Abstract
Plasma HDL levels are inversely associated with atherosclerosis. Inbred mouse strains differ in plasma HDL levels and susceptibility to atherosclerosis. Atherosclerosis-susceptible C57BL/6J mice possess plasma HDL levels 2-fold lower than atherosclerosis-resistant FVB/NJ mice. Polymorphisms have been previously identified between the two mouse strains in the major HDL apolipoproteins, ApoA-I and ApoA-II, which may affect their function on HDL. To begin to understand the HDL differences, we here report on a detailed comparison of the lipid-associated functions of the two mouse ApoA-I proteins. We demonstrate that these polymorphisms significantly alter the protein self-association properties, the ability of the proteins to clear lipid micelles from solution, and their binding affinity for mature mouse HDL. The changes in lipid binding do not appear to alter the ability of the protein to promote cholesterol efflux from cells or the formation of nascent HDL from primary hepatocytes. These apolipoprotein polymorphisms do not change the rate at which HDL protein or cholesterol are catabolized in vivo. Although the presence of the polymorphisms in ApoA-I alters important factors in HDL formation, the basis for the differences in the HDL plasma levels observed in the various mouse strains is more complex and requires additional investigation.
Keywords: cholesterol efflux, lipid binding, alpha helix, high density lipoprotein
Cardiovascular disease remains one of the leading causes of death worldwide (1). The progression of atherosclerosis is a major contributor to cardiovascular disease development (2). Although not fully understood, plasma lipoproteins play an important role in atherosclerosis, with plasma HDL cholesterol levels being inversely correlated with risk of cardiovascular disease (3). Much of our current knowledge of atherosclerosis comes from the use of atherosclerosis-susceptible animal models and particularly mouse models. Numerous inbred mouse strains display varying degrees of susceptibility to atherosclerotic lesion development, although the causes of these differences are unclear (4). Because wild-type (WT) mice do not develop atherosclerotic lesions, these comparisons were made using mice fed a cholate-containing diet high in fat and cholesterol in order to induce lesion formation. However, this diet also induces an inflammatory response. Since then, several studies have shown further strain differences in atherosclerosis susceptibility in apolipoprotein (Apo)E and LDL-receptor knockout mice, the most commonly used genetic models of atherosclerosis (5, 6). Most atherosclerosis studies have been performed in the C57BL/6 (C57) strain, which appears to be the most sensitive of the mouse strains. Thus, in both genetic models, the C57BL/6 mouse strain develops lesions more than 5-fold greater in size than the atherosclerosis-resistant FVB/NJ (FVB) strain. These two mouse strains also differ in lipoprotein levels. In WT, ApoE, and LDL-receptor knockout mice, the FVB strain displays 2-fold higher levels of plasma HDL cholesterol (5, 6). The same is true in numerous other atherosclerosis-resistant strains (7). The genetic basis of increased HDL cholesterol and the resulting impact on atherosclerosis susceptibility in these mouse strains is not completely understood.
ApoA-I and ApoA-II make up greater than 90% of the protein component of HDL, with ApoA-I being the predominant protein (∼70%) (8). These two proteins exhibit several sequence differences between the C57 and the FVB strains. The 240 amino acid ApoA-I of the atherosclerosis-susceptible C57 mouse possesses two differences in amino acid sequence from the FVB ApoA-I (C57 to FVB: Q225K, V226A) (9, 10). These two sequence differences are the only known naturally occurring polymorphisms in inbred mouse ApoA-I.
ApoA-I plays a significant role in HDL formation, and its absence results in at least a 70% reduction in plasma HDL levels (11). ApoA-I interacts with the ATP-binding cassette transporter ABCA1, by which ApoA-I receives cholesterol and phospholipid that is effluxed from the cell (12). ApoA-I is composed of 10 amphipathic α-helices, with helices 9 and 10 being responsible for the initial binding of human ApoA-I to lipid and its association with the ABCA1 transporter (13, 14). The two amino acids that differ between C57 and FVB ApoA-I are adjacent to each other in the center of helix 10. Mutations in helix 10 of human ApoA-I have been shown to decrease the rate of clearance of phospholipid in solution and the efflux rate of cholesterol from cultured macrophages (14). The Nichinan variant of human ApoA-I lacks E235 in helix 10 and is associated with decreased ABCA1-mediated cholesterol efflux from macrophages and decreased plasma HDL levels (15). It has been shown that the tertiary structure of lipid-free human ApoA-I is made up of two domains: an N-terminal α-helix bundle from residues 1–187 (through helix 7) and a C-terminal less organized region (helices 8–10), which is the major lipid-binding region of the molecule (16, 17). Recent work has suggested that the mouse ApoA-I is similarly made up of the N- and C-terminal domains, but, unlike the human ApoA-I, the N-terminal domain is more effective than the C-terminal in lipid binding and cholesterol efflux from macrophages (18–20). Thus, the role of the mouse C-terminal domain in ApoA-I function remains unclear. In light of this, it is important to understand whether the naturally occurring changes in the C-terminal domain of FVB ApoA-I significantly alter the lipid interactions of the protein, thereby potentially altering the protein function.
Here, we report the first in depth study of the role of these polymorphisms on lipid binding, cholesterol efflux, hepatic HDL production, and HDL clearance. The C-terminal polymorphisms result in significant differences in lipid binding capability and HDL association. However, the sequence differences alone do not appear to alter the cholesterol efflux capabilities of the lipid-free ApoA-I or of the mature HDL particle, nor do they appear to be solely responsible for plasma HDL cholesterol levels.
MATERIALS AND METHODS
All chemicals were purchased from Sigma Chemical Co. (St. Louis. MO) unless otherwise noted. C57BL/6J or FVBN/J mice were purchased from Jackson Laboratory (Bar Harbor, ME). Mice were bred, and the pups were weaned at 21–28 days and maintained on chow diet #7913 from Harlan Labs (Indianapolis, IN). Female mice were used for all experiments described. All procedures performed on the mice were in accordance with National Institute of Health and institutional guidelines.
HDL isolation
Plasma was obtained from fasted or nonfasted anesthetized mice. HDL was isolated from plasma by FPLC (21) or density gradient centrifugation. Density gradient centrifugation was done by bringing plasma volume to 1 ml in NaBr solution adjusted to a final density of 1.300 g/ml. On top of this were layered density solutions of 1.21 g/ml (3.2 ml), 1.063 g/ml (3.6 ml), 1.019 g/ml (3.2 ml), and 1.006 g/ml (1 ml), and the samples were spun for 22 h at 178,000 g. The resulting sample was fractionated manually into 20–30 fractions of equal volume. Whole plasma FPLC fractions and density gradient fractions were analyzed for protein (Bio-Rad Laboratories Inc., Hercules, CA), total cholesterol and triglyceride (Roche Diagnostics, Indianapolis, IN), and free cholesterol and phospholipid (Wako Chemicals, Richmond, VA). Fractions containing HDL were then combined and dialyzed/concentrated into PBS using a 100 Kd cut-off Centricon concentrator (Millipore, Billerica, MA) and stored at 4°C.
HDL apolipoprotein levels were analyzed using two methods. First, FPLC HDL fractions were analyzed by SDS–PAGE, and Coomassie-stained bands were quantitated using Fluorchem v2.0 analysis software. For the second method, HDL from 10 animals of each strain was isolated by density centrifugation, and the resulting HDL fractions were combined for each mouse strain. Density gradient centrifugation was performed again on this isolated HDL for further purification. The peak fraction was determined by cholesterol analysis of each gradient fraction, and the apolipoproteins in this fraction were identified and quantitated by proteomics analysis as described below (22). 3H-cholesterol-labeled HDL was generated in nonfasted C57 or FVB mice by oral gavage of 200 µl 3H-cholesterol (Perkin-Elmer, Waltham, MA) in olive oil at a concentration of 10 µCi/ml. After 8 h, mice were terminally bled, and HDL was isolated from plasma by density gradient centrifugation as described above.
Proteomic analysis of the HDL particles
Isolated HDL particles (10 μg protein) were adjusted to a final concentration of 100 μg/ml with digestion buffer (0.1% Rapigest; Waters, 50 mM Tris buffer, pH 8.0). Samples were reduced, alkylated, and digested at 37°C with two aliquots of trypsin (1:50, w/w, trypsin/protein; Promega), first for 2 h and then overnight. Proteolysis was stopped by adding HCl (final concentration, 50 mM). The samples were incubated for 45 min at 37°C to hydrolyze the Rapigest detergent and then clarified by centrifugation. The supernatants were dried under vacuum and resuspended in 5% acetonitrile (0.3% acetic acid) for mass spectrometric analysis. Tryptic digests (2 μg protein) were injected onto a trap column (Paradigm Platinum Peptide Nanotrap, 0.15 × 50 mm; Michrom Bioresources, Inc.), desalted for 5 min with 1% acetonitrile/0.1% formic acid (50 μl/min), eluted onto an analytical reverse-phase column (0.15 × 150 mm, Magic C18AQ, 5 μm, 200 A; Michrom Bioresources, Inc.), and separated at a flow rate of 1 μl/min over 180 min, using a linear gradient of 5% to 35% buffer B (90% acetonitrile, 0.1% formic acid) in buffer A (5% acetonitrile, 0.1% formic acid). Mass spectra were acquired in the positive ion mode using electrospray ionization in a linear ion trap mass spectrometer (LTQ; Thermo Electron Corp., San Jose, CA) with data-dependent acquisition (one MS survey scan followed by MS/MS scans of the eight most abundant peaks in the survey scan). For protein identification, MS/MS spectra were matched against the human International Protein Index database (23), using the SEQUEST (v 2.7) search engine with fixed Cys carbamidomethylation and variable Met oxidation modifications. The mass tolerance for precursor ions was 2.5 kDa, and the SEQUEST default tolerance was accepted for product ions (24). SEQUEST results were further validated with PeptideProphet and ProteinProphet, using an adjusted probability of ≥0.90 for peptides and ≥0.95 for proteins (25, 26). Each charge state of a peptide was considered a unique identification. Relative abundance of the proteins was expressed in terms of spectral counts (number of indentified MSMS spectra for peptides derived from a given protein) (27).
Recombinant apolipoprotein synthesis
Mature mouse ApoA-I cDNA was generated for each mouse strain by performing RT-PCR on total liver RNA for each mouse strain that had been isolated using Qiagen RNeasy mini kit (Qiagen, Valencia, CA). The 5′ primer used for cloning contained a BamHI site and the 3′ primer contained an XhoI site, allowing for subcloning into the pET28c+ vector (Novagen, Gibbstown, NJ). The resulting construct contained a poly-His site followed by T7 site directly upstream of the ApoA-I site and was transformed into the bacterial strain ER2566 (New England Biolabs, Ipswich, MA). Protein expression and purification was carried out as described previously (28). This was done for C57 and FVB ApoA-I mice, yielding recombinant His6-T7-ApoA-I proteins, referred to herein as T7-ApoA-I, differing only in the QV to KA amino acid polymorphism found between C57 and FVB ApoA-I. Purity of the isolated apolipoproteins was assessed by SDS-PAGE with Coomassie stain. ApoA-I was found to be greater than 90% pure for both mouse strains.
Peptide synthesis and purification
The mouse C57 and FVB ApoA-I helix 9/10 peptides (mouse ApoA-I amino acids 206–240) were synthesized manually by solid phase methods using standard t-butoxycarbonyl chemistry: NH2- PALEDLRHSL10MPMLETLKTQ20VQSVIDKASE30TLT –COOH (C57) or NH2- PALEDLRHSL10MPMLETLKTK20AQSVIDKASE30TLT –COOH (FVB). Peptides were prepared using preloaded (phenylacetamido)methyl resins (AnaSpec, Fremont, CA). Amino acids were deprotected with trifluoroacetic acid and coupled for 10 min with 0.5 M 2-(1H-benzotriazole-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate (Peptides International, Louisville, KY). Peptides were cleaved from the resin with hydrogen fluoride in the presence of p-cresol and purified using a reverse-phase, C18 preparative HPLC column (Rainin Dynamax) with an acetonitrile gradient in 0.1% trifluoroacetic acid at 60°C. Peptide purity was greater than 95% by analytical HPLC (Rainin C18 column). The molecular masses of the peptides were verified with electrospray ionization and matrix-assisted laser desorption ionization time-of-flight mass spectrometry. After purification, all peptides were lyophilized and stored at −20°C. The ApoA-I mimetic peptide 4F-KVEPLRA-4F (IHS) contains tandem 4F peptides linked by the 7 amino acid interhelical sequence found between helices 4 and 5 in human apoA-I. Synthesis of the IHS peptide has been described previously (29).
ApoA-I self-association
Recombinant T7-ApoA-I (0.33 mg/ml) in PBS was incubated for 3 h on ice with 3.5 mM bis(sulfo-succinimidyl)suberate (Pierce, Rockford, IL). The cross-linking reaction was quenched with the addition of 6× SDS-PAGE sample buffer. The samples were run on a 10% SDS-PAGE gel alongside noncrosslinked T7-ApoA-I and Biorad All Blue Precision Plus prestained protein standard (Biorad, Hercules, CA).
Dimyristoyl-phosphatidylcholine clearance
Dimyristoyl-phosphatidylcholine powder (DMPC) (Avanti Polar Lipids, Alabaster, AL) was dissolved in chloroform and dried under airflow while vortexing. The DMPC film was resuspended in PBS at 1 mg/ml and vortexed vigorously for 1 min to prepare multilamellar vesicles. DMPC multilamellar vesicles (0.25 mg/ml final concentration) were added to each well of a 96-well plate containing a final concentration range of 0.0–0.2 mg/ml of each recombinant protein or synthetic peptide tested. The decrease in absorbance over time at OD490 was measured every 15 s for 1 h on a BioTek MicroQuant microplate reader with the initial OD490 reading normalized to 1. The fraction cleared over 10 min was plotted against protein concentration and fit to the Michaelis-Menten equation to derive Vmax and Km kinetic values for each protein. To determine the size of the cleared particles, 6× native dye (100 mM Tris pH 6.8, 0.1% [w/v]) bromophenol blue, 6% glycerol final concentration) was added to the cleared samples and run overnight on a 3–36% nondenaturing PAGE gel alongside native molecular weight marker (GE Healthcare, Piscataway, NJ).
ApoA-I binding to HDL
Recombinant T7-ApoA-I (1 µg) was incubated at with varying concentrations of mouse HDL (0–2.5 μg protein) in PBS to measure the affinity of each ApoA-I isoform for HDL. ApoA-I was mixed with HDL and incubated at room temperature. After 30 min at 22°C, samples were loaded onto a 0.7% agarose gel in Tricine-Ca2+-Lactate (pH 8.4) and run for 2 h at 100V. The contents of the gel were then transferred onto Immobilon for Western blot with anti-T7-HRP. Lipid-free and HDL-bound T7-ApoA-I were quantitated from the blot using FluorChem v2.0 Spot Denso software. The total T7-ApoA-I signal was used to calculate the weight (μg) of bound and unbound T7-ApoA-I based off of the known total weight loaded onto each gel.
Surface plasmon resonance
The affinity of recombinant ApoA-I proteins possessing each polymorphism for C57 or FVB HDL was measured using surface plasmon resonance (SPR) on a BIAcore 3000 biosensor. Each recombinant protein was coupled to a CM5 sensor chip via amine coupling to carboxyl moieties on the dextran surface of the chip. The HDL analyte in HBS buffer (10 mM HEPES, 0.15 M NaCl, 50 µM EDTA, pH 7.4) was injected at 25°C at concentrations ranging from 50 nM to 1000 nM at a flow rate of 20 µl/min for 3 min with a dissociation time of 3 min. The kinetics of binding were analyzed based on the response difference between the immobilized T7-ApoA-I flow cell and the control flow cell using BiaCore BIAevaluation software using a global fit of 1:1 binding with drifting baseline.
ApoA-I dissociation from HDL
HDL was combined in PBS with varying concentrations of the 43-amino acid ApoA-I mimetic peptide IHS to a final concentration of 50 µg/ml HDL. Samples were shaken for 1 h at 37°C and immediately loaded in native loading dye onto a 3–36% nondenaturing polyacrylamide gel and run at 200 V for 4 h. The gel was then transferred to PVDF Immobilon (Millipore, Billerica, MA). After blocking with 5% dry milk, the Immobilon was probed with rabbit-anti-mouse ApoA-I antibody, followed by goat-anti-rabbit HRP-linked antibody and ECL Western detection reagent (ThermoScientific, Rockford, IL). HDL-bound and lipid-free ApoA-I bands were quantitated using FluorChem v2.0 Spot Denso software.
J774 cholesterol efflux
3H-cholesterol efflux from J774 cells was performed as described previously (29) with the following modifications. J774 cells were loaded overnight with DMEM + 1% FBS + 25 mg/ml Acetylated LDL + 2 µg/ml Sandoz ACAT inhibitor + 1µCi/ml 3H-cholesterol in the presence of 0.3 mM 8-(4-chlorophenyl-thio)-cAMP (CPT-cAMP). The next day, the cells were washed 3× with PBS, and DMEM containing varying amounts of recombinant T7-ApoA-I, synthetic peptide, or purified HDL was added. After 4 h, the media was removed from the cells, and media and cells were extracted and counted as described previously (29).
ApoA-I binding to J774 cells
J774 cells were incubated 15 h with 0.3 mM 8-(4-chlorophenyl-thio)-cAMP (CPT-cAMP) (Sigma Aldrich, St. Louis, MO). The cells were then washed 2× with PBS and incubated for 2 h at 37°C with varying concentrations of recombinant mouse T7-ApoA-I in serum-free DMEM. The cells were washed 2× with PBS and scraped into PBS+0.5% SDS, and the protein content of each sample measured using a microBCA protein kit (Pierce, Rockford, IL). Equal protein amounts of each sample were mixed with SDS-PAGE loading buffer and run on a 14% SDS-PAGE gel, transferred to Immobilon, and probed with rabbit-anti-mouse ApoA-I antibody. ApoA-I levels were quantitated using FluorChem Spot Denso software. ABCA1 levels from the same samples were run on a 4–12% gradient SDS-PAGE, transferred to Immobilon, and probed with rabbit-anti-mouse ABCA1 antibody (Novus Biologicals, Littleton, CO).
rHDL cholesterol esterification assay
rHDL was generated from C57 or FVB T7-ApoA-I using phosphatidylcholine, T7-ApoA-I, 3H-cholesterol, and nonlabeled cholesterol (molar ratio 200:1.2:1:1). Lipids were vortexed for 5 min in lecithin-cholesterol acyltransferase (LCAT) buffer (10 mM Tris, 0.14 M NaCl, 1 mM EDTA, pH 7.4), and 1.72 mg Na Cholate was added. The solution was revortexed until it was clear. T7-ApoA-I was added and briefly vortexed. Samples were dialyzed against LCAT buffer overnight at 4°C, and the resulting rHDL was used in the cholesterol esterification assay. Varying levels of rHDL for each T7-ApoA-I were mixed with 10 μl freshly isolated serum from an ApoA-I-deficient mouse (C57 background) in a final volume of 200 μl. Samples were incubated for 1 h at 37°C, and the esterification reaction was stopped with the addition of 1 vol isopropanol. Lipids were extracted by the addition of 8 vol hexane, dried, and resuspended in 50 μl hexane:isopropanol (8:1) containing 2 mg/ml each of nonlabeled cholesterol and cholesteryl ester as internal standards. Samples were run on thin layer chromatography Silica Gel IB2-F sheets (J.T. Baker, Phillipsburg, NJ) using hexane:ethyl ether:acetic acid (70:30:1) as a running solvent, and cholesterol and cholesteryl ester spots were cut out into Instafluor scintillation fluid (Perkin Elmer, Waltham, MA) and counted for radioactivity as described previously (29).
Primary hepatocytes
Primary hepatocytes were prepared from female C57 or FVB mice. The liver was perfused first with 100 ml Krebs Ringer + 20 mM glucose + 0.1 mM EGTA, followed by 100 ml Krebs Ringer + 20 mM glucose + 1.4 mM CaCl2 + 25 mg collagenase (Sigma #C5138) + 1 g BSA. The perfused liver was excised from the mouse and gently disrupted in DMEM +10% FBS using a glass pipette. The resulting cell suspension was filtered through a 70 µm cell strainer and washed once in DMEM + 10% FBS by centrifugation 5 min at 50 × g. The pellet was resuspended in 6 ml DMEM + lipoprotein-deficient FBS (LPDS, 2.5 mg/ml protein), and 6 ml 100% Percoll was added to this. This resuspension was centrifuged for 20 min at 50 g to pellet only live hepatocytes. The resulting pellet was washed two more times in DMEM + LPDS. The washed cell pellet was resuspended in Williams E + 2.5 mg/ml LPDS + 1% penicillin/streptomycin/glutamine (MediaTech, Herndon, VA), and cells (>80% viability by trypan blue exclusion) were plated on collagen-coated plates at 5.0 × 105 cells/35 mm dish.
Primary hepatocyte cholesterol/phospholipid efflux
Four hours after isolation, the primary hepatocyte media was changed to fresh Williams E media + 10% FBS + 1% penicillin/streptomycin/glutamine + 1 µCi/ml 3H-cholesterol or 1 µCi/ml 3H-choline (Perkin-Elmer, Waltham, MA). The following day, the cells were washed three times with PBS, and Williams E media + 2.5 mg/ml LPDS was added. Media and cells were harvested at various time points over 24 h. The media was used for lipid extraction (29) or density gradient centrifugation. ApoA-I-associated 3H-cholesterol or phospholipid was immunoprecipitated from media by an overnight rotating incubation at 4°C with rabbit-anti-mouse-ApoA-I antibody followed by an overnight rotating incubation at 4°C with protein-A-Sepharose (Sigma #P3391). The immunoprecipitate was washed three times with PBS and transferred to scintillation vials, and the 3H-cholesterol/phospholipid was dissociated from the immunoprecipitate by the addition of 1 ml isopropanol. After allowing for the isopropanol to evaporate, 3H-cholesterol or 3H-phospholipid in each sample was counted as previously described (29).
In vivo ApoA-I and HDL clearance
In vivo clearance of recombinant T7-ApoA-I was measured in C57 and FVB mice. Recombinant C57 or FVB T7-ApoA-I in PBS was injected retro-orbitally into each mouse, and an initial bleed was taken to measure the starting T7-ApoA-I plasma concentration. Subsequent bleedings were done over the course of 30 h, and the resulting plasma was diluted 1:3 in PBS. Equal volumes of diluted plasma were analyzed by Western blot using an HRP-linked anti-T7 antibody (Invitrogen, Grand Island, NY). The T7 signal for each plasma sample was quantitated using FluorChem v2.0 Spot Denso software. Plasma samples were also run on a 4–36% nondenaturing acrylamide gel and analyzed by anti-T7 Western blot to determine the lipid association of the injected recombinant T7-ApoA-I.
In vivo clearance of C57 and FVB HDL was measured using two methods to determine ApoA-I and HDL cholesterol clearance from the plasma. ApoA-I−/−/ApoE−/− mice on a C57 background were generated previously (30). Nonfasted mice were injected via the retro-orbital sinus with unlabeled or 3H-cholesterol-labeled HDL (generated as described above) derived from WT C57 or FVB mice and bled at various time points over the course of 36 h. At the final time point, the mice were terminally bled, and the plasma was subjected to differential centrifugation to isolate lipoproteins as described above. Plasma samples and lipoprotein fractions were analyzed for ApoA-I by Western blot (nonlabeled HDL injection) or 3H-cholesterol (3H-cholesterol-labeled HDL injection) by scintillation counting.
RESULTS
HDL composition
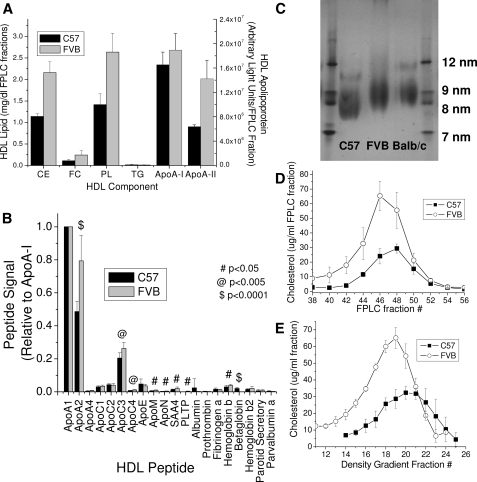
Figure 1 shows a comparison of the properties of the HDL isolated from C57 and FVB mice. C57 and FVB HDL have equivalent ApoA-I levels, whereas ApoA-II levels are ∼2-fold greater in the FVB HDL (Fig. 1A). This observation is similar to the findings of Doolittle et al. (9, 31) when comparing the C57 HDL with Balb/c HDL, which possesses the same ApoA-I and ApoA-II polymorphisms as the FVB mouse. To further characterize the proteins found on the HDL from the two mouse strains, proteomic analysis was done on C57 and FVB HDL that had been purified 2× by density gradient centrifugation. Again, the greatest difference in apoprotein was found for ApoA-II (Fig. 1B). There were significant increments in the minor protein components of the FVB HDL-associated proteins. HDL phospholipid and cholesterol levels were ∼2-fold greater in the FVB HDL as compared with the C57 HDL (Fig. 1A). Most of the difference in cholesterol levels was due to cholesteryl ester. The differences in HDL phospholipid and cholesterol at similar levels of ApoA-I suggested the FVB mouse possessed greater numbers of HDL particles, with fewer ApoA-I molecules per particle versus the C57 mouse, or a larger HDL particle, with more cholesterol and phospholipid per particle. Analysis of HDL from the two mouse strains on a native PAGE (Fig. 1C) or by FPLC (Fig. 1D) shows that the FVB HDL is indeed a larger particle than the C57 and comparable to the Balb/c HDL. This larger FVB HDL is less dense as evidenced by density gradient centrifugation (Fig. 1E).
Fig. 1.
HDL from FVB mice are enriched in cholesterol, phospholipid, and ApoA-II when compared with C57 mice. HDL isolated from each mouse strain was evaluated for (A) major lipid and apoprotein content (by SDS-PAGE), (B) total apoprotein content (by mass spectral quantitation), (C) lipoprotein size by native gel electrophoresis, or (D) lipoprotein size by FPLC fractionation of whole plasma followed by cholesterol analysis, and (E) lipoprotein density by density gradient centrifugation as described in Materials and Methods. Data points show the mean ± SD of triplicate samples (A, D, and E), 10 samples (B), or four samples (C).
ApoA-I properties
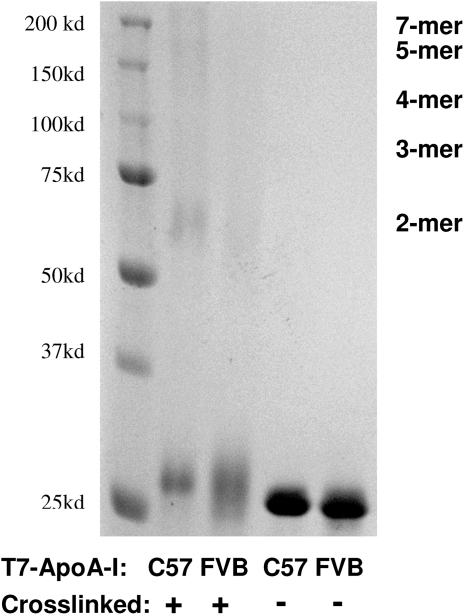
The recombinant C57 and FVB T7-ApoA-I proteins were compared in their degree of self-association. We had noted during the preparation of the recombinant proteins that the C57 T7-ApoA-I would not stay in solution at concentrations greater than 0.5 mg/ml, whereas the FVB T7-ApoA-I was soluble to nearly 1 mg/ml. Upon cross-linking the solubilized proteins to themselves, a significant portion of the C57 protein was found as oligomers, whereas the FVB protein was mainly still monomeric, indicating a greater degree of self-association for the C57-T7ApoA-I (Fig. 2).
Fig. 2.
Recombinant C57 T7-ApoA-I self-associates to a greater degree than FVB T7-ApoA-I. Recombinant T7-ApoA-I (0.33 mg/ml) was cross-linked with BS3 (3.5 mM) for 3 h on ice and run on a 10% SDS-PAGE gel alongside noncrosslinked T7-ApoA-I. Sizes of expected cross-linked multimers are indicated.
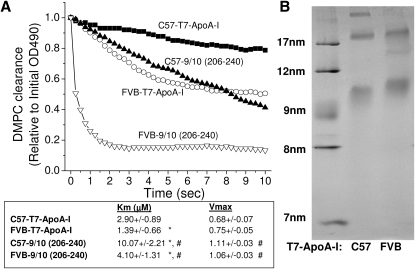
Several methods were used to assess whether the sequence differences between the C57 and FVB ApoA-I altered the lipid-binding characteristics of the proteins. The ability of the recombinant ApoA-I proteins to clear multilamellar vesicles of DMPC from solution was used as a measure of lipid solubilizing capability (Fig. 3). Recombinant FVB T7-ApoA-I cleared micelles of DMPC from solution more rapidly than the C57 T7-ApoA-I. The helix 10 portion of the ApoA-I protein is the site of the two polymorphisms between C57 and FVB ApoA-I. Previous studies have shown that synthetic peptides of human ApoA-I helix 9/10 are better lipid acceptors and stabilizers of ABCA1 than helix 10 alone (13). As such, synthetic peptides of the 9/10 helices of mouse ApoA-I were made possessing the C57 or FVB ApoA-I polymorphisms. When the synthetic peptides were used to clear DMPC, the FVB peptide cleared the solution more rapidly than the C57 peptide. The clearance kinetics of the recombinant ApoA-I and the 9/10 peptides were measured at various concentrations of each, showing the FVB ApoA-I has a significantly lower Km than the C57 protein but a similar Vmax value (Fig. 3A). Similar results were found for the FVB 9/10 peptide when compared with the C57 9/10 peptide. The recombinant ApoA-I proteins had lower Km values than their 9/10 helix peptide counterparts, suggesting an important role of the full-length mouse ApoA-I molecule in lipid solubilization. When the T7-ApoA-I-DMPC cleared solutions were run on a nondenaturing gel, the C57 and FVB T7-ApoA-I proteins were found to form lipoprotein particles that differed in size. Most notably, the FVB-T7-ApoA-I formed an HDL-sized particle larger than the C57, in accordance with what is seen in vivo (Fig. 3B).
Fig. 3.
Recombinant FVB ApoA-I and helix 9/10 synthetic peptide clear DMPC multilamellar vesicles more rapidly than C57 ApoA-I and 9/10 peptide, with FVB-ApoA-I forming larger HDL-sized particles. A: Multilamellar vesicles of DMPC (final concentration, 0.25 mg/ml) were added to 0–0.2 mg/ml C57 or FVB T7-ApoA-I or ApoA-I-9/10 synthetic peptide, and the absorbance at 490 nm was followed over the course of 1 h and normalized to the initial absorbance reading. Representative curves at 0.05 mg/ml are shown (minimum of five samples each). Percent turbidity of DMPC remaining at 10 min was plotted against A-I or 9/10 peptide concentration and fit to Michaelis-Menten kinetics. * P < 0.05 compared with C57. # P < 0.05 compared with corresponding T7-ApoA-I. B: Nondenaturing PAGE gel of 0.08 mg/ml T7-ApoA-I-cleared DMPC vesicles after 1 h clearance.
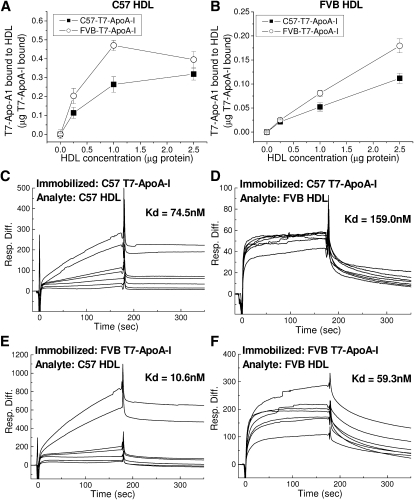
Agarose electrophoretic separation and surface plasmon resonance (SPR) were carried out to analyze the binding affinity of C57 or FVB HDL particles for recombinant C57 or FVB T7-ApoA-I (Fig. 4). Using agarose gel separation, varying levels of HDL were coincubated with recombinant T7-ApoA-I and run on an agarose gel to distinguish lipid-free ApoA-I from that which had bound to HDL during the course of incubation. FVB T7-ApoA-I was found to bind C57 and FVB HDL better than C57 T7-ApoA-I, with FVB and C57 T7-ApoA-1 proteins having a higher affinity for C57 HDL than for FVB HDL (Fig. 4A, B). SPR was also used to determine the binding kinetics of HDL to each recombinant protein. Fig. 4CndashF shows representative comparisons of C57 or FVB HDL binding to each T7-ApoA-I at HDL concentrations from 50 to 1,000 nM. The SPR binding data agree with the results obtained using agarose gel separation, with C57 and FVB HDL showing a higher total binding to FVB-T7-ApoA-I. The Kd values show that the FVB T7-ApoA-I has a higher affinity for HDL than the C57 T7-ApoA-I. Again, as with the agarose gel separation method, both ApoA-I isoforms display a greater affinity for the smaller C57 HDL than for the FVB HDL.
Fig. 4.
Recombinant C57 and FVB T7-ApoA-I bind C57 and FVB HDL with varying affinities. A, B:) Association of soluble T7-ApoA-I with HDL. Recombinant T7-ApoA-I (1 μg) was incubated with varying amounts of (A) C57 or (B) FVB HDL at 22°C for 30 min. Free and bound T7-ApoA-I were separated on a 0.7% agarose gel, transferred to Immobilon, and probed by anti-T7 Western blot for quantitation. Data points show the mean ± SD of triplicate samples. C–F: Kinetics of binding of C57 and FVB HDL to recombinant T7-ApoA-I as measured by SPR. A control blank (flow cell 1), C57 T7-ApoA-I (flow cell 2), or FVB T7-ApoA-I (flow cell 3) were bound to the CM5 sensor chip surface by amine coupling to carboxyl moieties on the dextran surface of the chip and 50, 100, 200, 300, 400, 800, and 1000 nM concentrations of HDL were flowed over the chip surface for a 3 min association time followed by a 3 min dissociation time. Representative sensograms at all HDL concentrations are shown here as the response difference between the immobilized T7-ApoA-I flow cell and the control flow cell. Binding kinetics were determined using a global fit based upon 1:1 binding with drifting baseline using the concentrations of analyte as described in Materials and Methods to arrive at the given Kd values.
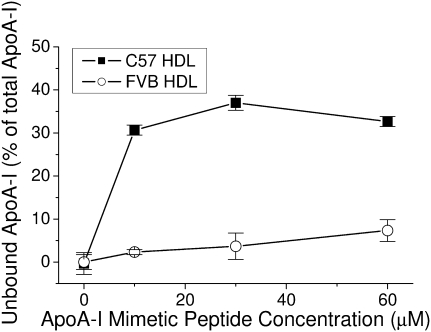
There is the possibility that the recombinant protein might interact with the HDL particle in a manner different from that of the endogenous protein. As such, the affinity of the endogenous protein for its own HDL particle was analyzed using an ApoA-I mimetic peptide (IHS) developed in our laboratory, which is able to dissociate ApoA-I from HDL in a concentration-dependent manner (29). In agreement with the results of the experiments using recombinant T7-ApoA-I, the endogenous FVB ApoA-I remained bound to its HDL at higher concentrations of the IHS peptide than C57 ApoA-I, which was easily dissociated from its HDL (Fig. 5).
Fig. 5.
Endogenous C57 ApoA-I is more readily dissociated from its HDL than is FVB ApoA-I. A mimetic peptide known to dissociate endogenous ApoA-I from its HDL was incubated at varying concentrations with C57 or FVB HDL as described in Materials and Methods. The levels of dissociation of endogenous ApoA-I were compared between the two mouse HDLs. Data points show the mean ± SD of triplicate samples.
Influence of polymorphism on cholesterol efflux
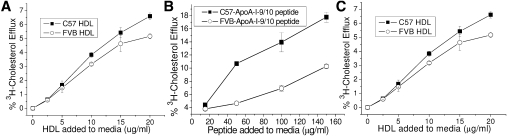
Because the polymorphisms in the ApoA-I peptides resulted in altered lipid binding, the ability of the proteins to efflux cholesterol from J774 macrophages was tested. Despite the greater lipid binding of the FVB T7-ApoA-I, the C57 T7-ApoA-I showed a slightly greater efflux capability when compared with FVB (Fig. 6A). The C57 9/10 peptide displayed a much greater ability to efflux cholesterol from J774 macrophages than the FVB 9/10 peptide (Fig. 6B). Additionally, the whole ApoA-I protein is a substantially more effective efflux acceptor than its 9/10 peptide counterpart, although the 9/10 peptide Vmax is probably higher. Holo-HDL isolated from each mouse strain was used in a similar cholesterol efflux study to determine whether the whole particles differ in their cholesterol efflux capabilities. The cholesterol efflux from J774 cells to HDL derived from each mouse strain is slightly greater for the C57 HDL than the FVB, similar to the results for the lipid-free T7-ApoA-I (Fig. 6C).
Fig. 6.
The differences in C57 or FVB apolipoprotein-mediated efflux of cholesterol from J774 macrophages are dependent on the type of acceptor. J774 macrophages were loaded with acetylated-LDL and labeled with 3H-cholesterol in the presence of an ACAT inhibitor and CPT-cAMP as described in Materials and Methods. Efflux of 3H-cholesterol to varying concentrations of C57 or FVB (A) recombinant T7-ApoA-I, (B) synthetic ApoA-I helix 9/10 peptide, or (C) HDL were measured. Data points show the mean ± SD of triplicate samples.
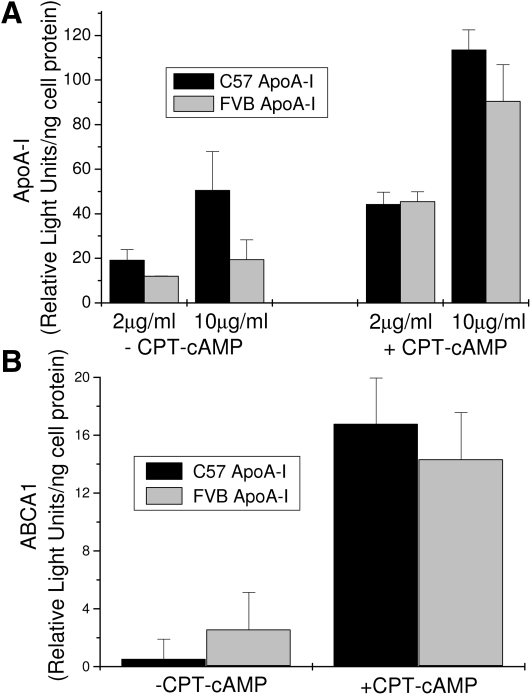
The binding of T7-ApoA-I to J774 macrophages was measured with and without the up-regulation of ABCA1 expression by CPT-cAMP (Fig. 7A). The binding of ApoA-I to the macrophages increases when ABCA-I expression is increased, as expected. However, although there is some indication that the C57 form of ApoA-I has slightly better binding to the macrophages, the differences are not significant (P > 0.05). These differences reflect the minor differences in cholesterol efflux seen for the ApoA-I proteins in Fig. 6. ApoA-I is known to stabilize the cellular ABCA1 levels by preventing degradation of ABCA1 (32, 33). The differences in the ApoA-I variants did not significantly (P > 0.05) affect the cellular levels of ABCA1, whereas the preincubation of the macrophages with CPT-cAMP dramatically increased ABCA1 protein levels as expected (Fig. 7B).
Fig. 7.
T7-ApoA-I binding to J774 macrophages with and without CPT-cAMP induction of ABCA1 expression. J774 macrophages were incubated 15 h with CPT-cAMP to induce ABCA1 expression. C57 or FVB recombinant T7-ApoA-I was then incubated with cells for 2 h and removed by PBS wash. A: T7-ApoA-I binding was measured by western blot of the T7-ApoA-I remaining associated with the cell protein as described in Materials and Methods. B: ABCA1 levels postincubation with 2 μg/ml C57 or FVB recombinant T7-ApoA-I as measured by ABCA1 Western blot of cell protein as described in Materials and Methods. Data points show the mean ± SD of triplicate samples.
Lecithin cholesterol acyltransferase activation
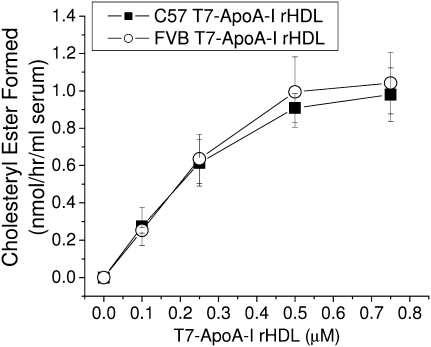
The bulk of the cholesterol found in the HDL of the two mice was found in its esterified form (Fig. 1A). ApoA-I is known to activate LCAT, which converts free cholesterol to cholesteryl ester (34). When rHDL made from the two recombinant T7-ApoA-I proteins were tested for LCAT activation, C57 and FVB T7-ApoA-I activated LCAT to similar extents (Fig. 8).
Fig. 8.
Recombinant HDL (rHDL) made from C57 and FVB T7-ApoA-I yield equivalent cholesterol esterification rates. rHDL particles made from phosphatidylcholine, recombinant T7-ApoA-I, 3H-cholesterol, and nonlabeled cholesterol (molar ratio, 200:1.2:1:1) were used to measure cholesterol esterification rates using serum from ApoA-I-deficient mouse as a source of LCAT during a 1 h incubation as described in Materials and Methods. Data points show the mean ± SD of four samples.
Apoprotein A-1 metabolism
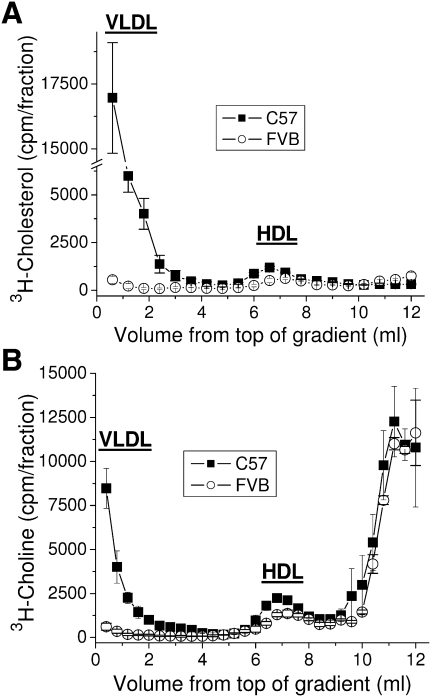
The liver is the main site of ApoA-I production as well as nascent HDL formation (35). Primary hepatocytes were prepared from both mouse strains, and the endogenous ApoA-I and HDL cholesterol secretion levels were analyzed. No difference was found for ApoA-I secretion levels for all time points tested (data not shown). The difference in HDL cholesterol secretion from the FVB hepatocytes favors the C57 HDL when compared with the C57 hepatocytes over 24 h, although the difference is small (Fig. 9A). Similar results were seen for shorter time points (data not shown). Interestingly, the C57 hepatocytes secrete the bulk of their cholesterol and phospholipid in association with particles in the density range of VLDL. The secretion of HDL phosphatidylcholine follows the same pattern as the HDL cholesterol (Fig. 9B). Thus, the altered lipid binding of the polymorphisms in ApoA-I does not appear to affect the secretion of cholesterol or phospholipids from hepatocytes to newly formed HDL particles. Additionally, the density of the HDL particles formed from these hepatocytes is slightly less for the C57 hepatocytes, unlike what is seen in vivo.
Fig. 9.
HDL secreted from FVB hepatocytes are no more dense and do not possess more cholesterol or phospholipids than HDL secreted from C57 hepatocytes. Primary hepatocytes were isolated from mature female C57 or FVB mice as described in Materials and Methods. After loading with (A) 3H-cholesterol or (B) 3H-choline cells were washed and incubated for 24 h. Media was harvested and separated into lipoprotein fractions by density gradient ultracentrifugation. Data points show the mean ± SD of triplicate samples.
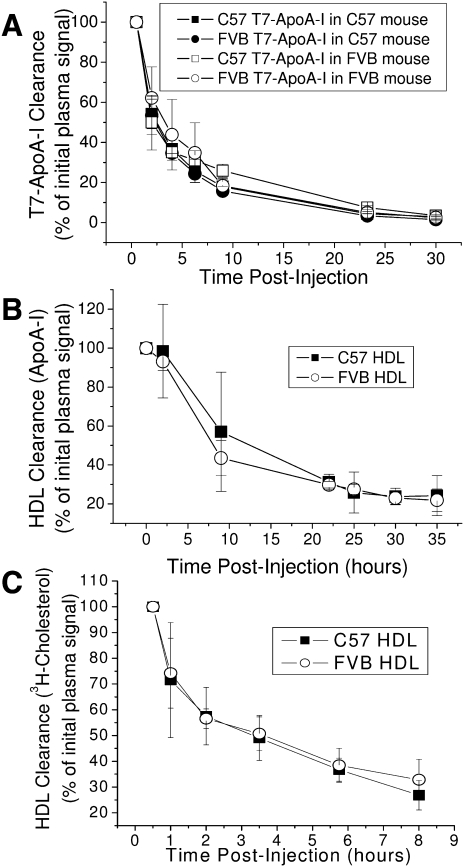
The data from cultured cells indicate the altered lipid binding resulting from the polymorphisms in ApoA-I does not significantly affect the secretion of cholesterol from hepatocytes to newly formed HDL particles or the efflux of cholesterol from macrophages to mature HDL. Given the different rates of dissociation from HDL observed for the two ApoA-I isoforms, it is plausible that the mature C57 HDL is catabolized more rapidly than the FVB HDL or that the altered lipid affinity of the apolipoproteins results in greater loss of lipid in the plasma. Recombinant C57 and FVB T7-ApoA-I were separately injected into WT C57 and FVB mice, and the disappearance of the T7 tag was followed over time (Fig. 10A). The clearance of recombinant protein from the plasma was the same regardless of ApoA-I type or strain background. This occurred even though the recombinant T7-ApoA-I rapidly associated with the endogenous HDL, as evidenced by its appearance in the HDL-sized portion of a nondenaturing gel, even for the early time points (data not shown). The clearance rates of the recombinant proteins were also the same after injection into ApoA-I−/−/ApoE−/− C57 mice (data not shown), which possess no detectable HDL (30). To determine if clearance of the holo-C57 or FVB HDL particle was the same, HDL purified from each mouse strain was injected into ApoA-I−/−/ApoE−/− C57 mice (Fig. 10B). The disappearance of ApoA-I over time in the plasma of these mice was equivalent despite the polymorphism differences in the associated apolipoproteins. The clearance rates were also similar when lipid-free recombinant ApoA-I was injected (data not shown). When 3H-cholesterol-labeled HDL from each mouse strain was injected into these ApoA-I−/−/ApoE−/− mice, the rate of disappearance of labeled cholesterol in the plasma was the same between the C57- and FVB-derived HDL (Fig. 10C).
Fig. 10.
C57 and FVB HDL ApoA-I and HDL cholesterol are cleared from the plasma at similar rates. A: C57 or FVB WT mice were injected retro-orbitally with recombinant C57 or FVB T7-ApoA-I and bled at various time points. Plasma from each time point was analyzed for T7 tag by Western blot. B and C: ApoA-I−/−/ApoE−/− mice on the C57 background were injected with (B) unlabeled or (C) 3H-cholesterol-labeled HDL obtained from C57 or FVB mice. Mice were bled at various time points and the plasma from each time point was analyzed for (B) ApoA-I levels by antimouse ApoA-I Western blot, or (C) 3H-cholesterol radioactive counts/min. Data points show the mean ± SD of triplicate samples.
DISCUSSION
Various inbred mouse strains demonstrate different susceptibilities to the development of atherosclerosis when fed an atherogenic diet or when genetically modified by removing ApoE or LDL receptor. These mice also have different plasma lipoprotein levels. Although the HDL of C57BL6 mice has been extensively studied, in part because this is an atherosclerosis susceptible mouse strain, the HDL of the FVB mouse has not been as widely examined. C57 and FVB mice differ in the level and size of HDL, with FVB mice having higher plasma levels and larger-sized HDL. Even when normalized for ApoA-I content, the FVB HDL has more cholesterol, cholesteryl ester, and phospholipid than the C57 HDL; as a result, FVB HDL is larger and less dense than C57 HDL. We here demonstrate that the two amino acid sequence variations in the C-terminal helix of C57 and FVB ApoA-I, the major HDL apolipoprotein, cause this protein to display several different properties, mostly affecting lipid-binding affinities.
We here report several approaches characterizing the differences in interaction of the two ApoA-I mouse proteins with lipid and with HDL. Each of these approaches yields results that are in agreement with one another, all indicating that the FVB ApoA-I variant is greater in its lipid binding and HDL association/affinity than the C57 ApoA-I variant. The data are especially intriguing given the work of Tanaka et al. (18), in which the lipid-binding activities of the N-terminal (residues 1–186) and C-terminal (residues 187–240) domains of lipid-free mouse (C57) ApoA-I are contrasted with those of human ApoA-I. In their study, most of the lipid binding of mouse A-I was found in the N-terminal region, whereas the C-terminal domain alone revealed almost no lipid binding. On the other hand, a hybrid molecule containing the human N-terminal fragment and the murine C-terminal domain exhibited improved lipid binding over the human N-terminal domain alone, suggesting that the murine C-terminal domain did afford an increment in lipid binding. They suggested that the initial lipid binding region of mouse A-I is most likely not in that C-terminal domain. It is worth noting that these are recombinant proteins, and the C-terminal domain includes helix 8 as well as 9 and 10. It is known that there are functional interactions among individual helices (13). Our results of lipid binding of the helix 9/10 peptides indicate a clear ability of these peptides to bind lipid and clear DMPC vesicles in the case of C57 and FVB 9/10 peptide, with FVB-9/10 being nearly as effective as the C57 full-length ApoA-I (Fig. 3, kinetic data). Natarajan and colleagues have observed lipid binding and cholesterol efflux, with synthetic 9/10 helices corresponding to the human sequences (13). No previous studies of the murine 9/10 helices have been done.
The mechanisms by which the polymorphisms confer greater lipid binding and HDL association on the FVB ApoA-I are not clear. The finding that the C57 apoprotein is more prone to self-association may indicate a conformational hindrance to the initial binding/association to lipid. However, the very nature of the protein when it is already in association with HDL appears to be altered, as evidenced by the ease with which the C57 protein is dissociated from its own HDL, whereas the FVB protein is difficult to dissociate from its HDL (Fig. 5). This may in part explain our HDL binding data done by SPR and the agarose separation assay, in which both variants of ApoA-I show a greater affinity for C57 HDL than for FVB. Given the limited surface area on an HDL particle, the ease with which the endogenous ApoA-I is dissociated is a key factor in the amount of recombinant A-I that can associate with the particle. In the case of the FVB HDL in which the endogenous A-I protein is tightly associated, the recombinant protein is less able to compete with it for binding to the HDL.
The functional properties of the two ApoA-I proteins that were assessed here did not reflect the differences in lipid association between the two proteins. First, the two ApoA-I proteins are not very different in promoting cholesterol efflux from cAMP-treated J774 cells, with C57 ApoA-I and HDL slightly more effective than their FVB counterparts. The fact that the efflux using the 9/10 peptides greatly accentuated this difference in favor of the C57 peptide further indicates that the increased lipid binding capabilities of the FVB ApoA-I does not result in an increase in ABCA1-mediated lipid efflux to this protein. There are numerous examples in the literature in which alterations in helix 10 of the human ApoA-I molecule results in decreased ABCA1-mediated cholesterol efflux (14, 15, 20, 36, 37). Macrophages and other cells that express ABCA1 have been found to possess two distinct ApoA-I binding sites: a low-capacity binding site on the ABCA1 protein itself, which interacts with ApoA-I at several locations on the ApoA-I molecule, and a high-capacity binding site within the membrane lipid domain generated by ABCA1 (37). The C-terminal helix 10 of human ApoA-I is largely responsible for the interaction of the protein with these lipid domains, and there is a positive correlation between the ability of different ApoA-I molecules to solubilize DMPC vesicles and their ability to promote ABCA1-mediated cholesterol efflux (37). The fact that this is not occurring in our studies is an interesting finding. Kono et al. (15) found that the human ApoA-I Nichinan E235 deletion mutant, which solubilizes DMPC vesicles less effectively than wild-type human ApoA-I, did not differ from the wild-type in its ability to promote cholesterol efflux from J774 macrophages. However, there was a significant difference in efflux from BHK overexpressing human ABCA1. They suggested that the lack of effect in the J774 cells was due to the interaction of human ApoA-I with mouse ABCA1. In our case, both ApoA-I and ABCA1 are from mouse. Whether this means the sensitivity of mouse ABCA1 cholesterol efflux activity to helix 10 variations is overall less than that of human ABCA1 remains unknown but may reveal important clues to the mechanisms of ABCA1 function. That the binding of C57 and FVB ApoA-I to J774 differed (albeit not significantly) in favor of the C57 ApoA-I suggests the affinity of ApoA-I for DMPC or HDL does not in these assays necessarily reflect its cellular binding affinity. We expected the FVB ApoA-I protein to show greater affinity for the high-capacity lipid binding site generated by ABCA1, but this is not evident from our results, suggesting the ABCA1-mediated lipid binding of ApoA-I is not directly correlated with the general lipid affinity of the ApoA-I protein variant.
The secretion of HDL cholesterol and phospholipid from primary hepatocytes is slightly greater for the C57 hepatocytes than for FVB hepatocytes. These results are contrary to what one would expect if nascent HDL formation was driving the differences in plasma HDL cholesterol and phospholipid found in these two strains of mice. However, these results do agree with the J774 efflux experiments, particularly the efflux to the 9/10 peptide, which is more effective when the C57 variant is used. This effect is apparently not significant in vivo in terms of regulation of the HDL phospholipid and cholesterol levels found in the two mice. Because the formation of nascent HDL by hepatocyte is ABCA1 dependent, these results with cells from the two strains seem to be in accord with the above results on the binding of ApoA-I to ABCA1 on J774 cells (i.e., that there is little distinction between the two native ApoA-I proteins). The binding to ABCA1 in J774 cells was performed with recombinant ApoA-I containing the T7 tag, whereas the hepatocyte production of HDL probably involving ABCA1 interaction is a reflection of the property of endogenous ApoA-I. In other words, with respect to interaction with ABCA1, there does not appear to be a difference between the recombinant T7-tagged protein and the natural apoprotein secreted from hepatocytes
Given the greater affinity of the FVB ApoA-I for HDL, we expected this difference to translate into a lower rate of catabolism of the FVB HDL because the ApoA-I protein should be more rapidly catabolized in vivo once dissociated from the HDL lipid core. This was not seen in vivo as the rates of catabolism of the two ApoA-I proteins and HDLs were similar for protein and cholesterol.
Taken together, these data indicate that, despite the variations in structure and physical properties of the two apolipoproteins found in HDL, neither the formation of a more cholesterol-rich nascent HDL particle in the FVB mouse nor the level of cholesterol efflux from peripheral cells such as macrophages accounts for the differences in level and size of the two HDL particles found at steady state in the plasma. Additionally, there appear to be no differences in the catabolism of HDL from the two mice. This underlines the complexity of HDL structure and function and suggests a complex protein-protein and/or protein-lipid interaction among the components of HDL. The manner in which the structural and physical differences observed between the two ApoA-I variants in turn affect their in vivo ApoA-I/HDL function remains to be determined. That these polymorphisms are likely functionally relevant is highlighted not only by their differences in self-association and lipid binding but also by their conservation among inbred mouse strains. To our knowledge, of the more than 40 strains examined, the Q225K, V226A mutations are the only mutations found, and they are roughly equally distributed among the strains (9, 38). Additionally, these mutations are uniquely associated, with no strains examined possessing solely Q225K or solely V226A.
One potential role may be in the interaction of these ApoA-I variations with polymorphic variants of the other major apolipoprotein found on HDL, ApoA-II. Between the C57 and FVB strains, there are three major polymorphisms in ApoA-II: E18D, V24M, and V36A (C57 to FVB). ApoA-I levels are similar between the FVB and C57 mice, but ApoA-II levels are several-fold higher in the FVB mouse (31) (Fig. 1). There is strong evidence in the literature associating ApoA-II levels with HDL particle size and cholesterol levels (8, 39–41). However, the way in which the ApoA-I and ApoA-II polymorphisms interact may be important as well. The presence of ApoA-II on recombinant HDL has been recently shown to alter the conformation of ApoA-I on the HDL particle (42). Although some data suggest that ApoA-II prevents dissociation of ApoA-I from HDL, there is evidence showing that the addition of ApoA-II displaces ApoA-I from HDL (43, 44). In this case, it is therefore possible that the greater affinity of FVB ApoA-I for HDL may allow the greater levels of ApoA-II to exist on the HDL particle without resulting in dissociation of the ApoA-I protein. That the FVB ApoA-I can alone form a larger recombinant HDL particle (Fig. 3B) may be significant in determining the levels to which other apoproteins, such as ApoA-II, can interact with the HDL particle. The interaction of the two major HDL apoproteins in modulating HDL composition and size merits further study. The role of the polymorphisms in the interactions of the two apoproteins may be important in HDL formation and stability. Additionally, Cavigiolio et al. (45) have shown that the exchange of ApoA-I between lipid-free and lipid-bound states may provide a means by which dysfunctional HDL are generated, further emphasizing the significance of ApoA-I affinity for HDL.
Another potential role for the increased HDL affinity of the FVB mouse may be during conditions of acute inflammation. During the acute phase response, levels of serum amyloid A increase dramatically and are found in the plasma associated with HDL-sized lipoproteins, whereas ApoA-I may dissociate from HDL and be catabolized, reducing plasma levels of ApoA-I HDL (46). Suzuki et al. (47) have recently shown that HDL is protective in mice infected with Salmonella typhimurium, preventing increases in plasma levels of interferon-β. The greater HDL affinity of one ApoA-I protein over another may play a role in the response to an inflammatory challenge. This is especially interesting given the finding that when macrophages from various inbred mouse strains were tested for susceptibility to bacillus anthracis lethal factor, four of the five mice known to possess the C57 ApoA-I/ApoA-II variants were resistant, whereas all of those known to possess the FVB ApoA-I/ApoA-II variants were susceptible (48). This finding merits further investigation.
In conclusion, we have characterized variants of ApoA-I that are found in inbred mouse strains that differ in HDL size and cholesterol. The variants are altered in the initial lipid binding domain of the apolipoprotein. These two amino acid variations alter the DMPC clearance and HDL binding capability of the apolipoprotein with no effect on the ability of the protein to promote cellular cholesterol efflux, HDL synthesis, or HDL catabolism. The physiological significance of the polymorphisms remains unknown but points to complex interactions between this and other apoproteins as well as with the lipids with which they are associated.
Acknowledgments
The authors thank Dr. Geoffrey Wool for providing the ApoA-I mimetic peptide and Dr. Elena Solomaha for assistance in conducting the SPR experiments.
Footnotes
Abbreviations:
- acetyl-CoA
- cholesterol-acyltransferase
- Apo
- apolipoprotein
- CPT-cAMP
- 8-(4-chlorophenyl-thio)-cAMP
- DMPC
- dimyristoyl-phosphatidylcholine
- IPTG
- isopropyl-β-1-D-thiogalactopyranoside
- LCAT
- lecithin-cholesterol acyltransferase
- LPDS
- lipoprotein-deficient serum
- SPR
- surface plasmon resonance
This work was supported by NHLBI (HL68661), the LeDucq Foundation, a National Institutes of Health Cardiovascular Training Grant, and The American Heart Association Postdoctoral Fellowship (T.J.S.). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health or other granting agencies.
REFERENCES
- 1.Thom T., Haase N., Rosamond W., Howard V. J., Rumsfeld J., Manolio T., Zheng Z-J., Flegal K., O'Donnell C., Kittner S., et al. 2006. Heart disease and stroke statistics–2006 update: a report from the American Heart Association Statistics Committee and Stroke Statistics Subcommittee. Circulation. 113: e85–e151 [DOI] [PubMed] [Google Scholar]
- 2.Perper J. A., Kuller L. H., Cooper M. 1975. Arteriosclerosis of coronary arteries in sudden, unexpected deaths. Circulation. 52: III27–III33 [PubMed] [Google Scholar]
- 3.Boden W. E. 2000. High-density lipoprotein cholesterol as an independent risk factor in cardiovascular disease: assessing the data from Framingham to the Veterans Affairs High Density Lipoprotein Intervention Trial. Am. J. Cardiol. 86: 19L–22L [DOI] [PubMed] [Google Scholar]
- 4.Paigen B., Morrow A., Brandon C., Mitchell D., Holmes P. 1985. Variation in susceptibility to atherosclerosis among inbred strains of mice. Atherosclerosis. 57: 65–73 [DOI] [PubMed] [Google Scholar]
- 5.Dansky H. M., Charlton S. A., Sikes J. L., Heath S. C., Simantov R., Levin L. F., Shu P., Moore K. J., Breslow J. L., Smith J. D. 1999. Genetic background determines the extent of atherosclerosis in ApoE-deficient mice. Arterioscler. Thromb. Vasc. Biol. 19: 1960–1968 [DOI] [PubMed] [Google Scholar]
- 6.Teupser D., Persky A. D., Breslow J. L. 2003. Induction of atherosclerosis by low-fat, semisynthetic diets in LDL receptor-deficient C57BL/6J and FVB/NJ mice: comparison of lesions of the aortic root, brachiocephalic artery, and whole aorta (en face measurement). Arterioscler. Thromb. Vasc. Biol. 23: 1907–1913 [DOI] [PubMed] [Google Scholar]
- 7.Svenson K. L., Von Smith R., Magnani P. A., Suetin H. R., Paigen B., Naggert J. K., Li R., Churchill G. A., Peters L. L. 2007. Multiple trait measurements in 43 inbred mouse strains capture the phenotypic diversity characteristic of human populations. J. Appl. Physiol. 102: 2369–2378 [DOI] [PubMed] [Google Scholar]
- 8.Schultz J. R., Verstuyft J. G., Gong E. L., Nichols A. V., Rubin E. M. 1993. Protein composition determines the anti-atherogenic properties of HDL in transgenic mice. Nature. 365: 762–764 [DOI] [PubMed] [Google Scholar]
- 9.Puppione D. L., Yam L. M., Bassilian S., Souda P., Castellani L. W., Schumaker V. N., Whitelegge J. P. 2006. Mass spectral analysis of the apolipoproteins on mouse high density lipoproteins. Detection of post-translational modifications. Biochim. Biophys. Acta. 1764: 1363–1371 [DOI] [PubMed] [Google Scholar]
- 10.Strausberg R. L., Feingold E. A., Grouse L. H., Derge J. G., Klausner R. D., Collins F. S., Wagner L., Shenmen C. M., Schuler G. D., Altschul S. F., et al. 2002. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc. Natl. Acad. Sci. USA. 99: 16899–16903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Plump A. S., Azrolan N., Odaka H., Wu L., Jiang X., Tall A., Eisenberg S., Breslow J. L. 1997. ApoA-I knockout mice: characterization of HDL metabolism in homozygotes and identification of a post-RNA mechanism of apoA-I up-regulation in heterozygotes. J. Lipid Res. 38: 1033–1047 [PubMed] [Google Scholar]
- 12.Zannis V. I., Chroni A., Krieger M. 2006. Role of apoA-I, ABCA1, LCAT, and SR-BI in the biogenesis of HDL. J. Mol. Med. 84: 276–294 [DOI] [PubMed] [Google Scholar]
- 13.Natarajan P., Forte T. M., Chu B., Phillips M. C., Oram J. F., Bielicki J. K. 2004. Identification of an apolipoprotein A-I structural element that mediates cellular cholesterol efflux and stabilizes ATP binding cassette transporter A1. J. Biol. Chem. 279: 24044–24052 [DOI] [PubMed] [Google Scholar]
- 14.Panagotopulos S. E., Witting S. R., Horace E. M., Hui D. Y., Maiorano J. N., Davidson W. S. 2002. The role of apolipoprotein A-I helix 10 in apolipoprotein-mediated cholesterol efflux via the ATP-binding cassette transporter ABCA1. J. Biol. Chem. 277: 39477–39484 [DOI] [PubMed] [Google Scholar]
- 15.Kono M., Tanaka T., Tanaka M., Vedhachalam C., Chetty P. S., Nguyen D., Dhanasekaran P., Lund-Katz S., Phillips M. C., Saito H. 2010. Disruption of the C-terminal helix by single amino acid deletion is directly responsible for impaired cholesterol efflux ability of apolipoprotein A-I Nichinan. J. Lipid Res. 51: 809–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saito H., Dhanasekaran P., Nguyen D., Holvoet P., Lund-Katz S., Phillips M. C. 2003. Domain structure and lipid interaction in human apolipoproteins A-I and E, a general model. J. Biol. Chem. 278: 23227–23232 [DOI] [PubMed] [Google Scholar]
- 17.Ajees A. A., Anantharamaiah G. M., Mishra V. K., Hussain M. M., Murthy H. M. K. 2006. Crystal structure of human apolipoprotein A-I: insights into its protective effect against cardiovascular diseases. Proc. Natl. Acad. Sci. USA. 103: 2126–2131 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Tanaka M., Koyama M., Dhanasekaran P., Nguyen D., Nickel M., Lund-Katz S., Saito H., Phillips M. C. 2008. Influence of tertiary structure domain properties on the functionality of apolipoprotein A-I. Biochemistry. 47: 2172–2180 [DOI] [PubMed] [Google Scholar]
- 19.Koyama M., Tanaka M., Dhanasekaran P., Lund-Katz S., Phillips M. C., Saito H. 2009. Interaction between the N- and C-terminal domains modulates the stability and lipid binding of apolipoprotein A-I. Biochemistry. 48: 2529–2537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vedhachalam C., Chetty P. S., Nickel M., Dhanasekaran P., Lund-Katz S., Rothblat G. H., Phillips M. C. 2010. Influence of apolipoprotein (Apo) A-I structure on nascent high density lipoprotein (HDL) particle size distribution. J. Biol. Chem. 285: 31965–31973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reardon C. A., Blachowicz L., White T., Cabana V., Wang Y., Lukens J., Bluestone J., Getz G. S. 2001. Effect of immune deficiency on lipoproteins and atherosclerosis in male apolipoprotein E-deficient mice. Arterioscler. Thromb. Vasc. Biol. 21: 1011–1016 [DOI] [PubMed] [Google Scholar]
- 22.Vaisar T., Pennathur S., Green P. S., Gharib S. A., Hoofnagle A. N., Cheung M. C., Byun J., Vuletic S., Kassim S., Singh P., et al. 2007. Shotgun proteomics implicates protease inhibition and complement activation in the antiinflammatory properties of HDL. J. Clin. Invest. 117: 746–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kersey P. J., Duarte J., Williams A., Karavidopoulou Y., Birney E., Apweiler R. 2004. The International Protein Index: an integrated database for proteomics experiments. Proteomics. 4: 1985–1988 [DOI] [PubMed] [Google Scholar]
- 24.Yates J. R., Eng J. K., McCormack A. L., Schieltz D. 1995. Method to correlate tandem mass spectra of modified peptides to amino acid sequences in the protein database. Anal. Chem. 67: 1426–1436 [DOI] [PubMed] [Google Scholar]
- 25.Keller A., Nesvizhskii A. I., Kolker E., Aebersold R. 2002. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74: 5383–5392 [DOI] [PubMed] [Google Scholar]
- 26.Nesvizhskii A. I., Keller A., Kolker E., Aebersold R. 2003. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75: 4646–4658 [DOI] [PubMed] [Google Scholar]
- 27.Liu H., Sadygov R. G., Yates J. R. 2004. A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal. Chem. 76: 4193–4201 [DOI] [PubMed] [Google Scholar]
- 28.Carnemolla R., Ren X., Biswas T. K., Meredith S. C., Reardon C. A., Wang J., Getz G. S. 2008. The specific amino acid sequence between helices 7 and 8 influences the binding specificity of human apolipoprotein A-I for high density lipoprotein (HDL) subclasses: a potential for HDL preferential generation. J. Biol. Chem. 283: 15779–15788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wool G. D., Reardon C. A., Getz G. S. 2008. Apolipoprotein A-I mimetic peptide helix number and helix linker influence potentially anti-atherogenic properties. J. Lipid Res. 49: 1268–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cabana V. G., Feng N., Reardon C. A., Lukens J., Webb N. R., de Beer F. C., Getz G. S. 2004. Influence of apoA-I and apoE on the formation of serum amyloid A-containing lipoproteins in vivo and in vitro. J. Lipid Res. 45: 317–325 [DOI] [PubMed] [Google Scholar]
- 31.Doolittle M. H., LeBoeuf R. C., Warden C. H., Bee L. M., Lusis A. J. 1990. A polymorphism affecting apolipoprotein A-II translational efficiency determines high density lipoprotein size and composition. J. Biol. Chem. 265: 16380–16388 [PubMed] [Google Scholar]
- 32.Martinez L. O., Agerholm-Larsen B., Wang N., Chen W., Tall A. R. 2003. Phosphorylation of a pest sequence in ABCA1 promotes calpain degradation and is reversed by ApoA-I. J. Biol. Chem. 278: 37368–37374 [DOI] [PubMed] [Google Scholar]
- 33.Wang N., Chen W., Linsel-Nitschke P., Martinez L. O., Agerholm-Larsen B., Silver D. L., Tall A. R. 2003. A PEST sequence in ABCA1 regulates degradation by calpain protease and stabilization of ABCA1 by apoA-I. J. Clin. Invest. 111: 99–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parks J. S., Li H., Gebre A. K., Smith T. L., Maeda N. 1995. Effect of apolipoprotein A-I deficiency on lecithin:cholesterol acyltransferase activation in mouse plasma. J. Lipid Res. 36: 349–355 [PubMed] [Google Scholar]
- 35.Eggerman T. L., Hoeg J. M., Meng M. S., Tombragel A., Bojanovski D., Brewer H. B. 1991. Differential tissue-specific expression of human apoA-I and apoA-II. J. Lipid Res. 32: 821–828 [PubMed] [Google Scholar]
- 36.Vedhachalam C., Liu L., Nickel M., Dhanasekaran P., Anantharamaiah G. M., Lund-Katz S., Rothblat G. H., Phillips M. C. 2004. Influence of ApoA-I structure on the ABCA1-mediated efflux of cellular lipids. J. Biol. Chem. 279: 49931–49939 [DOI] [PubMed] [Google Scholar]
- 37.Vedhachalam C., Ghering A. B., Davidson W. S., Lund-Katz S., Rothblat G. H., Phillips M. C. 2007. ABCA1-induced cell surface binding sites for ApoA-I. Arterioscler. Thromb. Vasc. Biol. 27: 1603–1609 [DOI] [PubMed] [Google Scholar]
- 38.Lusis A. J., Taylor B. A., Wangenstein R. W., LeBoeuf R. C. 1983. Genetic control of lipid transport in mice. II. Genes controlling structure of high density lipoproteins. J. Biol. Chem. 258: 5071–5078 [PubMed] [Google Scholar]
- 39.Warden C. H., Hedrick C. C., Qiao J. H., Castellani L. W., Lusis A. J. 1993. Atherosclerosis in transgenic mice overexpressing apolipoprotein A-II. Science. 261: 469–472 [DOI] [PubMed] [Google Scholar]
- 40.Weng W., Breslow J. L. 1996. Dramatically decreased high density lipoprotein cholesterol, increased remnant clearance, and insulin hypersensitivity in apolipoprotein A-II knockout mice suggest a complex role for apolipoprotein A-II in atherosclerosis susceptibility. Proc. Natl. Acad. Sci. USA. 93: 14788–14794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hedrick C. C., Castellani L. W., Warden C. H., Puppione D. L., Lusis A. J. 1993. Influence of mouse apolipoprotein A-II on plasma lipoproteins in transgenic mice. J. Biol. Chem. 268: 20676–20682 [PubMed] [Google Scholar]
- 42.Gauthamadasa K., Vaitinadin N. S., Dressman J. L., Macha S., Homan R., Greis K. D., Silva R. A. G. D. 2012. Apolipoprotein A-II mediated conformational changes of apolipoprotein A-I in discoidal high density lipoproteins. J. Biol. Chem.; Epub ahead of Print [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lagocki P. A., Scanu A. M. 1980. In vitro modulation of the apolipoprotein composition of high density lipoprotein. Displacement of apolipoprotein A-I from high density lipoprotein by apolipoprotein A-II. J. Biol. Chem. 255: 3701–3706 [PubMed] [Google Scholar]
- 44.Rye K-A., Wee K., Curtiss L. K., Bonnet D. J., Barter P. J. 2003. Apolipoprotein A-II inhibits high density lipoprotein remodeling and lipid-poor apolipoprotein A-I formation. J. Biol. Chem. 278: 22530–22536 [DOI] [PubMed] [Google Scholar]
- 45.Cavigiolio G., Geier E. G., Shao B., Heinecke J. W., Oda M. N. 2010. Exchange of apolipoprotein A-I between lipid-associated and lipid-free states: a potential target for oxidative generation of dysfunctional high density lipoproteins. J. Biol. Chem. 285: 18847–18857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Khovidhunkit W., Kim M-S., Memon R. A., Shigenaga J. K., Moser A. H., Feingold K. R., Grunfeld C. 2004. Effects of infection and inflammation on lipid and lipoprotein metabolism: mechanisms and consequences to the host. J. Lipid Res. 45: 1169–1196 [DOI] [PubMed] [Google Scholar]
- 47.Suzuki M., Pritchard D. K., Becker L., Hoofnagle A. N., Tanimura N., Bammler T. K., Beyer R. P., Bumgarner R., Vaisar T., de Beer M. C., et al. 2010. High-density lipoprotein suppresses the type I interferon response, a family of potent antiviral immunoregulators, in macrophages challenged with lipopolysaccharide. Circulation. 122: 1919–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roberts J. E., Watters J. W., Ballard J. D., Dietrich W. F. 1998. Ltx1, a mouse locus that influences the susceptibility of macrophages to cytolysis caused by intoxication with Bacillus anthracis lethal factor, maps to chromosome 11. Mol. Microbiol. 29: 581–591 [DOI] [PubMed] [Google Scholar]