Summary
flhE belongs to the flhBAE flagellar operon in Enterobacteria, whose first two members function in Type III secretion (T3S). In Salmonella enterica, absence of FlhE affects swarming, but not swimming, motility. Based on a chance observation of a ‘green’ colony phenotype of flhE mutants on pH indicator plates containing glucose, we have established that this phenotype is associated with lysis of flagellated cells in an acidic environment created by glucose metabolism. The flhE mutant phenotype of Escherichia coli is similar overall to that of S. enterica but is seen in the absence of glucose and, unlike in S. enterica, causes a substantial growth defect. flhE mutants have a lowered cytoplasmic pH in both bacteria, indicative of a proton leak. GFP reporter assays indicate that the leak is dependent on the flagellar system, is present before the T3S system switches to secretion of late substrates, and gets worse after the switch and upon filament assembly, leading to cell lysis. We show that FlhE is a periplasmic protein that co-purifies with flagellar basal bodies. FlhE may act as a plug or a chaperone to regulate proton flow through the flagellar T3S system.
Introduction
Bacterial flagella enable individual swimming motility through liquid or group swarming motility over a surface (Macnab, 1996, Harshey, 2003, Kearns, 2010). Flagellar biogenesis has been best studied in Salmonella enterica (Macnab, 2003, Chevance & Hughes, 2008, Harshey, 2011). The function of most genes involved in this process is now largely understood. In this work, we report on the function of flhE, one of the few flagellar genes with an unassigned role. This gene is found mainly in Enterobacteria (Liu & Ochman, 2007), where it is co-expressed with flhBA genes, which encode two major components of the Type III secretion (T3S) system. Absence of FlhE does not affect swimming motility in S. enterica (Minamino et al., 1994), but has been reported to eliminate swarming motility (Stafford & Hughes, 2007).
Flagellum assembly requires over 60 proteins. The flagellum consists of three major parts: a membrane-embedded basal body, an external hook, and a filament (Fig. 1; see farthest structure on the right). The basal body is composed of a stationary stator and a moving rotor. Force generating units called Mot proteins comprise the stator, and the basal rotor is made of proteins that constitute the cytoplasmic C-ring mounted on a membrane MS ring that is continuous with a periplasmic rod (Fig. 1; see inset). The rod penetrates the peptidoglycan layer to exit through a pore formed by two rings - P and L. Externally, the rod connects to the hook and filament. Two hook-associated proteins, or HAPs, form a short hook-filament junction. The filament is several cell lengths long, is composed of thousands of subunits of a single protein, and is held in place by a capping protein at the tip.
Fig. 1.
Expression and assembly of S. enterica flagellar proteins. Only structures/proteins relevant to this study are indicated. See text for details. OM, outer membrane; PG, peptidoglycan; IM, inner membrane; N and C, amino- and carboxy-terminal regions of secreted substrate.
Flagellar genes are expressed in a hierarchy of three temporal classes (Chevance & Hughes, 2008) (Fig. 1; flow diagram at bottom). The class I flhDC operon encodes two transcriptional regulators that direct σ70-dependent transcription from class II promoters, which control genes for assembly of the hook-basal body (HBB) substructure as well as two regulators - σ28 (FliA) and its inhibitor FlgM. Completion of the HBB signals export of FlgM, freeing σ28 to transcribe class III promoters, which control genes for the filament, the stator proteins, as well as the chemosensory pathway. The P ring (FlgI) and L ring (FlgH) proteins are secreted by the general (Sec) secretion pathway, but a T3S apparatus located at the base of the MS ring uses PMF-dependent secretion for export of all proteins that make up the rod, hook and filament (Minamino & Namba, 2008, Paul et al., 2008) (Fig. 1, inset). This apparatus consists of six integral membrane proteins (FlhA, FlhB, FliO, FliP, FliQ, FliR), three soluble components that form the ATPase complex for substrate delivery (FliI, FliH, FliJ), and four cytoplasmic proteins that act as substrate-specific chaperones (FlgN, FliA, FliS, FliT). FlhA and FlhB have substantial cytoplasmic domains, which along with the C-ring play an important role in docking the FliI ATPase (Erhardt & Hughes, 2010, Konishi et al., 2009). FlhA is involved in the early stage of export, and FlhB is important for the late stage. A recent mutagenesis study of FlhA has assigned it a PMF-driven export function (Hara et al., 2011). FlhB also acts as an export switch to control late flagellar protein export, as described below. The roles of FliOPQR are not yet understood.
Among proteins that are required for the structure and assembly of the rod and hook, two key players – FliK and FlhB - determine the rod-hook length and the switch to late secretion, respectively (Fig. 1). Absence of FliK abrogates hook-length control, resulting in polyhooks (Hirano et al., 1994). FliK is secreted intermittently to serve as a molecular ruler (Ferris & Minamino, 2006, Shibata et al., 2007, Erhardt et al., 2011). Interaction of the FliK N-terminus (FliKN) with the assembled hook cap (FlgD) is proposed to elicit a pause in FliK secretion. If the C-terminus of FliK (FliKC) is near the C-terminus of FlhB (FlhBC) when the pause occurs, which happens only when the hook reaches a length of 55 nm, their interaction causes auto-cleavage of FlhB, flipping the switch to a late secretion mode. FliK-dependent proteolytic cleavage of FlhB first promotes FlgM secretion, relieving the repression of σ28. Efficient secretion of all late protein substrates requires dedicated chaperones. During this process, the hook cap is discarded and successively replaced first by the HAPs FlgK and FlgL, and then by the capping protein FliD. Filament subunits (either FliC or FljB) are added beneath FliD. In the absence of either the HAP proteins or FliD, filament assembly fails, and flagellin subunits are secreted into the medium (Homma et al., 1984).
FlhE was identified in our laboratory as a protein whose absence reduces swarming motility in S. enterica (Butler et al., 2010). Prior to this, there was a report that a null mutation in flhE abolishes swarming, with no apparent effect on flagellar assembly or swimming behavior (Stafford & Hughes, 2007). FlhE contains an N-terminal signal sequence for export into the periplasm via the Sec pathway (Minamino et al., 1994, Stafford & Hughes, 2007). This sequence was shown to be essential for its swarming phenotype (Stafford & Hughes, 2007). A plausible role for FlhE might be related to the secretory functions of FlhA and/or FlhB, because flhE is co-transcribed with flhBA (Stafford & Hughes, 2007). In a study hinting at such a role, PL-ring-defective mutants, which are arrested for rod growth, were observed to switch to late secretion in suppressors bearing null alleles of flhE (Hirano et al., 2009). We show in the present study that absence of FlhE lowers the cytoplasmic pH in both S. enterica and Escherichia coli, impairs growth in E. coli, and causes filament assembly-dependent cell lysis in both bacteria. Our data implicate a role for FlhE in regulating proton flow through the PMF-driven flagellar secretion system.
Results
Absence of FlhE affects swarming, but not swimming, motility in S. enterica
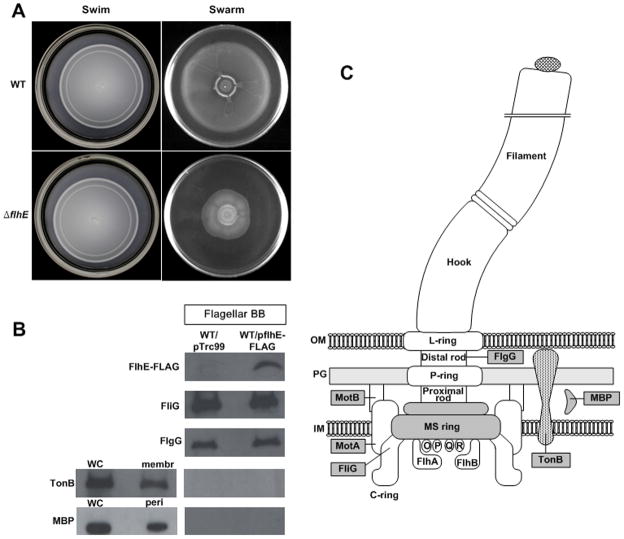
The swimming and swarming phenotypes of a flhE mutant compared to wild-type S. enterica are shown in Fig. 2A. As noted earlier (Minamino et al., 1994), there was no effect of the mutation on swimming; however, there was ~50% inhibition of swarming. An earlier report found complete inhibition of swarming in a flhE mutant (Stafford & Hughes, 2007). The difference in the extent of swarming inhibition could be due to differences in the parental strains used or in the growth conditions; swarming is also sensitive to parameters such as humidity and the commercial source of the agar (Harshey, 2003). Consistent with the earlier report (Stafford & Hughes, 2007), however, the swarming bacteria were not defective in flagella synthesis (see below).
Fig. 2.
Motility phenotype of a flhE mutant and localization of FlhE in S. enterica. A. Swimming and swarming motility of wild-type and flhE mutant (ST004) of S. enterica. Cultures were inoculated at the center of 0.3 % agar plates for swimming and 0.6 % agar plates for swarming. Plates were incubated at 37°C for 6 h. B. Western blots detecting the presence of FlhE-FLAG in isolated flagellar basal bodies. C-ring protein FliG and rod protein FlgG were detected with antibodies specific to these proteins. The bottom two panels show controls that follow the location of the membrane protein TonB and periplasmic maltose-binding protein (MBP) in either whole cells (WC), membranes (membr), the periplasm (peri) or basal body (BB) preparations, using antibodies to these proteins. All lanes contained ~3 μg of protein. C. Schematic showing location of the flagellar and membrane proteins shown in B.
FlhE is a periplasmic protein detectable in flagellar basal body preparations
The flhE gene encodes a 130-amino-acid protein. The N-terminal 16 amino acids of FlhE display features typical of a cleavable signal sequence. Full-length and cleaved forms of the protein were identified in maxicell labeling experiments in S. enterica, consistent with secretion of FlhE into the periplasm by the general secretory pathway (Minamino et al., 1994).
To determine the location of FlhE in flagellated S. enterica cells, FlhE-FLAG, a functional epitope-tagged variant of FlhE, was used. When the tagged protein was expressed from its chromosomal location, the flhE defect was fully complemented but the FLAG signal was weak, as expected due to the low mRNA levels of the flhBAE operon (Wang et al., 2004). When FlhE was expressed from a plasmid, cell fractionation experiments showed that it was present in the cytoplasmic and periplasmic fractions, but not in the membrane fraction (data not shown). Isolated basal body preparations showed the presence of FlhE using FLAG antibodies when the plasmid expressed FlhE-FLAG, but not in the vector control (Fig. 2B). The recovery of intact basal bodies was confirmed by detection of two proteins integral to it - the cytoplasmic C-ring protein FliG and the rod protein FlgG (see cartoon on right). Cross-contamination of the flagellar preparation from membrane or periplasmic fractions was assessed by probing for the presence of the cytoplasmic membrane protein TonB and the periplasmic maltose-binding protein (MBP) in these preparations. These proteins were not detected. In summary, these data show that the periplasmic protein FlhE is also included within the basal body.
flhE mutants are green on pH indicator plates, a phenotype associated with cell lysis Salmonella geneticists routinely use ‘green’ plates in transduction experiments to distinguish phage P22-carrying or P22-free transductants as green versus yellow colonies, respectively. Green plates contain LB media with ~0.8% glucose and the pH indicator dyes aniline blue and alizarin yellow. In the case of P22 lysogens, lysis is required to observe the green color (Smith & Levine, 1967). Other mutants that show a green phenotype, for example recA strains, are also associated with cell death and lysis. Although the molecular basis of the color-lysis association is not known, it is generally believed that anything that weakens the membrane integrity to cause solute leakage and cell lysis, gives green colonies.
We observed that even in the absence of exposure to P22, flhE mutant colonies were green on these plates (Fig. 3A). When propagated on green swarm plates, the whole-cell (WC) suspension of the flhE mutant is clearly green compared to wild-type (Fig. 3B). When centrifuged (3000 g, 5 min), the green material forms a layer on top of the cell pellet. The yellow and green layers in the flhE mutant pellet were both seen to consist of cells under the microscope. Aniline blue is known to bind to different macromolecules: nucleic acids, glucans, and hydrophobic proteins (Evans, 1984, Hough et al., 1985, Kippert & Lloyd, 1995). We surmise that the green color is the result of binding of the dyes to some cellular fraction either released from lysed cells or bound by dye that can permeate the dead cells (likely peptidoglycan, because spotting this macromolecule on the plates gave rise to green spots; not shown). Several tests for cell lysis were conducted and are shown in panels C-E. First, there was a 10% reduction in colony forming units (CFUs) in the flhE mutant compared to wild-type (Fig. 3C). Second, live-dead staining, where live cells are stained green and dead cells red, revealed a similar proportion (10%) of dead cells in the flhE mutant (Fig. 3D). Finally, agarose gel electrophoresis of supernatants of cultures, prepared as in Fig. 3B, showed the presence of released genomic DNA only in the flhE mutant (Fig. 3E). We conclude that the green color on the dye-containing plates arises from cell lysis within the flhE mutant colony. We surmise that cells in the green layer in Fig. 3B are less dense because of loss of their cytoplasmic content.
Fig. 3.
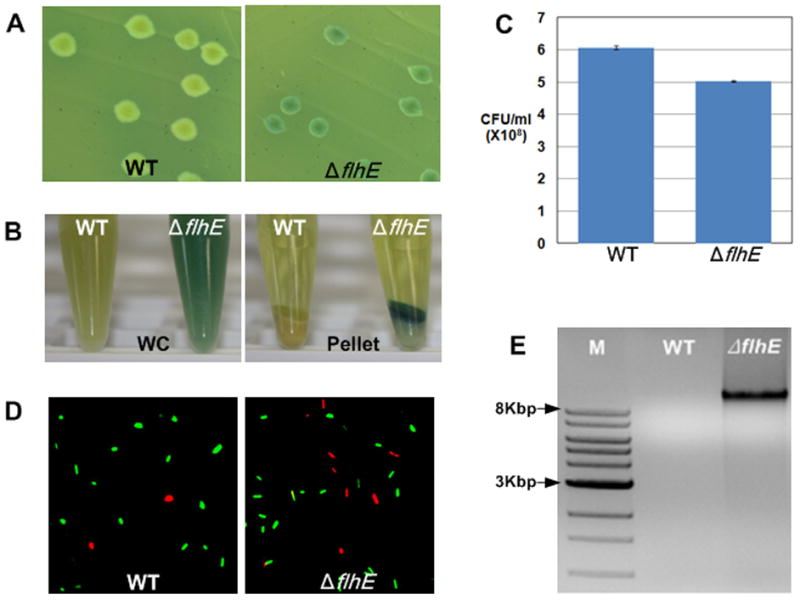
S. enterica flhE mutant colonies are green on pH indicator plates and show cell lysis. A. Color phenotype of wild-type and flhE (ST004) colonies on green plates. B. Color of cell suspensions of the strains in A, isolated from green swarm plates after 6 h of growth. WC, whole cell suspension; Pellet, low-speed centrifugation of WC showing a clear supernatant and yellow/green pellets. C. Colony forming units (CFU) of strains isolated as in B. D. Live-Dead stain used to visualize cell death in strains isolated as in B. E. Genomic DNA in the supernatants from B, electrophoresed on 0.8% agarose gels and stained with ethidium bromide. DNA size markers are indicated. See Experimental Procedures for details.
Cell lysis in S. enterica flhE mutants requires glucose
Glucose is a common component of both green plates and swarm plates. To test if glucose is needed for the cell-lysis phenotype, S. enterica wild-type and flhE mutant strains were grown on green swarm plates with and without glucose. The genomic DNA-release assay showed cell lysis only in the presence of glucose in the flhE mutant (Fig. 4A). Similar results were observed when arabinose was substituted for glucose.
Fig. 4.
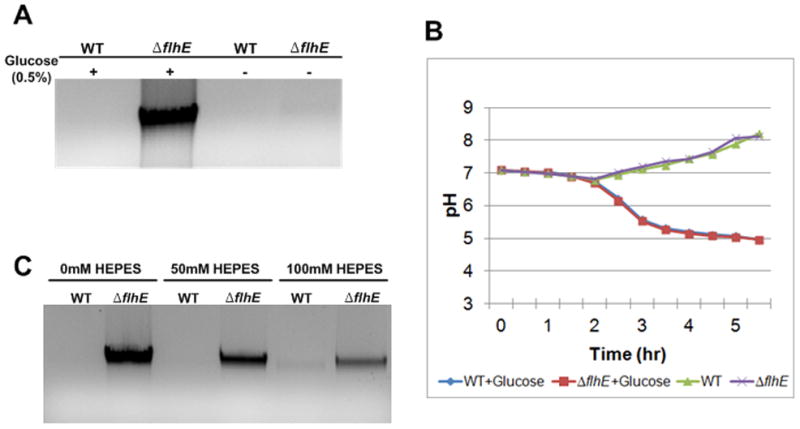
Cell lysis in a S. enterica flhE mutant depends on added glucose, whose metabolism lowers external pH. A. Agarose gel assay for lysis as in Fig. 3. B. External pH of indicated strains grown in LB medium with and without the addition of glucose, measured as described in Experimental Procedures. C. Effect of increasing HEPES buffer on cell lysis, measured as in A. The buffer was added to green swarm plates, and cells were harvested at 6 h. HEPES buffered well only at concentration over 200 mM. However, at these higher concentrations it was not well-tolerated by wild-type cells, which began to lyse. At 100 mM, the pH of the culture was ~0.5 units higher than the unbuffered culture.
Sugar metabolism in bacteria generates acids (Stokes, 1956), which lowers the pH of the growth medium, as seen in a broth culture in Fig. 4B. To test if it is the lowered pH that is responsible for cell lysis, the medium was initially buffered with HEPES to prevent pH changes. Increasing buffer concentration resulted in decreasing cell lysis (Fig. 4C), supporting this notion (HEPES concentrations higher than 100 mM were toxic, in that they induced lysis in wild-type cells). We conclude that FlhE provides a protective function against acidic pH in S. enterica. In experiments described below, we have used the green colony phenotype and/or genomic DNA release assay to dissect the timing and requirements for FlhE function.
Filament assembly is essential for cell lysis in flhE mutants
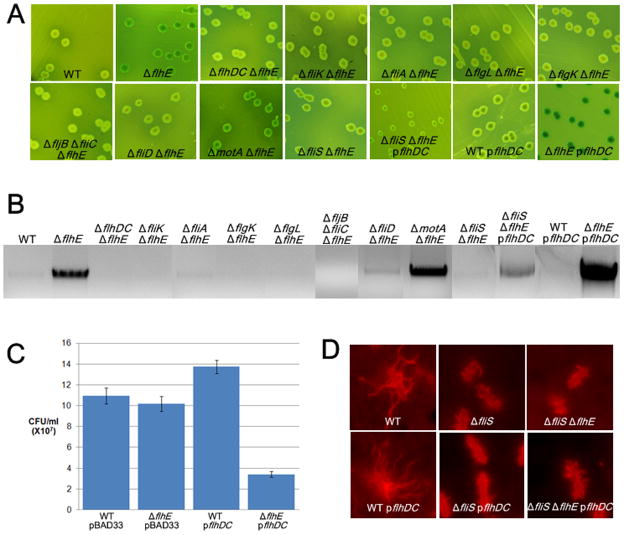
The green colony phenotype of the S. enterica flhE mutant provides a convenient handle for isolating second-site color suppressor mutants. A large-scale Tn10dCm transposon mutagenesis of the flhE strain yielded 3.4% yellow suppressors (i.e. wild-type colony color; see Experimental Procedures). A majority of these suppressors were non-motile, suggesting that the flagellar system was important for the green phenotype. Of those that were motile, none had wild-type levels of motility. Suppressor mutants displaying ~ 40–80% of parent motility were sequenced (Table S1). Of these, a majority (16/27) mapped between lrhA and yfbQ genes. LrhA is a positive regulator of flhDC in E. coli (Lehnen et al., 2002). Given that the mutants had diminished motility, we assume that the insertion diminished lrhA expression. When complemented with flhDC on a plasmid (pflhDC), these mutants regained full motility and turned green (data not shown), as did the mdo and rcs mutants known to downregulate flagellar gene expression in S. enterica (Toguchi et al., 2000, Wang et al., 2007). A consensus emerged that the green color depended on increased numbers of flagella.
To determine whether FlhE function is associated with a particular step of flagellar biogenesis, we systematically examined mutations in key flagellar genes in the flhE mutant background, only a subset of which is shown in Fig. 5. flhDC, fliK, fliA, flgK/L, fliC-fljB, and fliD mutants were yellow (Fig. 5A), and showed no lysis (genomic DNA release) on agarose gels (Fig. 5B), whereas the motA mutant was green and showed lysis. fliK and fliA mutants are defective in class 3 gene expression (see Fig. 1), whereas flgK/L, fliC-fljB and fliD mutants are proficient in both class 3 transcription and protein export but cannot assemble an intact filament (see Fig. S1). The motA mutant can assemble flagella but cannot power their rotation. We conclude that filament assembly, but not rotation, is essential for cell lysis.
Fig. 5.
Filament assembly, number, and length, but not rotation, are essential for cell lysis in a S. enterica flhE mutant. A. Color phenotype on green plates of mutants defective in various steps of flagellar assembly. In strains with pflhDC, arabinose (0.8%) was substituted for glucose in the medium to induce gene expression. Strains used from left to right (not including those with plasmids): WT 14028, ST004, ST496, ST121, ST214, ST210, ST137, ST212, ST146, ST157, ST442. B. Agarose gel electrophoresis of flhE mutants shown in A, propagated on green swarm plates, in which arabinose was substituted for glucose in strains with pflhDC. C. Cell viability of wild-type and flhE mutant strains complemented with pflhDC; 0.2% arabinose added as inducer. D. Flagellar staining of indicated strains with Texas Red coupled anti-IG antibody to FljB antibody.
We had noticed that when pflhDC was introduced into yellow suppressor mutants shown in Table S1, they not only regained higher levels of motility, but their colonies became darker green compared to the flhE mutant parent. Also, the green color of the flhE mutant (but not the wild-type parent) turned darker green in the presence of pflhDC (Fig 5A, compare last two panels on row 2). This darkening of the green color was accompanied by dramatically increased cell lysis as measured by both the genomic DNA-release assay (Fig. 5B) and the CFU assay (Fig. 5C). Lysis depended on filament assembly under these conditions as well i.e. all of the yellow mutants shown in Fig. 5A remained yellow in the presence of pflhDC (not shown). Under the inducing conditions used, pflhDC increases flagella numbers ~ 2–3 fold. These results show that not only were filaments important for cell lysis, but that higher numbers of flagella caused more lysis.
To explore this result further, we introduced a fliS mutation eliminating the filament-protein chaperone into the flhE strain. This mutant is reported to make short filaments (Yokoseki et al., 1995, Auvray et al., 2001) (Fig. 5D). The flhE fliS double mutant was yellow (Fig. 5A), and showed no lysis (Fig. 5B) unless the strain also contained pflhDC, in which case the number of short filaments doubled (Fig. 5D). Under those conditions lysis increased modestly compared to the flhE strain with pflhDC (Fig. 5C). We conclude that filament length is important for lysis and that increased filament numbers increase cell lysis.
Absence of FlhE shows filament assembly-dependent cell lysis even when flagella grow in the periplasm
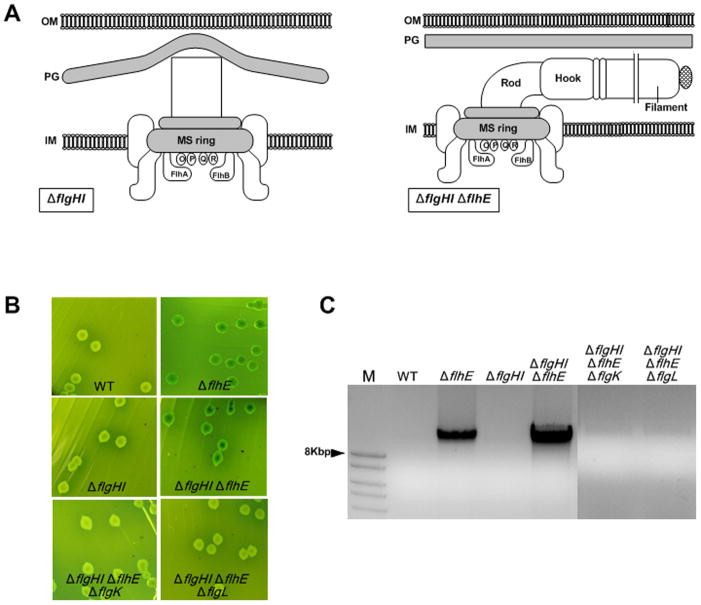
In order to gain insight into FlhE function by a different method, we monitored differences in gene expression profiles between wild-type and flhE mutant strains by microarray experiments. The data showed a slight upregulation of colanic acid biosynthesis genes in the S. enterica flhE mutant compared to wild-type (Table S2). Upregulation of these genes can be indicative of stress on the cell envelope activating, via the outer membrane protein RcsF, the Rcs signaling pathway (Laubacher & Ades, 2008). In gram-negative bacteria, stress sensors monitor permeability of the outer membrane, folding of envelope proteins, and energization of the inner membrane (Raivio, 2005, Rowley et al., 2006). To test whether the filament-dependent cell lysis was due to stress on the outer membrane from the point of exit of the filament, we repeated the experiments described above in a S. enterica PL-ring mutant (flgHI) background. Flagella do not exit the outer membrane in this situation, and they grow inside the periplasm when combined with a flhE mutation (Chevance et al., 2007) (Fig. 6A). Here too, the green color and lysis depended on absence of FlhE and the presence of an intact filament (Fig. 6B, C). flgK and flgL (HAP) mutants, which lack the hook-filament junction proteins, secrete flagellin subunits but fail to assemble a flagellum (see Fig. S1). The flhE flgHI mutant was observed to be more susceptible to lysis compared to the flhE mutant alone (Fig. 6C); indeed, induction of pflhDC in the flhE flgHI background was lethal. We conclude that filament-dependent lysis is independent of either external or periplasmic location of the flagella.
Fig. 6.
Cell lysis of flhE mutants in a S. enterica PL-ring mutant background. A. Cartoon showing impairment of rod growth in a flgHI mutant (lacking P and L rings); deletion of flhE relieves this defect, allowing filaments to assemble in the periplasm (Hirano et al., 2009). B. Color phenotype of wild-type and flgHI mutants defective in flhE and/or flgK, flgL on green plates. Strains used from left to right: 14028, ST004, ST453, ST454, ST478, ST482. C. Agarose gel electrophoresis of mutants shown in B.
Tsr-dependent tumbling response indicates lowered cytoplasmic pH in the flhE mutant
Experiments in Fig. 4 show that the cell lysis phenotype of the flhE mutant depends on a lower external pH. Because flhA, which is co-transcribed in flhE, is implicated in PMF-driven protein export (Hara et al., 2011), we wondered if these proteins function in a common pathway such that absence of FlhE might cause a proton leak. Acidification of the cytoplasm is perceived as a repellent (tumble-inducing or CW rotation-producing) signal by the Tsr chemoreceptor (Kihara & Macnab, 1981, Repaske & Adler, 1981). We therefore used this assay to assess whether the cytoplasm was acidified in the flhE mutant when the external pH was lowered. Because cells adapt rapidly to attractants and repellents, our experimental design incorporated our observation that upregulation of flhDC increases cell lysis (Fig. 5). pflhDC is under PBAD promoter control, and is thus induced by arabinose. Addition of arabinose has the dual effect of lowering the external pH and increasing flagella. Wild-type and tsr strains with and without the flhE mutation and harboring pflhDC were videotaped and their run-tumble bias monitored at 0 min (no inducer) and 30 min after arabinose addition, as described in Experimental Procedures. The data are shown in Fig. 7. In the flhE mutant, tumble frequency increased in the tsr+ (compare 7A and B) but not in the tsr strain (compare Fig. 7C and D). Based on this assay, we conclude that absence of FlhE leads to acidification of the cytoplasm.
Fig. 7.
Swimming behavior of flhE mutants with and without chemoreceptor Tsr. S. enterica wild-type, ΔflhE (ST004), tsr (RH2858), and tsr flhE (ST557) strains containing either the vector pBAD33 alone, or pflhDC, were induced for flhDC expression with addition of arabinose, and their swimming trajectories were observed and recorded at 0 min and 30 min after induction, as described in Experimental Procedures. ‘Normal bias’ refers to cells that swam straight for a distance of 25 μm without a tumble, which is the wild-type bias of cells in an isotropic medium.
Intracellular pH monitored by GFP reporter plasmids in S. enterica and E. coli
The chemoreceptor-based assay described above can only be performed after the switch to late or class 3 flagellar gene expression has taken place, because both the filament and the chemoreceptors are class 3 gene products (see Fig. 1). To test cytoplasm acidification by a second method, as well as to determine if the acidification is seen prior to the class 3 switch, a GFP reporter plasmid (GFPmut3b) that reports effectively on pH changes in the cytoplasm was used (Kitko et al., 2009). The data are shown in Fig. 8. The fluorescence emission spectra at various intracellular pHs, when equilibrated to the external pH using sodium benzoate, respond to changes in pH, as previously reported (Fig. 8A). These data were used to generate a standard curve correlating internal pH with fluorescence intensity (Fig. 8B). To test whether the internal pH of the flhE mutant was lower than that of its wild-type parent, the cultures were suspended in LB buffered to pH 5.5, similar to the pH measured during growth in glucose (Fig. 4B). Under these conditions, the cytoplasmic pH of the flhE mutant was seen to be lower than wild-type (Fig. 8C), supporting the results from the chemoreceptor/repellent assay shown in Fig. 7. However, attempts to measure pH differences in mutants stalled at various steps in the flagellar biogenesis pathway, gave variable results (showing differences between the mutants on some days and not on others), and were inconclusive.
Fig. 8.
Measurement of cytoplasmic pH in S. enterica by Green Fluorescent Protein Fluorimetry. A. Excitation spectra of cytoplasmic GFPmut3b as a function of pH. Wild-type S. enterica cells containing the GFP plasmid were suspended in M63 minimal-media adjusted to pH values varying from 5.5–7.5. Internal pH was equilibrated with external pH by addition of 30 mM sodium benzoate. B. Standard curve of cytoplasmic pH as a function of fluorescence signal (sum of 480nm to 510 nm). See Experimental Procedures for details. The error bars represent standard deviation of the mean (n = 3). C Wild-type and flhE (ST004) S. enterica strains were resuspended in media adjusted to pH 5.5, and GFP excitation spectra were recorded without addition of benzoate. Fluorescence measurements were converted to pH units using the standard curve (B) as described in Experimental Procedures. AU, arbitrary units.
Salmonella and E. coli have somewhat different acid resistance mechanisms (Foster, 2004). We therefore turned to E. coli, both to verify the S. enterica results and to test if the GFP pH reporter gave more reliable data in E. coli. In contrast to S. enterica, the E. coli flhE mutant grew poorly (Fig. 9A), and formed tiny colonies on LB plates. This mutant also showed a smaller swim-colony diameter and was completely defective in swarming (Fig. S2A). The growth defect is sufficient to explain the poor swim/swarm phenotypes of this mutant. Faster-growing variants, all of which were non-motile, accumulated readily, indicating that the growth defect of E. coli flhE strain is also related to the flagellar system. The E. coli flhE mutant behaved similarly to the S. enterica flhE mutant on green plates and in agarose-gel assays for cell lysis, except that these phenotypes were independent of added glucose (Fig. S2 B, C). As in S. enterica, the phenotypes depended on filament assembly. For example, in fliK and flgL mutant backgrounds, the flhE mutant failed to show growth defects (Fig. 9A; only fliK mutant shown) or release of genomic DNA (data not shown). GFPmut3b reporter assays were more reproducible in E. coli (Fig. 9B), although here too we experienced day-to-day variations in the absolute pH values. However, the difference in pH between the strains measured on the same day is statistically significant (p-value < 0.005 for pH difference between wild-type and flhE mutant). As with S. enterica, the flhE mutation lowered the cytoplasmic pH in E. coli. The lower pH was associated with the flhE mutation in both the fliK and flgL mutant backgrounds. These data indicate that the cytoplasm is acidified prior to the switch to late secretion (fliK flhE), is still seen after the switch (flgL flhE), and gets worse upon filament addition (flhE). Because the former two mutants neither lyse nor show growth defects (Fig. 9A), acidification, in and of itself, is not responsible for lysis. How filament assembly contributes to lysis is not clear.
Fig. 9.
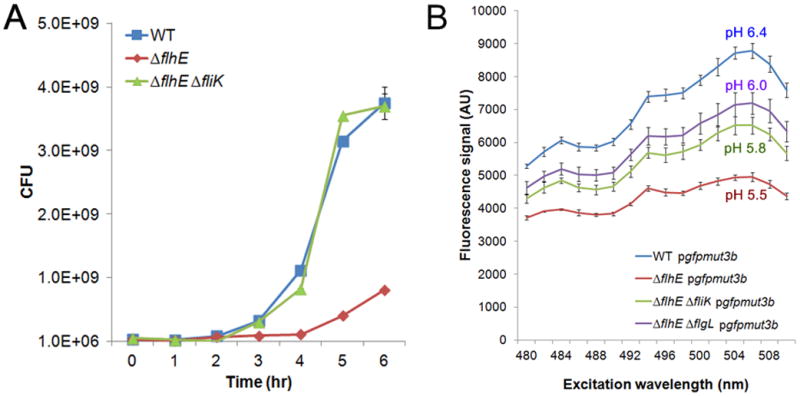
Measurement of growth rates and cytoplasmic pH of E. coli flhE mutants. A. Growth curves in LB broth of the indicated E. coli strains, expressed as colony forming units (CFUs). Strains used: wild-type RP437, ST839, ST907. B. GFPmut3b excitation spectra in wild-type E. coli and its flhE mutant derivatives, measured and converted to pH units as described in Fig. 8C. Strains as in A, with the addition of ST964.
Discussion
This study links FlhE to flagellar T3S function in S. enterica and E. coli. Based on flagella-specific growth defects and acidification of the cytoplasm in the absence of FlhE, our data suggest a role for FlhE in regulating proton flow through the T3S system.
Acidification of the cytoplasm vs cell lysis
In the absence of FlhE, a lowering of cytoplasmic pH is indicated by two different assays: a Tsr-dependent repellent response to low pH (Fig. 7) and changes in the fluorescence intensity of a GFP reporter (Figs. 8 and 9). We interpret these data to indicate a proton leak in the absence of FlhE. The leak must be through the flagellar T3S channels, because it depends on the flagellar system (Fig. 9). We note that proton leaks have been reported through the flagellar stator proteins in several motB mutants (Hosking et al., 2006). The flhE mutant phenotype we report is independent of Mot proteins (Fig. 5).
Overall, the flhE phenotypes were similar in S. enterica and E. coli. For example, flhE mutants in both bacteria were impaired for swarming (Fig. 2 and S2), were green on pH indicator plates (Figs. 3 and S2), showed filament-dependent cell lysis (Figs. 5, 6, 9, S1 and S2), and had lower cytoplasmic pH (Figs. 8, 9). However, there were important differences. For example, these phenotypes were glucose-dependent in S. enterica (Fig. 4), but were not glucose-dependent in E. coli (Fig. S2). The E. coli flhE mutant grew poorly in LB media in contrast to the S. enterica mutant (Figs. 4, 9). Thus, E. coli is more sensitive to the loss of flhE. This could be due to subtle differences in the T3S systems of the two bacteria, or to better tolerance of acid stress in Salmonella.
GFP reporter assays in E. coli indicate that the proton leak is present before the switch to late secretion and gets worse after the switch and upon filament addition (Fig. 9). However, cells do not lyse until filaments are added in both S. enterica (Figs. 5, 6 and S1) and E. coli (Figs. 9 and S2). Thus, cell-lysis is a filament-dependent, post-acidification event. We do not fully understand why this is so. We cannot attribute filament-induced lysis to the viscous load known to be exerted on rotating external filaments, because neither rotation nor an external location of the filaments is required (Figs. 5 and 6). That the lysis phenotype is heightened when flagella grow in the periplasm, where they are not anchored firmly in the peptidoglycan layer (Fig. 6), leads us to propose that, when the internal pH is low, the added stress of long filaments pulling on the inner membrane weakens the membrane further, and exacerbates the proton leak to cause solute efflux and lysis.
Non-viability/cell death of only 10% of cells in the S. enterica flhE mutant (Fig. 3) may reflect the fraction of cells that have the critical number of flagella needed to trigger lysis. We suggest that the flhE mutation affects swarming more than swimming because the fraction of cells with more flagella tends to lead the swarming pack and, being disproportionally affected, interferes with movement of cells that follow; in addition, the swarming environment is more aerobic than when swimming under the agar (Wang et al., 2004), and produces more lysis (data not shown).
Our data provide an explanation for the observation that absence of FlhE induces the late secretion switch in a PL-ring mutant background (Hirano et al., 2009). The flagellar secretion-specificity switch is coupled to HBB completion (Chilcott & Hughes, 2000, Chevance & Hughes, 2008), but it has been reported to occur prematurely, i.e. prior to HBB completion, in null mutations in flhE (Aldridge et al., 2006, Chevance et al., 2007, Hirano et al., 2009). Two events must take place for the flagellar T3S system to change secretion specificity: interaction of FliKC with FlhBC and cleavage of FlhBC. Although FlhB can autocleave spontaneously in vitro (Ferris et al., 2005), FliK interaction is thought to stimulate the cleavage in vivo. We propose that acidification of the cytoplasm that accompanies absence of FlhE, switches secretion specificity by changing the conformation of FlhB and/or proteins like FlhA/Flk that have been proposed to protect FlhB until its timely interaction with FliKC (Hirano et al., 2009).
FlhE function
Acidification of the cytoplasm in the absence of FlhE may imply a ‘proton plug’ or ‘lid’ function for FlhE. Such a function has been proposed for a periplasmic segment of the stator protein MotB, which together with MotA, forms a transmembrane proton channel (Hosking et al., 2006). The plug region was identified as a 20 amino acid segment of MotB, which, when deleted or mutated, caused a massive influx of protons, acidifying the cytoplasm without significantly depleting the proton motive force. The growth inhibition associated with the plug mutation was surmised to be a result of the acidification itself or some following consequence, such as potassium or water efflux from the cells. Under those conditions, cell lysis also depends on filament assembly (Fig. S3).
An alternative function for FlhE is that of a chaperone. Dedicated cytoplasmic chaperones are common in T3S systems, where they help maintain their substrates in a secretion-competent state, delivering them to secretion machinery in manner designed to facilitate travel through the T3S apparatus in an unfolded or partially folded manner (Galan & Wolf-Watz, 2006, Chevance & Hughes, 2008, Minamino & Namba, 2008). Periplasmic chaperones have been identified in assembly of the structural elements of flagella (Nambu & Kutsukake, 2000), needle complexes (Schuch & Maurelli, 2001) and pili (Sauer et al., 2000), where they are thought to bind, stabilize, and cap interactive surfaces of subunits until they are assembled. The MS ring scaffold has to accommodate six different T3S transport proteins, all correctly juxtaposed to regulate timely export. It is possible that FlhE assists in optimizing their packing arrangement, ensuring a tight seal.
A plug/lid role for FlhE fits in with the observation that the phenotype in Salmonella is manifest when the external pH is low. Such a role would tie in nicely with acquisition of the flhE gene mainly by the Enterics, as these bacteria experience low pH during transit through the stomach. However, in E. coli, the flhE mutant phenotype does not depend on lowering of the external pH. Furthermore, the presence of flhE in some non-enterics such as Azotobacter, Chromobacter, and Ralstonia, in which flhE is not part of the flhBA operon and displays only a 28–38% homology to the flhE gene in the enterics (Stafford & Hughes, 2007), also suggests that a role for FlhE as a chaperone is plausible. Although not much is known about the nature and assembly of flagellar secretion apparatus or of the proton channels that drive secretion (Minamino & Namba, 2008, Paul et al., 2008), it is attractive to think that FlhE might play a role in promoting optimal interactions among components of the T3S apparatus.
Experimental Procedures
Bacterial growth conditions, strain construction and reagents
Strains and plasmids are listed in Table 1. Bacteria were grown in 2% L-broth (LB) base unless otherwise stated. Swim plates were made with 0.3% Bacto agar, and swarm plates with 0.6% (S. enterica) or 0.5% (E. coli) Eiken agar (Eiken Chemical, Tokyo, Japan) supplemented with 0.5% glucose (Wang et al., 2004, Wang et al., 2005, Mariconda et al., 2006). Both swim and swarm plates were inoculated in the center of the plate with 8 μl of an overnight culture. All experiments were at 37°C for S. enterica and 30°C for E. coli (Parkinson, 1978). Green plates were prepared with 2% LB supplemented with 0.86% glucose, aniline blue (0.67 g/liter), alizarin yellow (0.065 g/liter), and either 1.5% Bacto agar (hard agar) or 0.5% Eiken agar (swarm agar; glucose concentration was 0.5% in these plates). Photographs of colonies growing on green plates were taken by Canon PowerShot SD1000. Antibiotics used in this study were ampicillin (100 μg/ml), chloramphenicol (30 μg/ml), kanamycin (50 μg/ml), and tetracycline (12.5 μg/ml).
Table 1.
Strains and plasmids used.
| Strain | Genotypea | Source (ref.) |
|---|---|---|
| Salmonella | ||
| 14028 | Wild-type ATCC strain | Mariconda et al. (2006) |
| ST005 | flhE::kan | This study |
| ST004 | ΔflhE | This study |
| ST084 | flhE::FLAG | This study |
| QW215 | flhDC::tetRA | Wang et al. (2007) |
| ST496 | flhDC::tetRA flhE::kan | This study |
| RH2153 | SJW/fliA::kan | Joe Mireles |
| ST649 | fliA::kan | This study |
| ST214 | fliA::kan ΔflhE | This study |
| RH2217 | ΔfljB ΔfliC | Adam Toguchi |
| ST212 | ΔfljB ΔfliC flhE::kan | This study |
| ST155 | ΔfliD | This study |
| ST146 | ΔfliD flhE::kan | This study |
| TH2955 | LT2/fliK77::Tn10 | Kelly Hughes |
| ST120 | fliK77::Tn10 | This study |
| ST121 | fliK77::Tn10 ΔflhE | This study |
| ST209 | ΔflgL | This study |
| ST210 | ΔflgL ΔflhE | This study |
| TH2929 | LT2/flgK563::Tn10 | Kelly Hughes |
| ST136 | flgK563::Tn10 | This study |
| ST137 | flgK563::Tn10 ΔflhE | This study |
| ST450 | flgHI::kan | This study |
| ST453 | ΔflgHI | This study |
| ST454 | ΔflgHI ΔflhE | This study |
| ST478 | flgHI::kan ΔflhE flgK563::Tn10 | This study |
| ST482 | flgHI::kan ΔflhE ΔflgL | This study |
| QW178 | ΔmotA | Wang et al. (2005) |
| ST157 | ΔmotA flhE::kan | This study |
| QW180 | ΔmotB | Wang et al. (2005) |
| ST580 | ΔmotB fliA::kan | This study |
| ST441 | fliS::kan | This study |
| ST442 | fliS::kan ΔflhE | This study |
| RH2858 | Δtsr | Susana Mariconda |
| ST557 | Δtsr flhE::kan | This study |
| RH2202 | Δhin (flagellar phase variation locked for fljB expression) | Adam Toguchi |
| RH2204 | Δhin (flagellar phase variation locked for fliC expression) | Adam Toguchi |
| ST149 | Δhin (RH2202) flhE::kan | This study |
| ST150 | Δhin (RH2204) flhE::kan | This study |
| TH341 | F::Tn10dCm | Kelly Hughes |
| E. coli | ||
| RP437 | Wild-type | Parkinson (1978) |
| ST839 | flhE::kan | This study |
| ST893 | fliK::kan | This study |
| ST903 | ΔfliK | This study |
| ST907 | ΔfliK flhE::kan | This study |
| ST963 | flgL::tetRA | This study |
| ST964 | flgL::tetRA flhE::kan | This study |
| DH5α | Δ(lacIZYA-argF) Φ80dlac Δ(lacZ)M15 | New England Biolabs |
| Phage | ||
| P22 | HT12/4int103 | Mariconda et al. (2006) |
| Plasmid | Expressed protein | Resistance | Replication Origin | Induction | Source (ref.) |
|---|---|---|---|---|---|
| pNK972 | Tn10 transposase in pBR322 | Ampicillin | ColE1 | Constitutively active | Kelly Hughes |
| pAmCyan | Expression vector used for Tn10dCm insertion mapping | Ampicillin | pUC | IPTG | CLONTECH |
| pKD4 | Source for Kan cassette | Kanamycin | oriR6K gamma | N.A | Datsenko and Wanner (2000) |
| pKD46 | Lamda Red recombinase | Ampicillin | repA101ts & oriR101 | Arabinose | Datsenko and Wanner (2000) |
| pCP20 | FLP recombinase | Ampicillin & Chloramphenicol | repA101ts | Constitutively active | Datsenko and Wanner (2000) |
| pMMB1311 | Source for GFPmut3b | Ampicillin | oriC | Constitutively active | Kitko et. al. (2010) |
| pTrc99a | Expression vector | Ampicillin | pBR | IPTG | Amann et al. (1988) |
| pBAD33 | Expression vector | Chloramphenicol | pACYC | Arabinose | Guzman et al. (1995) |
| pBAD30 | Expression vector | Ampicillin | pACYC | Arabinose | Guzman et al. (1995) |
| pJM01 | FlhE-FLAG | Ampicillin | pBR | IPTG | This study |
| pJM37 | GFPmut3b | Ampicillin | pBR | IPTG | This study |
| pflhDC | FlhDC | Chloramphenicol | pACYC | Arabinose | Asaka Suzuki |
| pmotABΔ51-60 | MotA, MotBΔ51-60 | Ampicillin | pACYC | Arabinose | Hosking et al. (2006) |
Unless otherwise indicated (e.g. SJW/LT2), all S. enterica strains are derivatives of 14028, and all E. coli strains constructed in this study are derivatives of RP437. Δ and :: refer to deletion of, or insertion/substitution within, respectively, the indicated gene. Note that deletions created by the Datsenko & Wanner (2000) method leave behind a ‘scar’ sequence of ~80 base pairs.
Deletion of S. enterica and E. coli genes was achieved by the one-step mutagenesis procedure previously described (Datsenko & Wanner, 2000, Wang et al., 2005). All primers used are listed in Table S3. Initial deletions involved selection with a kanamycin (kan) or tetracycline (tetRA) cassette. In S. enterica, the region replaced in flhE covers 330 bp, starting from nucleotide (nt) 61 (1 refers to A of the start codon), in flgL, 896 bp from nt 51, in fliD, 1404 bp from nt 1, in fliS, 248 bp from nt 100, and in flgHI, 1670 bp from nt 61 in flgH to nt 70 in flgI. In E. coli, for flhE, the region replaced covers 330 bp from nt 61, for flgL 876 bp from nt 40, and for fliK 1028 from nt 61. Verification of deletions was achieved by DNA sequencing. Mutant combinations in S. enterica were constructed by P22 transduction. Curing the kan cassette was achieved by expression of the FLP recombinase encoded on pCP20 (Datsenko & Wanner, 2000).
Antibodies against E. coli FliG were provided by David Blair, S. enterica anti-FlgG antibody by May Macnab and E. coli anti-TonB antibody by Kathleen Postle. Anti-FljB antibody was purchased from Becton Dickinson, anti-MBP antibody from New England Biolabs, anti-FLAG antibody from Sigma, and Texas-Red conjugated anti-rabbit antibody from Molecular Probes.
Plasmids
Plasmids obtained from other laboratories are indicated in Table 1. Flagellar genes from S. enterica were amplified with PCR from the chromosomal DNA of appropriate bacterial strains and ligated into pTrc99a (Amann et al., 1988). Epitope tags such as FLAG were conjugated to the genes by PCR. For GFPmut3b expression, gene sequences were amplified with PCR from the plasmid pMMB1311 and moved to pTrc99a.
Isolation of intact flagella from cells
Bacterial flagella were isolated as previously described (Francis et al., 1994) with the following modifications. A liter of wild-type S. enterica cells containing pTrc99-FlhE-FLAG were grown at 37°C with shaking in LB supplemented with 10 μM IPTG and ampicillin. They were collected in late log phase (4 h time point) by centrifugation at 4000 g for 20 min, and resuspended with 100 ml of ice-cold sucrose solution (0.5 M sucrose, 0.1 M Tris-HCl, pH 8.0). All solutions added were the same as those in the original protocol. The pH was raised with 1 M NaOH to pH 10–12. The lysate was centrifuged as described, and the pellets were re-suspended in 10 ml of alkaline solution.
SDS-PAGE, flagellin staining and Western blots
All samples were suspended in 6X SDS-PAGE sample buffer before electrophoresis in 12% polyacrylamide gels, followed by transfer to membrane for Western blot analysis. For whole cell extracts, the bacterial suspension was diluted to an OD600 = 0.6. Immobilion-P membrane (Millipore) was used for Western blots, performed according to standard protocols (Sambrook et al., 1989) and developed using the Enhanced chemiluminescence (ECL) kit from Amersham.
To detect the secreted flagellin, cells were harvested from on green swarm agar plates with 1XPBS after 6 h and normalized to OD600 = 0.3. They were centrifuged at 3000 g for 5 min, the supernatant was mixed with 6X SDS-PAGE sample buffer and electrophoresed in 10% polyacrylamide gels. The gels were washed twice with deionized water and stained for 1h with SimpleBlue™ SafeStain (Invitrogen).
Measurement of protein content
Protein concentration was measured using the BioRad Protein Assay according to the manufacturer’s instructions. Protein bands obtained from the Western blots were quantitated by BioRad Gel Doc and Quantity One 4.4.0 software.
Live/Dead Staining
The stain kit was purchased from Invitrogen, and cells were stained according to the manufacturer’s specifications. The kit includes two nucleic acid stains: green-fluorescent STYO 9 and red-fluorescent propidium iodide (PI). STYO 9 labels both live and dead bacteria alike, whereas PI reduces STYO 9 stain intensity only after crossing damaged cellular membranes. Staining was performed as described (Butler et al., 2010) with the following modifications. Cells were grown on swarm agar plates for 6 h and harvested with 1X PBS and diluted OD600 = 0.6. To determine red/green cell numbers, at least 200 cells were counted for each strain analyzed.
Growth curves, cell counts, and agarose gel electrophoresis
The growth profiles of strains were measured as colony-forming units (CFUs). Overnight cultures were diluted 1:100 in fresh LB (50 ml) and 100 μl of the cultures were collected every 1 h for up to 6 h. Serial dilutions were made with 1X PBS and plated to determine CFUs. To measure CFUs on green swarm plates, cells were grown on these plates for 6 h, collected with 5 ml of 1X PBS, normalized to OD600 = 0.3, and CFUs were determined after serial dilution.
To detect the chromosomal DNA on green swarm agar, swarm cells were harvested from plates after 6 h and adjusted to similar OD600 readings for all strains. The whole cell (WC) suspension was centrifuged at 3000 g for 5 min to separate the cell pellet and supernatant. 15 μl of supernatant were mixed with 3 μl of loading dye, electrophoresed on a 0.8% agarose gel with a 1kbp DNA ladder (NEW ENGLAND BioLabs) and visualized by a BioRad Gel Doc. The position and identity of genomic DNA were confirmed independently by monitoring the migration position of isolated genomic DNA under the same conditions, as well as testing the DNA band for chromosomal markers by PCR.
pH measurements of bacterial cultures
S. enterica cultures were grown overnight in LB (pH 7.0), diluted 1:100 in fresh LB broth (100 ml) with and without the addition of 0.5% glucose, and incubated up to 6 h. 5 ml of culture was collected every 30 min and pH of the culture was measured by a UB-10 UltraBasic pH/mV Meter (Denver Instruments).
Transposon mutagenesis to isolate extragenic supressors of the flhE green phenotype
Phage P22 was grown on donor stain (TH341) carrying Tn10dCm (defective Tn10, lacking the transposase), and used to generate a random pool of Tn10dCm by infection of a wild-type S. enterica strain harboring pNK972 (plasmid expressing the functional transposase gene of Tn10). The CmR transductants were pooled together, P22 lysates were prepared from the pool, and the Tn10dCm insertions were transduced into the S. enterica flhE::kan strain ST005, selecting on green plates supplemented with chloroamphenicol and kanamycin. Out of 26,827 colonies screened, 915 were yellow. 84% (771/915) of yellow mutants were non-motile. The remaining 144 motile mutants were re-tested by moving the Tn10dCm into a clean flhE::kan strain by P22 transduction to confirm the yellow phenotype. Although all the transductants were still motile, only 37 were truly yellow. These yellow motile mutants could be categorized into three classes based on their swimming speed: compared to the parent strain, the motilities of class I –class III mutants were >80%, 40–80%, and <40%, respectively. The Tn10dCm insertion sites of the class I and II mutant subtypes were mapped as follows: Chromosomal DNA from the mutants was purified by Wizard® Genomic DNA purification kit (Promega), partially fragmented by Sau3AI, and then ligated to a BamHI site in the high-copy number plasmid pAmCyan (Clontech) containing AmpR. Ligation mixtures were electroporated into E. coli DH5α. Plasmid DNA from CmR/AmpR transformants was purified using a Qiagen mini-prep kit and sequenced using vector primers from both sides of the insert (Table S3). The data are shown in Table S1; the gene descriptions are from (McClelland et al., 2001).
Microarray experiments
Transcription profiles of S. enterica wild-type and flhE mutant strains propagated on swarm plates for 3 h were obtained by microarray experiments performed as described (Wang et al., 2004).
Repellent response measured by monitoring trajectories of swimming cells
S. enterica wild-type, flhE, tsr, and tsr flhE mutant strains containing pBAD33-flhDC were used. Overnight cultures were diluted 1:100 in LB and shaken at 37°C until the cultures reached an OD600 of 0.6. A 10 μl aliquot (pre-induction) was sampled before arabinose was added (0.2% v/v) to induce flhDC expression and a second sample was taken after 30 min. Cells were observed by phase-contrast microscopy and recorded up to 10 min on an external Sony video recording device with Windows Movie Maker software. Three biological replicates were prepared for swim-motility analysis, and 90 to 200 cells were observed under each condition. To measure reversal frequencies, the straight-swimming distance was defined as ten-times the length of a bacterial cell (25 μm). Cells that swam straight without a tumble in this distance were categorized as having a wild-type or ‘normal’ bias. The swimming distance of each cell was measured manually on the computer screen.
Measurement of cytoplasmic pH with GFP reporter plasmid
Cytoplasmic pH was measured using GFPmut3b, a pH-sensitive green fluorescent protein, as described (Wilks & Slonczewski, 2007, Kitko et al., 2009). A standard pH curve was generated using a wild-type strain containing pTrc99a-GFPmut3b. An overnight culture grown at 37°C was diluted 1:100 in fresh potassium-modified LB (LBK [pH7.5]), which was buffered with 20 mM homopiperazine-N, N′-bis-2-(ethanesulfonic acid) (HOMOPIPES) and supplemented with 40 μM IPTG and ampicillin. Cells were grown to OD600 = 0.6 and pelleted at 4800 g for 4 min. They were resuspended to an OD600 of 0.4 in M63 minimal-medium (1.5% casein hydrolysate, 0.8% glycerol, and 50 mM HOMOPIPES). The pH of M63 medium was adjusted with KOH to pH 5.5 to 7.5. To equilibrate cytoplasmic pH with external pH, 30 mM sodium benzoate was added to the cultures.
Measurement of cytoplasmic pH of experimental samples was performed as described above, except that cultures were grown LBK buffered with 20 mM HOMOPIPES pH 5.5, the external pH attained by cultures growing in the presence of glucose (Fig. 4B), and sodium benzoate was omitted. Cultures were kept on ice prior to recording the GFPmut3b excitation spectra.
Fluorescence measurement of pH reporter plasmids
The excitation spectra of GFPmut3b were recorded using a PTI Quanta Master Model C scanning spectrofluorometer. Three ml of cell suspension were placed into a Bio-Rad VersaFluor cuvette with a path length of 10 mm. Excitation was measured from 480 to 510 nm (slit width of 2 nm), using an emission wavelength of 545 nm (slit width of 20 nm) (Wilks & Slonczewski, 2007). Three biological replicates were measured. To determine the standard curve correlating internal pH with fluorescence intensity, GFPmut3b intensities at pH 5.5, 6.0, 6.5, 7.0, and 7.5 were obtained. The intensities at pH5.5 and 7.5 were fitted to a linear equation (y = mx + b, where y is the GFPmut3b intensity, x is pH value, m is slope, and b is y-intercept) to obtain the slope (m = 4.75 × 104) and y-intercept (b = − 1.9 × 105). The equation (GFPmut3b signal intensity = 4.75 × 104 × pH - 1.9 × 105) was used to convert the signal intensities (sum of 480 nm to 510 nm) obtained from experimental samples to pH units.
Flagella staining and Microscopy
Flagellar filaments were stained as described (Attmannspacher et al., 2008) and cells were observed as reported previously (Paul et al., 2010).
Supplementary Material
Acknowledgments
We are grateful to Marvin Whiteley’s lab for allowing us the use of their microscope facility, to Joan Slonczewski for GFPmut3b, and to John Partridge for careful reading of the manuscript. This work was supported by a National Institutes of Health grant GM 57400.
References
- Aldridge P, Karlinsey JE, Becker E, Chevance FF, Hughes KT. Flk prevents premature secretion of the anti-sigma factor FlgM into the periplasm. Mol Microbiol. 2006;60:630–643. doi: 10.1111/j.1365-2958.2006.05135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann E, Ochs B, Abel KJ. Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli. Gene. 1988;69:301–315. doi: 10.1016/0378-1119(88)90440-4. [DOI] [PubMed] [Google Scholar]
- Attmannspacher U, Scharf BE, Harshey RM. FliL is essential for swarming: motor rotation in absence of FliL fractures the flagellar rod in swarmer cells of Salmonella enterica. Mol Microbiol. 2008;68:328–341. doi: 10.1111/j.1365-2958.2008.06170.x. [DOI] [PubMed] [Google Scholar]
- Auvray F, Thomas J, Fraser GM, Hughes C. Flagellin polymerisation control by a cytosolic export chaperone. J Mol Biol. 2001;308:221–229. doi: 10.1006/jmbi.2001.4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler MT, Wang Q, Harshey RM. Cell density and mobility protect swarming bacteria against antibiotics. Proc Natl Acad Sci U S A. 2010;107:3776–3781. doi: 10.1073/pnas.0910934107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevance FF, Hughes KT. Coordinating assembly of a bacterial macromolecular machine. Nature Rev Microbiol. 2008;6:455–465. doi: 10.1038/nrmicro1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevance FF, Takahashi N, Karlinsey JE, Gnerer J, Hirano T, Samudrala R, Aizawa S, Hughes KT. The mechanism of outer membrane penetration by the eubacterial flagellum and implications for spirochete evolution. Genes Dev. 2007;21:2326–2335. doi: 10.1101/gad.1571607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chilcott GS, Hughes KT. Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica Serovar Typhimurium and Escherichia coli. Microbiol Mol Biol Rev. 2000;64:694–708. doi: 10.1128/mmbr.64.4.694-708.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erhardt M, Hughes KT. C-ring requirement in flagellar type III secretion is bypassed by FlhDC upregulation. Mol Microbiol. 2010;75:376–393. doi: 10.1111/j.1365-2958.2009.06973.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erhardt M, Singer HM, Wee DH, Keener JP, Hughes KT. An infrequent molecular ruler controls flagellar hook length in Salmonella enterica. EMBO J. 2011;30:2948–2961. doi: 10.1038/emboj.2011.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans EA, Hoyne PA, Stone BA. Characteristics and specificity of the interaction of a fluorochrome from aniline blue (sirofluor) with polysaccharides. Carbohydrate Polymers. 1984;4:215–230. [Google Scholar]
- Ferris HU, Furukawa Y, Minamino T, Kroetz MB, Kihara M, Namba K, Macnab RM. FlhB regulates ordered export of flagellar components via autocleavage mechanism. J Biol Chem. 2005;280:41236–41242. doi: 10.1074/jbc.M509438200. [DOI] [PubMed] [Google Scholar]
- Ferris HU, Minamino T. Flipping the switch: bringing order to flagellar assembly. Trends Microbiol. 2006;14:519–526. doi: 10.1016/j.tim.2006.10.006. [DOI] [PubMed] [Google Scholar]
- Foster JW. Escherichia coli acid resistance: tales of an amateur acidophile. Nature Rev Microbiol. 2004;2:898–907. doi: 10.1038/nrmicro1021. [DOI] [PubMed] [Google Scholar]
- Francis NR, Sosinsky GE, Thomas D, DeRosier DJ. Isolation, characterization and structure of bacterial flagellar motors containing the switch complex. J Mol Biol. 1994;235:1261–1270. doi: 10.1006/jmbi.1994.1079. [DOI] [PubMed] [Google Scholar]
- Galan JE, Wolf-Watz H. Protein delivery into eukaryotic cells by type III secretion machines. Nature. 2006;444:567–573. doi: 10.1038/nature05272. [DOI] [PubMed] [Google Scholar]
- Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara N, Namba K, Minamino T. Genetic characterization of conserved charged residues in the bacterial flagellar type III export protein FlhA. PLoS One. 2011;6:e22417. doi: 10.1371/journal.pone.0022417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harshey RM. Bacterial motility on a surface: many ways to a common goal. Annu Rev Microbiol. 2003;57:249–273. doi: 10.1146/annurev.micro.57.030502.091014. [DOI] [PubMed] [Google Scholar]
- Harshey RM. New insights into the role and formation of flagella in Salmonella. In: Powollik S, editor. Salmonella: From Genome to Function. Caister Academic Press; UK: 2011. pp. 163–186. [Google Scholar]
- Hirano T, Mizuno S, Aizawa S, Hughes KT. Mutations in flk, flgG, flhA, and flhE that affect the flagellar type III secretion specificity switch in Salmonella enterica. J Bacteriol. 2009;191:3938–3949. doi: 10.1128/JB.01811-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T, Yamaguchi S, Oosawa K, Aizawa S. Roles of FliK and FlhB in determination of flagellar hook length in Salmonella typhimurium. J Bacteriol. 1994;176:5439–5449. doi: 10.1128/jb.176.17.5439-5449.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homma M, Fujita H, Yamaguchi S, Iino T. Excretion of unassembled flagellin by Salmonella typhimurium mutants deficient in hook-associated proteins. J Bacteriol. 1984;159:1056–1059. doi: 10.1128/jb.159.3.1056-1059.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosking ER, Vogt C, Bakker EP, Manson MD. The Escherichia coli MotAB proton channel unplugged. J Mol Biol. 2006;364:921–937. doi: 10.1016/j.jmb.2006.09.035. [DOI] [PubMed] [Google Scholar]
- Hough T, Bernhardt P, Knox RB, Williams EG. Applications of fluorochromes to pollen biology. II. The DNA probes ethidium bromide and Hoechst 33258 in conjunction with the callose-specific aniline blue fluorochrome. Stain Technol. 1985;60:155–162. doi: 10.3109/10520298509113906. [DOI] [PubMed] [Google Scholar]
- Kearns DB. A field guide to bacterial swarming motility. Nature Rev Microbiol. 2010;8:634–644. doi: 10.1038/nrmicro2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara M, Macnab RM. Cytoplasmic pH mediates pH taxis and weak-acid repellent taxis of bacteria. J Bacteriol. 1981;145:1209–1221. doi: 10.1128/jb.145.3.1209-1221.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kippert F, Lloyd D. The aniline blue fluorochrome specifically stains the septum of both live and fixed Schizosaccharomyces pombe cells. FEMS Microbiol Lett. 1995;132:215–219. doi: 10.1111/j.1574-6968.1995.tb07836.x. [DOI] [PubMed] [Google Scholar]
- Kitko RD, Cleeton RL, Armentrout EI, Lee GE, Noguchi K, Berkmen MB, Jones BD, Slonczewski JL. Cytoplasmic acidification and the benzoate transcriptome in Bacillus subtilis. PLoS One. 2009;4:e8255. doi: 10.1371/journal.pone.0008255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi M, Kanbe M, McMurry JL, Aizawa S. Flagellar formation in Cring-defective mutants by overproduction of FliI, the ATPase specific for flagellar type III secretion. J Bacteriol. 2009;191:6186–6191. doi: 10.1128/JB.00601-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laubacher ME, Ades SE. The Rcs phosphorelay is a cell envelope stress response activated by peptidoglycan stress and contributes to intrinsic antibiotic resistance. J Bacteriol. 2008;190:2065–2074. doi: 10.1128/JB.01740-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehnen D, Blumer C, Polen T, Wackwitz B, Wendisch VF, Unden G. LrhA as a new transcriptional key regulator of flagella, motility and chemotaxis genes in Escherichia coli. Mol Microbiol. 2002;45:521–532. doi: 10.1046/j.1365-2958.2002.03032.x. [DOI] [PubMed] [Google Scholar]
- Liu R, Ochman H. Origins of flagellar gene operons and secondary flagellar systems. J Bacteriol. 2007;189:7098–7104. doi: 10.1128/JB.00643-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macnab RM. Flagella and motility. In: Neidhardt FC, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. Vol. 1. ASM Press; Washington, D. C: 1996. pp. 123–145. [Google Scholar]
- Macnab RM. How bacteria assemble flagella. Annu Rev Microbiol. 2003;57:77–100. doi: 10.1146/annurev.micro.57.030502.090832. [DOI] [PubMed] [Google Scholar]
- Mariconda S, Wang Q, Harshey RM. A mechanical role for the chemotaxis system in swarming motility. Mol Microbiol. 2006;60:1590–1602. doi: 10.1111/j.1365-2958.2006.05208.x. [DOI] [PubMed] [Google Scholar]
- McClelland M, Sanderson KE, Spieth J, Clifton SW, Latreille P, Courtney L, Porwollik S, Ali J, Dante M, Du F, Hou S, Layman D, Leonard S, Nguyen C, Scott K, Holmes A, Grewal N, Mulvaney E, Ryan E, Sun H, Florea L, Miller W, Stoneking T, Nhan M, Waterston R, Wilson RK. Complete genome sequence of Salmonella enterica Serovar Typhimurium LT2. Nature. 2001;413:852–856. doi: 10.1038/35101614. [DOI] [PubMed] [Google Scholar]
- Minamino T, Iino T, Kutuskake K. Molecular characterization of the Salmonella typhimurium flhB operon and its protein products. J Bacteriol. 1994;176:7630–7637. doi: 10.1128/jb.176.24.7630-7637.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamino T, Namba K. Distinct roles of the FliI ATPase and proton motive force in bacterial flagellar protein export. Nature. 2008;451:485–488. doi: 10.1038/nature06449. [DOI] [PubMed] [Google Scholar]
- Nambu T, Kutsukake K. The Salmonella FlgA protein, a putativeve periplasmic chaperone essential for flagellar P ring formation. Microbiology. 2000;146(Pt 5):1171–1178. doi: 10.1099/00221287-146-5-1171. [DOI] [PubMed] [Google Scholar]
- Parkinson JS. Complementation analysis and deletion mapping of Escherichia coli mutants defective in chemotaxis. J Bacteriol. 1978;135:45–53. doi: 10.1128/jb.135.1.45-53.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul K, Erhardt M, Hirano T, Blair DF, Hughes KT. Energy source of flagellar type III secretion. Nature. 2008;451:489–492. doi: 10.1038/nature06497. [DOI] [PubMed] [Google Scholar]
- Paul K, Nieto V, Carlquist WC, Blair DF, Harshey RM. The c-di-GMP binding protein YcgR controls flagellar motor direction and speed to affect chemotaxis by a “backstop brake” mechanism. Mol Cell. 2010;38:128–139. doi: 10.1016/j.molcel.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raivio TL. Envelope stress responses and Gram-negative bacterial pathogenesis. Mol Microbiol. 2005;56:1119–1128. doi: 10.1111/j.1365-2958.2005.04625.x. [DOI] [PubMed] [Google Scholar]
- Repaske DR, Adler J. Change in intracellular pH of Escherichia coli mediates the chemotactic response to certain attractants and repellents. J Bacteriol. 1981;145:1196–1208. doi: 10.1128/jb.145.3.1196-1208.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley G, Spector M, Kormanec J, Roberts M. Pushing the envelope: extracytoplasmic stress responses in bacterial pathogens. Nature Rev Microbiol. 2006;4:383–394. doi: 10.1038/nrmicro1394. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 1989. [Google Scholar]
- Sauer FG, Mulvey MA, Schilling JD, Martinez JJ, Hultgren SJ. Bacterial pili: molecular mechanisms of pathogenesis. Curr Opin Microbiol. 2000;3:65–72. doi: 10.1016/s1369-5274(99)00053-3. [DOI] [PubMed] [Google Scholar]
- Schuch R, Maurelli AT. MxiM and MxiJ, base elements of the Mxi-Spa type III secretion system of Shigella, interact with and stabilize the MxiD secretin in the cell envelope. J Bacteriol. 2001;183:6991–6998. doi: 10.1128/JB.183.24.6991-6998.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata S, Takahashi N, Chevance FF, Karlinsey JE, Hughes KT, Aizawa S. FliK regulates flagellar hook length as an internal ruler. Mol Microbiol. 2007;64:1404–1415. doi: 10.1111/j.1365-2958.2007.05750.x. [DOI] [PubMed] [Google Scholar]
- Smith HO, Levine M. A phage P22 gene controlling integration of prophage. Virology. 1967;31:207–216. doi: 10.1016/0042-6822(67)90164-x. [DOI] [PubMed] [Google Scholar]
- Stafford GP, Hughes C. Salmonella typhimurium flhE, a conserved flagellar regulon gene required for swarming. Microbiology. 2007;153:541–547. doi: 10.1099/mic.0.2006/002576-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes JL. Enzymatic aspects of gas formation by Salmonella. J Bacteriol. 1956;72:269–275. doi: 10.1128/jb.72.2.269-275.1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toguchi A, Siano M, Burkart M, Harshey RM. Genetics of swarming motility in Salmonella enterica Serovar Typhimurium: critical role for lipopolysaccharide. J Bacteriol. 2000;182:6308–6321. doi: 10.1128/jb.182.22.6308-6321.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Frye JG, McClelland M, Harshey RM. Gene expression patterns during swarming in Salmonella typhimurium: genes specific to surface growth and putative new motility and pathogenicity genes. Mol Microbiol. 2004;52:169–187. doi: 10.1111/j.1365-2958.2003.03977.x. [DOI] [PubMed] [Google Scholar]
- Wang Q, Suzuki A, Mariconda S, Porwollik S, Harshey RM. Sensing wetness: a new role for the bacterial flagellum. EMBO J. 2005;24:2034–2042. doi: 10.1038/sj.emboj.7600668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Zhao Y, McClelland M, Harshey RM. The RcsCDB signaling system and swarming motility in Salmonella enterica Serovar Typhimurium: dual regulation of flagellar and SPI-2 virulence genes. J Bacteriol. 2007;189:8447–8457. doi: 10.1128/JB.01198-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilks JC, Slonczewski JL. pH of the cytoplasm and periplasm of Escherichia coli: rapid measurement by green fluorescent protein fluorimetry. J Bacteriol. 2007;189:5601–5607. doi: 10.1128/JB.00615-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoseki T, Kutsukake K, Ohnishi K, Iino T. Functional analysis of the flagellar genes in the fliD operon of Salmonella typhimurium. Microbiology. 1995;141(Pt 7):1715–1722. doi: 10.1099/13500872-141-7-1715. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.