyl sulfate. Biochimie. 1984 Nov-Dec;66(11-12):693–700. doi: 10.1016/0300-9084(84)90258-x. [DOI] [PubMed] [Google Scholar]
- Chen Y. M., Zhu Y., Lin E. C. The organization of the fuc regulon specifying L-fucose dissimilation in Escherichia coli K12 as determined by gene cloning. Mol Gen Genet. 1987 Dec;210(2):331–337. doi: 10.1007/BF00325702. [DOI] [PubMed] [Google Scholar]
- Diekmann S. Temperature and salt dependence of the gel migration anomaly of curved DNA fragments. Nucleic Acids Res. 1987 Jan 12;15(1):247–265. doi: 10.1093/nar/15.1.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorman C. J., Barr G. C., Ni Bhriain N., Higgins C. F. DNA supercoiling and the anaerobic and growth phase regulation of tonB gene expression. J Bacteriol. 1988 Jun;170(6):2816–2826. doi: 10.1128/jb.170.6.2816-2826.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drlica K. Biology of bacterial deoxyribonucleic acid topoisomerases. Microbiol Rev. 1984 Dec;48(4):273–289. doi: 10.1128/mr.48.4.273-289.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Filutowicz M. Requirement of DNA gyrase for the initiation of chromosome replication in Escherichia coli K-12. Mol Gen Genet. 1980 Jan;177(2):301–309. doi: 10.1007/BF00267443. [DOI] [PubMed] [Google Scholar]
- Franco R. J., Drlica K. DNA gyrase on the bacterial chromosome. Oxolinic acid-induced DNA cleavage in the dnaA-gyrB region. J Mol Biol. 1988 May 5;201(1):229–233. doi: 10.1016/0022-2836(88)90449-4. [DOI] [PubMed] [Google Scholar]
- Gasser S. M., Laroche T., Falquet J., Boy de la Tour E., Laemmli U. K. Metaphase chromosome structure. Involvement of topoisomerase II. J Mol Biol. 1986 Apr 20;188(4):613–629. doi: 10.1016/s0022-2836(86)80010-9. [DOI] [PubMed] [Google Scholar]
- Gellert M., Mizuuchi K., O'Dea M. H., Itoh T., Tomizawa J. I. Nalidixic acid resistance: a second genetic character involved in DNA gyrase activity. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4772–4776. doi: 10.1073/pnas.74.11.4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellert M., Mizuuchi K., O'Dea M. H., Nash H. A. DNA gyrase: an enzyme that introduces superhelical turns into DNA. Proc Natl Acad Sci U S A. 1976 Nov;73(11):3872–3876. doi: 10.1073/pnas.73.11.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilson E., Perrin D., Clement J. M., Szmelcman S., Dassa E., Hofnung M. Palindromic units from E. coli as binding sites for a chromoid-associated protein. FEBS Lett. 1986 Oct 6;206(2):323–328. doi: 10.1016/0014-5793(86)81005-5. [DOI] [PubMed] [Google Scholar]
- Goldstein E., Drlica K. Regulation of bacterial DNA supercoiling: plasmid linking numbers vary with growth temperature. Proc Natl Acad Sci U S A. 1984 Jul;81(13):4046–4050. doi: 10.1073/pnas.81.13.4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins C. F., Dorman C. J., Stirling D. A., Waddell L., Booth I. R., May G., Bremer E. A physiological role for DNA supercoiling in the osmotic regulation of gene expression in S. typhimurium and E. coli. Cell. 1988 Feb 26;52(4):569–584. doi: 10.1016/0092-8674(88)90470-9. [DOI] [PubMed] [Google Scholar]
- Hill C. W., Harnish B. W. Inversions between ribosomal RNA genes of Escherichia coli. Proc Natl Acad Sci U S A. 1981 Nov;78(11):7069–7072. doi: 10.1073/pnas.78.11.7069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamoto F., Tani S. Diversity of regulation of genetic transcription. Nat New Biol. 1972 Dec 6;240(101):172–175. doi: 10.1038/newbio240172a0. [DOI] [PubMed] [Google Scholar]
- Innis M. A., Tokunaga M., Williams M. E., Loranger J. M., Chang S. Y., Chang S., Wu H. C. Nucleotide sequence of the Escherichia coli prolipoprotein signal peptidase (lsp) gene. Proc Natl Acad Sci U S A. 1984 Jun;81(12):3708–3712. doi: 10.1073/pnas.81.12.3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovanovich S. B., Lebowitz J. Estimation of the effect of coumermycin A1 on Salmonella typhimurium promoters by using random operon fusions. J Bacteriol. 1987 Oct;169(10):4431–4435. doi: 10.1128/jb.169.10.4431-4435.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavenoff R., Bowen B. C. Electron microscopy of membrane-free folded chromosomes from Escherichia coli. Chromosoma. 1976 Dec 16;59(2):89–101. doi: 10.1007/BF00328479. [DOI] [PubMed] [Google Scholar]
- Kikuchi Y., Yoda K., Yamasaki M., Tamura G. The nucleotide sequence of the promoter and the amino-terminal region of alkaline phosphatase structural gene (phoA) of Escherichia coli. Nucleic Acids Res. 1981 Nov 11;9(21):5671–5678. doi: 10.1093/nar/9.21.5671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkegaard K., Wang J. C. Mapping the topography of DNA wrapped around gyrase by nucleolytic and chemical probing of complexes of unique DNA sequences. Cell. 1981 Mar;23(3):721–729. doi: 10.1016/0092-8674(81)90435-9. [DOI] [PubMed] [Google Scholar]
- Kohara Y., Akiyama K., Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987 Jul 31;50(3):495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- Kreuzer K. N., Alberts B. M. Site-specific recognition of bacteriophage T4 DNA by T4 type II DNA topoisomerase and Escherichia coli DNA gyrase. J Biol Chem. 1984 Apr 25;259(8):5339–5346. [PubMed] [Google Scholar]
- Liu L. F., Wang J. C. Supercoiling of the DNA template during transcription. Proc Natl Acad Sci U S A. 1987 Oct;84(20):7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockshon D., Morris D. R. Sites of reaction of Escherichia coli DNA gyrase on pBR322 in vivo as revealed by oxolinic acid-induced plasmid linearization. J Mol Biol. 1985 Jan 5;181(1):63–74. doi: 10.1016/0022-2836(85)90324-9. [DOI] [PubMed] [Google Scholar]
- Lodge J. K., Kazic T., Berg D. E. Formation of supercoiling domains in plasmid pBR322. J Bacteriol. 1989 Apr;171(4):2181–2187. doi: 10.1128/jb.171.4.2181-2187.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzel R., Gellert M. Regulation of the genes for E. coli DNA gyrase: homeostatic control of DNA supercoiling. Cell. 1983 Aug;34(1):105–113. doi: 10.1016/0092-8674(83)90140-x. [DOI] [PubMed] [Google Scholar]
- Mizuuchi K., Fisher L. M., O'Dea M. H., Gellert M. DNA gyrase action involves the introduction of transient double-strand breaks into DNA. Proc Natl Acad Sci U S A. 1980 Apr;77(4):1847–1851. doi: 10.1073/pnas.77.4.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu S., Kato M., Kohara Y., Mizuno T. Insertion sequence IS5 contains a sharply curved DNA structure at its terminus. Mol Gen Genet. 1988 Nov;214(3):433–438. doi: 10.1007/BF00330477. [DOI] [PubMed] [Google Scholar]
- O'Connor M. B., Malamy M. H. Mapping of DNA gyrase cleavage sites in vivo oxolinic acid induced cleavages in plasmid pBR322. J Mol Biol. 1985 Feb 20;181(4):545–550. doi: 10.1016/0022-2836(85)90426-7. [DOI] [PubMed] [Google Scholar]
- Poulsen P., Bonekamp F., Jensen K. F. Structure of the Escherichia coli pyrE operon and control of pyrE expression by a UTP modulated intercistronic attentuation. EMBO J. 1984 Aug;3(8):1783–1790. doi: 10.1002/j.1460-2075.1984.tb02046.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saadi S., Maas W. K., Hill D. F., Bergquist P. L. Nucleotide sequence analysis of RepFIC, a basic replicon present in IncFI plasmids P307 and F, and its relation to the RepA replicon of IncFII plasmids. J Bacteriol. 1987 May;169(5):1836–1846. doi: 10.1128/jb.169.5.1836-1846.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar A., Rupp W. D. Physical map of the recA gene. Proc Natl Acad Sci U S A. 1979 Jul;76(7):3144–3148. doi: 10.1073/pnas.76.7.3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinden R. R., Pettijohn D. E. Chromosomes in living Escherichia coli cells are segregated into domains of supercoiling. Proc Natl Acad Sci U S A. 1981 Jan;78(1):224–228. doi: 10.1073/pnas.78.1.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. L., Cantor C. R. Purification, specific fragmentation, and separation of large DNA molecules. Methods Enzymol. 1987;155:449–467. doi: 10.1016/0076-6879(87)55030-3. [DOI] [PubMed] [Google Scholar]
- Smith C. L., Econome J. G., Schutt A., Klco S., Cantor C. R. A physical map of the Escherichia coli K12 genome. Science. 1987 Jun 12;236(4807):1448–1453. doi: 10.1126/science.3296194. [DOI] [PubMed] [Google Scholar]
- Smith C. L., Kolodner R. D. Mapping of Escherichia coli chromosomal Tn5 and F insertions by pulsed field gel electrophoresis. Genetics. 1988 Jun;119(2):227–236. doi: 10.1093/genetics/119.2.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. L., Kubo M., Imamoto F. Promoter-specific inhibition of transcription by antibiotics which act on DNA gyrase. Nature. 1978 Oct 5;275(5679):420–423. doi: 10.1038/275420a0. [DOI] [PubMed] [Google Scholar]
- Smith C. L., Oishi M. Early events and mechanisms in the induction of bacterial SOS functions: analysis of the phage repressor inactivation process in vivo. Proc Natl Acad Sci U S A. 1978 Apr;75(4):1657–1661. doi: 10.1073/pnas.75.4.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H. O., Birnstiel M. L. A simple method for DNA restriction site mapping. Nucleic Acids Res. 1976 Sep;3(9):2387–2398. doi: 10.1093/nar/3.9.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder M., Drlica K. DNA gyrase on the bacterial chromosome: DNA cleavage induced by oxolinic acid. J Mol Biol. 1979 Jun 25;131(2):287–302. doi: 10.1016/0022-2836(79)90077-9. [DOI] [PubMed] [Google Scholar]
- Steck T. R., Drlica K. Bacterial chromosome segregation: evidence for DNA gyrase involvement in decatenation. Cell. 1984 Apr;36(4):1081–1088. doi: 10.1016/0092-8674(84)90058-8. [DOI] [PubMed] [Google Scholar]
- Sugino A., Peebles C. L., Kreuzer K. N., Cozzarelli N. R. Mechanism of action of nalidixic acid: purification of Escherichia coli nalA gene product and its relationship to DNA gyrase and a novel nicking-closing enzyme. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4767–4771. doi: 10.1073/pnas.74.11.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata S., Higashitani A., Takanami M., Akiyama K., Kohara Y., Nishimura Y., Nishimura A., Yasuda S., Hirota Y. Construction of an ordered cosmid collection of the Escherichia coli K-12 W3110 chromosome. J Bacteriol. 1989 Feb;171(2):1214–1218. doi: 10.1128/jb.171.2.1214-1218.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torriani A. Alkaline phosphatase subunits and their dimerization in vivo. J Bacteriol. 1968 Oct;96(4):1200–1207. doi: 10.1128/jb.96.4.1200-1207.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster T. A., Gibson B. W., Keng T., Biemann K., Schimmel P. Primary structures of both subunits of Escherichia coli glycyl-tRNA synthetase. J Biol Chem. 1983 Sep 10;258(17):10637–10641. [PubMed] [Google Scholar]
- Worcel A., Burgi E. On the structure of the folded chromosome of Escherichia coli. J Mol Biol. 1972 Nov 14;71(2):127–147. doi: 10.1016/0022-2836(72)90342-7. [DOI] [PubMed] [Google Scholar]
- Wu H. Y., Shyy S. H., Wang J. C., Liu L. F. Transcription generates positively and negatively supercoiled domains in the template. Cell. 1988 May 6;53(3):433–440. doi: 10.1016/0092-8674(88)90163-8. [DOI] [PubMed] [Google Scholar]
- Yang Y., Ames G. F. DNA gyrase binds to the family of prokaryotic repetitive extragenic palindromic sequences. Proc Natl Acad Sci U S A. 1988 Dec;85(23):8850–8854. doi: 10.1073/pnas.85.23.8850. [DOI] [PMC free article] [PubMed] [Google Scholar]