Abstract
Bacterial pathogens manipulate host cells to promote pathogen survival and dissemination. We used a 22,571 human cDNA microarray to identify host pathways that are affected by the Salmonella enterica subspecies typhimurium phoP gene, a transcription factor required for virulence, by comparing the expression profiles of human monocytic tissue culture cells infected with either the wild-type bacteria or a phoP∷Tn10 mutant strain. Both wild-type and phoP∷Tn10 bacteria induced a common set of genes, many of which are proinflammatory. Differentially expressed genes included those that affect host cell death, suggesting that the phoP regulatory system controls bacterial genes that alter macrophage survival. Subsequent experiments showed that the phoP∷Tn10 mutant strain is defective for killing both cultured and primary human macrophages but is able to replicate intracellularly. These experiments indicate that phoP plays a role in Salmonella-induced human macrophage cell death.
Salmonellosis is caused by ingestion of contaminated food or water. The bacteria are resistant to the acidic environment of the stomach and usually invade or are phagocytosed by cells lining Peyer's patches in the small intestine (1). Salmonella enterica subspecies typhimurium replicates in macrophages; in mice, it is thought that either macrophages or dendritic cells carry the bacteria from the Peyer's patches to the adjacent lymph nodes, spleen, and liver (2). S. typhimurium can kill macrophages by a caspase-1-dependent mechanism that also releases proinflammatory cytokines (3–6). Caspase-1 activation correlates with the ability to colonize the lymph nodes, spleen, and liver in mice (7). In immunocompetent humans, S. typhimurium does not usually cause systemic disease, and bacterial replication is instead limited to the intestine, with resulting gastroenteritis (1). It is unclear why S. typhimurium causes different diseases in mice and humans. Whereas S. typhimurium can kill human macrophages, the role of macrophage cell death in human disease is not yet known.
S. typhimurium phoP∷Tn10 mutants are avirulent in mice (8). It is unknown whether phoP is required for human gastroenteritis, but S. typhi phoP∷Tn10 mutants fail to cause typhoid fever in humans (9). phoP is the DNA-binding partner of a two-component response regulatory system that is activated after the bacteria enter host cells and that regulates transcription of diverse bacterial genes. For instance, phoP activates transcription of genes such as pmrA/B, pmrE, pmrF, and pagP, whose products play a role in bacterial resistance to anti-microbial peptides by chemically modifying lipopolysaccharide (LPS; ref. 10). phoP also induces genes involved in magnesium transport (mgtA, mgtCB), and has been shown to play a role in bacterial resistance to bile (11, 12). In addition, phoP represses genes that are important for bacterial entry into epithelial cells, namely those in the Salmonella pathogenicity island 1 (SPI1) (13).
What is the host molecular response to the activities of phoP-regulated genes? It has been demonstrated that phoP expression affects host cell antigen processing and presentation, but the downstream mediators, in both the bacteria and the host, are unknown (14). Microarrays provide us with a new tool for identifying host molecular pathways that a virulence determinant affects by enabling comparative analysis of the host transcriptional response to infection with virulent and avirulent mutant bacteria. Here, we identify differentially expressed genes from a human monocytic cell line infected with either wild-type S. typhimurium or an isogenic phoP∷Tn10 mutant under conditions in which bacterial growth was comparable. Based on these data, we hypothesized that phoP plays a role in macrophage cell death. Subsequent cell biological experiments showed that, indeed, phoP∷Tn10 mutants are defective for killing human U-937 monocytes and peripheral blood mononuclear cells (PBMCs). Thus, the phoP regulatory system controls gene product(s) that promote the death of human macrophages.
Materials and Methods
Bacterial Strains and Growth Conditions.
Salmonella strains are derived from SL1344 (15). The phoP mutant (phoP∷Tn10TetR) has an insertion at the phoP locus (8). Strain P7G2 (spiA∷mTn5Km) has an insertion in the spiA gene, a putative outer membrane component of the SPI2 type III secretion system (16). BJ66 (orgA∷Tn10Tet) has an insertion in orgA, a gene within SPI1, and is unable to cause mouse macrophage cell death within 2 hr of infection (6). Bacteria were grown overnight in LB without agitation at 37°C, diluted 1:50 in LB and grown 3 hr without agitation at 37°C (17). Before inoculation, bacteria were harvested by centrifugation and resuspended in PBS (for the microarray experiments) or diluted into PBS (all other experiments).
We confirmed the phenotype of the phoP∷Tn10 mutant strain by showing that it is unable to induce a known phoP-dependent promoter (pmrE/pagA) in N-minimal media with 8 μM magnesium, conditions under which the wild-type strain induced a phoP-regulated pagA promoter fused to a green fluorescent protein (GFP) reporter 5-fold relative to expression in 2 mM magnesium (data not shown) (11). Secondly, the phoP∷Tn10 mutant strain fails to replicate in the mouse macrophage RAW264.7 cell line, as previously reported (8, 18, 19).
Tissue Culture Infections and Bacterial Replication Assays.
For the microarray experiments, 5.4 × 107 U-937 cells per T175 flask were treated with phorbol 12-myristate 13-acetate (PMA, 10 ng/ml) to promote differentiation and adherence. After 46 hr, there were ≈1.6 × 108 cells per flask, and the medium (RPMI 1640, 10% FCS) was replaced with fresh medium without PMA. At 48 hr, S. typhimurium was added at a multiplicity of infection (moi) of 4–12 colony-forming units (cfu) per cell. An equivalent volume of PBS was added to uninfected controls. Thirty minutes after inoculation, gentamicin (100 μg/ml) was added to kill extracellular bacteria. Two hours after inoculation, the medium was replaced with medium containing 10 μg/ml gentamicin. Adherent cells were harvested at 0.5, 1, 2, 3, and/or 4 hr (time 0 is the time at which bacteria were added) after infection by scraping in 2 ml PBS and centrifugating for 2 min at 200 × g. The cell pellet was frozen in liquid nitrogen.
For the bacterial replication assays, 1 × 105 cells were seeded into 24-well dishes and infected in parallel with the T175 flasks at an moi of 10. Adherent cells were collected at 2 and 24 hr, lysed with 1% Triton X-100 for 5 min with vigorous pipetting, and plated for cfu (6, 17). The replication assay was repeated on four independent occasions, each in triplicate.
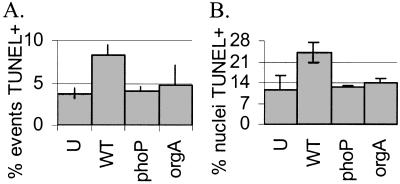
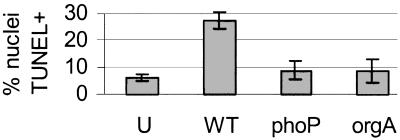
In all experiments, the bacteria were grown under conditions that allow for activity of the SPI1 type III secretion system, which secretes a cytotoxic protein(s) into host cells. However, by 4 hr, only a quarter of the wild-type-infected U-937 cells were dead as determined by terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling (TUNEL) staining (Fig. 3). Additionally, 4 hr after infection with wild-type Salmonella, we could consistently isolate reasonable amounts of high quality (as determined by distribution on an agarose gel) U-937 mRNA (data not shown).
Figure 3.
A phoP∷Tn10 mutant killed fewer U-937 macrophages than wild-type S. typhimurium by 4 hr after infection. (A) Suspended and adherent cells were harvested and processed for FACS. The percentage of TUNEL-positive events is shown. (B) Adherent U-937s were TUNEL and DAPI stained for fluorescence microscopy. U, uninfected; WT, wild-type; phoP, phoP∷Tn10 mutant; orgA, orgA∷Tn10 mutant.
RNA Isolation, Labeling, and Hybridization.
Cell pellets were thawed and homogenized in 2 ml Trizol (GIBCO/BRL, Germany) for 30 s with a Polytron PT 1200C (Kinematica, Littau-Lucerne, Switzerland), and total RNA was purified according to the manufacturer's instructions. Labeled cDNA was synthesized from 20 μg of RNA with an oligo(dT) primer and hybridized to a sequence-verified 22,571 spotted cDNA microarray on glass slides (20). The reference RNA we used was poly(A)+ RNA derived from a mixture of U-937 cells and the AGS human gastric tissue culture cell line (American Type Tissue Culture Collection). This reference RNA was labeled as previously described and hybridized to each array to allow for comparisons between different experiments (20). The hybridized slides were scanned and analyzed with a Gene Pix Scanner 4000A and the genepix program (Axon Instruments, Foster City, CA).
Data Analysis.
Data were analyzed with the Stanford University Microarray Database, Microsoft excel and access, and the significance analysis for microarrays (sam) program (http://www-stat-class.stanford.edu/SAM/SAMServlet; ref. 21). mRNAs for which at least 75% of the spots had a regression correlation greater than 0.6 were used in analysis (22). Missing data points were estimated with a K-Nearest-Neighbor imputator, where K equaled 10 (21).
To determine which mRNAs differed in the wild-type vs. uninfected samples, sam was run on the mean of the Log2 red/green normalized ratio where the time points from all experiments were grouped together. Arrays were paired according to experiment and time point. For example, the wild-type strain 2-hr time point from the third experiment was paired with the uninfected 2-hr time point from the third experiment. The differences in transcript levels between wild-type strain and phoP∷Tn10 mutant strain-infected samples, and the phoP∷Tn10 mutant strain vs. uninfected samples were calculated similarly. sam calculates a list of significant mRNAs and a false discovery rate (FDR), which is an estimate of the percentage of false positives. The relative levels of gene expression and standard errors reported in Tables 1 and 2 are the sam-generated ri and (si + so) values, respectively, where si is the standard deviation and so is an estimate of error (21). Data from all of the arrays used in this paper are available at http://genome-www4.stanford.edu/MicroArray/SMD/.
Table 1.
mRNAs that are more abundant in wild-type-infected and phoP∷Tn10 mutant-infected than in uninfected macrophages*
| Ratio† | SE | Ratio‡ | SE | |
|---|---|---|---|---|
| Inflammatory | ||||
| IL-8, C-X-C chemokine | 3.1 | 1.2 | 2.3 | 1.4 |
| GRO1, C-X-C chemokine§ | 7.0 | 1.2 | 8.6 | 1.3 |
| GRO1, C-X-C chemokine§ | 7.9 | 1.3 | 5.5 | 1.6 |
| GRO2, C-X-C chemokine | 7.2 | 1.3 | 6.5 | 1.6 |
| Mip-1-α, C-C chemokine | 4.1 | 1.3 | 2.3 | 1.9 |
| Mip-1-β, C-C chemokine | 7.8 | 1.5 | 3.9 | 2.2 |
| IL-1-β, cytokine§ | 4.8 | 1.3 | 6.4 | 1.5 |
| IL-1-β, cytokine§ | 4.3 | 1.3 | 3.5 | 2.0 |
| IL-23 p19 subunit, IL-6 family | 2.4 | 1.2 | 2.6 | 1.6 |
| Inhibin, beta A (activin A) | 3.5 | 1.2 | 4.4 | 1.4 |
| Prostaglandin-endoperoxide synthase 2 | 7.4 | 1.3 | 6.7 | 1.5 |
| Plasminogen activator inhibitor, type II | 3.4 | 1.2 | 3.2 | 1.5 |
| Transcription factor AP-1 subunit (Jun) | 3.0 | 1.2 | 2.1 | 1.7 |
| Transcription factor Spi-B | 3.1 | 1.2 | 2.1 | 1.4 |
| Cell death/survival | ||||
| NF-κB inhibitor (IKBA) (NKBI) | 3.3 | 1.3 | 2.6 | 1.7 |
| TNF receptor-associated factor 1 (TRAF1) | 2.4 | 1.2 | 2.5 | 1.4 |
| PRG1 (GLY96, DIF2, IEX1) | 3.7 | 1.3 | 3.2 | 1.6 |
| Other | ||||
| B-cell derived transcription factor | 2.0 | 1.3 | 2.0 | 1.6 |
| Connexin 26, gap junction protein | 6.0 | 1.3 | 4.0 | 1.5 |
| Connexin 26, gap junction protein | 3.5 | 1.2 | 2.2 | 1.9 |
| Superoxide dismutase 2, mitochondrial§ | 6.4 | 1.2 | 6.2 | 1.5 |
| Superoxide dismutase 2, mitochondrial§ | 6.7 | 1.2 | 4.9 | 1.6 |
| Transcription factor AP-2 alpha | 4.3 | 1.3 | 3.4 | 1.6 |
| Trinucleotide repeat containing 3§ | 2.6 | 1.2 | 2.8 | 1.4 |
| Trinucleotide repeat containing 3§ | 2.7 | 1.2 | 3.1 | 1.5 |
| Phorbol myristate acetate-induced protein | 5.6 | 1.3 | 4.5 | 1.5 |
| Similar to HMG-box transcription factor | 2.6 | 1.2 | 2.0 | 1.4 |
The false positive rate was 0.0% for the wild-type-infected vs. uninfected, and 9.3% for the phoP∷Tn10 mutant-infected versus uninfected.
Relative expression levels of wild-type-infected macrophages and uninfected macrophages.
Relative expression levels of phoP∷Tn10 mutant strain-infected macrophages and uninfected macrophages.
Two different cDNAs on the array represent the same gene.
Northern.
Northerns were performed on the same RNA samples used in the array experiments following standard protocols (23). Twenty micrograms of RNA were loaded per well. Blots were analyzed on a Molecular Dynamics PhosphorImager. GPD-1 (glycerol-3-phosphate dehydrogenase 1) was chosen as a loading control because the array data indicated that levels of this mRNA varied little between samples (data not shown). The Northern probes were identical to the cDNAs spotted onto the array.
Cell Death Assays.
For the lactate-dehydrogenase (LDH) release assays, 1 × 105 macrophages were seeded in 24-well dishes and infected with bacteria (moi 10) as described above. Twenty-four hours after infection, LDH release was measured with the Promega CytoTox 96 kit according to the instructions by using a Bio-Tek Instruments (Burlington, VT) plate reader at 490 nm. The LDH experiments were performed in triplicate on three separate occasions.
For the TUNEL-fluorescence-activated cell sorter (FACS) assays, 1 × 106 macrophages were seeded in 6-well dishes and infected with bacteria (moi 10) as described above. Twenty hours after infection, the media and two washes were collected and combined with cells lifted with 0.5 mM EDTA in PBS. Cells were harvested by centrifugation, fixed in 1% formaldehyde for 1 hr at room temperature, washed 3× with PBS, permeabilized in 0.25% Triton X-100 and 0.25% sodium citrate for 1–2 min at 4°C, washed 3× in PBS, and stained for TUNEL, according to kit instructions (Roche In Situ Cell Death Detection Kit, Germany). Cells were analyzed on a Becton Dickinson FACSCalibur and gated according to size. Samples were done in triplicate on three separate occasions, and a minimum of 800 events were counted per sample. TUNEL-positive cells were defined by comparing the amount of TUNEL staining to cells incubated in the reaction buffer without the enzyme.
For the fluorescence microscopy assay, 1 × 105 U-937s or PBMCs were harvested, seeded onto cover slips in 24-well dishes, and infected with bacteria (moi 10) as described above (24). Four hours after infection, cells were fixed in 1% formaldehyde, permeabilized in 1% T Triton X-100 and 1% sodium citrate for 1–2 min at 4°C, and stained with the TUNEL kit. Cover slips were mounted in media containing 1.5 μg/ml 4′,6-diamidino-2-phenylindole (DAPI; Vector Laboratories). Samples were prepared in triplicate on three separate occasions, and a minimum of 300 cells (as determined by DAPI staining and light microscopy) were counted per coverslip.
Results
phoP∷Tn10 Mutant Salmonella Replicate in U-937s.
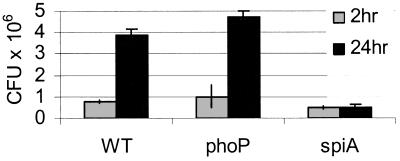
There are disparate reports on the ability of wild-type S. typhimurium to survive in PMA-treated U-937s (25, 26). We began our study by asking whether wild-type and phoP∷Tn10 mutant S. typhimurium strains replicate intracellularly under the experimental conditions used in our laboratory. We infected PMA-treated U-937 cells with wild-type and phoP∷Tn10 mutant Salmonella and measured bacterial cfu 2 and 24 hr after infection. Fig. 1 shows that, in U-937s, both the wild-type and phoP∷Tn10 mutant strains invaded and replicated, consistent with the results of Baker et al. (26). The U-937 cell line was not permissive for all Salmonella mutants; a spiA∷mTn5 mutant strain, which lacks part of the SPI2 type III secretion machinery required for intracellular replication in all known cell types, failed to replicate in U-937 cells by 24 h (Fig. 1; ref. 27).
Figure 1.
Wild-type and phoP∷Tn10 mutant Salmonella replicate in PMA-treated U-937s. At 2 (gray bars) and 24 (black bars) hr after infection, cells were lysed, and bacterial cfu were plated. WT, wild-type; phoP, phoP∷Tn10 mutant; spiA, spiA∷Tn5 mutant.
To confirm that the phoP∷Tn10 mutant bacteria maintained the tetracycline-marked transposon over the course of our experiments, performed in the absence of tetracycline, we assayed several individual colonies recovered after 24 hr of intracellular growth: 36 colonies of 36 tested were tetracycline resistant (data not shown). We conclude that the recovered bacteria retained the transposon and that the phoP∷Tn10 mutant can replicate at wild-type levels in PMA-treated U-937s.
Expression Profiles of Salmonella-Infected Vs. Uninfected Tissue Culture Cells.
We first profiled the transcriptional response of human tissue-culture macrophages to infection with wild-type Salmonella by comparing the expression profiles of wild-type-infected and uninfected U-937 cells by using a 22,571 human spotted cDNA microarray. PMA-activated U-937 macrophages were incubated with wild-type S. typhimurium or PBS for 30 min, and cells were harvested at 0.5, 1, 2, 3, and 4 hr after infection. Four independent experiments were performed. RNA was isolated, converted to labeled cDNA, and hybridized to the human arrays. Twenty-two of the RNA samples were independently labeled and hybridized to two different arrays to test reproducibility; the median correlation coefficient of the log2 red/green normalized ratio was 0.86 ± 0.18.
The data from wild-type-infected cells were compared with the uninfected control by using the sam program (see Methods). Time points were pooled to improve statistical significance. Sixty-eight mRNAs had a 2-fold or more difference in level. Fifty-five of the mRNAs had known or ascribable functions. Table 1 shows a subset of the mRNAs expressed at higher levels in wild-type Salmonella-infected cells that have known or putative functions. A complete version of Table 1 can be viewed in Table 3, which is published as supplemental data on the PNAS web site, www.pnas.org. An interesting set of genes in Table 1 includes those induced by the transcription factor NF-κB, such as the cytokine IL-1, the chemokine IL-8, an NF-κB inhibitor, and the growth regulator PRG1 (28, 29). The NF-κB pathway is induced in response to bacterial LPS, and its downstream mediators promote an acute inflammatory response and affect cell survival (28). Other inflammatory mediators induced by the infected macrophages included Jun, prostaglandin synthase, and inhibin A. These results agree well with previously published reports, in which individual gene products were assayed or smaller arrays were used, and reflect the ability of Gram-negative bacteria to stimulate innate immunity (30–32).
A phoP∷Tn10 Mutant Strain Elicits Expression of Many of the Same Macrophage mRNAs as Wild-Type Salmonella.
To identify host pathways that the phoP gene may affect, we examined the expression profiles of tissue culture macrophages infected with either a wild-type Salmonella strain or the phoP∷Tn10 mutant strain. RNA was harvested from U-937 cells infected with a wild-type Salmonella strain, a phoP∷Tn10 strain, or no bacteria, over replicate 4-hr time courses. A comparison between phoP∷Tn10 mutant Salmonella-infected and uninfected macrophages with the sam program revealed that the phoP∷Tn10 mutant strain elicited the expression of 36 mRNAs by 2-fold or more. The phoP∷Tn10 mutant strain elicited many of the same mRNAs as the wild-type bacteria, as 33 of the 36 mRNAs (91.7%) that were more highly expressed in phoP∷Tn10-infected macrophages were also more highly expressed in the wild-type-infected macrophages, relative to uninfected cells (Table 1). Also, all but one [macrophage inflammatory protein-1β (Mip-1β)] of the 33 mRNAs were induced to within 1 SD of the wild-type-infected macrophage expression levels. This analysis indicates that, overall, the inflammatory response of U-937 macrophages to wild-type Salmonella and the phoP∷Tn10 mutant strain is similar.
A phoP∷Tn10 Mutant Salmonella Fails to Elicit Host Cell Induction of Some Cell Death and Cell Cycle Genes.
On direct comparison of the wild-type strain and the phoP∷Tn10 mutant-infected macrophage data, we did not find any mRNAs whose abundance was lower in wild-type Salmonella-infected cells than in phoP∷Tn10 mutant-infected cells. However, we identified 34 mRNAs with expression levels that were 1.9-fold lower in phoP∷Tn10-infected macrophages than in wild-type-infected macrophages. Twenty-one of these mRNAs had known or putative functions (Table 2). Two of the known mRNAs affect antigen presentation, a process that phoP is known to interfere with (14). Cathepsin D is a lysosomal protease important for MHC peptide presentation (33). ILT3 is a cell surface molecule of the Ig superfamily that can both bind antigens and interfere with intracellular signaling to block cell activation by other surface molecules (34).
Table 2.
mRNAs that are more abundant in wild-type-infected than in phoP∷Tn10 mutant-infected macrophages*
| Ratio† | SE | |
|---|---|---|
| Inflammatory | ||
| CD9 antigen (p24) | 2.1 | 1.7 |
| E74-like factor 1 (ets domain transcription factor) | 2.5 | 1.5 |
| Leukocyte immunoglobulin-like receptor (ILT3) | 1.9 | 1.4 |
| Cathepsin D (lysosomal aspartyl protease) | 2.1 | 1.4 |
| Cell death/survival | ||
| STAT-induced STAT inhibitor 3 (SSI-3) | 2.2 | 1.5 |
| PIM-1 serine/threonine kinase | 2.0 | 1.5 |
| Tumor necrosis factor-α-induced protein 3 (A20) | 1.9 | 1.7 |
| Cell cycle | ||
| Cyclin-dependent kinase inhibitor 1B (p27, Kip1) | 2.0 | 1.5 |
| v-myb viral oncogene homolog-like 2 | 1.9 | 1.4 |
| Dihydrofolate reductase | 1.9 | 1.4 |
| Other | ||
| Thrombospondin 1; ECM, secreted, TGFB activator | 1.9 | 1.5 |
| Contactin 1, cell adhesion | 2.0 | 1.6 |
| Solute carrier family 15 (H+/peptide transporter) | 3.0 | 1.7 |
| Human rhotekin mRNA, Rho GTPase inhibitor | 2.2 | 1.6 |
| Carbonic anhydrase-related protein 10 | 2.6 | 1.5 |
| Ceruloplasmin (ferroxidase) copper scavanger | 1.9 | 1.4 |
| UDP glycosyltransferase 2 family, polypeptide B4 | 1.9 | 1.6 |
| Fatty acid binding protein 4, adipocyte | 2.6 | 2.2 |
| Fibroblast activation protein-α | 2.0 | 1.7 |
| α-methylacyl-CoA racemase | 2.3 | 1.6 |
| Suppression of tumorigenicity 14 | 2.1 | 1.7 |
False positive rate = 5.9%.
Relative expression levels of wild-type-infected macrophages and phoP∷Tn10 mutant-infected macrophages.
Nearly one-third of the twenty-one known genes that were differentially expressed in wild-type vs. phoP∷Tn10-infected monocytes are known to play roles in cell death, the cell cycle, or both (Table 2). SSI-3 (STAT-induced STAT inhibitor 3) encodes one of a family of proteins that suppress apoptosis via the signal transducers and activators of transcription (STAT) proteins, and SSI-3 itself has been shown to block cytokine-induced apoptosis (35, 36). PIM-1 is a kinase that protects against apoptosis and functions downstream of STAT family members (37–39). A20 is a zinc-finger protein that regulates cell death and limits inflammation by inhibiting NF-κB activity (40). The p27/Kip1 gene product promotes G1 cell cycle arrest in certain situations, such as upon Helicobacter pylori infection, that are followed by cell death (41, 42). The v-myb gene product is a member of the myb family of proteins, which are transcription factors that affect both cell cycle progression and cell death (43).
Northern Analysis of mRNA Samples.
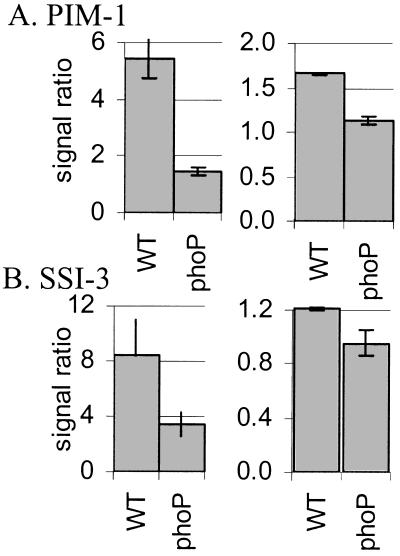
To confirm our microarray expression data by an independent method, we assayed by Northern analysis the relative abundance of two mRNAs that, according to the sam analysis, had different expression profiles in wild-type and phoP∷Tn10-infected macrophages. Fig. 2 shows the mRNA levels of SSI-3 and PIM-1, as quantified by array (Left) or Northern (Right) hybridization at the 2- through 4-hr time points. Whereas the Northern and array data show quantitative differences in relative expression levels, they agree that SSI-3 and PIM-1 are expressed at higher levels in wild-type-infected cells than in either phoP∷Tn10 mutant-infected or uninfected cells.
Figure 2.
Comparison of array and Northern data depicted as levels of mRNA in infected relative to uninfected macrophages. RNA from one of the array experiments was processed for Northern and probed with labeled PIM-1 or SSI-3 cDNA. (A) PIM-1 array (Left) and Northern (Right) data. (B) SSI-3 array (Left) and Northern (Right) data. The Northern data were normalized to a glycerol-3-phosphate dehydrogenase 1 (GPD-1) Northern. Data are expressed as the geometric mean of the bacterial-infected vs. uninfected macrophage ratios from the 2- to 4-hr time points. WT, wild-type-infected macrophages; phoP, phoP∷Tn10-infected macrophages.
The phoP∷Tn10 Mutant Salmonella Kills Fewer Tissue Culture Macrophages than Wild-Type Salmonella.
The array data showed that U-937 macrophages infected with the phoP∷Tn10 mutant do not express genes associated with cell death relative to that seen during wild-type infection. We therefore asked whether the phoP∷Tn10 mutation affects U-937 survival during Salmonella infection. We monitored cell death 4 hr after infection, by which time wild-type Salmonella infection is known to cause some macrophage cell death (data not shown; refs. 3 and 6). A TUNEL assay, which detects double-stranded breaks in chromosomal DNA, was used to quantify the percentage of dying cells. Fig. 3 shows the results of TUNEL staining monitored by FACS (Fig. 3A) or fluorescence microscopy (Fig. 3B). The results of both assays showed that about half as many macrophages were killed by infection with phoP∷Tn10 mutant Salmonella as with wild-type by 4 hr. Furthermore, the phoP∷Tn10 mutant strain promoted as little cell death as an orgA∷Tn10 mutant strain, which is defective for SPI1-mediated host cell killing in mouse macrophages (6).
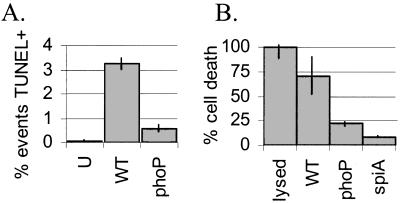
Macrophage cell death was also monitored at later times after inoculation to see whether the difference in the relative levels of wild-type and phoP∷Tn10 mutant Salmonella-mediated killing increased over the course of infection. We quantified the number of TUNEL-positive cells by FACS 20 hr after infection and found that wild-type-infected macrophages had approximately four times more TUNEL-positive events than phoP∷Tn10 mutant Salmonella (Fig. 4A). The number of TUNEL-positive events in wild-type-infected macrophages was lower at 20 hr than at 4 hr (Fig. 3A), likely because cells die and disintegrate throughout the 20-hr incubation. We used a different assay to measure cell death 24 hr after infection. The amount of the eukaryotic cytoplasmic enzyme LDH released into the media reflects the fraction of cells with damaged plasma membranes. Wild-type Salmonella-infected macrophages released approximately three times more LDH than phoP∷Tn10-infected macrophages (Fig. 4B). Thus, phoP is an important factor in Salmonella-induced U-937 cell death.
Figure 4.
A phoP∷Tn10 mutant killed fewer U-937 macrophages than wild-type S. typhimurium by 20 or 24 hr after infection. (A) Suspended and adherent cells were harvested and processed for FACS 20 hr after infection. The percentage of TUNEL-positive events is shown. (B) LDH release was measured 24 hr after infection. Results are presented as the percent cell death, where 100% is the amount of LDH in uninfected lysed cells. lysed, uninfected lysed; WT, wild-type; phoP, phoP∷Tn10 mutant; spiA, spiA∷Tn5 mutant.
phoP Is Important for Killing Human PBMCs.
To determine whether phoP plays a role in the death of primary human macrophages, we isolated PBMCs from healthy donors and counted the number of TUNEL-positive cells 4 hr after infection with wild-type and phoP∷Tn10 mutant Salmonella. We found that PBMCs, like the U-937 tissue culture cells, were less likely to die upon infection with the phoP∷Tn10 mutant bacteria than with wild-type Salmonella (Fig. 5).
Figure 5.
A phoP∷ Tn10 mutant strain is defective for killing human PBMCs. Adherent PBMCs were TUNEL and DAPI stained for fluorescence microscopy 4 hr after infection. U, uninfected; WT, wild-type; phoP, phoP∷Tn10 mutant; orgA, orgA∷Tn10 mutant.
Discussion
We used a spotted cDNA microarray to analyze Salmonella infection of the human monocytic U-937 cell line. It was not clear at the outset of these experiments whether it would be possible to distinguish between the response of tissue culture cells to wild-type S. typhimurium and avirulent phoP∷Tn10 mutant strains. Indeed, a recent study using a smaller array to compare the responses of macrophages incubated with live Salmonella vs. purified LPS did not detect any differences (31).
We first analyzed the expression profile of macrophages infected with wild-type bacteria relative to uninfected macrophages. Only 68 mRNAs of the 22,571 mRNAs on the array had significantly different levels of expression (Table 1). Most of the mRNAs elicited by wild-type Salmonella encode genes whose products function in the innate immune response. In addition, inflammatory-response mRNAs tended to be more abundant than genes that function in other processes. These results agree well with a recent study that examined expression of 588 mouse macrophage cDNAs by using a membrane-based array. However, our experiments identified a much lower percentage of differentially abundant mRNAs (0.3% vs. 13%), perhaps because our array was not specifically enriched for genes involved in immunity, a feature that enabled us to survey noninflammatory as well as inflammatory macrophage responses (31).
We then examined the molecular phenotype of phoP∷Tn10 mutant-infected macrophages. Over 90% of the macrophage mRNAs induced by the phoP∷Tn10 mutant Salmonella were also induced by the wild-type strain, indicating that, overall, both strains elicit similar inflammatory responses. These results are consistent with experiments showing that LPS isolated from a phoP∷Tn10 mutant strain stimulates monocyte innate immune responses in vitro (44).
Direct comparison of array data from phoP∷Tn10 mutant Salmonella-infected cells to wild-type-infected cells revealed 21 mRNAs of known or ascribable function that were expressed at higher levels in wild-type Salmonella-infected macrophages than in phoP∷Tn10 mutant-infected macrophages (Table 2). Two of these mRNA gene products affect antigen presentation, a process in which phoP has been implicated (14). Five of the mRNAs can play roles in cell death. Whereas the differences we observed in mRNA levels between wild-type- and phoP∷Tn10-infected macrophages were only 2-fold, it should be noted that relatively small changes in mRNA levels can significantly affect cell physiology. For example, small differences in either bcl-xL or survivin mRNA levels resulted in 4- to 5-fold increases in the number of dead tissue-culture breast carcinomal- or lung adenocarcinomal-derived cells, respectively (45, 46). Furthermore, we were most interested not in individual mRNAs that changed, but in groups of mRNAs that function in the same physiological process and were differentially expressed. Thus, based on the observation that five mRNAs that can play roles in cell death were differentially expressed between wild-type-infected macrophages and phoP∷Tn10-infected macrophages, we pursued the hypothesis that phoP affects cell death and demonstrated that phoP is required for Salmonella-induced human macrophage cell death (Figs. 3–5).
How does phoP mediate human macrophage cell death? During or shortly after S. typhimurium entry into host cells, the protein SipB is secreted into the host cell cytosol via the SPI1 type III secretion system (3, 4). SipB promotes macrophage cell death by cleaving and thus activating caspase-1, which triggers a cell death proteolytic cascade (4). A second type III secretion system, located in SPI2, has also been implicated in cell death. The SPI2 type III secretion system is activated after Salmonella have entered the phagosome and has been recently shown to promote mouse macrophage cell death by 12 h after infection (47). It is unknown which SPI2-secreted effecter(s) is involved. There are at least two ways by which phoP could mediate macrophage cell death via a type III secretion system. First, it is possible that phoP-regulated genes affect both SPI1 and SPI2 macrophage cell death, as a phoP-regulated outer membrane protein or LPS modification could be important for the translocation of effecter molecules by the type III secretion systems. Second, it has recently been shown that phoP positively regulates expression of srfJ. This gene encodes a protein homologous to eukaryotic lysosomal glucosyl ceramidases, which can play a role in host cell death. It has been proposed that the SrfJ protein is secreted into the cytosol of host cells, either by SPI2 or an alternative export system, where it could possibly play a role in host cell death (48). Finally, the phoP role in cell killing could also be independent of SPI1 and SPI2. For example, human macrophages may detect a phoP-regulated outer membrane protein or an LPS modification and respond by activating a cell death pathway.
What is the role of macrophage death in infection? In mice, cell death is correlated with establishing deep tissue infection, as caspase-1−/− mice are resistant to oral infection with S. typhimurium. The bacteria can colonize the Peyer's patches, but are impaired in disseminating to the mesenteric lymph nodes, liver, and spleen (7). Caspase-1-mediated cell death is accompanied by local inflammation, which results in the recruitment of inflammatory cells, and Salmonella may use macrophages and dendritic cells as a conduit to deeper tissue (2). It is unknown whether SPI2- or phoP-mediated cell death utilizes caspase-1 or promotes inflammation.
The experiments described in this report used microarrays to profile the transcriptional response of macrophages infected with Salmonella. It is not surprising that inflammatory genes constitute the major class of mRNAs that macrophages express in response to Salmonella. Indeed, the host reaction to most infectious agents is a generalized response mediated by the innate immune system and causing fever, malaise, and anorexia. In contrast, by directly comparing the expression profiles of wild-type and avirulent mutant Salmonella-infected macrophages, we dissected out a host pathway that a bacterial virulence determinant acts on. We then confirmed the relevance of this pathway with conventional cell biological methods. By using this approach, where two nearly identical pathogen strains are compared in a defined model system, we should be able to learn considerably more about the host pathways that individual virulence determinants affect. Thus, host microarrays will become particularly powerful tools for pathogenesis.
Supplementary Material
Acknowledgments
We thank Denise Monack, Nina Salama, Karen Guillemin, Karla Kirkegaard, Cathy Lee, and David Holden for critically reading the manuscript. We thank Ed Leonard, Nina Salama, Kaman Chan, and Julie Ross for help with PBMC experiments. We thank Rob Tibshirani, Steve Wagner, and John Storey for help with data analysis. C.S.D. is supported by an American Cancer Society PF-99-146-01-MBC grant. S.F. is supported by National Institutes of Health Grant AI26195.
Abbreviations
- PBMC
peripheral blood mononuclear cell
- PMA
phorbol 12-myristate 13-acetate
- moi
multiplicity of infection
- cfu
colony-forming unit
- TUNEL
terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling
- LDH
lactate-dehydrogenase
- DAPI
4′,6-diamidino-2-phenylindole
- LPS
lipopolysaccharide
- STAT
signal transducers and activators of transcription
- SSI-3
STAT-induced STAT inhibitor 3
References
- 1.Tsolis R M, Kingsley R A, Townsend S M, Ficht T A, Adams L G, Baumler A J. Adv Exp Med Biol. 1999;473:261–274. [PubMed] [Google Scholar]
- 2.Vazquez-Torres A, Jones-Carson J, Baumler A J, Falkow S, Valdivia R, Brown W, Le M, Berggren R, Parks W T, Fang F C. Nature (London) 1999;401:804–808. doi: 10.1038/44593. [DOI] [PubMed] [Google Scholar]
- 3.Chen L M, Kaniga K, Galán J E. Mol Microbiol. 1996;21:1101–1115. doi: 10.1046/j.1365-2958.1996.471410.x. [DOI] [PubMed] [Google Scholar]
- 4.Hersh D, Monack D M, Smith M R, Ghori N, Falkow S, Zychlinsky A. Proc Natl Acad Sci USA. 1999;96:2396–2401. doi: 10.1073/pnas.96.5.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Libby S J, Lesnick M, Hasegawa P, Weidenhammer E, Guiney D G. Cell Micro. 2000;2:49–58. doi: 10.1046/j.1462-5822.2000.00030.x. [DOI] [PubMed] [Google Scholar]
- 6.Monack D M, Raupach B, Hromockyj A E, Falkow S. Proc Natl Acad Sci USA. 1996;93:9833–9838. doi: 10.1073/pnas.93.18.9833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Monack D M, Hersh D, Ghori N, Bouley D, Zychlinsky A, Falkow S. J Exp Med. 2000;192:249–258. doi: 10.1084/jem.192.2.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miller S I, Kukral A M, Mekalanos J J. Proc Natl Acad Sci USA. 1989;86:5054–5058. doi: 10.1073/pnas.86.13.5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hohmann E L, Oletta C A, Killeen K P, Miller S I. J Infect Dis. 1996;173:1408–1414. doi: 10.1093/infdis/173.6.1408. [DOI] [PubMed] [Google Scholar]
- 10.Ernst R K, Guina T, Miller S I. J Infect Dis. 1999;179, Suppl. 2:S326–S330. doi: 10.1086/513850. [DOI] [PubMed] [Google Scholar]
- 11.Soncini F C, García Véscovi E, Solomon F, Groisman E A. J Bacteriol. 1996;178:5092–5099. doi: 10.1128/jb.178.17.5092-5099.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Velkinburgh J C, Gunn J S. Infect Immun. 1999;67:1614–1622. doi: 10.1128/iai.67.4.1614-1622.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Behlau I, Miller S I. J Bacteriol. 1993;175:4475–4484. doi: 10.1128/jb.175.14.4475-4484.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wick M J, Harding C V, Twesten N J, Normark S J, Pfeifer J D. Mol Microbiol. 1995;16:465–476. doi: 10.1111/j.1365-2958.1995.tb02411.x. [DOI] [PubMed] [Google Scholar]
- 15.Smith B P, Reina-Guerra M, Stocker B A, Hoiseth S K, Johnson E. Am J Vet Res. 1984;45:2231–2235. [PubMed] [Google Scholar]
- 16.Hensel M, Shea J E, Gleeson C, Jones M D, Dalton E, Holden D W. Science. 1995;269:400–403. doi: 10.1126/science.7618105. [DOI] [PubMed] [Google Scholar]
- 17.Lee C A, Falkow S. Proc Natl Acad Sci USA. 1990;87:4304–4308. doi: 10.1073/pnas.87.11.4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lindgren S W, Stojiljkovic I, Heffron F. Proc Natl Acad Sci USA. 1996;93:4197–4201. doi: 10.1073/pnas.93.9.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lundberg U, Vinatzer U, Berdnik D, von Gabain A, Baccarini M. J Bacteriol. 1999;181:3433–3437. doi: 10.1128/jb.181.11.3433-3437.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eisen M B, Brown P O. Methods Enzymol. 1999;303:179–205. doi: 10.1016/s0076-6879(99)03014-1. [DOI] [PubMed] [Google Scholar]
- 21.Tusher V, Tibshirani R, Chu G. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. . (First Published April 17, 2001; 10.1073/pnas.091062498) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iyer V R, Eisen M B, Ross D T, Schuler G, Moore T, Lee J C, Trent J M, Staudt L M, Hudson J, Boguski M S, et al. Science. 1999;283:83–87. doi: 10.1126/science.283.5398.83. [DOI] [PubMed] [Google Scholar]
- 23.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current Protocols in Molecular Biology. New York: Wiley; 1997. [Google Scholar]
- 24.Polesky A H, Ross J T, Falkow S, Tompkins L S. Infect Immun. 2001;69:977–987. doi: 10.1128/IAI.69.2.977-987.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwan W R, Huang X Z, Hu L, Kopecko D J. Infect Immun. 2000;68:1005–1013. doi: 10.1128/iai.68.3.1005-1013.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baker S J, Gunn J S, Morona R. Microbiology. 1999;145:367–378. doi: 10.1099/13500872-145-2-367. [DOI] [PubMed] [Google Scholar]
- 27.Cirillo D M, Valdivia R H, Monack D M, Falkow S. Mol Microbiol. 1998;30:175–188. doi: 10.1046/j.1365-2958.1998.01048.x. [DOI] [PubMed] [Google Scholar]
- 28.Ghosh S, May M J, Kopp E B. Annu Rev Immunol. 1998;16:225–260. doi: 10.1146/annurev.immunol.16.1.225. [DOI] [PubMed] [Google Scholar]
- 29.Schmidt W E, Arlt A, Trauzold A, Schafer H. Ann N Y Acad Sci. 1999;880:147–156. doi: 10.1111/j.1749-6632.1999.tb09517.x. [DOI] [PubMed] [Google Scholar]
- 30.Yamamoto Y, Klein T W, Friedman H. Infect Immun. 1996;64:3062–3068. doi: 10.1128/iai.64.8.3062-3068.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rosenberger C M, Scott M G, Gold M R, Hancock R E, Finlay B B. J Immunol. 2000;164:5894–5904. doi: 10.4049/jimmunol.164.11.5894. [DOI] [PubMed] [Google Scholar]
- 32.Svanborg C, Godaly G, Hedlund M. Curr Opin Microbiol. 1999;2:99–105. doi: 10.1016/s1369-5274(99)80017-4. [DOI] [PubMed] [Google Scholar]
- 33.Villadangos J A, Bryant R A, Deussing J, Driessen C, Lennon-Dumenil A M, Riese R J, Roth W, Saftig P, Shi G P, Chapman H A, et al. Immunol Rev. 1999;172:109–120. doi: 10.1111/j.1600-065x.1999.tb01360.x. [DOI] [PubMed] [Google Scholar]
- 34.Cella M, Dohring C, Samaridis J, Dessing M, Brockhaus M, Lanzavecchia A, Colonna M. J Exp Med. 1997;185:1743–1751. doi: 10.1084/jem.185.10.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morita Y, Naka T, Kawazoe Y, Fujimoto M, Narazaki M, Nakagawa R, Fukuyama H, Nagata S, Kishimoto T. Proc Natl Acad Sci USA. 2000;97:5405–5410. doi: 10.1073/pnas.090084797. . (First Published May 2, 2000; 10.1073/pnas.090084797) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Naka T, Matsumoto T, Narazaki M, Fujimoto M, Morita Y, Ohsawa Y, Saito H, Nagasawa T, Uchiyama Y, Kishimoto T. Proc Natl Acad Sci USA. 1998;95:15577–15582. doi: 10.1073/pnas.95.26.15577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lilly M, Sandholm J, Cooper J J, Koskinen P J, Kraft A. Oncogene. 1999;18:4022–4031. doi: 10.1038/sj.onc.1202741. [DOI] [PubMed] [Google Scholar]
- 38.Pircher T J, Zhao S, Geiger J N, Joneja B, Wojchowski D M. Oncogene. 2000;19:3684–3692. doi: 10.1038/sj.onc.1203684. [DOI] [PubMed] [Google Scholar]
- 39.Shirogane T, Fukada T, Muller J M, Shima D T, Hibi M, Hirano T. Immunity. 1999;11:709–719. doi: 10.1016/s1074-7613(00)80145-4. [DOI] [PubMed] [Google Scholar]
- 40.Lee E G, Boone D L, Chai S, Libby S L, Chien M, Lodolce J P, Ma A. Science. 2000;289:2350–2354. doi: 10.1126/science.289.5488.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sgambato A, Cittadini A, Faraglia B, Weinstein I B. J Cell Physiol. 2000;183:18–27. doi: 10.1002/(SICI)1097-4652(200004)183:1<18::AID-JCP3>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 42.Shirin H, Sordillo E M, Kolevska T K, Hibshoosh H, Kawabata Y, Oh S H, Kuebler J F, Delohery T, Weghorst C M, Weinstein I B, Moss S F. Infect Immun. 2000;68:5321–5328. doi: 10.1128/iai.68.9.5321-5328.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weston K. Curr Opin Genet Dev. 1998;8:76–81. doi: 10.1016/s0959-437x(98)80065-8. [DOI] [PubMed] [Google Scholar]
- 44.Guo L, Lim K B, Gunn J S, Bainbridge B, Darveau R P, Hackett M, Miller S I. Science. 1997;276:250–253. doi: 10.1126/science.276.5310.250. [DOI] [PubMed] [Google Scholar]
- 45.Olie R A, Simões-Wüst A P, Baumann B, Leech S H, Fabbro D, Stahel R A, Zangemeister-Wittke U. Cancer Res. 2000;60:2805–2809. [PubMed] [Google Scholar]
- 46.Simões-Wüst A P, Olie R A, Gautschi O, Leech S H, Häner R, Hall J, Fabbro D, Stahel R A, Zangemeister-Wittke U. Int J Cancer. 2000;87:582–590. doi: 10.1002/1097-0215(20000815)87:4<582::aid-ijc19>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 47.van Der Velden A W, Lindgren S W, Worley M J, Heffron F. Infect Immun. 2000;68:5702–5709. doi: 10.1128/iai.68.10.5702-5709.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Worley M J, Ching K H, Heffron F. Mol Microbiol. 2000;36:749–761. doi: 10.1046/j.1365-2958.2000.01902.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.