Abstract
RNA interference is a natural gene expression silencing system that appears throughout the tree of life. As the list of cellular processes linked to RNAi grows, so does the demand for tools to accurately measure RNAi dynamics in living cells. We engineered a synthetic RNAi sensor that converts this negative regulatory signal into a positive output in living mammalian cells thereby allowing increased sensitivity and activation. Furthermore, the circuit’s modular design allows potentially any microRNA of interest to be detected. We demonstrated that the circuit responds to an artificial microRNA and becomes activated when the RNAi target is replaced by a natural microRNA target (miR-34) in U2OS osteosarcoma cells. Our studies extend the application of rationally designed synthetic switches to RNAi, providing a sensitive way to visualize the dynamics of RNAi activity rather than just the presence of miRNA molecules.
Keywords: genetic switch, synthetic repressor, RNA interference, miR-34
Translational application of sophisticated synthetic devices for medicine and basic research is enabled by enhancing the synthetic biology toolkit with parts from mammalian cells.1,2 cellular mechanisms that play an important role in normal cell development and disease. Recently, functional synthetic gene switches have been successfully designed for applications in human health.3,4 Similar artificial networks could be designed to report important dynamic.
RNA interference (RNAi) is linked to essential processes including cell cycle progression5–7, cellular differentiation8, apoptosis9, and cancer development10 in myriad organisms. One class of small non-coding RNAs acts at the post-transcriptional level to inhibit translation from target messenger RNAs (mRNAs). MicroRNAs (miRNAs) are transcribed in the cell nucleus as long precursor molecules (pri-miRNA), processed into hairpin RNAs (pre-miRNA) by the protein Drosha11, and exported into the cytoplasm. Pre-miRNAs are cleaved, by the protein Dicer, into RISC-miRNA complex can then bind target mRNAs and inhibit translation.
Recent studies are beginning to connect the dynamics of miRNA expression with cellular and tissue phenotypes, advancing our knowledge of RNAi beyond a collection of data that show which miRNAs are present in a tissue at a fixed point in time. So far, time-course miRNA expression profiles have been created for different human developmental processes. The accumulation of a set of miRNAs including miR-1, miR-133a, miR-133b, and miR-206, has been observed during natural human fetal muscle development, as well as during artificial induction of muscle cell differentiation in situ.12 A key step in neural development is also linked to a dynamic miRNA expression profile, where differentiation-associated miRNAs accumulate and proliferation-regulating miRNAs decrease upon Schwann cell maturation.13 Periodic expression of some miRNAs appears to rely upon the circadian clock machinery14 and in some cases may regulate the clock itself.15 Rapid reporter systems that track closely with miRNA activity in real time will enable us to discover to what extent miRNA timing is linked to a biological purpose.
In the studies cited above, populations of cells or tissues were collected at specific time points and lysed for RNA analyses. Other recent studies have aimed to generate expression profiles in vivo by placing a fluorescence-producing gene (i.e., GFP) under the control of miRNA or short interfering RNA (siRNA) so that the loss of expression indicates RNAi activity.16–18 Using loss of signal as a proxy for RNAi has several disadvantages. Photo-bleaching or cell division in addition to RNAi can contribute to signal depletion, confounding the results of time-course experiments. Moreover, clear loss-of-signal can be delayed by the slow degradation of stable fluorescent proteins.19 Lastly, natural miRNAs with low activity levels may not efficiently knock down highly-expressed reporters. Ideally, RNAi reporters should be fast, sensitive, and produce a positive output.
We have engineered a synthetic reporter that converts RNAi-mediated gene silencing into a positive, visible signal on the order of hours in single living cells. We built a new genetic circuit based in part on the double repression scheme previously used by others.20 Thus far, such circuits have used strong synthetic gene promoters to create cell-type classifiers that detect high levels of miRNA.4 Our device extends the range RNAi detection for synthetic circuits. We engineered a constitutive human promoter that can be held in the initial inactivate state without relying an overwhelmingly strong repressor, giving the device the sensitivity to detect low levels of miRNA near the onset of RNAi. The double repression scheme places an output gene under the control of a repressor protein, which is targeted by a specific miRNA. This double repression approach links a gain of signal to RNAi activity. Thus, the observer can look for accumulation of signal above a negative background level, which can be seen more readily than loss of signal relative to positive background (Fig. 1A). The sooner the RNAi signal is seen, the more closely the reporter tracks with RNAi in real time, and the more useful the reporter is for live cell analysis.
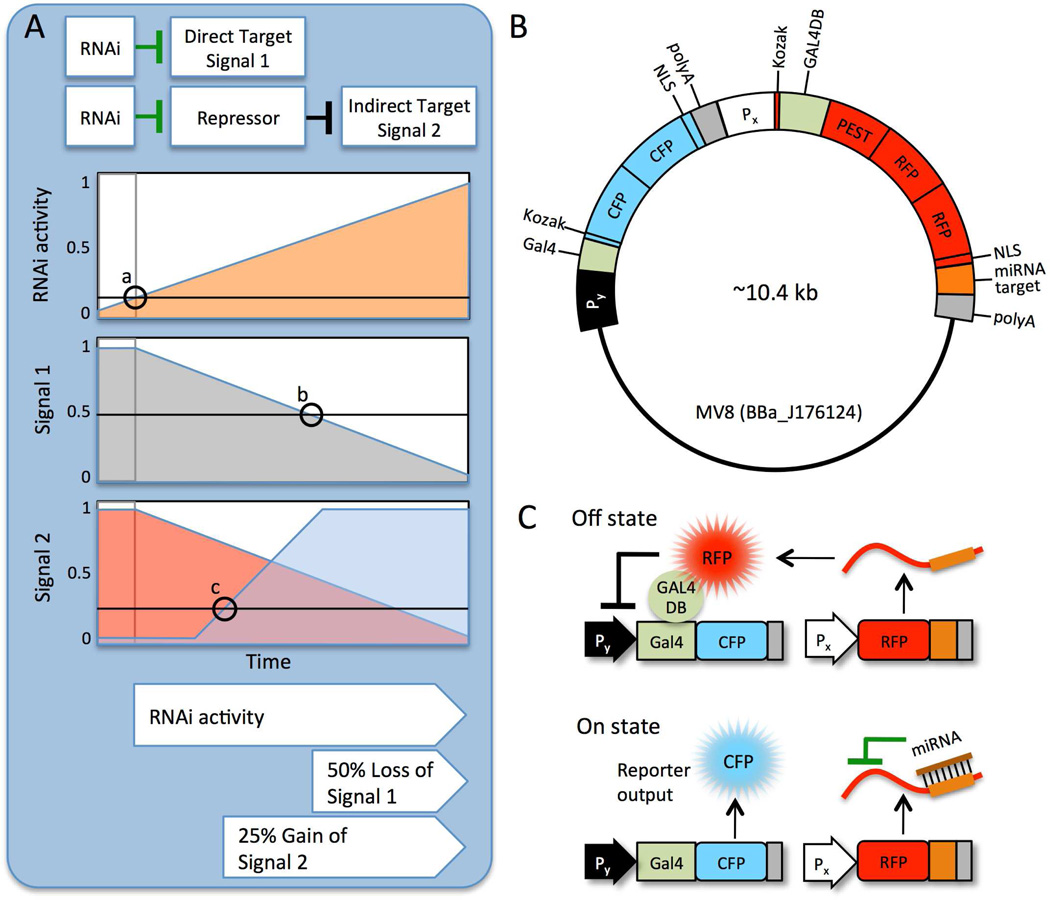
Figure 1. Schematic diagram of the mammalian miRNA reporter circuit.
(A) Qualitative comparison of two approaches for detecting RNAi, loss of signal vs. accumulation of signal. Signal 1 is directly repressed by RNAi. Signal 2 is repressed by a protein that is deactivated by RNAi. Once RNAi activity reaches an arbitrary threshold “a,” target protein degradation exceeds production. When Repressor concentration is insufficient to repress expression, Signal 2 accumulates (“c”) to a visible level sooner than Signal 1 is depleted by 50% (“b”). (B) General construct map. For all experiments, both genes were placed on the same vector. Py and Px = constitutive promoters driving expression of the CFP-Reporter and RFP-Repressor genes, respectively; NLS = nuclear localization sequence; CFP = AmCyan; RFP = mCherry; polyA = bovine growth hormone polyadenylation signal; PEST = protein degradation signal. (C) In the absence of miRNA, RFP-Repressor is expressed and inhibits CFP (Off). When a miRNA recognizes its specific target site within the 3’-UTR of the RFP-Repressor mRNA transcript, the Repressor protein is degraded and CFP is expressed (On).
Our genetic circuit has two states, which we qualitatively describe as “Off” and “On.” In the Off state, a cyan fluorescent signal is repressed. In the On state, the cyan fluorescent signal becomes activated. The circuit has a modular design (Fig. 1B), which in principle allows us to simply choose a corresponding target site for different miRNAs of interest could be readily switched to On in presence of a miRNA of interest. The Off state is maintained by a large double RFP-tagged protein (RFP-Repressor) that binds downstream of the CFP-Reporter promoter. The RFP-Repressor is placed under the control of miRNA such that miRNA expression leads to repressor silencing and activation of the reporter (Fig. 1C). Gene circuit components were optimized by identifying a combination of constitutive promoters for the CFP-Reporter and the RFP-Repressor that established the greatest ratio between the active cyan and repressed cyan states. A synthetic RNAi system (miR-luc) was used to test the performance of the device in cultured cells. We then added natural miRNA target sites to detect miR-34 in U2OS osteosarcoma cells. Our sensor system has produced the first evidence of cell-cycle-arrest associated miR-34 activity in unstressed U2OS cells.
RESULTS
Regulating transcription by steric hindrance from a fluorescent protein
The RFP-Repressor is a novel synthetic transcriptional repressor that binds near a promoter to occlude the transcriptional machinery through steric hindrance. Most of its bulk comes from two fluorophores (~54 kD); it does not rely upon a transcriptional repression domain to maintain gene repression. Thus, its promoter-silencing activity is essentially a function of repressor protein production and degradation, not epigenetic silencing effects. For instance, previously reported synthetic genetic switches utilize the Krüppel associated box (KRAB) repression domain.20,21 KRAB interacts with heterochromatin components such as HP1, which recruit chromatin modifying enzymes that form mitotically stable gene repression.22 We aimed to engineer fast reporter activation, where repression is reversed on the order of minutes or hours, before cell division occurs. Therefore we used steric hindrance, reminiscent of the TetR and LacI regulation systems23,24, to compete with PolII at the promoter and block transcriptional activation. Instead of tagging a full-length repressor with a fluorescent protein, we fused two mCherry monomers to the Gal4 DNA-binding domain from yeast to create a visible repressor. We included a PEST sequence motif to decrease repressor protein half-life when RNAi is active.
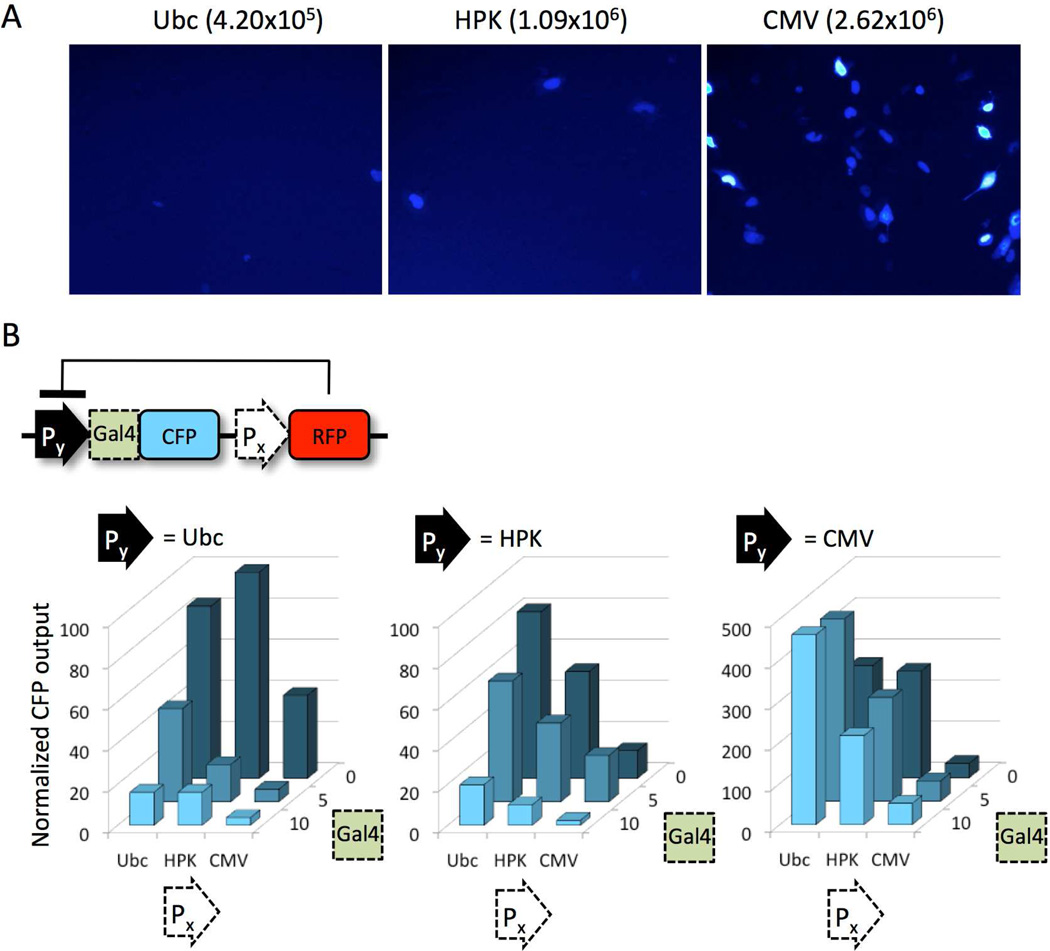
We optimized the Off state of the circuit by testing increasingly active constitutive promoters to drive RFP-Repressor expression. The relative strengths of the promoters we used are Ubiquitin C (Ubc) < human phosphoglycerate kinase (HPK) < cytomegalovirus (CMV), based on fluorescence microscopy and flow cytometry of cells transfected with CFP driven by each promoter (Fig. 2A). In constructs where a weak CFP promoter (Py = Ubc) had five Gal4 repressor binding sites (Gal4 = 5), we observed stronger CFP repression (relative to controls with zero binding sites, Gal4 = 0) as RFP-Repressor expression was increased (Fig. 2B); 0.8-, 2.5-, and 2.7- fold repression for RFP-Repressor promoters (Px) Ubc, HPK, and CMV, respectively. Switching the CFP promoter to HPK appeared to compromise the effectiveness of the repressor (0.46- and 0.44-fold repression for RFP-Repressor promoters Ubc and HPK). The CMV-driven CFP did not appear to respond to the repressor, compared to corresponding zero-Gal4 controls. Interestingly, the CMV-driven RFP-Repressor appears to decrease CFP levels when there are no repressor binding sites at the promoter of the CFP-Reporter (where Px = CMV and Gal4 = 0). This suggests that CMV on one plasmid may indirectly affect expression from the other plasmid, perhaps through competition for a fixed pool of transcription factors in the cell.
Figure 2. The off state can be tuned by increasing the concentration of RFP-Repressor at the output gene promoter.
(A) Fluorescence microscopy of cells carrying CFP driven by a Ubc, HPK, or CMV promoter. Numbers show total CFP intensity (mean CFP fluorescence in CFP-positive cells, multiplied by the frequency of CFP-positive cells) from flow cytometry. (B) RFP-Repressor expression levels were regulated by Ubc, HPK, and CMV promoters (Px), respectively. The local concentration of RFP-Repressor at the CFP promoter (Py) was modulated by inserting zero, 5, or 10 copies of RFP-Repressor binding sites (Gal4) downstream of the Py transcription start site. “Normalized CFP output” is the CFP intensity (mean CFP fluorescence in CFP/RFP-positive cells, multiplied by the frequency of CFP-positive cells) divided by the corresponding values shown in Fig. 2A (i.e., Py = Ubc values / Ubc 4.2×105; Py = HPK values / HPK 1.09×106; Py = CMV values / CMV 2.6×106). Each column is the average of triplicate flow cytometry experiments. Standard error is less than 25% of the CFP value with the exception of: 44% for Px = HPK, Py = Ubc, Gal4 = 5, and 31% for Px = CMV, Py = Ubc, Gal4 = 10. The arithmetic mean was used for all calculations.
We also considered the possibility that CMV might produce too much RFP-Repressor, the direct target of RNAi in this circuit, and overwhelm the natural silencing machinery. Therefore, we altered the binding sites to increase the local concentration of RFP-Repressor protein at the CFP-Reporter promoter. We found that doubling the number of repressor binding sites (from five to ten copies of Gal4) achieved greater repression (Fig. 2B). For instance, in constructs where a weak CFP promoter (Py = Ubc) was suppressed by an HPK-driven RFP-Repressor (Px = HPK), we observed 2.5- and 2.7-fold repression with 5 and 10 binding sites, respectively (Fig. 2B). Repression of HPK-driven CFP was 0.4- and 2.4-fold with 5 and 10 binding sites. We chose the HPK-CFP-Reporter construct since expression of CFP from the weak Ubc promoter was difficult to visualize via microscopy (Fig. 2A). This analysis allowed us to identify an optimal design (Px = HPK, Py = HPK, Gal4 = 10), where CFP repression can be acheived without high CMV-driven expression of the repressor.
The sensor is activated upon miRNA induction
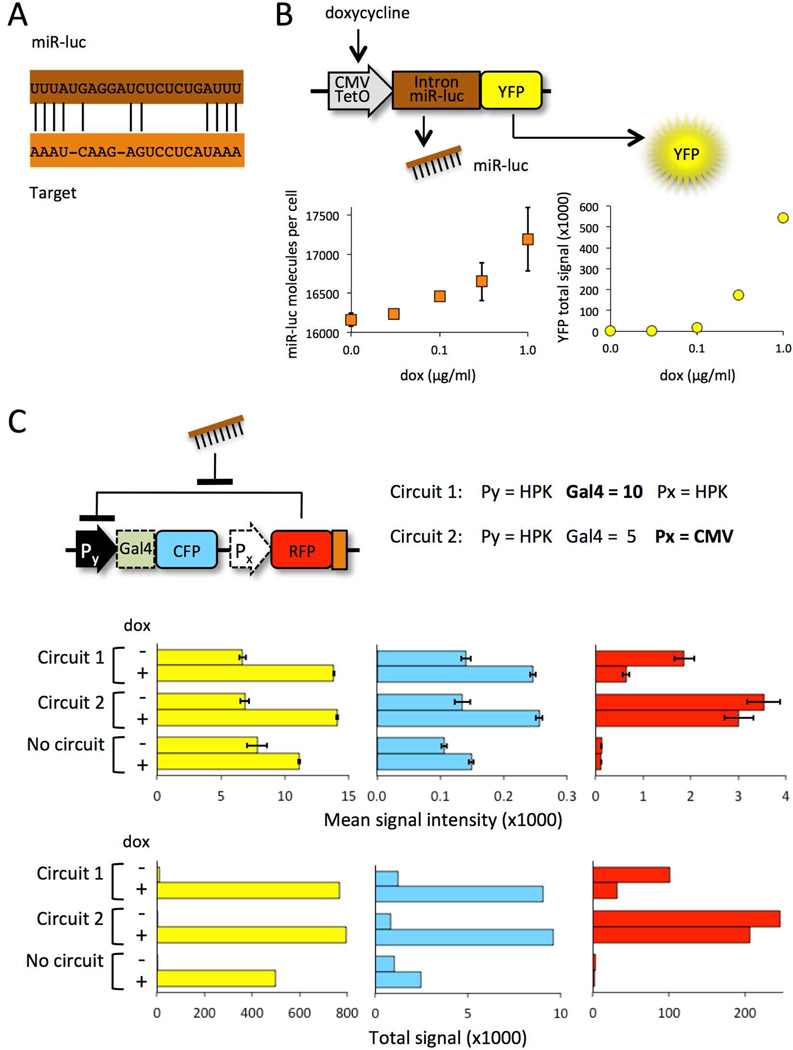
We demonstrated that the miR-sensor produces increased CFP signal in the presence of miRNA by using a drug-inducible orthogonal RNAi system. The JDS33 cell line25 carries a transgene that expresses luciferase miRNA (Luc-160126, referred to hereafter as “miR-luc”) when doxycycline (dox) is added to the cell culture medium. miR-luc has imperfect complementarity with the target sequences at the 3’ end of the RFP-Repressor gene (Fig. 3A), which is characteristic of natural miRNA regulation. Co-expressed visible YFP indicated the amount of miR-luc present, as confirmed by real time PCR analysis and flow cytometry (Fig. 3B). Therefore, the miR sensor should become activated when YFP is expressed.
Figure 3. The miRNA sensor responds to an artificial miRNA.
(A) YFP signal indicates the level of doxycycline-induced miR-luc in JDS33 cells. MiR-luc molecules per cell was measured by real time quantitiave RT-PCR. YFP signal was detected by flow cytometry analysis. YFP total signal = mean fluorescence × frequency of fluorescence signal. (B) Comparison of two miRNA sensor designs, one with ten binding sites for the RFP-Repressor (Circuit 1) vs. one with higher RFP-Repressor expression (Circuit 2). YFP, CFP, and RFP signal (left to right) was measured by flow cytometry. Top graphs: error bars show the standard error of the mean signal intensity. Bottom graphs: total signal (mean signal intensity × frequency of fluorescence signal). The arithmetic mean was used for all calculations.
We observed that the circuit becomes activated in the presence of miR-luc. Two different circuit configurations were compared. Circuit 1 had 10 Gal4 sites at the CFP-Reporter and an HPK (medium strength) promoter driving the RFP-Repressor. Circuit 2 had 5 Gal4 sites at the CFP-Reporter and a CMV (strong) promoter driving the RFP-Repressor (Fig. 3C). We transfected JDS33 cells with each circuit and treated the cells with 1 µg/ml dox for 48 hours. About 1.7×104 molecules of miR-luc per cell is present under these conditions (Fig. 3B). Shifts from low to higher CFP signal showed that both sensors became activated after dox-induced miR-luc expression (Fig. 3C). Activation of Circuit 1 (HPK-driven RFP-Repressor) was comparable to Circuit 2 (CMV-driven RFP-Repressor), thus a switch from Off to On does not require an over-expressed repressor.
A sensor designed to detect miR-34 is activated in human osteosarcoma cells over a short time scale
miRNA sensors carrying target sequences for the natural miRNA miR-34 become activated in the human bone cancer-derived cell line U2OS. miR-34 plays an important role in cell physiology. miR-34 silences cyclinD1 (CCDN1), cyclin dependent kinase 6 (CDK6), and B-cell lymphoma 2 (BCL2)9, which promote progression into mitosis and prevent apoptosis. Thus, miR-34 activity has a negative impact on cell proliferation and is expected to be diminished or absent in dividing cancer cells. miR-34 has been detected in U2OS at low or high levels, depending upon a proliferating or arrested growth status, respectively.27 We used our system to track miR-34 activity over a time interval of approximately one cell division (20 hours) in proliferating U2OS cells with potentially low levels of miR-34.
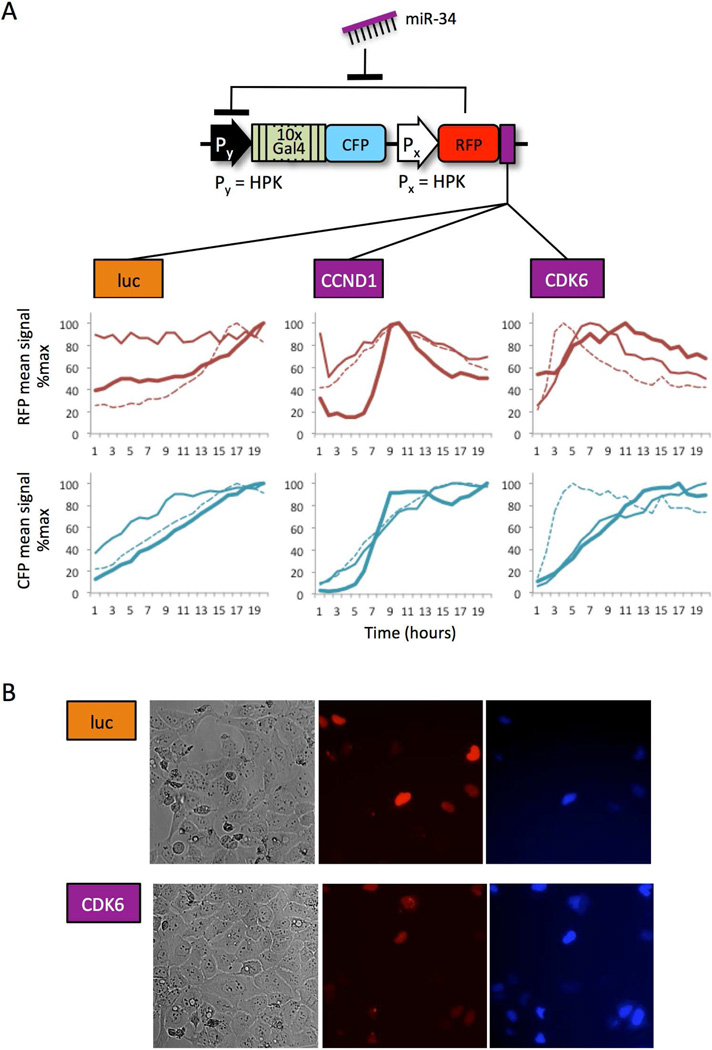
Sensors built with miR-34 target sites from CCND1 and CDK6, but not BCL2 showed switch-like activation in U2OS cells. All three sensors were constructed based upon the configuration for Circuit 1 (Fig. 3C), transfected into U2OS cells, and analyzed six hours later via live cell time-lapse microscopy. Circuit 1, without miR-34 target sites, was used as a negative control since it is not expected to become activated in U2OS cells that do not express artificial miR-luc. The negative control showed a gradual linear accumulation of both signals over the time frame analyzed. The BCL2 sensor behaved the same as the negative control construct (data not shown). In contrast, the CCDN1 and CDK6 sensors showed a rise and fall in RFP signal, accompanied by an increase in CFP production (Fig. 4A). The first derivatives of regression curves for CFP expression show higher peaks at earlier time points for the miR-34-regulated sensors (Figure S1). These results indicate that for time-course single cell analysis performed in under 20 hours, activated sensors can be distinguished by more rapid accumulation of CFP signal.
Figure 4. The behavior of natural microRNA sensors indicate miR-34 activity in U2OS cells.
(A) Live cells carrying one of three miR sensor circuits with different miR target sites (CCND1, CDK6, or luc) were imaged via fluorescence microscopy over a ~30 hour period. Each thick, thin, or dashed line corresponds to the same single cell in the RFP (top) and CFP (bottom) channel. RFP intensity and CFP output were measured as the mean signal of individual (within a single cell cycle). In cells where RFP-Repressor is targeted by the miR-34 silencer (CCND1 and CDK6), a rise and then fall in RFP signal is associated with faster accumulation of CFP output, compared to a negative control where RFP-Repressor is not targeted by miR-34 (luc). (B) The addition of a PEST tag to CFP reduces accumulation of CFP expression from a sensor, whereas the sensor that carries the CDK6 target is activated. Phase contrast, RFP, and CFP channels are shown from left to right.
Computational analysis of RNA hybridization suggests that the affinity of miR-34 for each of the targets may determine the differences in the behavior of the sensors we tested. We calculated the free energy of binding between CCND1, CDK6, and BCL2 target sites and each member of the miR-34 family (miR-34a, b, and c). Unresponsiveness of the BCL2 sensor might be a consequence of this target’s low affinity for miR-34, as indicated by relatively high free energies of the predicted miR-34-target hybrids (Fig. S2).
We also observed activation of a redesigned miR-34 sensor in which CFP was destabilized to reduce signal detected in the Off state. The accumulation of both RFP and CFP expression from the construct that was designed to be constitutively off (lacking miR-34 target sites) was unexpected, since our previous analyses indicated an inverse relationship between RFP and CFP (Fig. 4A). The time-course results may be due to some instability of the Off state before proteins reach steady state levels. We reduced accumulation of CFP signal in the inactive state by adding a PEST protein degradation tag to CFP (in circuit configuration Px = HPK, Py = HPK, Gal4 = 10). CFP destabilization does not appear to improve the signal to background ratio (0.4-fold, Fig. 4B) compared to the circuit with stable CFP (2.4-fold, Fig. 2B). However, cells carrying the miR-34 sensor with destabilized CFP did show greater CFP signal than a sensor that lacked the miR-34 target sites (Fig. 4B).
Conclusion
Our work presents two important advances for synthetic device engineering and applications. First, the RFP-Repressor may serve as a generally useful model for designing synthetic transcriptional regulators de novo. Other commonly used repressors (i.e., LacI and Tet) are derived from naturally occurring gene repression systems. To our knowledge, the RFP-Repressor is the first reported functional synthetic repressor that has been built entirely from protein modules that are not typically associated with transcriptional repression. Our results directly demonstrate that a functional repressor may be built from a protein designed simply to occupy sites immediately downstream of a constitutive promoter, and that its function depends upon the local concentration of repressors at the promoter.
Second, we have extended the application of the double repression circuit design to sense miRNAs previously detected at low levels in the proliferating cell state27, over a short time scale. miR sensor activation may be sensitive to low levels of miR-34, or activated sensors might signify the onset of miR-34 activity prior to subsequent cell cycle arrest. For medical applications in cancer therapy, our circuit design could be engineered to activate a cancer-killing gene near the onset of metastasis-associated miRNA activity. So far, we have expressed the switches from extra-chromosomal plasmids. Future applications that stably integrate the circuit into the genome will be useful for observing the stochasticity of miR-34 activity at the individual cell level, which is important for determining whether single cancer cells alter states to become more aggressive. Our work demonstrates the power and potential of modular rational design in extending the utility synthetic devices for research and medical applications.
METHODS
Constructs
DNA fragments with universal cloning sites (EcoRI, NotI, XbaI, SpeI, and PstI) were generated by PCR and assembled according to a modified BioBrick DNA assembly method28, using E. coli DH5-alpha maintained under standard conditions. All DNA fragment and annotated full-length construct sequences are freely available in the MIT Registry of Standard Biological Parts29 (See Table S1). The RFP-Repressor was constructed from a Gal4 DNA-binding domain followed by two copies of mCherry. The constitutive promoters Ubc, HPK and CMV were cloned upstream of five (5×Gal4) or ten (10×Gal4) copies of the Gal4 DNA binding sites to generate Gal4-regulated promoters. Complete constructs were excised from the V0120 high-copy cloning vector and inserted into modified pcDNA 3.1+ vectors. All plasmids were verified by restriction digests and DNA sequencing (Genewiz, Inc., Cambridge, MA) from the vector into the 5’ and 3’ ends of the inserts prior to transfection.
Cell culture and transfections
Cell line JDS33 is described by Shih et al.25 U2OS and JDS33 cells were grown in McCoy’s 5A medium supplemented with 10% tetracycline-free fetal bovine serum and 1% penicillin and streptomycin. Cells were grown at 37°C in a humidified CO2 incubator. For transfections, plasmid DNA was extracted and purified from E. coli using a QIAGEN miniprep kit. 1 µg plasmid DNA, 3 µl Lipofectamine LTX (Invitrogen), and 3 µl PLUS reagent in 190 µl Opti-MEM was added to ~2.5×105 cells per well (in a 12-well plate) in antibiotic-free growth medium.
Flow cytometry
Cells were grown in a 12-well plate (~5×105 per sample/ well), trypsinized, collected in growth medium, and washed and resuspended in 1× Dulbecco’s PBS. Flow cytometry was performed 18 hours after transfection using a BD Biosciences LSRII HTS-3 Laser high throughput sampler platform. For flow cytometry analyses to detect mCherry, YFP (Venus), and AmCyan, the following excitation lasers and filters were used: 594 nm Yellow/ DsRed (620/22), 488 nm Blue/FITC (520/50), and 405 nm Violet/ AmCyan (525/50). For every experiment, non-fluorescent U2OS cells were used to adjust the voltage to minimize autofluorescence signal in all three channels, and to create a 1:1 forward and side scatter ratio. Background was subtracted from signal by setting the threshold gate above the signal from non-fluorescent U2OS cells. Data was visualized using FACS Diva software, and statistically analyzed using FlowJo software.
Quantitative real-time reverse transcription PCR of miR-luc and YFP detection
JDS33 cells were cultured in 6-well plate (~1x106 per sample/ well) and treated with 0, 0.01, 0.03, 0.1, 0.3 or 1 µg/ml doxycycline to induce YFP and miR-luc expression for 48 hours. One set of samples was harvested for YFP detection via flow cytometry. A duplicate set was processed for RNA isolation. RNA was extracted with TRIzol Reagent (Invitrogen 15596-018) and purified according to the Invitrogen protocol. cDNA synthesis and PCR were designed as described in Raymond et al.30 cDNA synthesis (SuperScript III, Invitrogen 18080-051) was performed according to the manufacturer’s protocol with either 5 µg template RNA from the dox-treated JDS33 cells (Experimental) or 2 pmol synthesized “Input Control” RNA oligos (IDT DNA), plus 50 pmol miR-luc “tailed” primers (5’-catgatcagctgggccaagaaaatcagagag) instead of oligo dT in a final reaction volume of 20 µl. To normalize for Experimental RNA loading, oligo dT primers were used in separate reactions to generate Total cDNA for measuring GAPDH transcripts. Completed reactions were treated with 1 µl of RNaseH.
qRT-PCR was performed on Experimental cDNA, Input Control cDNA, or Total cDNA. Experimental and Input Control cDNA reactions contained 0.8 µl cDNA, 36.0 pmol primers (miR-luc forward 5’-tttatgaggatctctct; miR-luc reverse 5’-catgatcagctgggcca), and 7.5 µl SYBR Green Master Mix (Applied Biosystems) in a final volume of 15 µl. GAPDH reactions contained 0.8 µl Total cDNA and 2.25×10−3 pmol primers (GAPDH forward 5’-ccgcatcttcttttgcgtcgcc; GAPDH reverse 5’-accaggcgcccaatacgacc). For standardization, we used a synthesized DNA oligo template Standard that carries the full miR-luc PCR template sequence. qRT-PCR was performed as described above using 0.8 ul of 1.0×10−5, ×10−4, ×10−3, or ×10−2 uM synthesized DNA solution and 36.0 pmol primers (miR-luc forward and miR-luc reverse).
Fold change relative to the least abundant standard template was caluclated as 2^(CtStandard 1 - Ctsample) for all samples. DNA oligo molecules per reaction for Standards 1 through 4 (4.816×106, ×107, ×108, and ×109) was plotted against fold change to generate a standard curve. The line of best fit equation was used to calculate miR-luc molecules per reaction for the Experimental cDNA samples; these values were divided by Total cDNA GAPDH normalization values (2^(CtGAPDH sample / CtGAPDH 0 dox)) and adjusted to take into account 73% cDNA synthesis efficiency (as indicated by the Input Control values). “miR-luc molecules per cell” (Fig. 3A) was calculated as normalized Experimental miR-luc molecules per reaction divided by 8000 cells per reaction.
Single cell fluorescence assays
U2OS cells were cultured in 12-well glass-bottom plates and transiently transfected with miRNA sensor circuit constructs for 6 hrs. Phase contrast, red fluorescent, and cyan fluorescent images were collected at 1 hour intervals using a Nikon inverted microscope, controlled by Metamorph acquisition software. Mean fluorescence intensities of individual interphase nuclei were manually measured using ImageJ.
Supplementary Material
ACKNOWLEDGMENTS
We thank J. Moore for assistance with flow cytometry, Z. Xie and J. Lohmeuller for general advice, and J. Shih for JDS33 cells and the miR-luc sequences. This work was supported, in whole or in part, by National Institutes of Health Grants GM36373 (to PAS) and 1F32GM087860 (to KAH). FC was supported by University of Bologna. DF and AY were supported by the Harvard CSB (P50GM068763).
Footnotes
AUTHOR CONTRIBUTIONS
KAH and PAS conceptualized the project. KAH, FC, DF, and AY built and transfected constructs and performed flow cytometry. KAH and DF performed time-course microscopy. FC and AY designed and performed miR-luc RT-PCR. DF performed the RNAhybrid analysis. All authors drafted the manuscript.
SUPPORTING INFORMATION AVAILABLE
Supplemental material includes a detailed list of constructs as entries into the MIT Registry of Standard Biological Parts (Table S1), Figure S1, Figure S2, supplemental methods, and references. This information is available free of charge via the Internet at http://pubs.acs.org/.
REFERENCES
- 1.Greber D, Fussenegger M. Mammalian synthetic biology: engineering of sophisticated gene networks. J. Biotechnol. 2007;130:329–345. doi: 10.1016/j.jbiotec.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 2.Haynes KA, Silver PA. Eukaryotic systems broaden the scope of synthetic biology. J. Cell Biol. 2009;187:589–596. doi: 10.1083/jcb.200908138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ye H, Daoud-El Baba M, Peng R-W, Fussenegger M. A synthetic optogenetic transcription device enhances blood-glucose homeostasis in mice. Science. 2011;332:1565–1568. doi: 10.1126/science.1203535. [DOI] [PubMed] [Google Scholar]
- 4.Xie Z, Wroblewska L, Prochazka L, Weiss R, Benenson Y. Multi-Input RNAi-Based Logic Circuit for Identification of Specific Cancer Cells. Science. 2011;333:1307–1311. doi: 10.1126/science.1205527. [DOI] [PubMed] [Google Scholar]
- 5.Boyerinas B, Park S-M, Hau A, Murmann AE, Peter ME. The role of let-7 in cell differentiation and cancer. Endocr. Relat. Cancer. 2010;17:F19–F36. doi: 10.1677/ERC-09-0184. [DOI] [PubMed] [Google Scholar]
- 6.Lin S-Y, Johnson SM, Abraham M, Vella MC, Pasquinelli A, Gamberi C, Gottlieb E, Slack FJ. The C elegans hunchback homolog, hbl-1, controls temporal patterning and is a probable microRNA target. Developmental Cell. 2003;4:639–650. doi: 10.1016/s1534-5807(03)00124-2. [DOI] [PubMed] [Google Scholar]
- 7.Nimmo RA, Slack FJ. An elegant miRror: microRNAs in stem cells, developmental timing and cancer. Chromosoma. 2009;118:405–418. doi: 10.1007/s00412-009-0210-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Merkerova M, Belickova M, Bruchova H. Differential expression of microRNAs in hematopoietic cell lineages. Eur. J. Haematol. 2008;81:304–310. doi: 10.1111/j.1600-0609.2008.01111.x. [DOI] [PubMed] [Google Scholar]
- 9.Sun F, Fu H, Liu Q, Tie Y, Zhu J, Xing R, Sun Z, Zheng X. Downregulation of CCND1 and CDK6 by miR-34a induces cell cycle arrest. FEBS Lett. 2008;582:1564–1568. doi: 10.1016/j.febslet.2008.03.057. [DOI] [PubMed] [Google Scholar]
- 10.Lynam-Lennon N, Maher SG, Reynolds JV. The roles of microRNA in cancer and apoptosis. Biological Reviews. 2009;84:55–71. doi: 10.1111/j.1469-185X.2008.00061.x. [DOI] [PubMed] [Google Scholar]
- 11.Cullen B. Transcription and Processing of Human microRNA Precursors 10.1016/j.molcel.2004.12.002 : Molecular Cell | ScienceDirect.com. Molecular Cell. 2004 doi: 10.1016/j.molcel.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 12.Koutsoulidou A, Mastroyiannopoulos NP, Furling D, Uney JB, Phylactou LA. Expression of miR-1, miR-133a, miR-133b and miR-206 increases during development of human skeletal muscle. BMC Dev. Biol. 2011;11:34. doi: 10.1186/1471-213X-11-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gokey NG, Srinivasan R, Lopez-Anido C, Krueger C, Svaren J. Developmental Regulation of MicroRNA Expression in Schwann Cells. Mol. Cell. Biol. 2011 doi: 10.1128/MCB.06270-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang M, Lee J-E, Padgett RW, Edery I. Circadian regulation of a limited set of conserved microRNAs in Drosophila. BMC Genomics. 2008;9:83. doi: 10.1186/1471-2164-9-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng H-YM, Obrietan K. Revealing a role of microRNAs in the regulation of the biological clock. Cell Cycle. 2007;6:3034–3035. doi: 10.4161/cc.6.24.5106. [DOI] [PubMed] [Google Scholar]
- 16.De Pietri Tonelli D, Calegari F, Fei J-F, Nomura T, Osumi N, Heisenberg CP, Huttner WB. Single-cell detection of microRNAs in developing vertebrate embryos after acute administration of a dual-fluorescence reporter/sensor plasmid. Biotech. 2006;41:727–732. doi: 10.2144/000112296. [DOI] [PubMed] [Google Scholar]
- 17.Zhang X, Zabinsky R, Teng Y, Cui M, Han M. microRNAs play critical roles in the survival and recovery of Caenorhabditis elegans from starvation-induced L1 diapause. Proc. Natl. Acad. Sci. U.S.A. 2011;108:17997–18002. doi: 10.1073/pnas.1105982108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Varghese J, Lim SF, Cohen SM. Drosophila miR-14 regulates insulin production and metabolism through its target, sugarbabe. Genes & Development. 2010;24:2748–2753. doi: 10.1101/gad.1995910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Milo R, Jorgensen P, Moran U, Weber G, Springer M. BioNumbers--the database of key numbers in molecular and cell biology. Nucleic Acids Research. 2010;38:D750–D753. doi: 10.1093/nar/gkp889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rinaudo K, Bleris L, Maddamsetti R, Subramanian S, Weiss R, Benenson Y. A universal RNAi-based logic evaluator that operates in mammalian cells. Nature Biotechnology. 2007;25:795–801. doi: 10.1038/nbt1307. [DOI] [PubMed] [Google Scholar]
- 21.Kramer BP, Viretta AU, Baba MD-E, Aubel D, Weber W, Fussenegger M. An engineered epigenetic transgene switch in mammalian cells. Nature Biotechnology. 2004;22:867–870. doi: 10.1038/nbt980. [DOI] [PubMed] [Google Scholar]
- 22.Ryan RF, Schultz DC, Ayyanathan K, Singh PB, Friedman JR, Fredericks WJ, Rauscher FJ. KAP-1 corepressor protein interacts and colocalizes with heterochromatic and euchromatic HP1 proteins: a potential role for Kruppel-associated box-zinc finger proteins in heterochromatin-mediated gene silencing. Mol. Cell. Biol. 1999;19:4366–4378. doi: 10.1128/mcb.19.6.4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramos JL, Martinez-Bueno M, Molina-Henares AJ, Teran W, Watanabe K, Zhang X, Gallegos MT, Brennan R, Tobes R. The TetR family of transcriptional repressors. Microbiol. Mol. Biol. Rev. 2005;69:326–356. doi: 10.1128/MMBR.69.2.326-356.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lewis M. The lac repressor. Biol. C. R. 2005;328:521–548. doi: 10.1016/j.crvi.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 25.Shih JD, Waks Z, Kedersha N, Silver PA. Visualization of single mRNAs reveals temporal association of proteins with microRNA-regulated mRNA. Nucleic Acids Research. 2011;39:7740–7749. doi: 10.1093/nar/gkr456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chung KH. Polycistronic RNA polymerase II expression vectors for RNA interference based on BIC/miR-155. Nucleic Acids Research. 2006;34:e53–e53. doi: 10.1093/nar/gkl143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He C, Xiong J, Xu X, Lu W, Liu L, Xiao D, Wang D. Functional elucidation of MiR-34 in osteosarcoma cells and primary tumor samples. Biochemical and Biophysical Research Communications. 2009;388:35–40. doi: 10.1016/j.bbrc.2009.07.101. [DOI] [PubMed] [Google Scholar]
- 28.Phillips I, Silver P. DSpace@MIT : A New Biobrick Assembly Strategy Designed for Facile Protein Engineering. 2006 [Google Scholar]
- 29.Endy D. Foundations for engineering biology. Nat Cell Biol. 2005;438:449–453. doi: 10.1038/nature04342. [DOI] [PubMed] [Google Scholar]
- 30.Raymond CK, Roberts BS, Garrett-Engele P, Lim LP, Johnson JM. Simple, quantitative primer-extension PCR assay for direct monitoring of microRNAs and short-interfering RNAs. RNA. 2005;11:1737–1744. doi: 10.1261/rna.2148705. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.