Abstract
Hepatitis C virus (HCV) infects 3% of world population and is the leading cause of liver failure in the United States. A unique feature of HCV is that the viral particles are integral to very low density lipoprotein (VLDL)-derived lipoprotein particles. The virus is assembled into VLDL in hepatocytes, and released out of the cells together with VLDL. The virus then infects more hepatocytes by entering the cells through low density lipoprotein receptor, which mediates uptake of majorities of VLDL-derived lipoprotein particles. These observations suggest that HCV may belong to a novel class of viruses that is associated with VLDL. Understanding the relationship between HCV and VLDL metabolism may reveal new strategies to treat HCV infection.
Hepatitis C virus (HCV) infects more than 170 million people worldwide 1. According to the Centers for Disease Control and Prevention, 4.1 million Americans are estimated to be infected by HCV, 3.2 million of whom become chronically infected. These individuals account for most cases of liver failure in the United States 2. The most effective therapy for HCV infection involves inhibiting a HCV-encoded enzyme 3. However, the HCV genome rapidly acquires mutations that render drug resistance owing to the low fidelity of the viral replication machinery 4. Thus, these inhibitors must be combined with interferon in order to significantly improve treatment outcome of HCV infection. Because of the expense and severe side effects that accompany interferon treatment 5, the search for new strategies to treat HCV infection is merited.
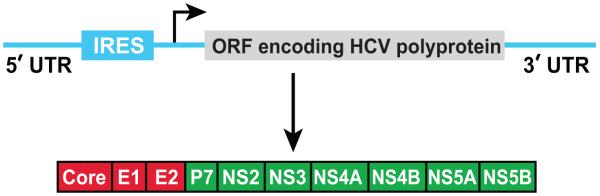
HCV is a single-stranded positive-sense RNA virus of the Flaviviridae family 6. The 9.6-kilobase HCV genome encodes a single polyprotein that is post-translationally processed into at least 10 structural and nonstructural (NS) proteins 7 (Figure 1). The amino-terminal one-third of the polyprotein encodes virion structural proteins: Core, E1 and E2. The remainder of the genome encodes NS proteins that are not found in viral particles but instead are required for replication and assembly of the virus. The NS3, NS4A, NS4B, NS5A and NS5B proteins, which are necessary and sufficient for replication of viral RNA 8, form a viral replication complex on endoplasmic reticulum (ER) membranes 9-12.
Figure 1.
Diagram of HCV genome. HCV genomic RNA contains 3′ and 5′-untranslated regions (UTR) that are required for viral replication. The 5′-UTR also contains an internal ribosomal entry site (IRES) that directs translation of a viral polyprotein, which is further proteolytically processed into 10 proteins. The viral structure and nonstructural protein are labeled in red and green, respectively.
Association of HCV with very low density lipoprotein (VLDL)
An intriguing feature of HCV is that the viral particles are found in complex with VLDL, which plays an important role in transporting cholesterol and triglyceride from the liver to peripheral tissues 13. VLDL contains a hydrophobic core of neutral lipids consisting of triglycerides and cholesteryl esters surrounded by a surface coat containing phospholipids, free cholesterol, and lipoproteins including apolipoprotein B (apoB) and apolipoprotein E (apoE) 14. HCV particles isolated from the serum of virus-infected patients exhibited a density similar to that of VLDL 15-17. Moreover, these particles were rich in triglyceride and contained apoB and apoE 15, 16. Recently, Merz et al developed a strain of HCV in which the E2 protein was tagged with a FLAG epitope, and purified the HCV virion produced from the human hepatoma Huh7 cells through affinity purification with anti-FLAG 18. The purified HCV virion appeared to contain more apoE than viral proteins at the surface of the particles 18. Lipidomic analysis revealed that cholesteryl esters comprised almost half of the total lipid content in the affinity-purified viral particles 18. In sharp contrast, the viral envelope derived from host cell membranes is predominantly composed of phospholipids. The triglyceride content in the purified viral particles were not determined owing to technical difficulty 18.
Electron microscopy (EM) analysis of HCV either isolated from patient serum or affinity-purified from the culture medium of virus-infected Huh7 cells revealed structures that contained lipid-rich cores resembling lipoprotein particles rather than the classical viral capsid structure 15, 18. These observations suggest that the viral genome and capsid may be hidden within the hydrophobic core of VLDL. This structure may allow HCV to evade B cell/antibody-mediated immune surveillance during circulation, thereby providing a plausible explanation as to why the viral infection cannot be effectively prevented by vaccination. In contrast to these studies, EM analysis revealed structures resembling enveloped viral particles in a fraction of culture medium enriched in HCV infectivity 19. Thus, HCV may exist as multiple forms, and more sophisticated EM analyses such as cryo-EM capable of visualizing structures within the hydrophobic cores of VLDL may be necessary to identify capsid structure of HCV.
Assembly of HCV-VLDL complex
The hepatic synthesis of VLDL requires generation of lipid droplets enriched in neutral lipids such as triglycerides and cholesteryl esters in the ER lumen 14. These lipid droplets are produced by reactions catalyzed by microsomal triglyceride transfer protein (MTP) 20-22. Although not formally demonstrated, apoE might also play an important role to generate these lipid droplets 23. Upon fusion with apoB, these lipid droplets can be secreted out of cells as nascent VLDL through exocytosis 14. In addition to generating lipid droplets in the ER lumen, MTP also stabilizes apoB during translation by transferring lipids to the nascent polypeptide chain of apoB 14, 22. In the absence of this lipid transfer, the secretion of apoB is blocked and the protein is rapidly degraded in cells 24, 25. VLDL secretion also requires hepatic synthesis of phosphatidylcholine (PC), the major phospholipid on the surface of the lipoprotein particles 26. In human hepatoma Huh7 cells, long chain acyl-CoA synthetase 3 (ACSL3)-mediated PC synthesis is required for secretion of apoB 27.
Proteomic analysis of ER membrane vesicles containing HCV RNA and viral replication complex composed of viral proteins NS3-NS5B revealed that these vesicles were enriched in apoB, apoE, MTP and ACSL3 12. The reason for co-localization of the HCV replication and VLDL assembly appears to lie in a requirement for co-assembly and secretion of VLDL and HCV particles. Thus, secretion of HCV virion from virus-infected Huh7 cells was inhibited when cells were treated with pharmacological inhibitors of MTP 12, 28, 29. Secretion of HCV was also inhibited in cells transfected with a siRNA targeting apoE 30 or ACSL3 27. The results regarding apoB are not consistent: knockdown of apoB was shown to block HCV secretion in two studies 12, 28 but had no effect on release of HCV virion in another study 29. This discrepancy is most likely caused by the different HCV infection system used in the studies. In the reports showing apoB was required for secretion of HCV, care was taken to ensure that HCV infection did not result in cellular apoptosis so that viral particles were only released through exocytosis. In contrast, the study showing the opposite result used a system known to cause apoptosis of virus-infected cells 31. Consequently, intracellular infectious HCV particles containing NS5A were released into culture medium from dying cells 29. Since apoB is not required to produce intracellular HCV particles 29, it is not surprising to observe apoB-independent production of infectious HCV particles in culture medium using this system.
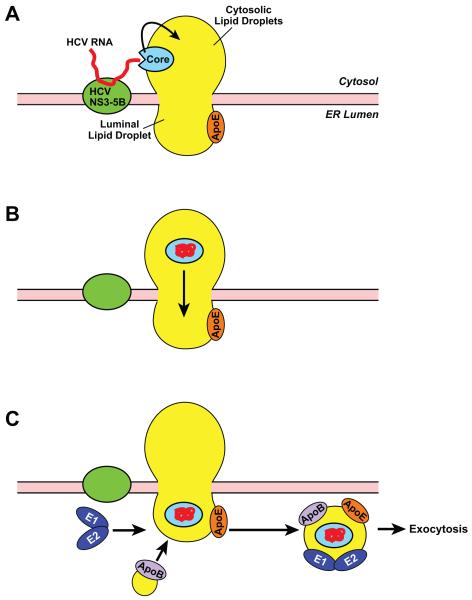
A puzzling question regarding assembly of HCV-VLDL complex is how viral genome synthesized at cytosolic face of the ER is transported across the membrane bilayers to reach ER lumen where it is packaged into VLDL. A clue to the question may come from a unique property of HCV-encoded capsid core protein. HCV core protein contains two domains: an NH2-terminal hydrophilic domain (D1) that binds viral RNA, and a COOH-terminal hydrophobic domain (D2) that interacts with neutral lipid 32, 33. In HCV-infected Huh7 cells, the majority of core proteins are localized at the surface of cytosolic lipid droplets that are in contact with ER membranes containing the HCV replication complex 34, which is also the site for VLDL assembly 12. Thus, HCV may replicate at an ER domain enriched in neutral lipids that can bud toward cytosol or lumen to form lipid droplets in both locations. A hypothetic model is proposed in Figure 2 to explain translocation of HCV capsid based on this localization. Core is targeted to cytosolic lipid droplets through its D2 domain, after it is cleaved from the viral polyprotein. The hydrophilic D1 domain is exposed to the cytosol, ready to accept viral RNA synthesized by the viral replication complex (Figure 2A). Once associated with viral RNA, core protein undergoes a conformational change so that hydrophilic residues that bind viral RNA are folded inside, whereas hydrophobic residues are exposed at the surface. This conformational change allows the core-RNA complex to become completely embedded in the hydrophobic core of lipid droplets (Figure 2B). The viral capsid-RNA complex then travels through the neutral lipid-rich ER membrane to reach lipid droplets in the ER lumen. The HCV-containing luminal lipid droplets then fuse with apoB, acquire two other lipoprotein-like viral structural proteins E1 and E2 35, and are secreted out of the cells through exocytosis (Figure 2C).
Figure 2.
A hypothetic model illustrating HCV assembly. HCV RNA is synthesized by HCV replication complex composed of viral proteins NS3-NS5B at the cytosolic face of the ER membranes. The viral RNA then binds to the core protein localized at the surface of the lipid droplets adjacent to the viral replication complex (A). Upon binding with viral RNA, core goes through a conformational change so that the core-RNA complex enters the core of cytosolic lipid droplets, allowing the complex to reach lipid droplets in the ER lumen by traveling through ER domains enriched in neutral lipids (B). The HCV-containing luminal lipid droplets fuse with apoB, acquire E1 and E2 heterodimer, and are secreted out of cells through exocytosis (C).
The model shown in Figure 2 predicts that HCV capsids are able to enter the hydrophobic core of lipid droplets. Although there have not been many studies characterizing localization of cellular proteins in hydrophobic cores of lipid droplets, such localization was reported through EM analysis 36, 37. Further studies are required to determine whether these host proteins facilitate translocation of HCV capsids. This model also predicts that HCV capsids travel across the membrane through neutral lipid-rich domain of the ER. ApoB has been reported to translocate from the ER lumen into the cytosol though a mechanism that involves lipid droplets 38. This observation implies that lipid droplets across ER membranes are continuous and proteins may be able to transport across ER membranes through lipid droplet intermediates. More imaging and biochemical analyses will be required to further validate the model shown in Figure 2.
Requirement of LDL receptor (LDLR) for HCV entry
Once released out of cells, HCV enters more hepatocytes for a new round of infection. Several receptors for HCV entry have been identified based on their interaction with E2 39, including CD81 and Scavenger receptor-B1 40-44. Proteins forming tight junction such as claudin-1 and occludin have also been implicated in HCV entry 45, 46. However, all of these receptors appear to function at later stages of viral entry since they are not required for HCV to bind to the cell surface 45, 47. This initial binding is at least partially mediated by LDLR, which plays a predominant role in acquiring VLDL-derived lipoprotein particles 48. It has been reported that cellular binding or uptake of HCV particles isolated from infected patients correlated with LDL receptor activity on cell surface 15, 49-51. LDLR was also required for infectious entry of HCV virion produced from Huh7 cells, and this entry depended on the interaction between the receptor and apoE on the viral particles 52. LDLR does not directly interact with viral proteins as the receptor was not required for entry of HCV pseudo particles, which were assembled by displaying HCV structural proteins E1 and E2 onto retroviral core particles that were not in complex with lipoproteins 53.
Mutations disrupting the function of the LDLR have been identified in human. These mutations produce autosomal dominant familial hypercholesterolemia, which affect 0.2% of world population 54. Affected individuals have elevated plasma levels of LDL-cholesterol, which causes premature coronary atherosclerosis. However, during evolution when dietary cholesterol was scant, these mutations may not produce such a severe phenotype but may actually protect these individuals from infection of HCV or HCV-related virus. An analysis comparing the frequency of HCV infection in people expressing normal LDL receptor versus those affected by familial hypercholesterolemia will be needed to test the hypothesis. This hypothesis may also be tested with mice containing human liver grafts, an animal model successfully used to study HCV infection 55-57. If this hypothesis is correct, mice grafted with human liver derived from patients affected by familial hypercholesterolemia are expected to resist the infection by HCV.
Treating HCV infection with drugs targeting VLDL metabolism
The dependency on VLDL for HCV life cycle offers opportunities to treat the viral infection with drugs targeting VLDL metabolism. Several MTP inhibitors have already been tested in clinical trials because of their ability to block VLDL secretion, thereby lowering the plasma levels of VLDL-triglycerides and LDL-cholesterol 58, 59. Long-term treatment with MTP inhibitors led to the accumulation of fat in livers thus hampering the drugs to be approved for treatment of hypercholesterolemia, which may require life-long administration in the case of familial hypercholesterolemia 58, 59. However, short-term treatment (up to several weeks) reduced the plasma level of VLDL with only minor adverse effects, which disappeared after drug removal 58. Since the standard treatment for HCV infection with drugs targeting the viral enzymes lasts for only about 12 weeks, MTP inhibitors may be combined with these drugs to treat HCV infection. MTP inhibitors also have the advantage in that they target a host protein rather than viral proteins so they are less likely to face the drug-resistance problem caused by mutations in the viral genome.
Another drug that inhibits VLDL assembly is an antisense RNA drug targeting apoB 60. Unlike MTP inhibitors, apoB antisense RNA lowered VLDL secretion in the absence of accumulation of fat in livers 61. However, in cultured cells knockdown of apoB by siRNA was less potent than MTP inhibitors to inhibit HCV production 12, 28. Thus, more studies are required to determine the efficacy of apoB antisense RNA on treatment of HCV infection in vivo.
Acknowledgments
I thank Russell DeBose-Boyd for his critical comments to the manuscript. JY is supported by research grants from the NIH (HL-20948, AI 090119).
Footnotes
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- (1).Chisari FV. Unscrambling hepatitis C virus-host interactions. Nature. 2005;436:930–2. doi: 10.1038/nature04076. [DOI] [PubMed] [Google Scholar]
- (2).Brown RS. Hepatitis C and liver transplantation. Nature. 2005;436:973–8. doi: 10.1038/nature04083. [DOI] [PubMed] [Google Scholar]
- (3).Schlutter J. Therapeutics: New drugs hit the target. Nature. 2011;474:S5–S7. doi: 10.1038/474S5a. [DOI] [PubMed] [Google Scholar]
- (4).Sylvestre D. Perspective: Recognizing resistance. Nature. 2011;474:S11. doi: 10.1038/474S11a. [DOI] [PubMed] [Google Scholar]
- (5).Feld JJ, Hoofnagle JH. Mechanism of action of interferon and ribavirin in treatment of hepatitis C. Nature. 2005;436:967–72. doi: 10.1038/nature04082. [DOI] [PubMed] [Google Scholar]
- (6).Appel N, Schaller T, Penin F, Bartenschlager R. From structure to function: new insights into hepatitis C virus RNA replication. J Biol Chem. 2006;281:9833–6. doi: 10.1074/jbc.R500026200. [DOI] [PubMed] [Google Scholar]
- (7).Lindenbach BD, Rice CM. Unravelling hepatitis C virus replication from genome to function. Nature. 2005;436:933–8. doi: 10.1038/nature04077. [DOI] [PubMed] [Google Scholar]
- (8).Lohmann V, Korner F, Koch JO, Herian U, Theilmann L, Bartenschlager R. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science. 1999;285:110–3. doi: 10.1126/science.285.5424.110. [DOI] [PubMed] [Google Scholar]
- (9).Egger D, Wolk B, Gosert R, Bianchi L, Blum HE, Moradpour D, Bienz K. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J Virol. 2002;76:5974–84. doi: 10.1128/JVI.76.12.5974-5984.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Gosert R, Egger D, Lohmann V, Bartenschlager R, Blum HE, Bienz K, Moradpour D. Identification of the Hepatitis C Virus RNA Replication Complex in Huh-7 Cells Harboring Subgenomic Replicons. J Virol. 2003;77:5487–92. doi: 10.1128/JVI.77.9.5487-5492.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Ye J, Wang C, Sumpter R, Jr., Brown MS, Goldstein JL, Gale M., Jr. Disruption of hepatitis C virus RNA replication through inhibition of host protein geranylgeranylation. Proc. Natl. Acad. Sci.USA. 2003;100:15865–70. doi: 10.1073/pnas.2237238100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Huang H, Sun F, Owen DM, Li W, Chen Y, Gale M, Jr., Ye J. Hepatitis C virus production by human hepatocytes dependent on assembly and secretion of very low-density lipoproteins. Proc. Natl. Acad. Sci.USA. 2007;104:5848–53. doi: 10.1073/pnas.0700760104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Ye J. Reliance of Host Cholesterol Metabolic Pathways for the Life Cycle of Hepatitis C Virus. PLoS Pathog. 2007;3:1017–22. doi: 10.1371/journal.ppat.0030108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Sundaram M, Yao Z. Recent progress in understanding protein and lipid factors affecting hepatic VLDL assembly and secretion. Nutr Metab. 2010;7:35–51. doi: 10.1186/1743-7075-7-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Andre P, Komurian-Pradel F, Deforges S, Perret M, Berland JL, Sodoyer M, Pol S, Brechot C, Paranhos-Baccala G, Lotteau V. Characterization of low- and very-low-density hepatitis C virus RNA-containing particles. J Virol. 2002;76:6919–28. doi: 10.1128/JVI.76.14.6919-6928.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Nielsen SU, Bassendine MF, Burt AD, Martin C, Pumeechockchai W, Toms GL. Association between Hepatitis C Virus and Very-Low-Density Lipoprotein (VLDL)/LDL analyzed in iodixanol density gradients. J Virol. 2006;80:2418–28. doi: 10.1128/JVI.80.5.2418-2428.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).André P, Perlemuter G, Budkowska A, Bréchot C, Lotteau V. Hepatitis C virus particles and lipoprotein metabolism. Semin Liver Dis. 2005;25:93–104. doi: 10.1055/s-2005-864785. [DOI] [PubMed] [Google Scholar]
- (18).Merz A, Long G, Hiet MS, Br++gger B, Chlanda P, Andre P, Wieland F, Krijnse-Locker J, Bartenschlager R. Biochemical and morphological properties of Hepatitis C virus particles and determination of their lipidome. J Biol Chem. 2011;286:3018–32. doi: 10.1074/jbc.M110.175018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Gastaminza P, Dryden KA, Boyd B, Wood MR, Law M, Yeager M, Chisari FV. Ultrastructural and biophysical characterization of hepatitis C virus particles produced in cell culture. J. Virol. 2010;84:10999–1009. doi: 10.1128/JVI.00526-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Wang Y, Tran K, Yao Z. The activity of microsomal triglyceride transfer protein is essential for accumulation of triglyceride within microsomes in McA-RH7777 cells. J Biol Chem. 1999;274:27793–800. doi: 10.1074/jbc.274.39.27793. [DOI] [PubMed] [Google Scholar]
- (21).Kulinski A, Rustaeus S, Vance JE. Microsomal triacylglycerol transfer protein is required for lumenal accretion of triacylglycerol not associated with apoB, as well as for apoB lipidation. J Biol Chem. 2002;277:31516–25. doi: 10.1074/jbc.M202015200. [DOI] [PubMed] [Google Scholar]
- (22).Hussain MM, Shi J, Dreizen P. Microsomal triglyceride transfer protein and its role in apoB-lipoprotein assembly. J Lipid Res. 2003;44:22–32. doi: 10.1194/jlr.r200014-jlr200. [DOI] [PubMed] [Google Scholar]
- (23).Fazio S, Yao Z. The enhanced association of apolipoprotein E with apolipoprotein B-containing lipoproteins in serum-stimulated hepatocytes occurs intracellularly. Arterioscler Thromb Vasc Biol. 1995;15:593–600. doi: 10.1161/01.atv.15.5.593. [DOI] [PubMed] [Google Scholar]
- (24).Yao Z, Tran K, McLeod RS. Intracellular degradation of newly synthesized apolipoprotein B. J Lipid Res. 1997;38:1937–53. [PubMed] [Google Scholar]
- (25).Ginsberg HN, Fisher EA. The ever-expanding role of degradation in the regulation of apolipoprotein B metabolism. J Lipid Res. 2009;50:S162–S166. doi: 10.1194/jlr.R800090-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Cole LK, Vance JE, Vance DE. Phosphatidylcholine biosynthesis and lipoprotein metabolism. Biochim Biophys Acta. 2011 doi: 10.1016/j.bbalip.2011.09.009. doi:10.1016/j.physletb.2003.10.071. [DOI] [PubMed] [Google Scholar]
- (27).Yao H, Ye J. Long chain acyl-CoA synthetase 3-mediated phosphatidylcholine synthesis is required for assembly of very low density lipoproteins in human hepatoma Huh7 Cells. J Biol Chem. 2008;283:849–54. doi: 10.1074/jbc.M706160200. [DOI] [PubMed] [Google Scholar]
- (28).Gastaminza P, Cheng G, Wieland S, Zhong J, Liao W, Chisari FV. Cellular determinants of hepatitis C virus assembly, maturation, degradation, and secretion. J Virol. 2008;82:2120–9. doi: 10.1128/JVI.02053-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Jiang J, Luo G. Apolipoprotein E but not B is required for the formation of infectious Hepatitis C virus particles. J Virol. 2009;83:12680–91. doi: 10.1128/JVI.01476-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Chang KS, Jiang J, Cai Z, Luo G. Human apolipoprotein E is required for infectivity and production of hepatitis C virus in cell culture. J Virol. 2007;81:13783–93. doi: 10.1128/JVI.01091-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Deng L, Adachi T, Kitayama K, Bungyoku Y, Kitazawa S, Ishido S, Shoji I, Hotta H. Hepatitis C virus infection induces apoptosis through a Bax-triggered, mitochondrion-mediated, Caspase 3-dependent pathway. J Virol. 2008;82:10375–85. doi: 10.1128/JVI.00395-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).McLauchlan Properties of the hepatitis C virus core protein: a structural protein that modulates cellular processes. J Viral Hepat. 2000;7:2–14. doi: 10.1046/j.1365-2893.2000.00201.x. [DOI] [PubMed] [Google Scholar]
- (33).Hope RG, Murphy DJ, McLauchlan J. The domains required to direct core proteins of Hepatitis C virus and GB Virus-B to lipid droplets share common features with plant oleosin proteins. J Biol Chem. 2002;277:4261–70. doi: 10.1074/jbc.M108798200. [DOI] [PubMed] [Google Scholar]
- (34).Miyanari Y, Atsuzawa K, Usuda N, Watashi K, Hishiki T, Zayas M, Bartenschlager R, Wakita T, Hijikata M, Shimotohno K. The lipid droplet is an important organelle for hepatitis C virus production. Nat Cell Biol. 2007;9:1089–97. doi: 10.1038/ncb1631. [DOI] [PubMed] [Google Scholar]
- (35).Icard V, Diaz O, Scholtes C, Perrin-Cocon L, Rami+¿re C, Bartenschlager R, Penin F, Lotteau V, Andre P. Secretion of Hepatitis C virus envelope glycoproteins depends on assembly of apolipoprotein B positive lipoproteins. PLoS ONE. 2009;4:e4233. doi: 10.1371/journal.pone.0004233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Robenek MJ, Severs NJ, Schlattmann K, Plenz G, Zimmer KP, Troyer D, Robenek H. Lipids partition caveolin-1 from ER membranes into lipid droplets: updating the model of lipid droplet biogenesis. FASEB J. 2004;18:866–8. doi: 10.1096/fj.03-0782fje. [DOI] [PubMed] [Google Scholar]
- (37).Robenek H, Robenek MJ, Troyer D. PAT family proteins pervade lipid droplet cores. J Lipid Res. 2005;46:1331–8. doi: 10.1194/jlr.M400323-JLR200. [DOI] [PubMed] [Google Scholar]
- (38).Ohsaki Y, Cheng J, Suzuki M, Fujita A, Fujimoto T. Lipid droplets are arrested in the ER membrane by tight binding of lipidated apolipoprotein B-100. J Cell Sci. 2008;121:2415–22. doi: 10.1242/jcs.025452. [DOI] [PubMed] [Google Scholar]
- (39).von Hahn T, Rice CM. Hepatitis C virus entry. J Biol Chem. 2008;283:3689–93. doi: 10.1074/jbc.R700024200. [DOI] [PubMed] [Google Scholar]
- (40).Pileri P, Uematsu Y, Campagnoli S, Galli G, Falugi F, Petracca R, Weiner AJ, Houghton M, Rosa D, Grandi G, Abrignani S. Binding of hepatitis C virus to CD81. Science. 1998;282:938–41. doi: 10.1126/science.282.5390.938. [DOI] [PubMed] [Google Scholar]
- (41).Scarselli E, Ansuini H, Cerino R, Roccasecca RM, Acali S, Filocamo G, Traboni C, Nicosia A, Cortese R, Vitelli A. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO J. 2002;21:5017–25. doi: 10.1093/emboj/cdf529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Bartosch B, Vitelli A, Granier C, Goujon C, Dubuisson J, Pascale S, Scarselli E, Cortese R, Nicosia A, Cosset FL. Cell entry of hepatitis C virus requires a set of co-receptors that include the CD81 tetraspanin and the SR-B1 scavenger receptor. J Biol Chem. 2003;278:41624–30. doi: 10.1074/jbc.M305289200. [DOI] [PubMed] [Google Scholar]
- (43).Koutsoudakis G, Herrmann E, Kallis S, Bartenschlager R, Pietschmann T. The level of CD81 cell surface expression is a key determinant for productive entry of hepatitis C virus into host cells. J Virol. 2007;81:588–98. doi: 10.1128/JVI.01534-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Kapadia SB, Barth H, Baumert T, McKeating JA, Chisari FV. Initiation of hepatitis C virus infection is dependent on cholesterol and cooperativity between CD81 and scavenger receptor B type I. J Virol. 2007;81:374–83. doi: 10.1128/JVI.01134-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Evans MJ, von Hahn T, Tscherne DM, Syder AJ, Panis M, Wolk B, Hatziioannou T, McKeating JA, Bieniasz PD, Rice CM. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature. 2007;446:801–5. doi: 10.1038/nature05654. [DOI] [PubMed] [Google Scholar]
- (46).Ploss A, Evans MJ, Gaysinskaya VA, Panis M, You H, Jong Y, Rice CM. Human occludin is a hepatitis C virus entry factor required for infection of mouse cells. Nature. 2009;457:882–6. doi: 10.1038/nature07684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Cormier EG, Tsamis F, Kajumo F, Durso RJ, Gardner JP, Dragic T. CD81 is an entry coreceptor for hepatitis C virus. Proc. Natl. Acad. Sci.USA. 2004;101:7270–4. doi: 10.1073/pnas.0402253101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Goldstein LJ, Brown SM. The low-density lipoprotein pathway and its relation to atherosclerosis. Annu Rev Biochem. 1977;46:897–930. doi: 10.1146/annurev.bi.46.070177.004341. [DOI] [PubMed] [Google Scholar]
- (49).Agnello V, Abel G, Elfahal M, Knight GB, Zhang QX. Hepatitis C virus and other Flaviviridae viruses enter cells via low density lipoprotein receptor. Proc. Natl. Acad. Sci.USA. 1999;96:12766–71. doi: 10.1073/pnas.96.22.12766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Wunschmann S, Medh JD, Klinzmann D, Schmidt WN, Stapleton JT. Characterization of hepatitis C virus (HCV) and HCV E2 interactions with CD81 and the low-density lipoprotein receptor. J Virol. 2000;74:10055–62. doi: 10.1128/jvi.74.21.10055-10062.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Molina S, Castet V, Fournier-Wirth C, Pichard-Garcia L, Avner R, Harats D, Roitelman J, Barbaras R, Graber P, Ghersa P, Smolarsky M, Funaro A, Malavasi F, Larrey D, Coste J, Fabre JM, Sa-Cunha A, Maurel P. The low-density lipoprotein receptor plays a role in the infection of primary human hepatocytes by hepatitis C virus. J Hepatol. 2007;46:411–9. doi: 10.1016/j.jhep.2006.09.024. [DOI] [PubMed] [Google Scholar]
- (52).Owen DM, Huang H, Ye J, Gale J. Apolipoprotein E on hepatitis C virion facilitates infection through interaction with low-density lipoprotein receptor. Virology. 2009;394:99–108. doi: 10.1016/j.virol.2009.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Bartosch B, Dubuisson J, Cosset FL. Infectious hepatitis C virus pseudo-particles containing functional E1-E2 envelope protein complexes. J Exp Med. 2003;197:633–42. doi: 10.1084/jem.20021756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Hobbs HH, Brown MS, Goldstein JL. Molecular genetics of the LDL receptor gene in familial hypercholesterolemia. Hum Mutat. 1992;1:445–66. doi: 10.1002/humu.1380010602. [DOI] [PubMed] [Google Scholar]
- (55).Meuleman P, Leroux-Roels G. The human liver-uPA-SCID mouse: A model for the evaluation of antiviral compounds against HBV and HCV. Antiviral Res. 2008;80:231–8. doi: 10.1016/j.antiviral.2008.07.006. [DOI] [PubMed] [Google Scholar]
- (56).Kneteman NM, Toso C. In vivo study of HCV in mice with chimeric human livers. Methods Mol Biol. 2009;510:383–99. doi: 10.1007/978-1-59745-394-3_29. [DOI] [PubMed] [Google Scholar]
- (57).Chayama K, Hayes CN, Hiraga N, Abe H, Tsuge M, Imamura M. Animal model for study of human hepatitis viruses. J Gastroenterol Hepatol. 2011;26:13–8. doi: 10.1111/j.1440-1746.2010.06470.x. [DOI] [PubMed] [Google Scholar]
- (58).Chandler CE, Wilder DE, Pettini JL, Savoy YE, Petras SF, Chang G, Vincent J, Harwood HJ., Jr. CP-346086: an MTP inhibitor that lowers plasma cholesterol and triglycerides in experimental animals and in humans. J Lipid Res. 2003;44:1887–901. doi: 10.1194/jlr.M300094-JLR200. [DOI] [PubMed] [Google Scholar]
- (59).Cuchel M, Bloedon LT, Szapary PO, Kolansky DM, Wolfe ML, Sarkis A, Millar JS, Ikewaki K, Siegelman ES, Gregg RE, Rader DJ. Inhibition of microsomal triglyceride transfer protein in familial hypercholesterolemia. N Engl J Med. 2007;356:148–56. doi: 10.1056/NEJMoa061189. [DOI] [PubMed] [Google Scholar]
- (60).Burnett JR. Drug evaluation: ISIS-301012, an antisense oligonucleotide for the treatment of hypercholesterolemia. Curr opin Mol Ther. 2006;8:461–7. [PubMed] [Google Scholar]
- (61).Thomas T, Ginsberg H. Development of apolipoprotein B antisense molecules as a therapy for hyperlipidemia. Curr Atheroscler Rep. 2010;12:58–65. doi: 10.1007/s11883-009-0078-7. [DOI] [PubMed] [Google Scholar]