Abstract
TOR (target of rapamycin) is a serine/threonine kinase, evolutionarily conserved from yeast to human, which functions as a fundamental controller of cell growth. The moderate clinical benefit of rapamycin in mTOR-based therapy of many cancers favors the development of new TOR inhibitors. Here we report a high throughput flow cytometry multiplexed screen using five GFP-tagged yeast clones that represent the readouts of four branches of the TORC1 signaling pathway in budding yeast. Each GFP-tagged clone was differentially color-coded and the GFP signal of each clone was measured simultaneously by flow cytometry, which allows rapid prioritization of compounds that likely act through direct modulation of TORC1 or proximal signaling components. A total of 255 compounds were confirmed in dose-response analysis to alter GFP expression in one or more clones. To validate the concept of the high throughput screen, we have characterized CID 3528206, a small molecule most likely to act on TORC1 as it alters GFP expression in all five GFP clones in an analogous manner to rapamycin. We have shown that CID 3528206 inhibited yeast cell growth, and that CID 3528206 inhibited TORC1 activity both in vitro and in vivo with EC50s of 150 nM and 3.9 μM, respectively. The results of microarray analysis and yeast GFP collection screen further support the notion that CID 3528206 and rapamycin modulate similar cellular pathways. Together, these results indicate that the HTS has identified a potentially useful small molecule for further development of TOR inhibitors.
TOR (Target of Rapamycin) proteins are ser/thr protein kinases phylogenetically conserved from yeast to man (1–3). Yeast possesses two TOR proteins that function in two distinct protein complexes, TOR complex 1 (TORC1) and TOR complex 2 (TORC2). TORC1 is sensitive to rapamycin and promotes protein synthesis and other anabolic processes, while inhibiting autophagy and other catabolic and stress-response processes (3). TORC2 is largely insensitive to rapamycin and appears to regulate spatial aspects of growth, such as cell polarity (4). While there are currently no known TORC2 specific inhibitors, TORC1 can be specifically inhibited with rapamycin, which has been used to characterize the TORC1 pathway in both mammals and budding yeast (2). With the aid of rapamycin, the yeast TORC1 pathway has been extensively investigated (see the comprehensive review on yeast TORC1 in (3)). Numerous distal readouts of the yeast TORC1 pathway and distinct signaling branches that are regulated by TORC1/rapamycin have been identified in budding yeast, including: 1) the RTG signaling pathway mediated by Rtg1p/Rtg3p that activates genes required for biosynthesis and homeostasis of glutamate and glutamine (5–8); 2) the nitrogen-discrimination pathway (NDP) mediated by Gln3p that activates genes enabling cells to import and catabolize poor nitrogen sources under nitrogen limitations (9, 10); 3) the stress-response pathway mediated by Msn2p/Msn4p that regulates the transcription response to a wide range of stressors (11); and 4) signaling that controls translation, such as ribosomal protein synthesis, translation initiation and mRNA turnover (12, 13). TORC1 regulates gene expression in these pathways mainly by controlling translocation of the transcription factors. The downstream effectors or substrates that link TORC1 activity to these readouts are not well understood. So far, only Sch9p kinase and Tap42p phosphatase have been identified as direct TORC1 substrates that mediate TOR signaling to its distal readouts (14, 15). More effectors and substrates need to be identified. Moreover, these signaling branches are not independent, but rather engage in substantial cross-talk while also interacting with other signaling pathways (16–18), thus constituting a complicated regulatory network. Therefore, there is an ongoing need to identify novel components and mechanisms in the TORC1 pathway as well as to isolate new chemical probes to delineate the TORC1 pathway.
The mammalian TOR cognate, mTOR has emerged as a therapeutic cancer target due to its central roles in controlling cell growth (1). Rapamycin (or its analogs) is a first generation TOR inhibitor that has shown promising results in preclinical pharmacological studies, but has not lived up to expectations in clinical trials (1, 19, 20). New mTOR inhibitors or novel chemicals that act in concert with rapamycin would be valuable (21, 22). Although more potent ATP-competitive mTOR inhibitors that target both mTORC1 and mTORC2 have been developed (23–25), small molecules that selectively and potently inhibit either TORC1 or TORC2 are lacking. These molecules are anticipated as the new generation of TOR inhibitors and are likely suitable for unveiling therapeutically relevant mechanisms (20).
Budding yeast has been a useful system for high throughput screening (HTS) and for drug target identification and mechanism discovery (26–28). More importantly, molecules identified from yeast screens have potential for translation into higher organisms (22, 29). Flow cytometry is a versatile high speed cell analysis method for proteomics and systems biology (30). HT flow cytometry (HTFC), such as HyperCyt®, enables the processing of 96- or 384-well plates in as little as 3 and 12 min, respectively. It is therefore well suited for large-scale cell screening and selection applications (31–34), such as budding yeast in suspension cell culture. Taking advantage of the yeast GFP collection which consists of 4,159 GFP tagged ORFs comprising 75% of the yeast proteome (35), we identified rapamycin-responsive GFP clones and conducted a multiplexed HTFC screen to search for compounds that alter GFP expression in five rapamycin-responsive GFP clones. These five GFP clones represent the readouts of four branches of the TORC1 signaling pathway (Figure 1A), and allows evaluation of compound activity on multiple branches simultaneously. We sought to identify molecules that functionally mimic rapamycin with distinct structure as well as molecules selective for individual branches that could target effectors in the TORC1 pathway or interfere with other non-TOR, cross-talk signaling mechanisms. These molecules would represent new chemical tools for delineation of the yeast TOR pathway or serve as potential drug leads for mTOR based therapies.
Figure 1.
Development of multiplexing strategies for HTFC screen of the yeast TORC1 pathway. A) Schematic diagram of the yeast TORC1 pathway showing four signaling branches mediated by various transcription factors and the distal readouts. B) Time-course analysis of GFP expression in rapamycin-responsive GFP clones. GFP clones were treated with 1% DMSO and 0.22 μM rapamycin respectively, and GFP signal was measured by flow cytometry at hourly intervals up to 3hrs. C, D) Multiplexing strategy. Five GFP clones were diluted to 0.5 OD600 mL−1 and differentially stained with Alexa Fluor® 405 and Alexa Fluor® 633 dyes according to the staining scheme (C). The multiplex staining was analyzed by flow cytometry (D). Five Distinct cell populations were distinguished in the dot plot, and each gate (1–5) represents an individual GFP clone (Left panel, 0 hr). The multiplex was stable for 3 hrs under normal yeast growth conditions (right panel, 3 hr) with a distinct population of unbarcoded daughter cells observed (gate 6). The GFP histogram of AGP1 cells (Gate 2) was displayed at the upper-right, showing no cross-contamination of different clones in the multiplex after 3 hrs.
Here we report the screening results from the Molecular Libraries Small Molecule Repository (MLSMR) consisting of ~320,000 compounds. We identified a small molecule CID 3528206 that alters GFP expression in all five GFP clones in an analogous manner to rapamycin and showed that this molecule behaved in a manner consistent with inhibition of yeast TORC1 in follow-up biochemical and cell-based assays.
RESULTS AND DISCUSSION
Identification of rapamycin-responsive yeast GFP clones
To determine the appropriate TORC1-regulated GFP clones for HTS, we evaluated the yeast GFP collection for rapamycin-responsive clones. A total of 106 GFP clones were identified as responsive to rapamycin in YPD media from the primary screen and 58 clones were confirmed (Supplemental Table S1). GFP fluorescence was down-regulated in 18 clones and up-regulated in 40 clones. Note that all down-regulated clones encode ribosomal proteins, which is in agreement with the fact that rapamycin inhibits protein synthesis. Twenty-six of the up-regulated clones encode proteins involved in RTG, NDP, SPS (36), and stress response pathways that are known to be regulated by the TORC1 pathway (Supplemental Table S1). Other rapamycin up-regulated clones encode proteins that have not yet been linked to the TORC1 pathway, including several uncharacterized ORFs (Supplemental Table S1). Characterization of these GFP clones may identify novel components in the yeast TORC1 pathway, and may prove valuable for evaluation of compounds identified from the subsequent HTS. Since YPD medium compromised the multiplexing staining protocol (see below), we retested 96 positive GFP clones from the primary screen in SC media. A total of 50 rapamycin-responsive GFP clones were confirmed in SC media (Supplemental Table S1). To determine the GFP clones for yielding good Z′ (>0.5), a statistical factor to assess the quality of HTS (37), we picked the ones that showed a robust response to rapamycin in SC media for a 3-hr time-course study (Figure 1B and data not shown). With DMSO treatment, the GFP signal in the CIT2, AGP1, MEP2 and LAP4 clones remained constant over time. With rapamycin treatment, the GFP signal in these clones increased greater than 3-fold at 3 hr compared to DMSO treatment. In contrast, the GFP signal in the RPL19A clone increased steadily with DMSO treatment but remained unchanged with rapamycin treatment, resulting in a 1.5-fold decrease of the GFP signal with rapamycin treatment compared to DMSO treatment. We selected these five GFP clones, CIT2, AGP1, MEP2, LAP4 and RPL19A representing four signaling branches of the yeast TORC1 pathway for multiplexed HTS (Figure 1A).
Development of a 5-plex strategy
Multiplexing is a powerful capability of flow cytometry and various strategies have been developed for beads, fixed and live cells (38). We created a live cell-based multiplex using “barcoding” to discriminate the different clones in the multiplex. We used Alexa Fluor 405® (Ex/Em: 400/424 nm) and/or Alexa Fluor® 633 (Ex/Em: 632/647 nm) to stain the yeast cells at two levels of fluorescence intensity and then interrogated the cells for changes in GFP expression (Ex/Em: 488/507 nm). The staining scheme was shown in Figure 1C. In Figure 1D (0 hr), we can readily distinguish five distinct cell populations (gates 1–5). To test the stability of the barcode, we analyzed the stained multiplex at hourly intervals up to 4 hrs under normal yeast growth condition (Figure 1D and data not shown). After 3 hrs incubation, these five cell populations remained distinguishable (Figure 1D, 3 hrs), and when gating on a discrete population, the resulting GFP histogram was a homogeneous population with negligible contamination from other clones. However, a distinct population (~50%) of new-born daughter cells was generated at 3 hrs (Figure 1D, 3 hrs, gate 6), and it dominated the culture (80%) after 4 hrs (data not shown). As only the stained cells (Mother cells) in gates 1–5 are analyzed, too many daughter cells result in insufficient number of mother cells for data analysis. To balance the duration of compound incubation and mother cell number, we selected 3 hrs for HTS.
Identification of compounds that modulate expression of GFP fusion genes
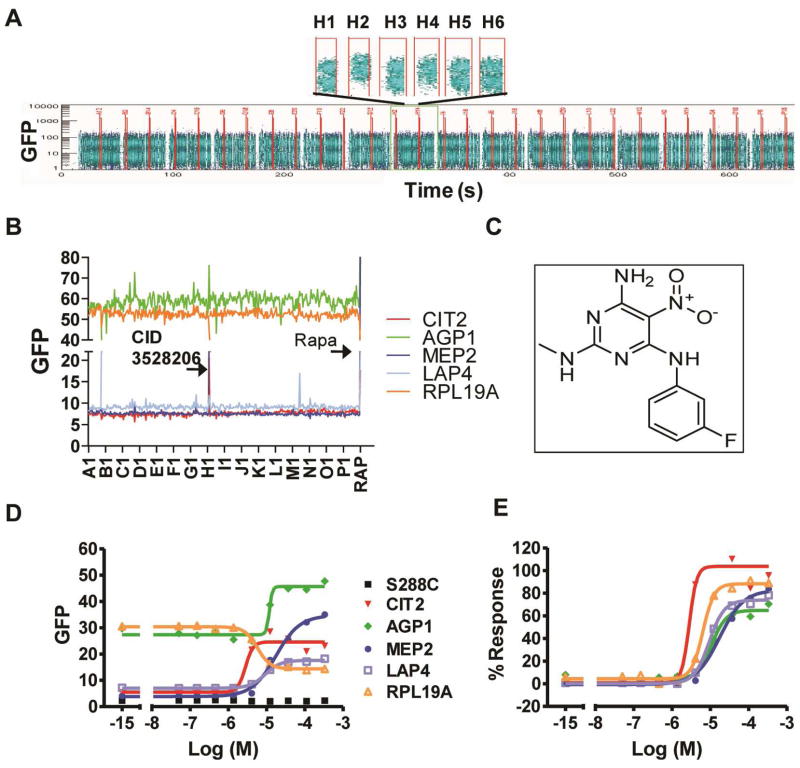
HTS was carried out in a 5-plex assay format in which CIT2, AGP1, MEP2, LAP4 and RPL19A GFP fusion clones were color-coded with Alexa Fluor® dyes and evaluated simultaneously with respect to the alteration of GFP expression. Gating based on FL6 and FL8 emission distinguished the five GFP clones as shown in Figure. 1D. The original screening data and the intensity of GFP signal of each clone in each well from the sample plate ML32337 is displayed in Figure 2A and 2B, respectively, showing the hit compound CID 3528206 in well H4. The average Z′ were all above 0.5 for each target clone (data not shown). Primary screening of the MLSMR (~320,000 compounds) in 5-plex format resulted in the identification of 210, 51, 1682, 1090 and 982 active compounds for CIT2, AGP1, MEP2, LAP4 and RPL19A clones, respectively (Table 1). The complete results from the multiplex screen are available on PubChem (PubChem summary AID 1908, http://pubchem.ncbi.nlm.nih.gov). A total of 255 active compounds were confirmed: 19, 19, 12, 205 and 102 actives for the CIT2, AGP1, MEP2, LAP4 and RPL19A clones, respectively (Table 1). We then analyzed the distribution of these 255 confirmed compounds among the five GFP clones. A total of 176 compounds selectively targeted one clone while 79 compounds targeted multiple clones, among which, 1, 2, 16 and 60 compounds altered GFP expression in five, four, three and two clones, respectively (Table 2). This distribution pattern suggests that we may have identified compounds that target either various signaling nods in the yeast TORC1 pathway or target pathways engaged in cross-talk with TORC1. Notably, a large number of hits (67 in total) target both LAP4 and RPL19A clones. We observed that dozens of LAP4 hits that were statistically inactive for RPL19A in the primary single-point screen were confirmed active for the RPL19A clone in the dose-response analysis. This observation may suggest the co-regulation of LAP4 and RPL19A branches.
Figure 2.
Confirmation of CID 3528206 as a hit from the multiplexed HTS. A) Screen shot of sample plate ML32337. Each block represents a group of 20 samples and 2 controls (one positive and one negative control), separated by blank wells. Each sample contains data for 5 targets. The negative control (DMSO, well H1), positive control (rapamycin, well H2) and a potential hit (well H4) were shown above. B) The GFP signal of each strain in each well of plate ML32337 was analyzed by HyperView® software. The arrows denote the hit in well H4 (CID 3528206) and the rapamycin positive control. C) Chemical structure of CID 3528206. D, E) Dose-dependent regulation of GFP expression in yeast GFP clones by CID 3528206. Raw median channel fluorescence of GFP signal (D) and percent response with respect to rapamycin (E) of each GFP clone were graphed.
Table 1.
Hits summary from the primary and confirmatory screening
| Targets | CIT2 | AGP1 | MEP2 | LAP4 | RPL19A |
|---|---|---|---|---|---|
| Primary hits 1 | 210 | 51 | 1682 | 1090 | 982 |
| Confirmed hits 2 | 19 | 19 | 12 | 205 | 102 |
A total of ~320, 000 compounds were screened.
A total of ~2,500 compounds were screened in single point confirmation. A total of 613 compounds were screened in dose response confirmation. A total of 255 hits were confirmed.
Table 2.
Distribution of confirmed hits
| # of hits | 1 | 2 | 1 | 4 | 1 | 4 | 6 | 5 | 3 | 2 | 50 | 6 | 7 | 2 | 128 | 33 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CIT2 | * | * | * | * | * | * | ||||||||||
| AGP1 | * | * | * | * | * | * | * | |||||||||
| MEP2 | * | * | * | * | * | |||||||||||
| LAP4 | * | * | * | * | * | * | * | * | * | * | * | |||||
| RPL19A | * | * | * | * | * | * | * | * |
active for the indicated GFP clones
While all these molecules are interesting and worthwhile pursuing, subsequent characterization and target identification remains challenging. For instance, the hits for LAP4-GFP clone may act on pathways including PKA, autophagy, stress response and TORC1 pathways. Since it is unlikely one can design a simple assay to clarify all these hits, we focused on the hit CID 3528206 that most likely acts on TORC1 as it alters GFP expression in all five GFP clones in an analogous manner to rapamycin. We are also characterizing the LAP4 hits to validate potential autophagy regulators (data not shown).
Purified CID 3528206 powder was obtained to confirm its activity in the dose-response assay (Figure 2D–E). CID 3528206 increased GFP expression in the CIT2, AGP1, MEP2 and LAP4 clones and decreased GFP expression in the RPL19A clone (Figure 2D). Its EC50 on all clones ranges from 3–13 μM. The percentage response of the clones to CID 3528206 varied from 40–100% compared to rapamycin (Figure 2E). This indicates that this small molecule represents a distinct chemical structure that may function like rapamycin (Figure 2C).
Compound CID 3528206 inhibits yeast cell growth, but is not toxic
As rapamycin arrests yeast cell growth (39, 40), we first determined the toxicity and effect of CID 3528206 on cell growth. Compared to DMSO treatment, CID 3528206 significantly inhibited cell growth at 20 μM (Figure 3A). The growth inhibition caused by CID 3528206 was dose-dependent (Figure 3B). To test if CID 3528206 is toxic to yeast cells, we evaluated yeast viability following treatment with 30 μM CID 3528206 by a standard colony forming unit (cfu) assay (Figure 3C). Similar to DMSO and rapamycin treatments, greater than 90% of the cells treated with 30 μM CID 3528206 were viable at all three time points analyzed. In contrast, a 4-hour treatment with the fungicidal compound amphotericin B reduced the corresponding cfu to levels below 0.5% when compared to control cells. These data indicate that CID 3528206 inhibits yeast cell growth, but is not toxic at a concentration of 30 μM.
Figure 3.
CID 3528206 inhibits yeast cell growth, but is not toxic. A) CID 3528206 inhibits yeast cell growth. BY4741 cells were treated with 1% DMSO, 0.22 μM rapamycin and 20 μM CID 3528206 (Cmpd) in duplicate, respectively. OD600 was measured at different time points. B) Dose-dependent inhibition of yeast cell growth by CID 3528206. BY4741 cells were treated with varying concentrations of CID 3528206 (Cmpd) or control compounds in duplicate. OD600 was measured after 5.5 hrs treatment. C) Yeast viability assay. BY4741 Cells were treated with 30 μM CID 3528206 (Cmpd), rapamycin (Rapa), amphotericin B (Amph) and DMSO in triplicate, respectively.OD600 and cfu were measured at different time points, and percent viability was calculated by cfus relative to the number of cells. D) CID 3528206 inhibited growth of wild-type cells but not the TORC1 bypass cells. A series of 10-fold dilutions of BY4741 and TORC1 bypass cells were spotted onto YPD plates containing indicated concentrations of CID 3528206 or control compounds, and grown for 3 days.
CID 3528206 inhibits yeast cell growth through modulation of TORC1
Rapamycin inhibits TORC1 activity allosterically when complexed with Fpr1p and thus exhibits a cytostatic effect on wild-type yeast cell growth. When the downstream TORC1 effectors are constitutively active, cells can bypass TORC1 activity and resist rapamycin treatment, such as with the TORC1 bypass cells bearing SCH9 2D3E and GLN3 KO alleles (41, 42). We tested whether the TORC1 bypass cells resist CID 3528206 treatment (Figure 3D). Consistent with the previous report, rapamycin inhibited wild-type cell growth but not TORC1 bypass cell growth at 0.2 μM (41, 42). CID 3528206 also inhibited wild-type cell growth but not TORC1 bypass cell growth at 30 μM, indicating that the growth inhibition effect of CID 3528206 is mediated by TORC1, and suggesting that CID 3528206 either acts, like rapamycin, directly on TORC1, or upstream of TORC1.
CID 3528206 inhibits TORC1 in vivo and in vitro
To investigate whether CID 3528206 inhibits TORC1, we first examined the in vivo phosphorylation of Sch9p (a direct substrate of TORC1) (42). Like rapamycin, CID 3528206 caused dose-dependent dephosphorylation of the C-terminal phosphorylation sites in Sch9p (Figure 4A). TORC1 activity was quantified in Figure 4B, and IC50 was calculated as 3.9 μM, which is consistent with the range of EC50s of all clones shown in Figure 2D. This result indicates that CID 3528206 inhibits TORC1 activity in vivo, suggesting that CID 3528206 acts on TORC1 or upstream. To ask if CID 3528206 targets TORC1 directly, we performed TORC1 in vitro kinase assay (Figure 4C, 4D). CID 3528206 completely inhibited Sch9p phosphorylation at 1 μM or higher concentrations. CID 3528206 also inhibited Kog1p auto-phosphorylation to the greatest extent (~70% inhibition) at 1 μM or higher concentrations. Dose-response curve-fitting showed that CID 3528206 inhibited both Sch9p and Kog1 phosphorylation with IC50s ~ 150 nM (Figure 4D). This result favors the possibility that CID 3528206 targets TORC1 directly rather than acting on its upstream components. Further identification of TORC1 mutants resistant to CID 3528206 could confirm this hypothesis. Note that the in vitro IC50 (150 nM) is 20-fold lower than the in vivo IC50 (3.9 μM), which may be due to cell permeability or efflux systems that result in a lower cellular concentration of CID 3528206.
Figure 4.
CID 3528206 directly inhibits TORC1. A, B) CID 3528206 inhibited TORC1 activity in vivo. Yeast cells expressing Sch9p-5HA were treated with DMSO vehicle or varying concentrations of CID 3528206 for 30 min. A) Western blots using anti-HA antibody to detect the extent of Sch9p C-terminal phosphorylation; B) The ratio of the hyperphosphorylated over the hypophosphorylated Sch9p was used to quantify TORC1 activity in vivo. C, D) CID 3528206 inhibited TORC1 activity in vitro. In vitro kinase assay using Sch9p as substrate was performed in triplicate (C) and quantified as Mean ± SD (D).
Genome-wide comparison of CID 3528206 with rapamycin on yeast gene expression
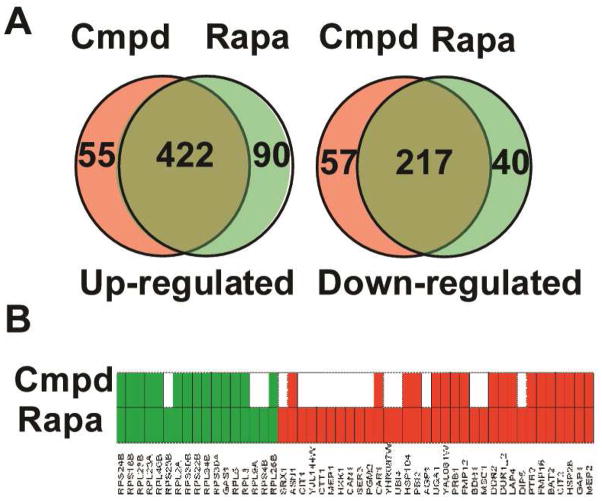
To compare the effect of CID 3528206 and rapamycin on yeast gene expression, we performed microarray analysis using Affymetrix® yeast gene chips (Figure 5A). CID 3528206 significantly altered the transcription of 751 yeast genes, among which 477 genes were up-regulated and 274 genes were down-regulated. Rapamycin significantly increased the transcription of 512 genes and decreased the transcription of 257 genes. Venn diagram analysis showed that 422 genes up-regulated by CID 3528206 (88.5%) were also up-regulated by rapamycin while 217 genes that were down-regulated by CID 3528206 (79.2%) were also down-regulated by rapamycin. Overall, CID 3528206 and rapamycin shared 85.1% regulated genes. Such a significant overlap in gene expression profiling indicates that CID 3528206 and rapamycin modulate similar cellular pathways. Since the mRNA levels do not necessarily reflect the protein levels, we took advantage of the rapamycin-responsive GFP clones identified in the yeast GFP collection to test whether CID 3528206 altered GFP expression in these clones (Figure 5B). The GFP signal in 14 out of 17 rapamycin-down-regulated clones was decreased by CID 3528206. Note that GFP expression in the other three rapamycin-down-regulated clones was also decreased by CID 3528206 to 0.67–0.72 fold, which is slightly higher than the 0.66-fold cut-off value. This result indicates that CID 3528206 down-regulated GFP expression in almost all rapamycin down-regulated clones. The GFP signal in 19 out of 33 (57.6%) rapamycin-up-regulated clones was up-regulated by CID 3528206. Overall, CID 3528206 altered GFP expression in 33 out of 50 (66.0%) rapamycin-responsive GFP clones. This significant overlap is in good agreement with the microarray data, and confirms the common cellular function of CID 3528206 and rapamycin.
Figure 5.
CID 3528206 shares common cellular function with rapamycin. A) Venn diagram analysis of the CID 3528206-regulated genes and rapamycin-regulated genes identified from microarray analysis. B) CID 3528206 altered GFP expression of rapamycin-responsive GFP clones. Fifty rapamycin-responsive GFP clones (Supplemental table S1) were grown in SC media and treated with DMSO, 30 μM CID 3528206 or 0.22 μM rapamycin respectively for 3 hrs in triplicate. GFP fluorescence was measured by flow cytometry. A 1.5-fold change in fluorescence was used as the cut-off value for up-regulated (Red) and down-regulated (Green) clones.
Preliminary structure-activity relationship (SAR) exploration
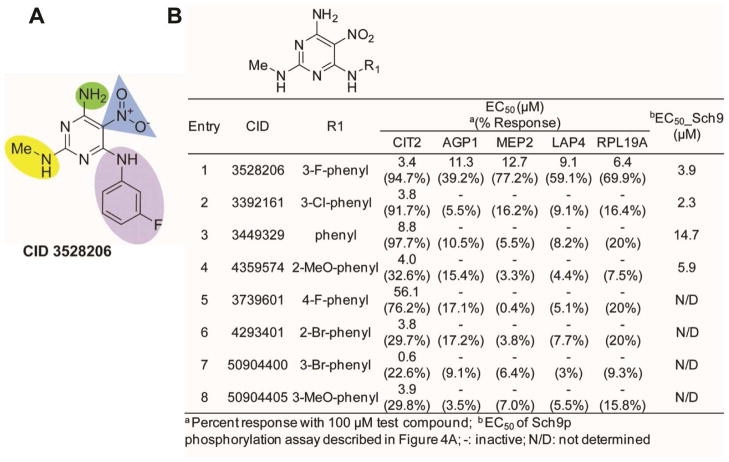
To explore the SAR optimization, we surveyed 57 analogs where the substitutions were focused on four regions of the scaffold (Figure 6A, shaded regions). Alteration of the free primary amino substituent (Figure 6A, green shaded region) or modification of the nitro group (Figure 6A, blue shaded region) was not tolerated. Modest changes in alkyl substituent of the methyl amine (Figure 6A, yellow shaded area) led to one active compound, but with an altered activity profile (data not shown). Interestingly, changes to the 3-fluorophenyl appendage (Figure 6A, purple shaded region) afforded a set of analogs that appeared to exclusively alter CIT2 GFP expression, unlike CID 3528206, which modulated GFP expression in all five GFP clones (Figure 6B). Surprisingly, these CIT2 branch-selective analogs also inhibited Sch9p phosphorylation in vivo (Figure 6B). Due to solubility challenges with some analogs in this series (data not shown), the possible link between physiochemical properties and CIT2 branch selectivity is being investigated with the pursuit of compounds with enhanced solubility.
Figure 6.
SAR optimization of CID 3528206. A) Shaded regions of targeted SAR optimization. B) Active analogs generated from changes to the 3-fluorophenyl appendage (purple shaded region). The results of dose-response and in vivo Sch9p phosphorylation assays are shown.
CONCLUSION
In summary, the flow cytometry based multiplexing HTS has successfully identified a yeast TORC1 inhibitor and provided a novel scaffold for further development of TOR inhibitors. Interestingly, preliminary SAR exploration on the single chemotype identified analogs exhibiting both rapamycin-like activity and CIT2/RTG branch selective activity. Further investigation of structural and biological basis of the analogs may reveal novel mechanisms in the yeast TORC1 pathway. As the TOR complexes and signaling pathways are highly conserved (2, 3), CID 3528206 may have potential for inhibiting mTORC1 activity. Characterization of the activity of CID 3528206 in other organisms and comprehensive SAR analysis are under way to elucidate the mechanism of action of CID 3528206.
METHODS
Yeast strains and reagents
Yeast strains and plasmids used in this study are listed in supplemental Table S2 and S3, respectively. The yeast GFP collection (Invitrogen) was screened as described previously (43).
The fluorescent dyes Alexa Fluor 405® and Alexa Fluor 633® as well as Pluronic F-68 were purchased from Invitrogen. Rapamycin was purchased from TOCRIS Biosciences.
Multiplexed HTS
The CIT2, AGP1, MEP2, LAP4 and RPL19A GFP fusion clones were grown separately overnight in 15 mL SC media at 30 °C to 0.5–1.0 OD600. Approximately 10 OD600 cells were washed with PBS buffer once and resuspended in 10 mL PBS buffer supplemented with 0.03% (v/v) Pluronic F-68. Different combinations of Alexa Fluor® 405 (0, 16 uL, 100 uL of 1 mg mL−1 DMSO stock solution) and Alexa Fluor® 633 (0, 4 uL, 100 uL of 1 mg mL−1 DMSO stock solution) dyes were added to cells and incubated for 45 min at room temperature. Cells were washed with PBS twice and resuspended in SC media. Stained cells were combined and diluted into fresh SC media at 0.5 OD600. 10 μL mixture was added to each well in 384-well plates in the sequence below: 1st, 5 μL SC media supplemented with 0.03% Pluronic F-68; 2nd, 100 nL compounds; 3rd, 5 μL mixed cells. Plates were incubated at 30 °C upside down for 3 hrs. The cells in the multiplex were interrogated for GFP expression levels using the established HTFC at UNMCMD (44). Flow cytometric light scatter and fluorescence emission at 530 ± 20 nm (FL1), 665 ± 10 nm (FL8) and 450 ± 25 nm (FL6) are collected. The resulting time-gated data files were analyzed with HyperView® software to determine compound activity in each well. A compound was considered active if the change in fluorescence was greater than 50% of DMSO-treated controls. The quality control statistic Z′ (37) for each GFP clone was calculated with the GFP fluorescence of rapamycin- and DMSO-treated controls. Active compounds were further confirmed in single point and dose-response analyses in a single-plex format. A counter-screen for green fluorescent compounds was also performed using a non-GFP parental strain S288c.
Yeast viability and cell growth assays
Colony forming unit (cfu) assay was performed to evaluate yeast viability. BY4741 cells were treated with 1% DMSO, 0.22 μM rapamycin, 30 μM CID 3528206, or 2.5 μg mL−1 amphotericin B in triplicate for 24 hrs. OD600 and cfu were measured at 4, 7 and 24 hrs. Percent viability was calculated by cfus relative to the number of cells in the culture. The TORC1 bypass (MP138-4C cell containing p1290) and BY4741 cells spot assay was performed as described (41, 42).
Sch9p in vitro and in vivo phosphorylation assays
Sch9p in-vitro phosphorylation assay was performed as described (41, 42). Sch9p in-vivo phosphorylation assay was performed using YL515 cells containing plasmids pRS413, pRS416 and pJU1058 in the chemical fragmentation analysis as described (41, 42).
Microarray analysis
Overnight cultures of BY4741 cells in YPD were diluted to 0.05 OD600 and allowed to grow to 0.2 OD600. Cells were treated with 0.22 μM rapamycin, 20 μM CID 3528206 and 1% DMSO vehicle control in duplicate for 3 hrs, respectively. Cells were collected and RNA was isolated with MasterPure® Yeast RNA purification kit (Epicentre). cDNA probes were generated from total RNA and used for hybridization to yeast gene chips (Affymetrix) at Keck-UNM Genomics Resource. The RMA algorithm of Expression Consol (v1.1, Affymetrix) was used to generate and normalize signal intensities. The significant analysis of microarray (SAM) (45) was performed with a false discovery rate (FDR) of 0.1 to identify the genes whose expressions are significantly up- or down-regulated by CID 3528206 or rapamycin compared to DMSO treatment. The microarray data (Accession number GSE33320) are available at http://www.ncbi.nlm.nih.gov/geo.
Supplementary Material
Acknowledgments
We thank S. Chavez, D. Perez, A. Evangelisti, M. Garcia, V. Salas, K. Gouveia, G. Pickett, E. Manzanilla, K. Niewola-Staszkowska, and S. Haesendonckx for technical assistance. This research was supported by NIH grants U54MH084690, U54HG005031 and R03 MH086450-01 and a grant from the Swiss National Foundation (CDV).
Footnotes
Supporting Information Available: This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Virgilio C, Loewith R. The TOR signalling network from yeast to man. Int J Biochem Cell Biol. 2006;38:1476–1481. doi: 10.1016/j.biocel.2006.02.013. [DOI] [PubMed] [Google Scholar]
- 3.De Virgilio C, Loewith R. Cell growth control: little eukaryotes make big contributions. Oncogene. 2006;25:6392–6415. doi: 10.1038/sj.onc.1209884. [DOI] [PubMed] [Google Scholar]
- 4.Kamada Y, Fujioka Y, Suzuki NN, Inagaki F, Wullschleger S, Loewith R, Hall MN, Ohsumi Y. Tor2 directly phosphorylates the AGC kinase Ypk2 to regulate actin polarization. Mol Cell Biol. 2005;25:7239–7248. doi: 10.1128/MCB.25.16.7239-7248.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sekito T, Thornton J, Butow RA. Mitochondria-to-nuclear signaling is regulated by the subcellular localization of the transcription factors Rtg1p and Rtg3p. Mol Biol Cell. 2000;11:2103–2115. doi: 10.1091/mbc.11.6.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Giannattasio S, Liu Z, Thornton J, Butow RA. Retrograde response to mitochondrial dysfunction is separable from TOR1/2 regulation of retrograde gene expression. J Biol Chem. 2005;280:42528–42535. doi: 10.1074/jbc.M509187200. [DOI] [PubMed] [Google Scholar]
- 7.Sekito T, Liu Z, Thornton J, Butow RA. RTG-dependent mitochondria-to-nucleus signaling is regulated by MKS1 and is linked to formation of yeast prion [URE3] Mol Biol Cell. 2002;13:795–804. doi: 10.1091/mbc.01-09-0473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Z, Sekito T, Epstein CB, Butow RA. RTG-dependent mitochondria to nucleus signaling is negatively regulated by the seven WD-repeat protein Lst8p. EMBO J. 2001;20:7209–7219. doi: 10.1093/emboj/20.24.7209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crespo JL, Daicho K, Ushimaru T, Hall MN. The GATA transcription factors GLN3 and GAT1 link TOR to salt stress in Saccharomyces cerevisiae. J Biol Chem. 2001;276:34441–34444. doi: 10.1074/jbc.M103601200. [DOI] [PubMed] [Google Scholar]
- 10.Bertram PG, Choi JH, Carvalho J, Ai W, Zeng C, Chan TF, Zheng XF. Tripartite regulation of Gln3p by TOR, Ure2p, and phosphatases. J Biol Chem. 2000;275:35727–35733. doi: 10.1074/jbc.M004235200. [DOI] [PubMed] [Google Scholar]
- 11.Beck T, Hall MN. The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature. 1999;402:689–692. doi: 10.1038/45287. [DOI] [PubMed] [Google Scholar]
- 12.Xiao L, Grove A. Coordination of Ribosomal Protein and Ribosomal RNA Gene Expression in Response to TOR Signaling. Curr Genomics. 2009;10:198–205. doi: 10.2174/138920209788185261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Powers T, Walter P. Regulation of ribosome biogenesis by the rapamycin-sensitive TOR-signaling pathway in Saccharomyces cerevisiae. Mol Biol Cell. 1999;10:987–1000. doi: 10.1091/mbc.10.4.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huber A, Bodenmiller B, Uotila A, Stahl M, Wanka S, Gerrits B, Aebersold R, Loewith R. Characterization of the rapamycin-sensitive phosphoproteome reveals that Sch9 is a central coordinator of protein synthesis. Genes Dev. 2009;23:1929–1943. doi: 10.1101/gad.532109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shamji AF, Kuruvilla FG, Schreiber SL. Partitioning the transcriptional program induced by rapamycin among the effectors of the Tor proteins. Curr Biol. 2000;10:1574–1581. doi: 10.1016/s0960-9822(00)00866-6. [DOI] [PubMed] [Google Scholar]
- 16.Soulard A, Cremonesi A, Moes S, Schutz F, Jeno P, Hall MN. The rapamycin-sensitive phosphoproteome reveals that TOR controls protein kinase A toward some but not all substrates. Mol Biol Cell. 2010;21:3475–3486. doi: 10.1091/mbc.E10-03-0182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pedruzzi I, Dubouloz F, Cameroni E, Wanke V, Roosen J, Winderickx J, De Virgilio C. TOR and PKA signaling pathways converge on the protein kinase Rim15 to control entry into G0. Mol Cell. 2003;12:1607–1613. doi: 10.1016/s1097-2765(03)00485-4. [DOI] [PubMed] [Google Scholar]
- 18.Vinod PK, Sengupta N, Bhat PJ, Venkatesh KV. Integration of global signaling pathways, cAMP-PKA, MAPK and TOR in the regulation of FLO11. PLoS One. 2008;3:e1663. doi: 10.1371/journal.pone.0001663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Benjamin D, Colombi M, Moroni C, Hall MN. Rapamycin passes the torch: a new generation of mTOR inhibitors. Nat Rev Drug Discov. 2011;10:868–880. doi: 10.1038/nrd3531. [DOI] [PubMed] [Google Scholar]
- 20.Guertin DA, Sabatini DM. The pharmacology of mTOR inhibition. Sci Signal. 2009;2:pe24. doi: 10.1126/scisignal.267pe24. [DOI] [PubMed] [Google Scholar]
- 21.Aghajan M, Jonai N, Flick K, Fu F, Luo M, Cai X, Ouni I, Pierce N, Tang X, Lomenick B, Damoiseaux R, Hao R, Del Moral PM, Verma R, Li Y, Li C, Houk KN, Jung ME, Zheng N, Huang L, Deshaies RJ, Kaiser P, Huang J. Chemical genetics screen for enhancers of rapamycin identifies a specific inhibitor of an SCF family E3 ubiquitin ligase. Nat Biotechnol. 2010;28:738–742. doi: 10.1038/nbt.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sarkar S, Perlstein EO, Imarisio S, Pineau S, Cordenier A, Maglathlin RL, Webster JA, Lewis TA, O’Kane CJ, Schreiber SL, Rubinsztein DC. Small molecules enhance autophagy and reduce toxicity in Huntington’s disease models. Nat Chem Biol. 2007;3:331–338. doi: 10.1038/nchembio883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Q, Wang J, Kang SA, Thoreen CC, Hur W, Ahmed T, Sabatini DM, Gray NS. Discovery of 9-(6-Aminopyridin-3-yl)-1-(3-(trifluoromethyl)phenyl)benzo[h][1,6]naphthyr idin-2(1H)-one (Torin2) as a Potent, Selective, and Orally Available Mammalian Target of Rapamycin (mTOR) Inhibitor for Treatment of Cancer. J Med Chem. 2011;54:1473–1480. doi: 10.1021/jm101520v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Q, Chang JW, Wang J, Kang SA, Thoreen CC, Markhard A, Hur W, Zhang J, Sim T, Sabatini DM, Gray NS. Discovery of 1-(4-(4-propionylpiperazin-1-yl)-3-(trifluoromethyl)phenyl)-9-(quinolin-3- yl)benzo[h][1,6]naphthyridin-2(1H)-one as a highly potent, selective mammalian target of rapamycin (mTOR) inhibitor for the treatment of cancer. J Med Chem. 2010;53:7146–7155. doi: 10.1021/jm101144f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feldman ME, Apsel B, Uotila A, Loewith R, Knight ZA, Ruggero D, Shokat KM. Active-site inhibitors of mTOR target rapamycin-resistant outputs of mTORC1 and mTORC2. PLoS Biol. 2009;7:e38. doi: 10.1371/journal.pbio.1000038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoon S, St Onge RP, Giaever G, Nislow C. Yeast chemical genomics and drug discovery: an update. Trends Pharmacol Sci. 2008;29:499–504. doi: 10.1016/j.tips.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 27.Giaever G. A chemical genomics approach to understanding drug action. Trends Pharmacol Sci. 2003;24:444–446. doi: 10.1016/S0165-6147(03)00225-6. [DOI] [PubMed] [Google Scholar]
- 28.Armour CD, Lum PY. From drug to protein: using yeast genetics for high-throughput target discovery. Curr Opin Chem Biol. 2005;9:20–24. doi: 10.1016/j.cbpa.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 29.Minic J, Sautel M, Salesse R, Pajot-Augy E. Yeast system as a screening tool for pharmacological assessment of g protein coupled receptors. Curr Med Chem. 2005;12:961–969. doi: 10.2174/0929867053507261. [DOI] [PubMed] [Google Scholar]
- 30.Nolan JP, Yang L. The flow of cytometry into systems biology. Brief Funct Genomic Proteomic. 2007;6:81–90. doi: 10.1093/bfgp/elm011. [DOI] [PubMed] [Google Scholar]
- 31.Young SM, Curry MS, Ransom JT, Ballesteros JA, Prossnitz ER, Sklar LA, Edwards BS. High-throughput microfluidic mixing and multiparametric cell sorting for bioactive compound screening. J Biomol Screen. 2004;9:103–111. doi: 10.1177/1087057103262335. [DOI] [PubMed] [Google Scholar]
- 32.Sklar LA, Carter MB, Edwards BS. Flow cytometry for drug discovery, receptor pharmacology and high-throughput screening. Curr Opin Pharmacol. 2007 doi: 10.1016/j.coph.2007.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Edwards BS, Oprea T, Prossnitz ER, Sklar LA. Flow cytometry for high-throughput, high-content screening. Curr Opin Chem Biol. 2004;8:392–398. doi: 10.1016/j.cbpa.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 34.Black CB, Duensing TD, Trinkle LS, Dunlay RT. Cell-based screening using high-throughput flow cytometry. Assay Drug Dev Technol. 2011;9:13–20. doi: 10.1089/adt.2010.0308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O’Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 36.Ljungdahl PO. Amino-acid-induced signalling via the SPS-sensing pathway in yeast. Biochem Soc Trans. 2009;37:242–247. doi: 10.1042/BST0370242. [DOI] [PubMed] [Google Scholar]
- 37.Zhang JH, Chung TD, Oldenburg KR. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 38.Krutzik PO, Nolan GP. Fluorescent cell barcoding in flow cytometry allows high-throughput drug screening and signaling profiling. Nat Methods. 2006;3:361–368. doi: 10.1038/nmeth872. [DOI] [PubMed] [Google Scholar]
- 39.Barbet NC, Schneider U, Helliwell SB, Stansfield I, Tuite MF, Hall MN. TOR controls translation initiation and early G1 progression in yeast. Mol Biol Cell. 1996;7:25–42. doi: 10.1091/mbc.7.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakashima A, Maruki Y, Imamura Y, Kondo C, Kawamata T, Kawanishi I, Takata H, Matsuura A, Lee KS, Kikkawa U, Ohsumi Y, Yonezawa K, Kamada Y. The yeast Tor signaling pathway is involved in G2/M transition via polo-kinase. PLoS One. 2008;3:e2223. doi: 10.1371/journal.pone.0002223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wanke V, Cameroni E, Uotila A, Piccolis M, Urban J, Loewith R, De Virgilio C. Caffeine extends yeast lifespan by targeting TORC1. Mol Microbiol. 2008;69:277–285. doi: 10.1111/j.1365-2958.2008.06292.x. [DOI] [PubMed] [Google Scholar]
- 42.Urban J, Soulard A, Huber A, Lippman S, Mukhopadhyay D, Deloche O, Wanke V, Anrather D, Ammerer G, Riezman H, Broach JR, De Virgilio C, Hall MN, Loewith R. Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol Cell. 2007;26:663–674. doi: 10.1016/j.molcel.2007.04.020. [DOI] [PubMed] [Google Scholar]
- 43.Davidson GS, Joe RM, Roy S, Meirelles O, Allen CP, Wilson MR, Tapia PH, Manzanilla EE, Dodson AE, Chakraborty S, Carter M, Young S, Edwards B, Sklar L, Werner-Washburne M. The proteomics of quiescent and nonquiescent cell differentiation in yeast stationary-phase cultures. Mol Biol Cell. 2011;22:988–998. doi: 10.1091/mbc.E10-06-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Edwards BS, Young SM, Ivnitsky-Steele I, Ye RD, Prossnitz ER, Sklar LA. High-content screening: flow cytometry analysis. Methods Mol Biol. 2009;486:151–165. doi: 10.1007/978-1-60327-545-3_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.