Summary
DNA methylation is highly dynamic during mammalian embryogenesis. It is broadly accepted that the paternal genome is actively depleted of 5-methyl cytosine at fertilization, followed by passive loss that reaches a minimum at the blastocyst stage. However, this model is based on limited data, and to date no base-resolution maps exist to support and refine it. Here, we generated genome-scale DNA methylation maps in mouse gametes and through post-implantation embryogenesis. We find that the oocyte already exhibits global hypomethylation, most prominently at specific families of long interspersed element-1 and long terminal repeat retro-elements, which are disparate between gametes and resolve to lower methylation values in zygote. Surprisingly, the oocyte contributes a unique set of Differentially Methylated Regions (DMRs), including many CpG Island promoter regions, that are maintained in the early embryo but are lost upon specification and absent from somatic cells. In contrast, sperm-contributed DMRs are largely intergenic and resolve to hypermethylation after the blastocyst stage. Our data provide a complete genome-scale, base-resolution timeline of DNA methylation in the pre-specified embryo, when this epigenetic modification is most dynamic, before returning to the canonical somatic pattern.
Introduction
Cytosine methylation in mammals is an epigenetic modification that is largely restricted to CpG dinucleotides and serves multiple critical functions, including stable repression of target promoters, maintaining genomic integrity, establishing parent-specific imprinting patterns, and silencing endogenous retrotransposon activity1,2. In somatic tissues, CpG methylation exhibits global patterns based on relative CpG density: CpG islands at housekeeping or developmental promoters are largely unmethylated, while non-regulatory CpGs distributed elsewhere in the genome are largely methylated1,3. This landscape is relatively static across all somatic tissues, where the majority of methylated CpGs are pre-established and inherited through cell division. Generally, only a small fraction of CpGs switch their methylation status as part of an orchestrated regulatory event4–7.
DNA methylation is much more dynamic during mouse germ-cell and pre-implantation development. The classical model postulates that at fertilization, a targeted, though widespread, catalytic process “actively” removes DNA methylation contributed by the paternal gamete. Recent evidence implicates a demethylation mechanism that transitions through a hydroxymethylated (hmC) intermediate that is catalyzed by the Tet3 member of the Tet family8,9. However, only a portion of hydroxylated targets appears to be actively catalyzed to complete demethylation, and the identity of these targets remains unknown10. Following this dramatic change in the zygote, there appears to be a passive loss of global DNA methylation levels that continues until the blastocyst stage, where the inner cell mass (ICM) that gives rise to the embryo proper is first specified (reviewed by Ref. 11). Recent evidence suggests this passive depletion may also be facilitated in part by Tet enzyme-mediated hydroxylation10. After specification of the ICM, the embryo implants into the uterine lining in concert with gastrulation, which is accompanied by a global remethylation of the genome that is believed to contribute to lineage restriction and the loss of cellular potency12,13.
Unfortunately, little is known on a quantitative, genome-wide scale about the specific dynamics of cytosine methylation during these earliest developmental stages14. The classical model is drawn from observations made using either global measurements, such as immunohistochemistry, or from limited analysis at individual loci using bisulfite sequencing11,12,15–22. Key questions about DNA methylation patterns in early development remain open, including which genomic features are specifically targeted, as well as the identities of Differentially Methylated Regions (DMRs) inherited from either gamete beyond known Imprint Control Regions (ICRs). Here, we use new genomic high-resolution methylation profiling strategies23,24 to gain insight into the underlying mechanisms and regulatory principles of DNA methylation as it functions in early mammalian development.
Results
Genome-scale methylation maps of murine embryogenesis
To generate a global and high-resolution view of early mammalian DNA methylation dynamics, we collected oocytes and sperm, as well as zygote, 2-, 4-, and 8- cell cleavage stage embryos, the inner cell mass (ICM) and E6.5/7.5 post-implantation embryos (Fig. 1a, Supplementary Fig. 1 and 2). All samples were extensively washed and purified to remove any somatic or gametic contaminants; maternal biasing from meiotic polar bodies (representing a 1x or 0.5x static genomic contaminant, respectively) was excluded by mechanical biopsy (Supplementary Fig. 1 and Supplementary Movie 1) and was further confirmed by assessing the paternal (129×1) to maternal (C57/B6×DBA) ratio of known single nucleotide polymorphisms (SNPs) (Supplementary Figs. 3 and 4). We generated reduced representation bisulfite sequencing (RRBS)4 libraries from each stage to provide a comprehensive timeline of DNA methylation patterns during early mouse embryogenesis.
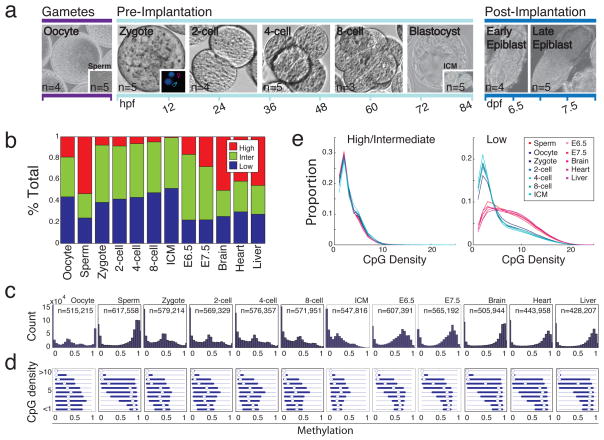
Figure 1. Global CpG methylation dynamics across early murine embryogenesis.
a. Schematic representation of samples isolated and purified for methylation analysis with replicate number (n) highlighted. hpf: hours post fertilization; dpf: days post fertilization.
b. Fraction of 100bp tiles with High (≥0.8, red), Intermediate (Inter, >0.2 and <0.8, green) and Low (≤0.2, blue) methylation values. Brain, heart and liver tissue are shown for adult comparisons.
c. Histogram of methylation values across 100bp tiles. n is the number of tiles for each stage.
d. Boxplots of methylation values at different local CpG densities highlight the difference between hypomethylated pre-implantation tissues and the adult pattern seen in sperm, post-implantation and somatic samples. Bulls-eye indicates the median, edges the 25th/75th percentile and whiskers the 2.5th/97.5th percentile.
e. CpG density of >0.2 methylation (left panel) and ≤0.2 methylation (right panel) tiles in stages that display somatic versus embryonic patterning (red and blue lines, respectively).
Compared to all other genome-wide profiling strategies currently available, RRBS is optimally suited for the low cell numbers that can be obtained from embryonic samples23–25. Within our range of 0.5–10ng genomic DNA, RRBS provides high sensitivity and reproducibility, and the expected genomic coverage (Supplementary Fig. 2). On average, we obtained the methylation status of 1,062,216 CpGs for comparative analysis (Supplementary Table 1). Unfortunately, bisulfite sequencing cannot distinguish between methyl- and hydroxymethyl- cytosine (hmC), and current methods for global profiling of hmC lack the sensitivity to investigate the pre-implantation stages in this study9,26–31. Thus, we cannot draw any definitive conclusions regarding the base resolution hmC distribution, though this modification has not yet been linked to a regulatory mechanism other than to potentiate demethylation32. Given this ambiguity, regions of high mC/hmC methylation, especially those retained over multiple time points, could still be expected to function as if methylated.
Global CpG methylation in the early embryo does not resemble somatic patterns
Current models postulate a phase of global hypomethylation during mammalian pre-implantation development that reaches a minimum at the morula/blastocyst stage. However, it is unknown which genomic regions are affected or how similar the embryonic methylation pattern is to adult1. To address these questions, we investigated the global dynamics of CpG methylation using 100bp tiles (Methods). Intriguingly, we found that oocytes are already dramatically and globally hypomethylated compared to sperm (0.32 median methylation in oocyte versus 0.83 in sperm, Supplementary Fig. 5). We examined the relative proportion of genomic regions at each stage falling into high (≥ 0.8), intermediate (>0.2 and <0.8) or low (≤0.2) methylated categories. Notably, oocyte methylation levels more closely resembled those of early embryonic time points than the levels in sperm, post-implantation embryos, or adult tissues (Fig. 1b). We also observed a gradual increase in the fraction of tiles that exhibit intermediate and low methylation values from oocytes to the ICM, which is consistent with loss of methylation over multiple cleavage divisions (Fig. 1b).
Sperm and post-implantation embryos show a strong inverse relationship between CpG density and methylation levels that is characteristic of somatic cells. In oocyte and pre-implantation samples, this dependence is weaker (Fig. 1c,d). In both pre- and post-implantation embryos, methylated CpGs (>0.2) tend to occur in low CpG density regions, as they do in somatic cells (Fig. 1e, left). However, the alternate relationship between higher CpG density and low methylation is not apparent in the oocyte or pre-implantation embryo (Fig. 1e, right). In summary, pre-implantation development represents a unique developmental period where methylation is differentially positioned and regulated before being restored and maintained in a somatic fashion.
Two major transitions in methylation levels during early development
We next searched for substantial changes in regional DNA methylation through development. For each pair of consecutive stages, we compared methylation levels of each tile and classified it as changed if the difference exceeded 0.2 and was significant according to an FDR-corrected t-test. The most dramatic changes in DNA methylation occurred during two developmental transitions: between sperm and the zygote and between the ICM and the post-implantation embryo (Fig. 2a). At each of these transitions, the majority of changes were unidirectional (Fig. 2b): a gross reduction upon fertilization (mean change = 0.47 decrease for 37% of regions examined) and massive remethylation from the ICM onwards (mean = 0.46 increase in methylation at 66% of tiles). Within E6.5 and E7.5 post-implantation embryos, the methylation levels at the majority of assayed tiles were stable or increased slightly (Fig. 2b). However, more subtle global changes, reflecting a gradual decrease in methylation, were observed from zygote/early cleavage through the 8-cell stage and into the ICM, where methylation levels reached their lowest observed values (Fig. 1b,c).
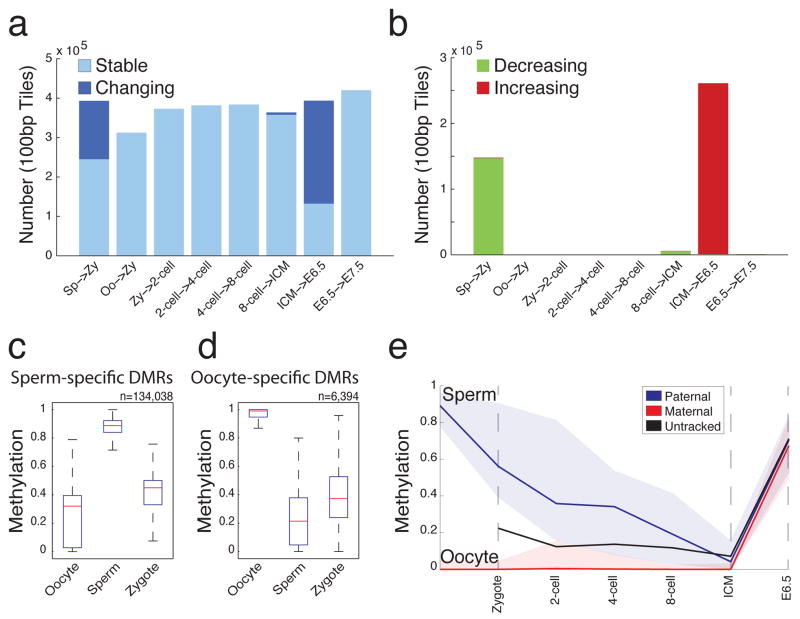
Figure 2. Major transitions in DNA methylation levels during early development.
a. 100bp tiles available for pairwise comparison across consecutive embryonic stages. Tiles that remain unchanged (stable) at the indicated transitions are shown in light blue. Tiles that change by greater than 0.2 and are significant by t-test are highlighted in dark blue.
b. 100bp tiles with increasing (red) or decreasing (green) methylation levels at each consecutive transition show that major transitions are largely unidirectional.
c. Boxplot of methylation levels for sperm-specific DMRs (n=134,038 tiles). Red line indicates the median, edges the 25th/75th percentile and whiskers the 2.5th/97.5th percentile.
d. Boxplot of methylation levels for oocyte-specific DMRs (n=6,394 tiles) as in (c).
e. 74 CpGs within sperm-specific DMR tiles (c) could be ascribed to paternal and maternal alleles and tracked across stages. Paternal CpG methylation values (blue line, median; colored space, 25th/75th percentile) exhibit marked decrease by the zygote stage while maternal CpG methylation (red line, median; colored space, 25th/75th percentile) remain unchanged. If untracked, these CpGs have an intermediate methylation value between those ascribed to a parent-of-origin (black line).
The oocyte defines the early methylation landscape
Active demethylation is expected to occur prior to pronuclear fusion or the completion of DNA synthesis11,33. When we compare methylation patterns between sperm and zygote, the majority of regions in the genome show reduced methylation in the zygote with few additional changes in 2-cell embryos (Fig. 2b). Interestingly, the vast majority of tiles that are methylated at significantly different levels between gametes show higher methylation levels in sperm than in oocyte and many are reduced to levels at or near those of the oocyte (Fig. 2c,d). Using SNPs, we confirmed this observation by tracking 74 CpGs that fell within these tiles and could be assigned paternal or maternal specific values. Zygotes displayed a decrease in paternal methylation in contrast to maternally contributed CpGs, which remained unmethylated (Fig. 2e). Zygotes isolated here are likely in earlier stages of S phase, such that either a passive, replication based mechanism could result in the synthesis of unmethylated, nascent DNA or DNA methylation could be removed by a targeted process10,34–36. The similarities in methylation levels between zygote and 2-cell, which represents one complete round of replication, argues that at least some observed demethylation is a consequence of targeted removal, but distinguishing between these two models may be complicated by the coupling of proposed Base-Excision repair mechanisms and DNA replication itself35.
In contrast, the few regions that are significantly hypermethylated in oocyte compared to sperm exhibit intermediate values in the zygote, suggesting a more direct inheritance of the allelic methylation state (Fig. 2d). The disparity in the zygotic resolution of regions that are differentially methylated between the gametes indicates that the oocyte largely reflects the zygotic/pre-implantation methylome and prescribes its architecture (Supplementary Fig. 6). Thus, the oocyte methylome, rather than the sperm methylome, appears to be more reflective of patterns in the early embryo.
Decreasing methylation at fertilization is most prominent at specific paternally hypermethylated repeat classes
Consistent with a demethylation model, we confirmed that the vast majority (96%) of tiles that are hypermethylated in sperm in our data set become less methylated in the zygote. Most of these tiles already exhibit lower methylation in the oocyte, such that additive effects could also explain more subtle decreases in many regions. Interestingly, tiles exhibiting the most extreme methylation changes during the sperm to zygote transition are enriched for Long Interspersed Elements (LINEs) (P<4.7×10−184, FDR<0.05, hypergeometric enrichment) (Fig. 3a, Supplementary Table 2). We directly estimated the methylation level for individual LINEs surveyed by RRBS at each stage and found that changes in these elements are markedly bimodal during the sperm to zygote transition, with 18% of LINEs reducing their methylation values by over 0.45 (Fig. 3a). By comparison, 10% of captured Long Terminal Repeat (LTR) retroelements exhibit similar levels of demethylation, but the distribution was not as clearly bimodal (Fig. 3b, Supplementary Table 3). Short Interspersed Elements (SINEs) are generally less methylated in sperm than other repeat classes, as has been noted in human37, and also exhibit dramatic shifts in their methylation values from sperm to early embryo, but without the apparent bimodality observed for LINE elements (Supplementary Fig. 7).
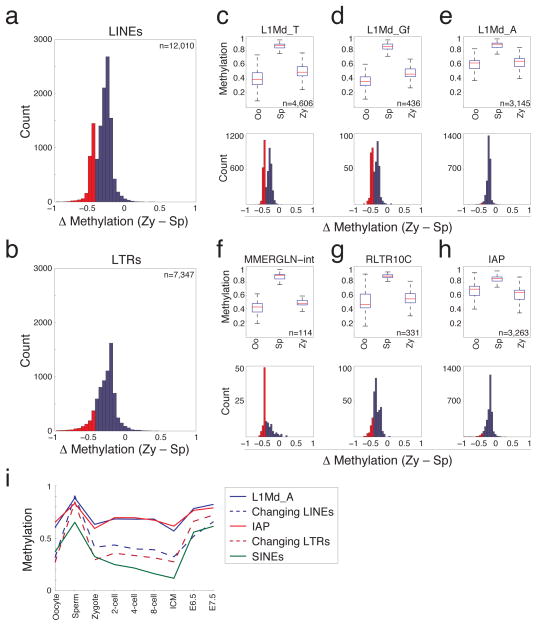
Figure 3. Specific families of LINE and LTR retroelements exhibit the most dramatic changes in the sperm to zygote transition.
a. Histogram of the difference in methylation levels, where negative values represent tiles decreasing from sperm to zygote, within LINE retroelement features that are captured by RRBS. 85% of the elements have a significant difference (P<0.04, FDR<0.05; t-test). The distribution is bimodal with 18% of elements displaying a change in methylation status by greater than 0.45 as highlighted in red.
b. Differences in methylation between sperm and zygote within annotated LTR retroelements. Compared to LINEs, a smaller fraction of elements appear regulated by DNA demethylation (61% significant, 10% of those sampled exhibiting changes greater than 0.45 as highlighted in red).
c–e. Boxplots of methylation levels in oocyte, sperm and zygote (top panels) as well as the distributions of change in methylation levels between sperm and zygote (bottom panels) for specific LINE-1 families, including those that are (c,d) or are not dynamic (e). Top panels: Red line indicates the median, edges the 25th/75th percentile and whiskers the 2.5th/97.5th percentile. Bottom panels: members of each family that are demethylated by greater than 0.45 are highlighted in red.
f–h. Boxplots of methylation levels in oocyte, sperm and zygote (top panels) and the distributions of change in methylation levels between sperm and zygote (bottom panels) for specific families of LTR containing retroelements, including MMERGLN (f), RLTR10C (g) and IAP elements (h). Top and bottom panels as in (c–e).
i. Mean methylation level for all elements of the L1Md_A LINE (solid blue line) and IAP LTR class (solid red line) that do not dramatically change contrasted by LINEs (dashed blue line) and LTR elements (dashed red line) that show the greatest loss at fertilization. SINE elements (green line) are less methylated in sperm than other repeat elements and are generally demethylated to oocyte levels.
Surprisingly, LINEs that changed most dramatically during the sperm to zygote transition largely consisted of two closely related families of L1 LINEs, L1Md_T and L1Md_Gf (Fig. 3c,d, P<4.7×10−184, P<7.9×10−6; hypergeometric enrichment test)38,39. Repeats from these families had the largest and most consistent decrease, while those from other equally represented families, such as L1Md_A elements, showed smaller changes upon fertilization and maintained higher methylation values in both oocyte and zygote (Fig. 3e, Supplementary Fig. 8). Similarly, several LTR families exhibit considerable loss of methylation within the zygote (Fig. 3f,g), while the Class II Intracisternal A-particles (IAPs, Fig. 3h) did not. The latter is consistent with the known retention of high methylation levels of IAPs throughout cleavage19.
Interestingly, during early development, all retrotransposons resolved identically, reaching minimal values at ICM before increasing to levels observed within somatic cells by E6.5/7.5 (Fig. 3i). Thus, repeat elements exist in a less methylated state primarily in the pre-implantation stages (Supplementary Fig. 7). Bisulfite sequencing cannot answer if methylated cytosines at these repeats are converted to hmCs before a subset is further targeted for complete demethylation. Some mCs may be targeted for active demethylation via this intermediate form, while the remaining mC/hmC residues may lose their methylation passively through cleavage, consistent with recent metaphase immunostaining results10.
Sperm and the oocyte contribute distinct genomic features as heritable DMRs
While loss of methylation is widespread, some epigenetic information must be differentially contributed from the two gametes, including known ICRs that maintain their allele-specific methylation pattern throughout embryogenesis40. We systematically searched for inherited DMRs contributed from either gamete, by applying linear regression to all tiles that had mean methylation ≥0.75 in one gamete and ≤0.25 in the other. We identified 376 oocyte-contributed DMRs with intermediate methylation levels in the zygote (P<0.047, FDR<0.05, ANOVA; linear regression residual <0.29, FDR<0.1, Fig. 4a) and 4,894 sperm-contributed DMRs (Fig. 4c). Notably, oocyte-contributed DMRs primarily reside in CpG island-containing promoters (Fig. 4b, Supplementary Table 4), whereas sperm-contributed DMRs were predominantly intergenic (Fig. 4d). The sperm- and oocyte-contributed DMRs also differed substantially in their relative CpG densities (Supplementary Fig. 9).
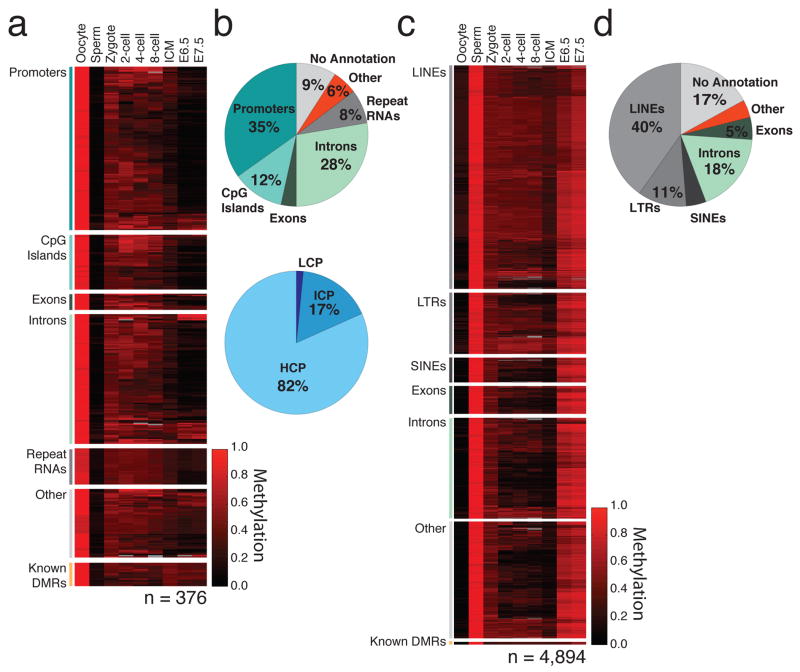
Figure 4. Differentially methylated regions represent discrete gamete specific feature classes.
a. Heatmap of methylation levels (black: 0; red: 1; grey: missing value) in 376 identified 100bp tiles (rows) that behave as oocyte-contributed DMRs in the zygote. Tiles are sorted by functional classes and clustered within each class. 15 known ICRs, shown at the bottom, behave similarly in the early embryo and retain intermediate methylation through implantation.
b. Genomic features (top) and promoters of different CpG densities (bottom) in oocyte-contributed DMRs. Top: oocyte DMRs are enriched for promoters. Bottom: most of the 105 promoters that overlap oocyte-contributed DMR tiles are high CpG density promoters containing CpG Islands (HCPs, light blue).
c. Heatmap of methylation levels (black: 0; red: 1; grey: missing value) in 4,894 identified 100bp tiles (rows) that behave as sperm-contributed DMRs in pre-implantation embryos. Tiles are sorted by functional classes (labels, left) and clustered within each class. Known DMRs contributed by sperm areat the bottom.
d. Genomic features in sperm-contributed DMRs are generally intergenic.
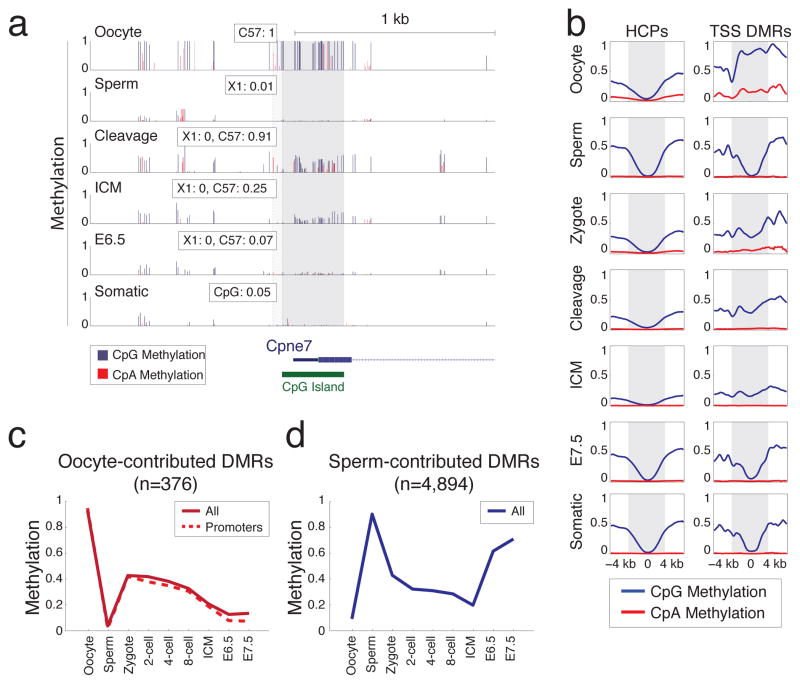
We next focused specifically on oocyte-contributed promoter DMRs, in part due to their unusual enrichment for high CpG containing promoters (HCPs). While this set had no clear functional enrichment, it did include the promoters of several interesting genes that are not expressed in later stages of oogenesis, such as the somatic isoform of Dnmt1, and Dnmt3b41–43, suggesting a repressive function for at least some DMRs. The use of genotyped strains allowed us to confirm that the methylation proximal to the CpG island promoter of Copine VII (Cpne7), another putative DMR, was directly inherited from the oocyte (Fig. 5a). As a set, oocyte contributed promoter DMRs retained intermediate methylation values from the zygote through the ICM, followed by resolution to hypomethylation in the specified embryo (Fig. 5b,c). Thus, CpG island methylation is transiently stabilized during cleavage divisions before re-establishing an unmethylated state around implantation. A distinct methylation pattern during pre-implantation development is also observed in sperm-contributed DMRs, which retain intermediate methylation values through the ICM, before being hypermethylated post-implantation at typical somatic levels (Fig. 5d).
Figure 5. DMRs resolve after cleavage to univalent hyper- or hypo- methylated values in a gamete-of-origin specific fashion.
a. Single CpG resolution methylation within 2kb of the Cpne7 promoter in gametes and across embryonic development (rows). Dark gray bar highlights the CpG island. A CpG proximal to this island can be tracked to a phase resolving SNP and this region is highlighted in light gray, with paternal (X1) and maternal (C57) methylation values highlighted as an inset for each trackable phase. Values for SNP methylation in “Cleavage” correspond exactly to those captured in the zygote. Blue bars: CpG methylation; Red bars: CpA methylation.
b. Composite plot of CpG (blue) and CpA (red) methylation for all HCPs (left) and for promoters that are specifically hypermethylated in oocytes (TSS DMRs, right). The region +/− 2kb of the TSS is marked in gray. Identified promoter DMRs contributed by the oocyte are hypermethylated around the periphery of the TSS and resolve to intermediate values throughout cleavage. An expected HCP methylation architecture is re-acquired for these DMRs around implantation.
c. Mean methylation levels and stage for oocyte-contributed DMRs in promoters (red, dashed line) versus our complete set (red, solid line).
d. Sperm-contributed DMRs (blue line) generally resolve to hypermethylation.
Notably, while RRBS is designed to enrich for CpG dinucleotides (6-fold), it does capture the other three non-CpG dinucleotides at normal frequencies. Of these, CpA is the predominant target for methylation in mouse and human44,45. Consistent with previous locus-specific observations46,47, we found that oocytes had the highest global CpA methylation level observed across pre-implantation development, and that this level decreased by ~50% in the zygote stage (Supplementary Fig. 10, Fig. 5b). This indicates that non-CpG methylation is inherited as part of the oocyte-contributed methylated alleles but is more rapidly lost.
Discussion
To better understand the regulation of methylation patterns during its most dynamic phase, we generated genome-scale maps of DNA methylation in both gametes and through the complete pre-implantation timeline. We find that methylation contributed by sperm to the zygote is most dramatically altered in retro-elements of specific families, while other elements remain more protected and retain higher methylation levels throughout development (Supplementary Fig. 11). The methylation status of the oocyte is a strong predictor of levels in the zygote, and regions that are already hypomethylated in the oocyte could explain much of the disparities between the early embryo and sperm. Possibly, the mechanism and targets of DNA demethylation during female gametogenesis could be similar to those at work during fertilization34. Regardless, the embryonic pattern more closely resembles that of the oocyte until the later stages of pre-implantation, where DNA methylation is further decreased.
In addition to classical ICRs, which exhibit parent-of-origin specific methylation maintained through adulthood, a substantial number of CpG island promoters are specifically hypermethylated in the oocyte, in agreement with a recent study25. Surprisingly, these regions retain intermediate values indicating differential allelic methylation before gradually decreasing through ICM specification and gastrulation, where somatic methylation patterns are re-established (Supplementary Fig. 11).
It remains to be investigated whether the diverse targets that exhibit low methylation levels during embryogenesis are the consequence of a single regulatory principle. LINE and LTR activity in the early embryo is associated with some of the earliest transcriptional events during zygotic genome activation. Targeted depletion by antisense oligonucleotides of the L1Md_T class as well as certain LTRs have demonstrated a general requirement for retrotransposon transcription for progression through cleavage48,49. These observations may also support data suggesting the elongation factor/histone acetyltransferase ELP3 is a component of the DNA demethylation machinery and could explain a tight relationship between demethylation and transcription-associated complexes50.
It is likely that current interest in hmC will spur technical improvements that will permit quantitative dissection of mC and hmC patterns, which will help answer remaining questions regarding Tet3’s universal necessity for conversion to unmethylated cytosines, as well as the effect hmC may have on Dnmt-mediated inheritance32. Tet3’s global conversion to hmC of the paternal genome does not appear to lead to equivalently dramatic demethylation based on the retention of bisulfite-detected signal. The feature-specific dynamics of DNA methylation at fertilization suggest that Tet3 and hmC may be required for targeted demethylation, as well as for driving a gradual hypomethylation over cleavage9,10. Further experiments will be required to characterize this division-dependent demethylation in more detail, and expand it to regions with lower GC content that are under-represented in RRBS. Importantly, other mechanisms must retain heritable methylation information because many targets display relative epigenetic stability from zygote onward and many of these features exhibit embryogenesis-specific methylation patterns.
In summary, our genome-scale single base resolution data provide improved understanding of the relationship and general regions exhibiting DNA demethylation at fertilization. This expands earlier models derived from immunohistochemistry-based observations and begins to address remaining open questions, setting the stage for future epigenetic studies in early mouse development.
Methods Summary
Gametes, cultured cleavage stage embryos, immunosurgically dissected ICM, and post-implantation embryos were isolated as described previously (see Methods). Samples were purified through sequential KSOM microdrops (Millipore) and polar body contaminants mechanically dissected using XY laser (HamiltonThorne) assisted biopsy (Supplementary Figure 1, Supplementary Movie 1). Reduced representation bisulfite libraries were generated from Proteinase K purified, MspI digested genomic DNA and sequenced using the Illumina Genome Analyzer II platform. Sequenced reads were aligned to the Mouse Genome Build 37 (mm9) using a custom computational pipeline that accounted for the strain identity of purified samples, which were either inbred or hybrid strains to provide adequate SNP tracking. Sampled cytosines covered ≥10x were used for single CpG analysis. Alternatively, single CpGs were incorporated into features taken from Ref 4 or into 100bp tiles using a 5x threshold; methylation levels reported for a sample is the average across replicates that met these threshold criteria. Tiles were considered changing between two samples if they exhibited a methylation difference ≥0.2 and statistical significance through a t-test after correction for multiple hypothesis testing (FDR < 0.05) using the Benjamini-Hochberg method. Enrichment for retrotransposon families used annotations from the RepeatMasker track of the UCSC genome browser. Novel DMRs were identified from a pool of 100bp tiles where one gamete had a mean methylation ≥0.75 and the other had a mean methylation ≤0.25. Linear regression applied to this set identified tiles that had zygotic methylation values that fell halfway between those of oocyte and sperm. SNPs between 129×1 paternal and BDF1 (C57/B6 × DBA2/J) maternal genomes were taken from Mouse Genome Informatics and used to assess relative maternal contamination as well as segregated by parent-of-origin to track allelic methylation values in DMRs and for sites exhibiting demethylation.
Data availability
RRBS data is deposited at the Gene Expression Omnibus under accession number GSE34864.
Methods
Preparation of Samples
Isolation of gametes, pre- and post-implantation embryos was performed using procedures described in detail elsewhere51. Briefly, 4–6 week old BDF1 female mice (Charles River) were injected with 5 IU of Pregnant Mare Gonadotropin (Sigma) followed 46h later by 5 IU Human Chorionic Gonadotropin (Sigma). Primed mice were then either directly used to collect oocytes or mated with 129×1 male mice (Jackson) to collect fertilized embryos. Twelve hours after final hormone injection, oocytes or zygotes were isolated from the ampulla under mineral oil and collected in hyaluronidase containing M2 medium (Millipore) drops to eliminate cumulus cells or spermatocyte contaminants. Oocytes were then depleted of somatic contaminants via progressive dilution through sequential drops of CO2 buffered, amino acid supplemented KSOM medium (Millipore) until no somatic contaminants were observed.
Embryos were cultured in KSOM until collection at progressive cleavage stages with isolation occurring within 6 hours of the first observed cleavage event for that stage. Zygotes were screened for the presence of visible pronuclei and subjected to XY Clone (Hamilton Thorne) laser assisted polar body biopsy using an 8μm bore piezo pipette (Humagen, Supplementary Figure 1 and Supplementary Movie 1). Clean cleavage stage embryos underwent an identical approach, with developmental progression unhindered by biopsy conducted at the 2-cell stage (Supplementary Figure 1). For each collection, batches of embryos were carefully screened to ensure each stage did not contain any abnormal embryos. Collection for zygotes was timed at ~10 hpf with fertilization assumed to occur 6–8 hrs after HCG injection, which was again confirmed by the relative synchronicity of the first cleavage division and by relative pronuclear stage. Biopsies were conducted in M2 media (Millipore) in batches of 5–10 embryos to reduce time on the micromanipulator stage. Before the final collection, cleaned and sorted samples were washed with Acid Tyrode’s solution (Sigma) to eliminate the zona pellucida and to deplete any residual somatic contaminants or polar bodies through a short series of additional washes.
The inner cell mass was collected from blastocysts flushed from the uteri of naturally mated mice 3.5 days after fertilization using M2 or DMEM followed by sequential washing in KSOM. The ICM itself was enriched from collected blastocysts by treating the embryo with rabbit anti-mouse serum (Sigma) before immunosurgical depletion of the trophectoderm using Guinea Pig Complement Serum (Sigma). Isolated ICMs were serially washed after isolation to remove contaminants (see Figure 1).
E6.5 and 7.5 embryos were isolated using mechanical dissection of the decidua from the uterine lining of mated mice. Samples were again serially washed and extra-embryonic tissues dissected from ICM-derived tissues using fine glass capillaries (see Figure 1).
Swimming sperm samples were isolated in BSA supplemented Human Tubule Fluid (Millipore) from the caudal epididymis of male mice within 5 days of a successful natural mating as scored by copulation plug. Sperm were incubated in buffered HTF as in in vitro fertilization for over 1 hr in part to reduce somatic contaminants and samples were scored for relative quality under 10x microscopy before snap freezing.
All samples were then collected at minimal volume and either snap frozen or immediately resuspended in DNA lysis buffer.
Preparation of Reduced Representation Bisulfite Sequencing Libraries
RRBS libraries were generated as described52–54. Briefly, DNA was isolated from snap frozen embryos in DNA lysis buffer (100mM Tris-HCl (pH 8.5), 5mM EDTA, 0.2% SDS, 200mM NaCl) supplemented with 300μg/mL Proteinase K (Invitrogen) followed by Phenol:Chloroform extraction, Ethanol precipitation and resuspension in EB buffer. Isolated DNA was then subjected to MspI digestion (NEB), end repair using Klenow 3′-5′ exo– (NEB) supplemented with GTP, meCTP, and ATP in a 1:1:10 ratio to facilitate 3′ A tailing, and ligation of standard adapters using ultraconcentrated 106 U T4 DNA ligase (NEB) and extended 20 hour ligation at 16°C. Size selection of 40–150 and 150–270bp fragments containing ligated adapter was conducted by extended gel electrophoresis using NuSieve 3:1 agarose (Lonza) and gel extraction (Qiagen) using 50ng dephosphorylated, sonicated E. coli DNA as a protective carrier and to increase overall yield. The isolated molecular weight fractions in a given RRBS library were then separately treated with sodium bisulfite using the Epitect® Bisulfite conversion and column purification system (Qiagen) with a modified conversion strategy as described52. Following clean up, the optimal, minimum PCR cycle number required to generate the final libraries was gauged using diagnostic PCRs for each library. Final libraries were then generated from the complete bisulfite converted pool and purified through a second round of gel electrophoresis. High- and low- molecular weight fragments were then either sequenced separately or pooled at a 2:1 ratio by mass to assume an equimolar representation of both size ranges. Libraries were then sequenced on an Illumina Genome Analyzer II before alignment and analysis. The sequencing reads were aligned to the Mouse Genome Build 37 (mm9) using a custom computational pipeline taking into account the strain background for each sample4,54. To supplement our data set we included sperm replicate 2 from Ref. 25 (SRA#ERP000689).
Estimating methylation levels
The methylation level of each sampled cytosine was estimated as the number of reads reporting a C, divided by the total number of reads reporting a C or T. Single CpG methylation levels were limited to those CpGs that had at least 10-fold coverage. For 100bp tiles, reads for all the CpGs that were covered more than 5-fold within the tile were pooled and used to estimate the methylation level as described for single CpGs. The CpG density for a given single CpG is the number of CpGs 50bp up- and downstream of that CpG. The CpG density for a 100bp tile is the average of the CpG density for all single CpGs used to estimate methylation level in the tile. CpA methylation levels were estimated in the same way as for CpG methylation.
The methylation level reported for a sample is the average methylation level across replicates. A replicate will contribute to the average only if it meets the coverage criteria within the replicate. Technical replicates were averaged before contributing to the sample average.
Genomic features
High density CpG promoters (HCP), intermediate density CpG promoters (ICP), low density CpG promoters (LCP), transcription start sites (TSS), CpG island, and DMR annotations were taken from Ref. 4. Promoters are defined as 1kb up and downstream of the TSS. LINE, LTR, and SINE annotations were downloaded from the UCSC browser (mm9) RepeatMasker tracks. Gene annotations were downloaded from the UCSC browser (mm9) refseq track. In each case, the methylation level of an individual feature is estimated by pooling read counts for all CpGs within the feature that are covered greater than 5-fold, and levels are only reported if a feature contains at least 5 CpGs with such coverage (in contrast to 100bp tiles where no minimum number of CpGs is required). A tile is annotated as a genomic feature if any portion of the tile overlaps with the feature and may be annotated by more than one feature (e.g. the same can be annotated as both a promoter and a gene).
Identification of tiles with changing methylation levels and their enrichments
A tile is considered changing if it both has a methylation difference ≥0.2 between two stages and is significant in a two sample t-test with unequal variance after correction for multiple hypothesis testing (FDR < 0.05) using the Benjamini-Hochberg method55. Enrichment p-values are from the hypergeometric distribution where the background is the number of tiles that have a methylation difference ≥0.2 and are corrected for multiple hypotheses at FDR < 0.05, based on the number of gene sets tested.
Identification of enriched retrotransposon families
The same procedure for identifying changing tiles was applied to the methylation levels of retrotransposon elements to identify changing elements. Enrichment for families was done using annotations from the RepeatMasker track of the UCSC genome browser.
Novel DMR identification
100bp tiles where one gamete had a mean methylation greater than 0.75 and the other gamete had a mean methylation of less than 0.25 were flagged as potential DMRs. Linear regression was used to identify tiles which had methylation levels in zygote which were halfway between the methylation levels in oocyte and sperm. Only tiles that had two replicates present in each time point were considered. Residuals were calculated as the mean of the differences between the model predictions and the data taking into account missing values. ANOVA was used to assign a p-value to each tile. A tile was considered a novel DMR if it had a residual in the tenth percentile of tiles tested and a significant p-value from ANOVA with a Benjamini-Hochberg FDR < 0.05. A residual in the tenth percentile corresponds to an FDR < 0.1 by a permutation test where zygote methylation values are shuffled for potential DMR tiles. In the pie charts (Fig. 5b,d), the genomic feature that covered the most novel tiles was reported first and then subtracted from the set before reporting the feature which covered the next largest number of tiles. This procedure was repeated until all tiles were categorized. The one exception was for oocyte-contributed DMRs where promoters were taken out before genes.
Identification of SNPs
An initial set of SNPs between 129×1 and BDF1 (C57/B6 × DBA2/J) was taken from Mouse Genome Informatics56. The set was filtered such that SNPs that fell into the following categories were removed: (1) SNPs that had inconsistent entries for the same position, (2) SNPs not trackable by RRBS (C/T or A/G), (3) SNPs between C57/B6 and DBA2/J, and (4) SNPs that were not covered by X1 and BDF1 in an in silico digest. The log odds ratio [log2(X1 count +0.01/C57 count+0.01)] was calculated for each SNP that was covered in the data set (n=786). SNPs that had at least 10x coverage with an average log odds ratio across all replicates between −5 and 5 and a Sperm X1 log odds ratio greater than 2 were considered of stringent quality (n=636) and used to assess both maternal bias and to serve as a general quality control metric for all libraries incorporated into the data set.
Parent-of-origin methylation tracking
The 636 SNPs identified above corresponded to 1674 CpG dinucleotides and were used to track allelic single CpG methylation. Reads were segregated into either X1 or BDF1 according to SNP type, and CpG methylation levels were called in the same manner described above. SNP normalized methylation values (Supplementary Fig. 4) are the average of the methylation values derived from each strain.
Supplementary Material
Acknowledgments
We would like to thank all the members of the Meissner and Regev labs. Manuel Garber, Nir Yosef, Jimmie Ye, Richard Koche, Christoph Bock, Rene Maehr and Dieter Egli for technical advice and discussion. All members of the Broad Sequencing Platform in particular Fontina Kelly and James Meldrim, Tim Fennel, Kathleen Tibbetts and Jennifer Fostel. We also thank Stuart Levine, Michael Gravina, and Kevin Thai from the MIT BioMicro Center. AR is an investigator of the Merkin Foundation for Stem Cell Research at the Broad Institute. This work was supported by the NIH Pioneer Award (5DP1OD003958), the Burroughs Wellcome Career Award at the Scientific Interface and HHMI (to AR), the Harvard Stem Cell Institute (to TSM) and the NIH (5RC1AA019317, U01ES017155 and P01GM099117), the Massachussetts Life Science Center and the Pew Charitable Trusts (to AM) and a Center for Excellence in Genome Science from the NHGRI (1P50HG006193-01, to AR and AM).
Footnotes
Author contributions
ZDS and AM conceived the study and ZDS, MMC, AM facilitated its design. ZDS collected samples and performed methylation profiling, MMC performed all analysis with assistance from TSM and ZDS. HG and AG provided critical technical assistance and expertise. ZDS, MMC, TSM, AR and AM interpreted the data. ZDS, MMC, and AM wrote the paper with the assistance of the other authors.
The authors declare no competing financial interests.
References
- 1.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 2.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33(Suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 3.Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 2008;9:465–476. doi: 10.1038/nrg2341. nrg2341 [pii] [DOI] [PubMed] [Google Scholar]
- 4.Meissner A, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weber M, et al. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- 6.Weber M, et al. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- 7.Ji H, et al. Comprehensive methylome map of lineage commitment from haematopoietic progenitors. Nature. 2010;467:338–342. doi: 10.1038/nature09367. nature09367 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wossidlo M, et al. 5-Hydroxymethylcytosine in the mammalian zygote is linked with epigenetic reprogramming. Nat Commun. 2011;2:241. doi: 10.1038/ncomms1240. ncomms1240 [pii] [DOI] [PubMed] [Google Scholar]
- 9.Gu TP, et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 2011;477:606–610. doi: 10.1038/nature10443. nature10443 [pii] [DOI] [PubMed] [Google Scholar]
- 10.Inoue A, Zhang Y. Replication-dependent loss of 5-hydroxymethylcytosine in mouse preimplantation embryos. Science. 2011;334:194. doi: 10.1126/science.1212483. science.1212483 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293:1089–1093. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- 12.Kafri T, et al. Developmental pattern of gene-specific DNA methylation in the mouse embryo and germ line. Genes Dev. 1992;6:705–714. doi: 10.1101/gad.6.5.705. [DOI] [PubMed] [Google Scholar]
- 13.Borgel J, et al. Targets and dynamics of promoter DNA methylation during early mouse development. Nat Genet. 2010;42:1093–1100. doi: 10.1038/ng.708. ng.708 [pii] [DOI] [PubMed] [Google Scholar]
- 14.Meissner A. Epigenetic modifications in pluripotent and differentiated cells. Nat Biotechnol. 2010;28:1079–1088. doi: 10.1038/nbt.1684. nbt.1684 [pii] [DOI] [PubMed] [Google Scholar]
- 15.Razin A, Shemer R. DNA methylation in early development. Hum Mol Genet. 1995;4:1751–1755. doi: 10.1093/hmg/4.suppl_1.1751. [DOI] [PubMed] [Google Scholar]
- 16.Monk M, Boubelik M, Lehnert S. Temporal and regional changes in DNA methylation in the embryonic, extraembryonic and germ cell lineages during mouse embryo development. Development. 1987;99:371–382. doi: 10.1242/dev.99.3.371. [DOI] [PubMed] [Google Scholar]
- 17.Rougier N, et al. Chromosome methylation patterns during mammalian preimplantation development. Genes Dev. 1998;12:2108–2113. doi: 10.1101/gad.12.14.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mayer W, Niveleau A, Walter J, Fundele R, Haaf T. Demethylation of the zygotic paternal genome. Nature. 2000;403:501–502. doi: 10.1038/35000656. [DOI] [PubMed] [Google Scholar]
- 19.Lane N, et al. Resistance of IAPs to methylation reprogramming may provide a mechanism for epigenetic inheritance in the mouse. Genesis. 2003;35:88–93. doi: 10.1002/gene.10168. [DOI] [PubMed] [Google Scholar]
- 20.Oswald J, et al. Active demethylation of the paternal genome in the mouse zygote. Curr Biol. 2000;10:475–478. doi: 10.1016/s0960-9822(00)00448-6. [DOI] [PubMed] [Google Scholar]
- 21.Santos F, Hendrich B, Reik W, Dean W. Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev Biol. 2002;241:172–182. doi: 10.1006/dbio.2001.0501. [DOI] [PubMed] [Google Scholar]
- 22.Kim SH, et al. Differential DNA methylation reprogramming of various repetitive sequences in mouse preimplantation embryos. Biochem Biophys Res Commun. 2004;324:58–63. doi: 10.1016/j.bbrc.2004.09.023. S0006-291X(04)02067-4 [pii] [DOI] [PubMed] [Google Scholar]
- 23.Bock C, et al. Quantitative comparison of genome-wide DNA methylation mapping technologies. Nat Biotechnol. 2010 doi: 10.1038/nbt.1681. nbt.1681 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harris RA, et al. Comparison of sequencing-based methods to profile DNA methylation and identification of monoallelic epigenetic modifications. Nat Biotechnol. 2010 doi: 10.1038/nbt.1682. nbt.1682 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smallwood SA, et al. Dynamic CpG island methylation landscape in oocytes and preimplantation embryos. Nat Genet. 2011 doi: 10.1038/ng.864. ng.864 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Davis T, Vaisvila R. High sensitivity 5-hydroxymethylcytosine detection in Balb/C brain tissue. J Vis Exp. 2011:2661, pii. doi: 10.3791/2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ficz G, et al. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473:398–402. doi: 10.1038/nature10008. nature10008 [pii] [DOI] [PubMed] [Google Scholar]
- 28.Szulwach KE, et al. Integrating 5-hydroxymethylcytosine into the epigenomic landscape of human embryonic stem cells. PLoS Genet. 2011;7:e1002154. doi: 10.1371/journal.pgen.1002154. PGENETICS-D-11-00676 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Williams K, et al. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011;473:343–348. doi: 10.1038/nature10066. nature10066 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu H, et al. Genome-wide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in transcriptional regulation in mouse embryonic stem cells. Genes Dev. 2011;25:679–684. doi: 10.1101/gad.2036011. 25/7/679 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu Y, et al. Genome-wide regulation of 5hmC, 5mC, and gene expression by Tet1 hydroxylase in mouse embryonic stem cells. Molecular cell. 2011;42:451–464. doi: 10.1016/j.molcel.2011.04.005. S1097-2765(11)00283-8 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Branco MR, Ficz G, Reik W. Uncovering the role of 5-hydroxymethylcytosine in the epigenome. Nat Rev Genet. 2011 doi: 10.1038/nrg3080. nrg3080 [pii] [DOI] [PubMed] [Google Scholar]
- 33.Santos F, Hendrich B, Reik W, Dean W. Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev Biol. 2002;241:172–182. doi: 10.1006/dbio.2001.0501. [DOI] [PubMed] [Google Scholar]
- 34.Hajkova P, et al. Genome-wide reprogramming in the mouse germ line entails the base excision repair pathway. Science. 2010;329:78–82. doi: 10.1126/science.1187945. 329/5987/78 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wossidlo M, et al. Dynamic link of DNA demethylation, DNA strand breaks and repair in mouse zygotes. Embo J. 2010;29:1877–1888. doi: 10.1038/emboj.2010.80. emboj201080 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Popp C, et al. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature. 2010;463:1101–1105. doi: 10.1038/nature08829. nature08829 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Molaro A, et al. Sperm methylation profiles reveal features of epigenetic inheritance and evolution in primates. Cell. 2011;146:1029–1041. doi: 10.1016/j.cell.2011.08.016. S0092-8674(11)00942-1 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Waterston RH, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 39.Goodier JL, Ostertag EM, Du K, Kazazian HH., Jr A novel active L1 retrotransposon subfamily in the mouse. Genome Res. 2001;11:1677–1685. doi: 10.1101/gr.198301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Edwardsa C, Ferguson-Smith A. Mechanisms regulating imprinted genes in clusters. Current Opinion in Cell Biology. 2007;19:281–289. doi: 10.1016/j.ceb.2007.04.013. [DOI] [PubMed] [Google Scholar]
- 41.Bestor TH. The DNA methyltransferases of mammals. Hum Mol Genet. 2000;9:2395–2402. doi: 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- 42.Hirasawa R, et al. Maternal and zygotic Dnmt1 are necessary and sufficient for the maintenance of DNA methylation imprints during preimplantation development. Genes Dev. 2008;22:1607–1616. doi: 10.1101/gad.1667008. 22/12/1607 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lucifero D, et al. Coordinate regulation of DNA methyltransferase expression during oogenesis. BMC Dev Biol. 2007;7:36. doi: 10.1186/1471-213X-7-36. 1471-213X-7-36 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ramsahoye BH, et al. Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc Natl Acad Sci U S A. 2000;97:5237–5242. doi: 10.1073/pnas.97.10.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ziller MJ, et al. Genomic Distribution and Inter-Sample Variation of Non-CpG Methylation across Human Cell Types. PLoS Genet. 2011;7:e1002389. doi: 10.1371/journal.pgen.1002389. PGENETICS-D-11-00694 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Haines T, Rodenhiser D, Ainsworth P. Allele-specific non-CpG methylation of the Nf1 gene during early mouse development. Dev Biol. 2001;240:585–598. doi: 10.1006/dbio.2001.0504. [DOI] [PubMed] [Google Scholar]
- 47.Tomizawa S, et al. Dynamic stage-specific changes in imprinted differentially methylated regions during early mammalian development and prevalence of non-CpG methylation in oocytes. Development. 2011;138:811–820. doi: 10.1242/dev.061416. dev.061416 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Beraldi R, Pittoggi C, Sciamanna I, Mattei E, Spadafora C. Expression of LINE-1 retroposons is essential for murine preimplantation development. Mol Reprod Dev. 2006;73:279–287. doi: 10.1002/mrd.20423. [DOI] [PubMed] [Google Scholar]
- 49.Kigami D, Minami N, Takayama H, Imai H. MuERV-L is one of the earliest transcribed genes in mouse one-cell embryos. Biol Reprod. 2003;68:651–654. doi: 10.1095/biolreprod.102.007906. [DOI] [PubMed] [Google Scholar]
- 50.Okada Y, Yamagata K, Hong K, Wakayama T, Zhang Y. A role for the elongator complex in zygotic paternal genome demethylation. Nature. 2010;463:554–558. doi: 10.1038/nature08732. nature08732 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nagy A. Manipulating the mouse embryo: a laboratory manual. 3. Cold Spring Harbor Laboratory Press; 2003. [Google Scholar]
- 52.Smith ZD, Gu H, Bock C, Gnirke A, Meissner A. High-throughput bisulfite sequencing in mammalian genomes. Methods. 2009;48:226–232. doi: 10.1016/j.ymeth.2009.05.003. S1046-2023(09)00111-X [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gu H, et al. Preparation of reduced representation bisulfite sequencing libraries for genome-scale DNA methylation profiling. Nature protocols. 2011;6:468–481. doi: 10.1038/nprot.2010.190. nprot.2010.190 [pii] [DOI] [PubMed] [Google Scholar]
- 54.Gu H, et al. Genome-scale DNA methylation mapping of clinical samples at single-nucleotide resolution. Nat Methods. 2010;7:133–136. doi: 10.1038/nmeth.1414. nmeth.1414 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B. 1995;57:289–300. [Google Scholar]
- 56.Blake JA, Bult CJ, Kadin JA, Richardson JE, Eppig JT. The Mouse Genome Database (MGD): premier model organism resource for mammalian genomics and genetics. Nucleic Acids Res. 2011;39:D842–848. doi: 10.1093/nar/gkq1008. gkq1008 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RRBS data is deposited at the Gene Expression Omnibus under accession number GSE34864.