Abstract
Many bacteria use large modular enzymes for the synthesis of polyketide and peptide natural products. These multidomain enzymes contain integrated carrier domains that deliver bound substrates to multiple catalytic domains, requiring coordination of these chemical steps. Non-Ribosomal Peptide Synthetases (NRPSs) load amino acids onto carrier domains through the activity of an upstream adenylation domain. Our lab recently determined the structure of an engineered two-domain NRPS containing fused adenylation and carrier domains. This structure adopted a domain swapped dimer that illustrated the interface between these two domains. To continue our investigation, we now examine PA1221, a natural two-domain protein from Pseudomonas aeruginosa. We have determined the amino acid specificity of this new enzyme and used domain specific mutations to demonstrate that loading the downstream carrier domain within a single protein molecule occurs more quickly than loading of a non-fused carrier domain inter-molecularly. Finally, we have determined crystal structures of both the apo- and holo-PA1221 protein, the latter using a valine-adenosine vinylsulfonamide inhibitor that traps the adenylation-carrier domain interaction. The protein adopts a similar interface to that seen with the prior adenylation-carrier protein construct. A comparison of these structures with previous structures of multidomain NRPSs suggests that a large conformational change within the NRPS adenylation domains guides the carrier domain into the active site for thioester formation.
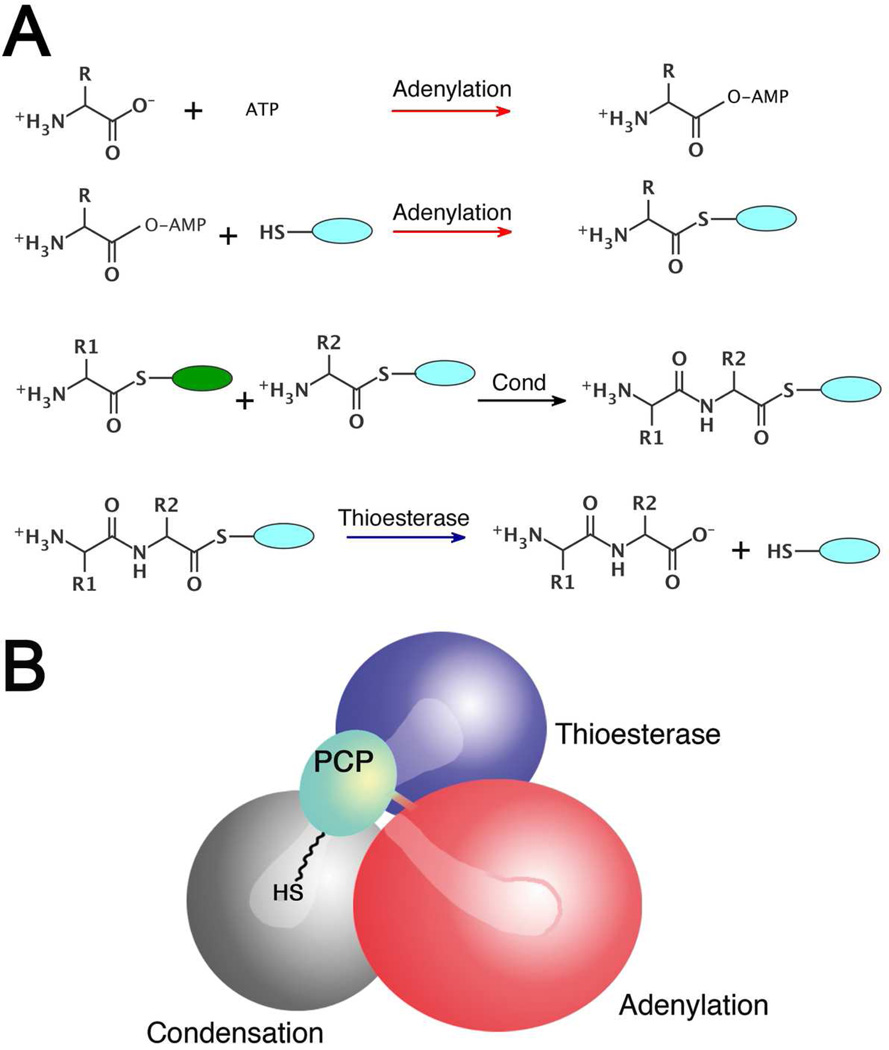
Non-ribosomal Peptide Synthetases (NRPSs) are modular enzymes that contain multiple catalytic domains joined as a single, multidomain protein. These domains function in a coordinated fashion to direct the synthesis of the peptide product (1). During synthesis, the amino acid and peptide substrates are bound covalently to a peptidyl carrier protein (PCP) domain, which is post-translationally modified to contain a phosphopantetheine cofactor on a conserved serine residue (2). Amino acids are first loaded onto the PCP by an upstream adenylation domain, and then delivered to upstream or downstream condensation domains, where peptide bond formation occurs. Final release of peptide is catalyzed by a terminal thioesterase domain. This catalytic strategy requires that a typical PCP domain of an internal module must visit at least three catalytic domain active sites (Figure 1A) to enable a) initial loading by an upstream adenylation domain, b) peptide bond formation by an upstream condensation domain, and c) peptide off-loading or hydrolysis by a downstream condensation or thioesterase domain (3). Structural studies of single- and multi-domain NRPS proteins, as well as studies that use molecular and biochemical tools to probe the impact of mutations at the interface or linkers between domains, are providing insights into the domain interfaces and structural elements that enable the conformational flexibility necessary to deliver the nascent peptide to the catalytic domains.
Figure 1.
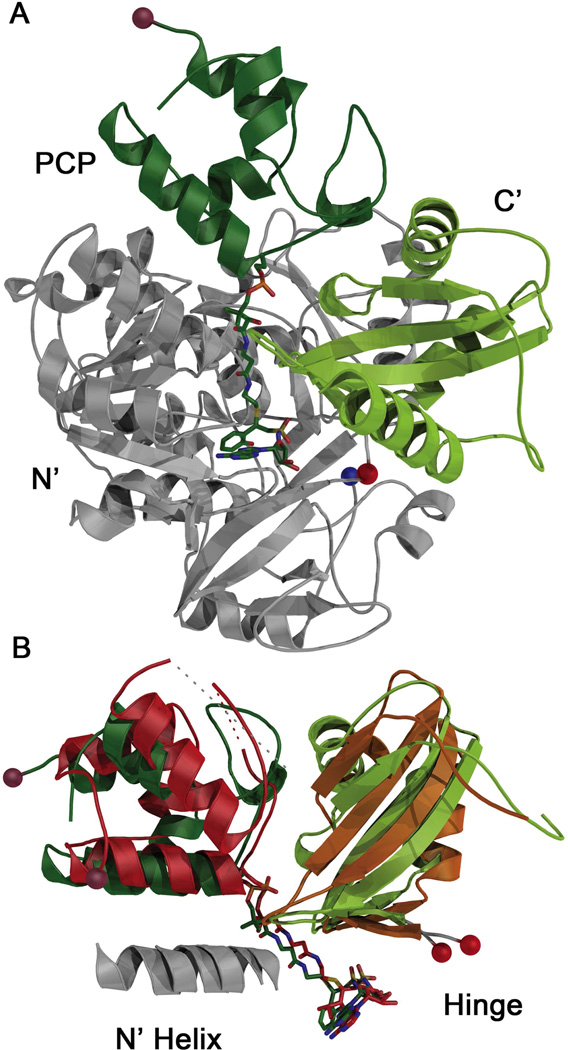
Multidomain catalysis of peptide synthesis by the NRPSs. (A) Reactions catalyzed by the most common NRPS catalytic domains. The adenylation domain catalyzes a two step reaction to first adenylate the amino acid and then covalently load the downstream carrier domain, represented by the blue oval. The pantetheine cofactor is represented by the thiol SH. The domain alternation hypothesis suggests a 140° rotation of a small C-terminal subdomain within the adenylation domain is used to adopt the catalytic conformations for the two partial reactions. The condensation domain catalyzes peptide bond formation, transferring the loaded amino acid from an upstream carrier domain (green) to form the dipeptide on the downstream carrier (blue). The terminal thioesterase domain catalyzes thioester hydrolysis, releasing the peptide. (B) Schematic representation of SrfA-C highlighting the structural problem of the domain rearrangements required to enable the phosphopantetheine cofactor to reach the neighboring active sites. The schematic represents the domain orientation observed in the SrfA-C structure (PDBID:2VSQ). The condensation domain is grey, the adenylation domain is red, the PCP domain is light blue, and the thioesterase is dark blue.
Individual NRPS domain structures have been determined, including adenylation (4–7), condensation (8), peptidyl carrier protein (9), and thioesterase (10) domains. Additionally, structures of multi-domain NRPSs have been determined that identify how the PCP interacts with each of the catalytic domains. A structure of the intermolecular interaction between the adenylation and PCP domains of the enterobactin system demonstrates how the PCP interacts with the adenylation domain for amino acid loading (11). The interaction between the PCP and the thioesterase domain has been determined both by NMR and X-ray crystallography (12, 13), with the crystal structure of the holo-PCP providing clear insights into the pantetheine binding site. Exciting insights into PCP-Condensation domain interactions were provided by SrfA-C, a complete terminal module containing the condensation, adenylation, PCP, and thioesterase domains (14). In this structure, the phosphopantetheinylation site of the PCP, mutated to an alanine to facilitate crystallization, was located ~16 Å from the catalytic histidine of the condensation domain, roughly the length of the phosphopantetheine arm (Figure 1B). However, the PCP domain is 57 and 43 Å from the active sites of the adenylation and thioesterase domains, demonstrating that a substantial rearrangement is necessary to deliver the PCP and peptide substrate to the adjacent domains (14).
The NRPS adenylation domains belong to the ANL superfamily of adenylating enzymes (15). Members of this family are 450–600 residues in length and contain two subdomains. The ANL enzymes all carry out two partial reactions at the same active site, an initial adenylation reaction and a second reaction that is either a thioester-forming reaction or, in the case of luciferase, a multi-step oxidative reaction. These two partial reactions require changes to the active site that are achieved through domain rotation of the smaller C-terminal subdomain. Upon completion of the adenylation half reaction, the C-terminal subdomain undergoes a ~140° rotation to present a different conserved region that positions the pantetheine moiety to the acyl-adenylate for thioester formation (16). The SrfA-C adenylation domain adopted a conformation that approximates the adenylate-forming conformation, with the two subdomains more open relative to the catalytic conformation. The rotation of the C-terminal subdomain, as demonstrated for ANL superfamily members (16–19), may provide one necessary domain rearrangement required of modular NRPSs to coordinate delivery of the downstream PCP domain (14, 15).
Our lab recently determined the structure of the adenylation-carrier protein interaction using the intermolecular module from enterobactin biosynthesis. We created a fusion protein between the self-standing adenylation domain EntE (20) and the acyl carrier protein from EntB (21). This EntE-B protein was crystallized using a mechanism-based inhibitor that mimics the adenylate intermediate, reacting covalently with the thiol of the incoming phosphopantetheine (11). Similar inhibitors have been used in the structural characterization of thioester bond formation in SUMO E1 (22) and the ANL adenylating enzyme o-succinylbenzoate-CoA synthetase (23). The crystal structure of EntE-B demonstrates the regions of the PCP that interact with the adenylation domain. In particular, the 10 residues that precede the pantetheinylation site (loop 1) and the helix that follows this position (helix 2) form the interaction surface for the adenylation domain. Although the EntE-B chimeric protein was designed to elucidate the intra-molecular interactions observed in a natural two-domain adenylation-PCP protein, the protein crystallized as a dimer of intermolecular interactions, where the phosphopantetheine and PCP of chain A were directed into the active site of EntE chain B, while the PCP of chain B was donated back to the adenylation domain of chain A. These inter-molecular interactions were validated by directed mutagenesis experiments to improve the ability of an EntE homolog to recognize EntB. Guided by the EntE-B crystal structure, changes to several surface residues of this homolog, BasE, improved catalytic efficiency with the EntB carrier domain by as much as 50-fold (11). This structure therefore provides insight into the EntE and EntB interaction and, presumably, the intermolecular transfer interface of other NRPS adenylation and PCP domains.
To continue our investigation of adenylation-PCP interactions, we identified natural two-domain proteins that may serve as suitable targets for structural and functional investigation. The human pathogen Pseudomonas aeruginosa contains a biosynthetic operon encoded by the PA1221-PA1211 genes1. Although the product of this pathway is unknown, this operon contains several of the hallmarks of a novel NRPS cluster, including a di-domain adenylation-PCP protein, PA1221, and self-standing condensation, thioesterase, and adenylation domains encoded by PA1220, PA1219, and PA1215, respectively. Additionally, the operon contains a potential Major Facilitator Superfamily (MFS) transporter (24), encoded by PA1212. We present here structural and functional analysis of PA1221. We have identified the prefered substrate of PA1221 and demonstrate that intra-molecular loading occurs more quickly than acylation of a PCP domain on a separate polypeptide molecule. The structures of both the apo- and holo-PA1221 proteins adopt the thioester-forming conformation. In the presence of a mechanism-based inhibitor, the PCP domain is observed interacting with the adenylation domain in a conformation that is very similar to that seen with the EntE-EntB structure (11).
Materials and Methods
Cloning, Expression, and Purification
The gene encoding PA1221 was cloned from Pseudomonas aeruginosa strain PAO1 into pET15b containing a N-terminal 5X His-tag and TEV protease recognition site (25). PA1221 was expressed in BL21(DE3) cells grown at 37°C to an OD600 of 0.6 followed by induction with 500 µM IPTG and overnight incubation at 16°C. Cells were lysed by sonication in 100 mM Tris, pH 7.5, 500 mM NaCl, 30 mM imidazole, 1 mM MgCl2, 15% glycerol, 0.2 mM TCEP, and protease inhibitor cocktail (Sigma), and passed over a 5 ml Ni2+ HiTrap Chelating HP column (GE Healthcare). The column was washed with 5 column volumes of lysis buffer and eluted with lysis buffer containing 300 mM imidazole. Fractions containing PA1221 were pooled and dialyzed (with one change) overnight against lysis buffer lacking imidazole with TEV protease at a 1:80 ratio to remove the 5x His-tag (25). The dialyzed protein was then passed over the Ni2+ HiTrap Chelating HP column and the flow-through fractions containing untagged PA1221 were collected. PA1221 was concentrated with a 50 kDa molecular weight cutoff concentrator (Millipore) to 7.5 mg/mL before flash freezing in liquid N2 (26). One liter of cell culture yielded ~15 mg of pure protein.
The K499L and S553A mutations were generated with the QuikChange II Site-Directed Mutagenesis kit (Agilent) following the manufacturer’s protocol. Mutant enzymes were purified as described for wild-type enzyme.
Phosphopantetheinylation of PA1221
Purified PA1221 was phosphopantetheinylated (27, 28) by incubation with 10 nM of the non-specific phosphopantetheinyl transferase Sfp, 12.5 mM MgCl2, and 1 mM CoA in lysis buffer for 50 minutes at 22°C. Following phosphopantetheinylation, Sfp and CoA were removed from the reaction sample by buffer exchange through a 50 kDa molecular weight cut-off concentrator (Millipore) 5 times.
Analysis of the PA1221 adenylation reaction
PA1221 substrate preference was assessed utilizing the radiolabeled pyrophosphate exchange assay with amino acids, 2-aminobenzoic acid, 4-chlorobenzoic acid, and acetate as performed previously (20, 29). In brief, 1 µM of PA1221 was incubated with 2 mM ATP, 0.2 mM Na4PPi, 5 mM amino acid or carboxylate substrate, and 0.150 µCi 32P-Na4PPi (Perkin-Elmer) in reaction buffer (50 mM HEPES pH 7.5, 100 mM NaCl, and 1 mM MgCl2) for 10 min at 37°C. The reaction was quenched with 1.2 % (w/v) activated charcoal, 100 mM unlabeled Na4PPi, and 350 mM perchloric acid. The charcoal was collected by centrifugation and washed twice with H2O. After washing, the charcoal was resuspended in 0.5 mL of H2O and transferred to scintillation vials. Radiolabel incorporated into nucleotide was quantified with a Packard Tri-Carb 1900 TR Liquid Scintillation Counter (Perkin-Elmer). PA2412 was purified as described previously (30) and included in the PA1221 adenylation reactions at 0.8-, 1.0-, and 1.2-fold molar ratios using the 32P-PPi exchange assay.
Analysis of loading of the PA1221 adenylation domain
Inter- and intra-molecular loading were assessed through 3H-valine incorporation into PA1221. We first monitored the reaction at 37°C using 2 mM valine, 0.45 µCi 3H-Valine, and 1 mM ATP. These initial reactions contained 10 µM of wild-type or mutant PA1221 for reactions containing a single enzyme. In the reaction with the combination of the two compensatory mutants, the reactions contained 5 µM of each mutant. Because of the rapid rate of acylation of the wild-type protein at 37°C, and because of the significant thioester hydrolysis rate (t½ ~40 min at pH 7.5 at 25°C, (31)), a second series of reactions was set up and carried out on ice.
The time course included reaction buffer (100 mM Tris, pH 7.5, 250 mM NaCl, 1 mM MgCl2, 15 % glycerol, and 0.2 mM TCEP) with 12 µM of WT protein or 12 µM of each mutant protein, 250 µM valine, 0.6 µCi 3H-Valine, and initiated with 0.8 mM ATP. Reactions were quenched with 20% TCA at 0, 5, 10, 15, 30, 45, 60, 90, and 120 minutes. Protein pellets were washed 3 times with acetone and then resuspended in 8 M urea and transferred to vials containing scintillation fluid for counting in the Packard Tri-Carb 1900 TR Liquid Scintillation Counter (Perkin-Elmer).
Inhibitor Synthesis
The valine-adenosine vinylsulfonamide (Val-AVS) inhibitor was synthesized in four steps from N-tert-butoxycarbonyl-L-valine Weinreb amide S1 in 45% overall yield. S1 was reduced with lithium aluminum hydride to the corresponding aminoaldehyde S2 that was immediately olefinated with diphenylphosphoryl methylsulfonamide S3 to afford vinylsulfonamide S4. Mitsunobu coupling of S4 with protected adenosine derivative S5 furnished S6. Global deprotection of S6 with 80% aqueous TFA yielded Val-AVS, which was purified by recrystallization from methanol and diethylether. Detailed experimental procedures are provided in the Supporting Information. Data for Val-AVS: mp 148–150 °C; −14.3 (c 0.1, MeOH); 1H NMR (600 MHz, CD3OD) δ 0.94 (d, J = 7.2 Hz, 3H), 1.00 (d, J = 7.2 Hz, 3H), 2.02–2.05 (m, 1H), 3.32–3.34 (m, 2H), 3.79 (t, J = 7.8 Hz, 1H), 4.23–4.26 (m, 1H), 4.35–4.37 (m, 1H), 4.79 (t, J = 5.4 Hz, 1H), 5.95 (d, J = 6.0 Hz, 1H), 6.60 (dd, J = 15, 7.2 Hz, 1H), 6.77 (d, J = 15 Hz, 1H), 8.28 (s, 1H), 8.32 (s, 1H); 13C NMR (125 MHz, CD3OD) δ 18.0, 19.0, 32.3, 45.8, 58.0, 72.9, 74.8, 85.7, 91.6, 121.3, 135.6, 137.3, 143.3, 149.9, 150.9, 153.1; HRMS (ESI+) calcd for C16H26N7O5S [M + H]+ 428.1711, found 428.1726 (error 3.5 ppm).
Crystallization of apo- and holo-PA1221
Crystallization conditions for apo-PA1221 in the presence of 1 mM valine and 1 mM ATP were identified from sparse matrix screening using hanging-drop vapor diffusion at 4°C. Final crystals of apo-PA1221 at 7.5 mg/ml were obtained using a mother liquor of 50 mM MES, pH 6.0, 100 mM glycine, and 1.5–4 % polyethylene glycol (PEG) 8000. Crystals were carefully removed from skin-encased drops and cryoprotected in equilibrated reservoir solution with 6–20 % (2R,3R)-(−)-butanediol. Crystals grew within 24 hr; SDS-PAGE analysis of protein crystals showed there was no proteolysis of the PA1221 protein.
Crystals of holo-PA1221 grew in the presence of the mechanism based inhibitor Val-AVS. The inhibitor was dissolved in 100 % DMSO to a final concentration of 26 mM and added directly to 10.2 mg/ml holo-PA1221 at a 1.2-fold molar excess over protein. The inhibitor and protein were kept on ice for 30 minutes prior to setting up the crystallization experiment. Crystals grew at 4°C via hanging-drop vapor diffusion using a cocktail of 50 mM HEPES, pH 8.5, and 20 % methyl ether PEG 8000. Crystals grew initially as clusters from which individual plates could be isolated and cryoprotected in equilibrated reservoir solution supplemented with 8 % (2R,3R)-(−)-butanediol.
Data Collection, Processing, and Refinement
Data for the crystal of apo-PA1221 were collected at SSRL beamline 9–2 using the BluIce software package (32). HKL2000 (33) was used to index and scale the data. The structure was solved by molecular replacement using the full-length phenylalanine-activating adenylation domain of Gramicidin synthetase A (PheA) (PDB: 1AMU, (5)) as a search model with MOLREP from the CCP4 package (34). It was immediately obvious that the C-terminal domain adopted the thioester-forming conformation and it was removed from the model. The C-terminal subdomain was then manually modeled, followed by iterative rounds of model building and TLS refinement in COOT (35) and both Refmac5 (36) and PHENIX (37). The model was validated and checked using MolProbity (38) throughout model building and refinement.
Data for the holo-PA1221 co-crystallized with Val-AVS were also collected at SSRL beamline 9–2; the data were processed with iMOSFLM (39) and SCALA (40). The model of the adenylation domain from apo-PA1221, residues 16–510, lacking the cofactor was used as a molecular replacement search model for holo-PA1221 using MOLREP. All non-protein atoms were removed from the model. Residues 520–602 of the PCP domain were modeled manually. The structure of holo-PA1221 was built and refined as with the apo-PA1221 structure.
The ligands for both structures were generated using Jligand (34) and chirality manually restrained according to previous deposited phosphopantetheine and AMP definitions. Structure factors and coordinates for both the apo (4DG8) and holo (4DG9) structures have been deposited with the Protein Data Bank.
Results
Substrate Specificity and Activity of the PA1221 Protein
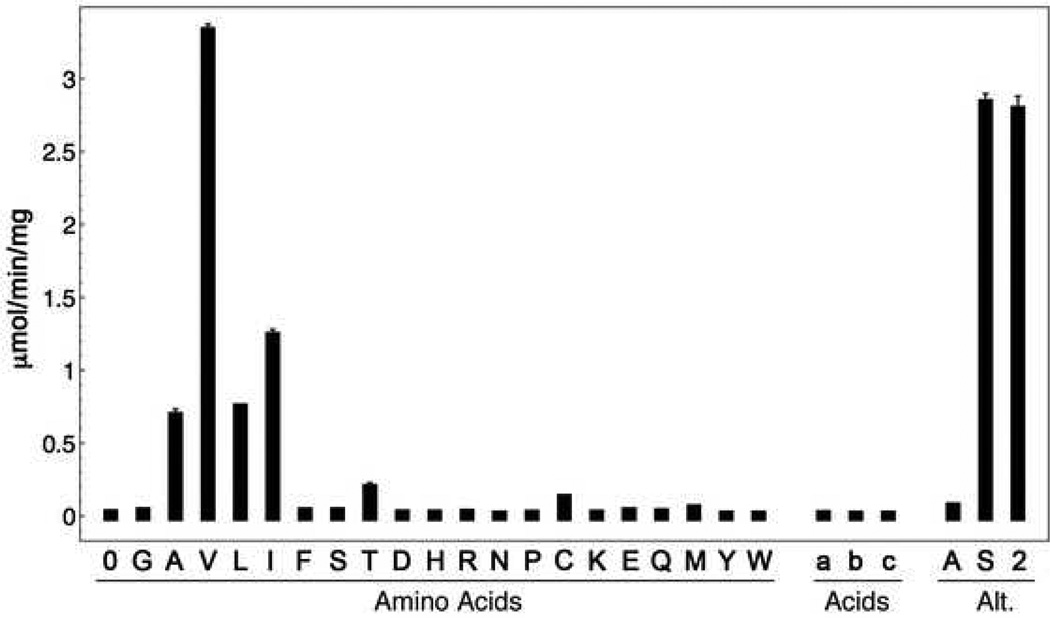
We first determined the substrate specificity of the PA1221 adenylation domain. Bioinformatic analysis using the amino acid residues likely to form the substrate-binding pocket (41, 42) predicted a preference for small, aliphatic amino acids. We set up a broad spectrum of amino acids, as well as 4-chlorobenzoate, 2-aminobenzoate, and acetate, to cover a range of potential substrates. Valine resulted in the highest activity, with Ile, Leu, and Ala utilized as substrates at a lower rate (Figure 2).
Figure 2.
Substrate specificity of PA1221. Specificity was tested utilizing the 32P-PPi exchange assay. Amino acids tested are reported as single letter code and background activity, lacking a carboxylate substrate, is labeled “0”. Additional acids were tested and labeled a, b, and c, representing acetate, 4-chlorobenzate, and 2-amino-benzoate, respectively. Three other assays were performed in a series of alternate experiments (Alt.). Two mutant enzymes were tested with valine as the amino acid substrate, the K499L A10 adenylation domain mutant (A) and the Ser533Ala phosphopantetheinyl attachment site mutant (S). Finally, the impact of 1.2-fold molar excess of the MbtH-like protein PA2412 is shown in the final bar (2).
Two mutant enzymes were produced to analyze intra- vs inter-molecular substrate loading (see below). We tested the role of Lys499, a conserved catalytic residue from the A10 motif, for its involvement in the adenylation reaction. Analysis of adenylation activity with the K499L mutant resulted in roughly 1% of the WT activity in the presence of 5 mM valine, consistent with other A10 mutants belonging to the ANL superfamily (43). We also made the S553A mutant, altering the PCP domain site of cofactor addition to make an obligate apo-proteion. The S553A mutant incorporated radio-labeled pyrophosphate at similar levels to WT (Figure 2) showing that adenylate-forming activity is independent of cofactor addition to the PCP.
The P. aeruginosa MbtH-Like protein PA2412 has no effect on adenylation activity
Many NRPS biosynthetic clusters contain a small, ~70 residue protein that enhances the adenylation activity of NRPS adenylation domains (44, 45). Members of the MbtH-like family of proteins contain a conserved hydrophobic patch on one surface of the protein (30) that has been shown to be important for this enhanced adenylation activity (46). P. aeruginosa contains a single MbtH-like protein that is upregulated with the biosynthetic operon for the peptide siderophore pyoverdine. Because MbtH-like proteins have been shown to act on multiple NRPS clusters within one organism (47, 48), we tested the effects of PA2412 on the adenylation activity of PA1221. PA2412 did not enhance the PA1221 adenylation reaction (Figure 2).
Structural characterization of apo-PA1221
We subjected apo-PA1221 protein to crystallization trials in the presence and absence of different combinations of nucleotides and amino acids. In the presence of 1 mM ATP and valine, apo-PA1221 crystallized in P43212 space group with one molecule in the asymmetric unit. The structure was obtained by molecular replacement using the full-length PheA as a search model. The initial electron density deteriorated at Asp417, the hinge residues joining the N- and C-terminal subdomains, indicating that PA1221 was not in the adenylate-forming conformation adopted by PheA. The C-terminal subdomain was therefore removed and rebuilt manually. The final model refined to an R and Rfree of 18.6 and 23.4 %, respectively (Table 1) and includes residues 15–510 and 243 water molecules. No electron density was observed for the PCP domain, residues 512–618. SDS-PAGE analysis of comparable crystals showed no protolysis had occurred. The C-terminal loop containing the catalytic Lys499 is poorly ordered and residues Ala496 through Leu500 are not included in the final model.
Table 1.
Crystallographic Data Collection and Refinement Statistics.
| Data Collection | apo-PA1221 | holo-PA1221 |
|---|---|---|
| Beamline | SSRL BL 9-2 | SSRL BL 9-2 |
| Wavelength (Å) | 0.9795 | 0.9795 |
| Space Group | P43212 | C2221 |
| Unit cell a, b, c (Å), | 92.56, 92.56, 164.39 | 53.19, 231.18, 122.05 |
| Asymmetric unit | 1 molecule | 1 molecule |
| Resolution range | 30.0-2.15 | 61.0-2.55 |
| No. Obs. | 275,595 | 97,138 |
| No. unique reflections | 36,771 | 25,006 |
| Completeness % (outer) | 92.83 (91.3) | 100 (99.4) |
| I/σ (outer) | 12.7 (2.2) | 9.4 (2.2) |
| Rmerge (%) (outer) | 8.7 (49) | 11.4 (57.1) |
|
Structure Refinement | ||
| Resolution range | 29.38-2.15 | 61.03-2.55 |
| R-factor (%) | 18.6 | 18.2 |
| Rfree (%) | 23.4 | 23.8 |
| No. protein/solvent atom | 3780/243 | 4389/77 |
| RMSD bond distances (Å) | 0.009 | 0.008 |
| RMSD bond angles (°) | 1.27 | 1.36 |
| Average B-factor (Å2) | ||
| Protein, Solvent, Ligands | 38.1, 37.8, 26.3 | 40.6, 32.6, 27.7 |
|
Ramachandran analysis | ||
| Preferred (%) | 97.6 | 95.6 |
| Allowed (%) | 1.8 | 3.5 |
| Outliers (%) | 0.2 | 0.2 |
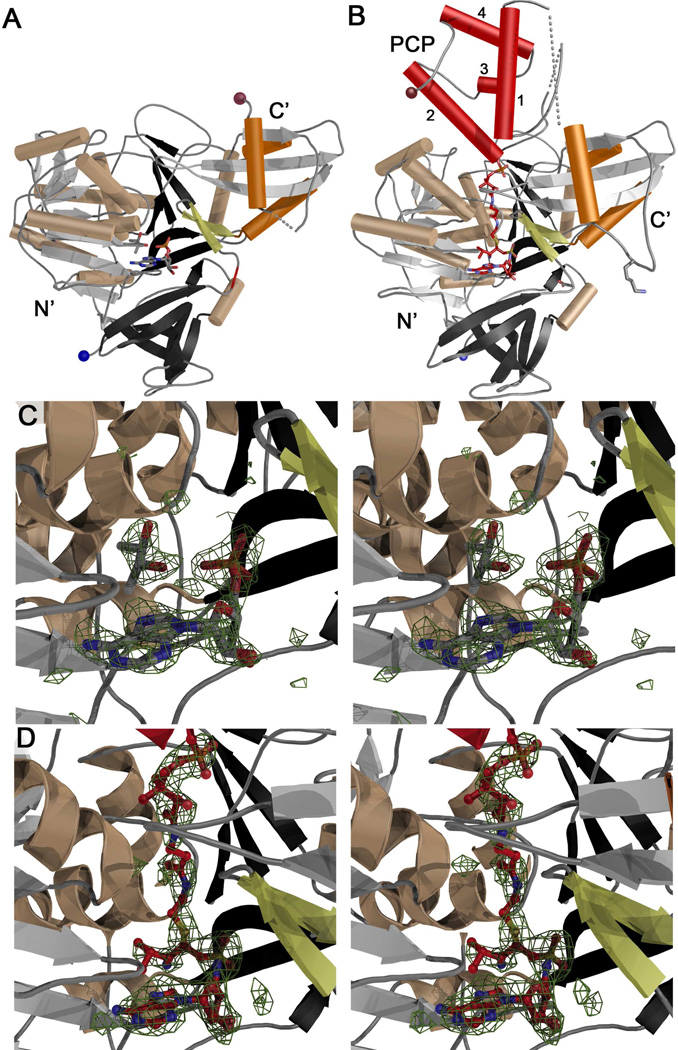
The structure of the PA1221 adenylation domain is similar to prior structures of ANL enzymes (Figure 3A). The N-terminal domain aligns with PheA with an RMS displacement of 1.5 Å over 323 Cα atoms of homologous residues. The C-terminal domain similarly aligns with an RMS displacement of 1.4 Å over 68 Cα atoms. The active site contained AMP bound in the nucleotide binding pocket. Electron density was observed near the AMP in the acyl-binding pocket (Figure 3C). We modeled this density as a molecule of valine, glycine, and (2R,3R)-(−)-butanediol, and found that the latter, which was used as the cryoprotectant, to refine most optimally. A second molecule of cryoprotectant was modeled in density close to Trp256.
Figure 3.
Structures of apo- and holo-PA1221. (A) Ribbon diagram of apo-PA1221 with N-terminal domain colored grey, black, and wheat, the C-terminal subdomain highlighted with orange helices. The A8 loop, an antiparallel sheet that immediately follows the hinge at Asp417, is shown in yellow and the N- and C-termini are indicated with blue and brown spheres. (B) Ribbon diagram of holo-PA1221, colored as in panel A with the PCP colored red and the four helices labeled 1–4. Lys499, the catalytic lysine from the A10 motif, is shown in stick representation. The same loop, Ala496-Leu500, is disordered in the apo-structure and is shown with a dotted line in panel A. (C) Stereo image of apo-PA1221 active site with AMP and (2R,3R)-(−)-butanediol and Fo-Fc difference density contoured at 3 σ. (D) Stereo image of holo-PA1221 phosphopantetheine tunnel and active site with phosphopantetheine and Val-AVS with Fo-Fc difference density contoured at 3 σ. Both electron density maps were created with coefficients calculated prior to inclusion of ligands in the model.
The PA1221 adenylation domain contains two subdomains, the larger N-terminal subdomain residues 16–417 and the smaller C-terminal subdomain 418–510. Asp417 is the “hinge residue” that has been shown to exhibit the primary conformational change to accommodate the two catalytic conformations (18). Ten conserved motifs, labeled A1–A10, have been described for this enzyme family (5, 15). Aside from the partial disordering of motif A10, PA1221 contains no deviations from these conserved motifs, which appear to play similar roles as observed previously.
Structural characterization of holo-PA1221
The structure of apo-PA1221 did not allow us to observe the PCP domain (Figure 3A). We therefore employed the use of a vinyl-sulfonamide inhibitor (49) designed to capture the incoming phosphopantetheine moiety of the carrier domain upon reaction with the adenylate mimic. The holo-PA1221 co-crystallized with the inhibitor in C2221 space group with one molecule in the asymmetric unit. The structure was solved using molecular replacement with apo-PA1221 as the search model. The holo-PA1221 protein also adopted the thioester-forming conformation. In this crystal form the PCP domain was resolved (Figure 3B) and built through residue 602, out of 618. The model refined to an R and Rfree of 18.2 and 23.8 %, respectively (Table 1). Several loops show weak electron density. An N-terminal loop encompassing residues Thr134-Cys141 is weakly ordered. The linker that joins the adenylation domain to the PCP is disordered from residue 511–519. Within the PCP domain, the regions that interact with the adenylation domain, residues Gly550-Ser563, exhibit the best electron density compared to the regions distal to the domain interface. After the fourth helix at residues Asp584-Arg593, the chain adopts the start of a fifth helix at Pro597-Leu601, which packs against the first helix of the PCP domain. The final 13 residues of PA1221 are disordered.
Ser553, the phosphopantetheine attachment site, had continuous positive difference density reaching from the serine side chain to the active site located in the interface between the adenylation N- and C-terminal subdomains (Figure 3D). The phosphopantetheine and the Val-AVS inhibitor were modeled into the positive Fo-Fc difference density as one molecule, reflecting the covalent reaction of the thiol with the vinylsulfonamide linker.
Active site interactions of the adenylation domain
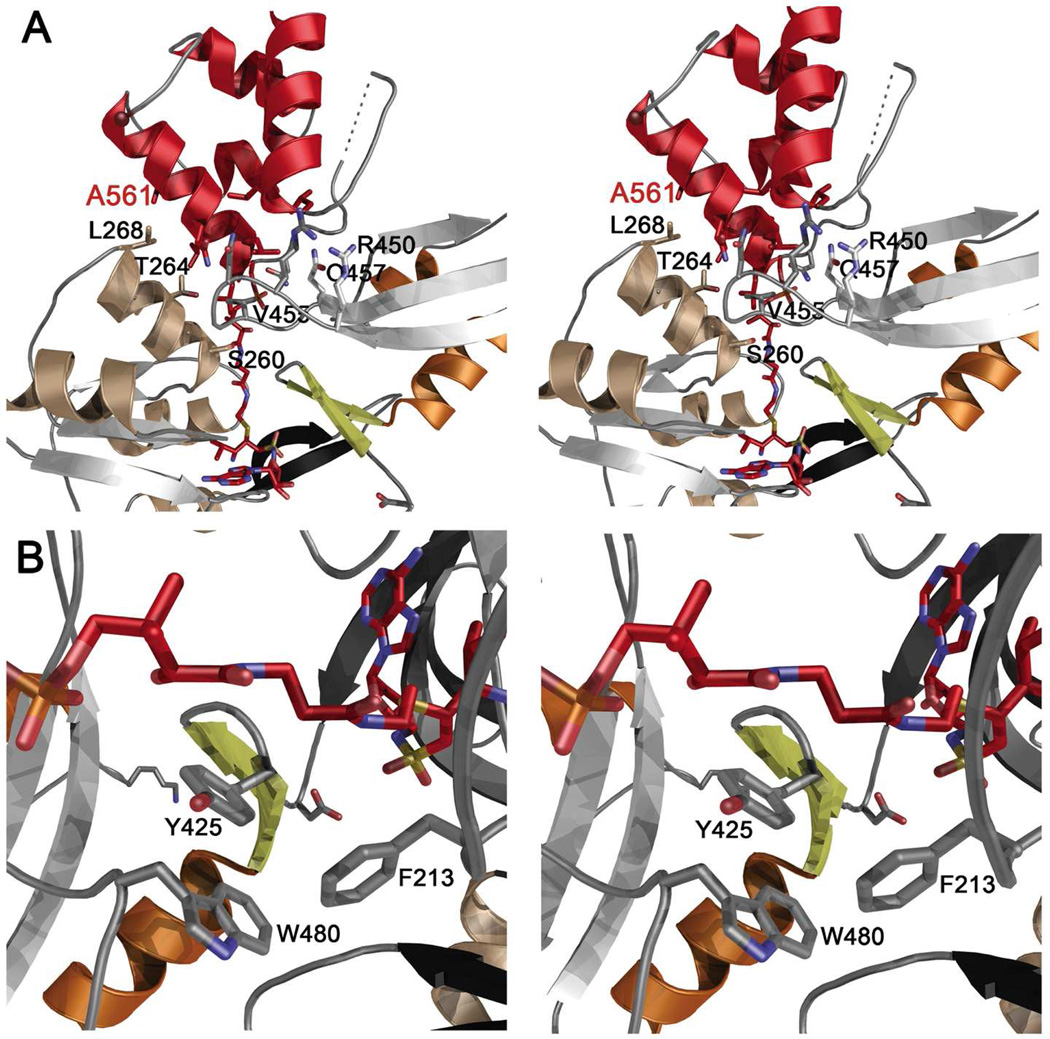
The crystal structure of the enzyme bound to Val-AVS provides a detailed view of the enzyme active site (Figure 4), illustrating the interactions with the nucleotide, amino acid substrate, and phosphopantetheine moieties. The adenine binds in a hydrophobic pocket formed on one side by the aromatic side chains of Tyr307 and Phe412, and on the other side by the main chain of Gly283-Asp284-Ile285. The ribose hydroxyls both interact with the side chain of Asp400 of the A7 motif. The sulfonamide nitrogen interacts with the side chain of Lys421, a residue on the turn that follows the hinge residue of the adenylation domain. The valine amine interacts with the side chain of Asp214 and the main chain carbonyl of Thr314, which is the only outlier in the Ramachandran analysis in both structures. The strained torsion angles of Thr314 (Φ = 74, Ψ = −91) may help orient the Asp214 side chain. As observed previously (11, 15, 50, 51), the phosphopantetheine makes limited interactions with the protein. The amide nitrogens hydrogen bond to the carbonyls of Asn423 and Gly424, residues on the A8 loop that follows the hinge. There are no interactions between the adenylation domain and the phosphate from the cofactor.
Figure 4.
Stereorepresentations of holo-PA1221. (A) Ribbon diagram of the interaction between the Adenylation and PCP domains. The PCP is shown in red, while the adenylation domain is colored as in Figure 3B. Several residues from the adenylation domain are labeled in black to orient the viewer. The PCP residue Ala561 is labeled red; this residue is located on helix 2. The residues that form the hydrophobic interface with this helix include Thr264 and Leu268. Leu 261 and Leu265 are shown but not labeled for clarity. Val455 is shown interacting with Leu554 and Leu555, which are not labeled. Residues that contribute to the hydrogen bonding network include Arg450, Arg452, Asn453, and Gln457 are shown. These residues interact primarily with main chain atoms of the PCP. (B) Phe213, the conserved aromatic residue of the A4 motif, is stabilized by interactions with aromatic residues Tyr425 and Trp480 of the C-terminal domain. Rotation of Phe213 opens the phosphopantetheine tunnel allowing proper binding of the pantetheine for thioester formation.
After the first structural elucidation of an NRPS adenylation domain, several groups analyzed the residues that form the amino acid binding pocket and noted the correlation between the identity of these residues and the substrate specificity (41, 52). We examined the valine binding pocket to compare to the prior predictions (Figure S1 of Supporting Information). Generally, the Stachelhaus (41) substrate-specificity conferring code (DALWLGGTFK) agrees well with the active-site residues in the model. This original prediction was based on the large phenylalanine substrate of PheA (5). Analysis of the PA1221 structure demonstrates, however, that several of the predicted residues are greater than 6 Å from the valine and therefore may be more susceptible to substitution without impacting substrate specificity. In particular, Leu218, Leu280, and Gly306, the structurally equivalent residues to Trp239, Ile299, and Ala322 of PheA, are removed from the binding pocket and likely do not contribute to the specificity except through potential “second shell” interactions.
A conserved aromatic residue forms the A4 motif of the ANL adenylating enzymes (15). This residue is positioned over the adenylate in the adenylate-forming conformation and rotates away to clear the phosphopantetheine tunnel in the thioester-forming conformation. This rotation appears to be controlled by the residues present in the A8 motif that interact with the aromatic side chain of the A4 residue. In PA1221, the A4 residue, Phe213, interacts with Tyr425 and Trp480, which are both located on the C-terminal subdomain (Figure 4B). This hydrophobic interaction mimics the direct hydrogen bonding interaction observed in other ANL enzymes between an A4 histidine residue and a glutamic acid from the A8 loop (17). Other important residues on the C-terminal subdomain that interact with the N-terminal domain include Lys456, which hydrogen bonds to Asp284, and Arg426, which forms a salt bridge with Glu311.
Functional analysis of intra- vs. inter-molecular loading with the PA1221 protein
Because the EntE-B protein crystallized as a domain-swapped dimer with the PCP domain from one chimeric protein molecule donated to the adenylation domain of a partner (11), we were sensitive to the possibility that the PA1221 protein could also adopt such a dimeric interaction. In the structure of holo-PA1221, the linker that joins the adenylation and PCP domains is disordered. Within one crystallographic asymmetric unit, the C-terminus of the adenylation domain (Glu510) is 17 Å from the start of the carrier domain (Leu520). Glu510 is 24 Å from the Leu520 residue of a symmetry-related molecule. Thus, while the distance of the intra-molecular interaction was shorter, the nine disordered residues from 511 to 519 could span the 17 or 24 Å to form either the intra- or the inter-molecular interaction.
To investigate whether PA1221 preferred to load the PCP domain intra-molecularly rather than inter-molecularly, we created two mutants of PA1221. The first mutated enzyme, K499L, contained a substitution in the conserved A10 catalytic lysine and was designed to be defective for the adenylation partial reaction. The second mutation, S553A, replaced the pantetheinylation site Ser553 with an alanine. We tested both enzymes in the PPi-exchange assay (Figure 2) and demonstrated that the K499L mutant was defective for the adenylation reaction while the S553A mutant retained activity for the first partial reaction.
We then asked whether these proteins were competent for loading with 3H-valine (Figure 5A). The wild-type PA1221 was able to load valine in an ATP-dependent manner. Neither the A10 mutant K499L nor the PCP mutant S553A could load valine. We then combined the two mutant proteins in equal amounts and demonstrated that 3H-valine could be loaded. This combination of the two mutant proteins showed that the active adenylation domain of the S553A mutant could react inter-molecularly to load the pantetheine of K499L mutant enzyme (Figure 5A).
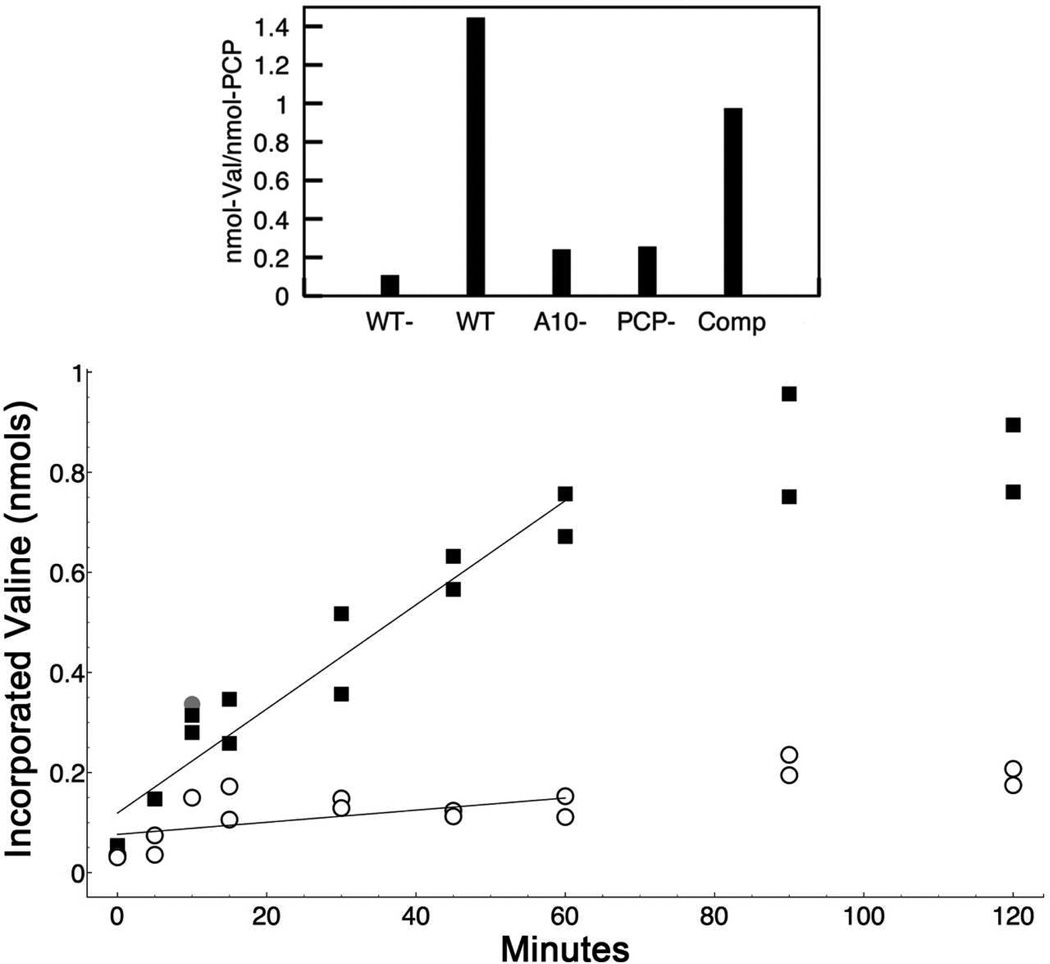
Figure 5.
Functional analysis of intra- vs. inter-molecular loading with the PA1221 protein. The top panel shows the initial loading of 3H-Valine onto PA1221 at 37°C. The individual bars represent averages of two assays with wild-type enzyme in the absence (WT-) and presence (WT) of ATP, the K499L mutant (A10-), the S553A mutant (PCP-) and a co-incubation reaction containing equal amounts of the K499L and the S553A mutant enzymes (Comp). Results are expressed as nmol valine incorporated per nmol of functional PCP as the experiment with the compensatory mutants used equal amounts of total protein, or half as much of each functional domain. In the bottom panel, the reaction was monitored on ice, slowing the reaction to observe differences between the wt and the combination of the K499L and S553A mutant enzymes. Charging with 3H-valine was monitored over two hours. Loading of the WT enzyme is represented by the filled squares. The inter-molecular reaction, forced through the reaction containing equal amounts of the two compensatory mutants, K499L and S553A, is represented by the open circle. All data points, reflecting duplicate reactions at each time point, are shown. The filled grey circle is an anomalous data point from the compensatory mutant experiment that was omitted from the linear regression plot to better represent the rate of incorporation. Results are expressed as nmol of valine incorporated into the 3 nmol of holo-PCP domain used in both experiments.
Detailed kinetic analysis is challenged by the fact that for the intra-molecular activity of the wild-type protein, we can only achieve a single turnover per protein molecule. We therefore used a time course to assess loading (Figure 5B). The protein was monitored on ice, slowing the reaction sufficiently to allow analysis. The wild-type reaction contained 12 µM holo-PA1221. A second reaction contained the K499L and S553A mutant proteins at 12 µM each (24 µM total protein concentration). This combination of compensating mutant enzymes showed activity however the rate of the reaction of the wild-type protein was nearly 10 times greater. This result showed that the intra-molecular loading occurs faster than the obligate inter-molecular loading (Figure 5B). Thus, the PA1221 adenylation domain prefers to load the fused PCP domain within a single protein chain.
The activity and the distance analysis suggests that the asymmetric unit represents a single protein molecule with a disordered linker between the adenylation and PCP domains. However, until a structure with a linker containing visible electron density is determined, we cannot strictly rule out the possibility that the current structure is an inter-molecular dimer, as observed with the EntE-B structure (11).
Interactions between the adenylation and PCP domains
The rotation of the C-terminal subdomain into the thioester-forming conformation allows for the PCP to interact with both the N- and C-terminal regions of the adenylation domain (Figure 4A). There are two regions that form these interactions. One region is mostly hydrophobic while the second is a network of hydrogen bonding interactions. The hydrophobic patch of interactions occurs between helix 2 from the PCP and Ala259-Leu268 that form a helix from the N-terminal subdomain. Along PCP helix 2, Leu554, Leu557, and Ala561, interact with Leu261, the methyl group of Thr264, Leu265, and Leu268 from the N-terminal subdomain. In the SrfA-C structure, helix 2 of the PCP is aligned parallel to a helix within the condensation domain. Thus, helix 2 adopts parallel helical stacking interactions to interact with both the adenylation and condensation domains. Also contributing to this hydrophobic interaction are Leu554 and Leu555 at the beginning of helix 2 of the PCP and Val455 in the C-terminal subdomain.
The hydrogen bonding interactions occur mostly between loop 1 of the PCP and the C-terminal subdomain. These interactions reside mostly along loop 1 of the PCP and the loop connecting β-strands 3 and 4 in the C-terminal subdomain. Arg450 and Arg452 of the C-terminal subdomain interact with the main chain carbonyls of Glu533-Gly536. The side chain of Gln457 hydrogen bonds with the main chain carbonyls of Leu535 and Gly550. Finally, the side chain of Asn552, located just before the phosphopantetheine attachment Ser553, forms a hydrogen bond with the main chain carbonyl of Lys456. An aspartic acid at this residue was proposed to be a critical interaction between the EntE adenylation domain with the EntB PCP (11).
Discussion
We present here the crystallographic structure of an NRPS adenylation domain bound to a PCP domain in a functional interaction. The structure, like the structure of the engineered EntE-B chimeric protein (11), was determined through the use of a vinylsulfonamide inhibitor (49) that mimics the adenylate intermediate and reacts covalently with the incoming thiol from the phosphopantetheine cofactor. The inhibitor was critical for trapping the PCP domain as the structure of the apo-PA1221 protein lacking the inhibitor resulted in a carrier domain that could not be observed in the electron density. The inhibitor provides a means to trap the conformationally flexible protein and enable crystallization. The structure of the enzyme inhibitor complex also provides exquisite insights into the active site. Comparison of the PA1221 structure with other multidomain NRPS enzymes illustrates the large-scale conformational changes that are necessary to deliver the carrier protein to neighboring catalytic domains (53).
The C-terminal subdomain of the PA1221 adenylation domain adopts the expected thioester-forming conformation for interactions with the PCP domain. The C-terminal subdomain of both the apo and holo forms of the protein superimpose well, except for residues Arg450-Gln457. This loop between the two longest β-strands of the C-terminal domain moves to accommodate helix 2 of the PCP domain and the phosphopantetheine in the holo structure.
Immediately following the hinge at residue Asp417 is a two-stranded antiparallel β-sheet (motif A8). In the adenylate-forming conformation, this motif is directed away from the active site while, in the thioester-forming conformation, it points into the active site. In this conformation, seven of the ten residues on this loop play important roles, including interactions with the adenylate analog (Gln419, Lys421, and Arg426), the phosphopantetheine moiety (Asn423 and Gly424), and the N-terminal subdomain (Asn423, Tyr425, Arg426, and Asp428). This loop therefore plays a major role in the architecture of the thioester-forming conformation.
Comparison of the holo-PA1221 structure to the EntE-B interface
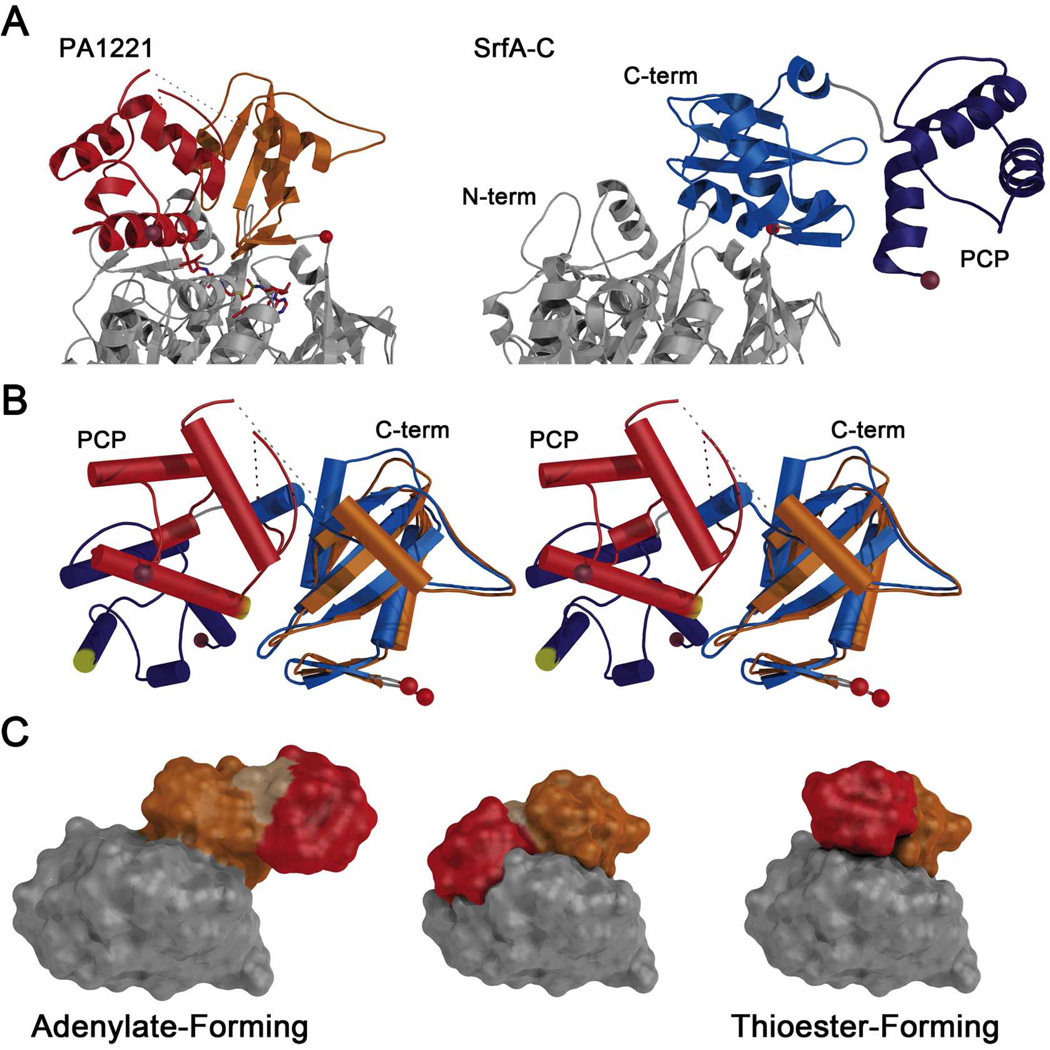
We compared the structure of PA1221 to the structure of the interaction between the EntE adenylation domain and the EntB carrier protein domain (Figure 6). These two domains were genetically fused to form a chimeric protein (11). The C-terminal subdomains of EntE and the PA1221 adenylation domain exhibit 26 % sequence identity over 105 residues. The EntB and the PA1221 PCP domains exhibit 20 % identity over 66 core residues. Nonetheless, the orientation of the carrier domains is very similar in the two adenylation-PCP complexes. There are small differences in the nature of several of the structural elements in the two structures. For example, helix 1 is longer by one turn in the PA1221 structure and a portion of loop 1 that joins helices 1 and 2 adopts a different orientation. However, it is clear that the main elements of the interface are remarkably conserved. Helix 2 of the PCP stacks against a helix from the N-terminal domain. The loop that precedes the phosphopantetheine attachment site forms multiple interactions with the C-terminal subdomain of the adenylating proteins. In both the PA1221 and EntE-B structures, the adenylation domains adopt the thioester-forming conformation to recognize their partner PCP; the relative orientation of the PCP to the catalytic domain is the same in both structures.
Figure 6.
Comparison of holo-PA1221 and EntE-B. (A) The interaction of the EntE-B protein (PDBID:3RG2) with N-terminal domain in grey, C-terminal subdomain is green, and PCP in forest green. The N- and C-termini, as well as the hinge residue, are shown with blue, brown, and red spheres respectively. The orientation is similar to the PA1221 orientation in Figure 3 (B) C-terminal subdomains and PCP domains of holo-PA1221 and EntE-B structurally aligned by least squares fitting of N-terminal domains, which are not shown for clarity. The N-terminal domain helices that interact with helix 2 of the PCP are shown in grey for both PA1221 and EntE-B and labeled N’ helix.
Adenylation Domain Alternation and NRPS Domain Choreography
The shared conformational states of EntE-B and PA1221 now allows comparison of the structures of the PA1221 protein and the SrfA-C termination module (14) to provide insights into the necessary conformational dynamics that could govern NRPS biosynthesis (Figure 7). In the SrfA-C structure, the condensation domain and adenylation N-terminal subdomain share a substantial interface that is not believed to change in different catalytic states. The adenylation C-terminal subdomain, the PCP, and the thioesterase domains likely move relative to this ordered “platform” to facilitate the necessary domain interactions (14). Unlike the more compact PA1221 structure, the SrfA-C adenylation domain adopts an extended conformation (Figure 7A) that positions the PCP away from the adenylation active site, allowing it to approach the condensation domain.
Figure 7.
Comparison of holo-PA1221 with SrfA-C adenylation and PCP domains. (A) The PA1221 and SrfA-C proteins were aligned on the basis of the N-terminal subdomain for comparison of the overall organization of C-terminal subdomains and PCPs. Where the (left) PA1221 carrier domain packs against the adenylation domains, the SrfA-C protein (right) extends the PCP to interact with the neighboring condensation domain. (B) The SrfA-C (blue) and PA1221 (orange and red) C-terminal subdomain and PCPs were aligned on the basis of the central sheet in the C-terminal subdomain. A stereo representation illustrates the differences in rotations of the PCP domain relative to the C-terminal subdomain. The pantetheine attachment site at the start of helix 2 of the PCP is highlighted in yellow. (C) Schematic model for the role of domain alternation in the movement of modular PCPs. Each panel illustrates the SrfA-C adenylation and PCP domains. The N- and C-terminal subdomains are grey and orange; the PCP is red. The adenylate-forming model is the orientation of the experimentally determined adenylation and PCP domains from 2VSQ. The middle model was created by superposing the C-terminal subdomain and PCP of 2VSQ as a rigid body onto PA1221, using the C-terminal subdomains for alignment. In this model, the PCP and N-terminal subdomains overlap, illustrating that a second component of rotation is needed to adopt the assumed thioester-forming model (right panel), which was created by superimposing both the SrfA-C C-terminal subdomain and the PCP onto the similar domain orientation in PA1221.
Members of the ANL superfamily of enzymes (15, 51, 54) use a 140° domain rotation of the C-terminal subdomain to present different residues to the same active site to carry out both steps of the reaction (15, 18). NRPS adenylation domains adopt both conformations as well. The self-standing adenylation domain DltA has been crystallized in both conformations (4, 55) and mutations at opposing faces of the C-terminal domain of the self-standing adenylation domain EntE impair only one of the two partial reactions (21). The latest multidomain structures of SrfA-C, approximating the adenylate-forming conformation, and EntE-B and PA1221, in the thioester-forming orientation, confirm the existence of both states in multi-domain NRPSs.
We asked whether the rotation of the adenylation C-terminal subdomain directly guides the PCP into the adenylation active site by determining if the C-terminal subdomain and the PCP move as a rigid body. We aligned fragments of SrfA-C and PA1221, composed of the C-terminal subdomain and the PCP, using the central sheet of the C-terminal subdomains as the basis for the alignment (Figure 7B). This shows that the relative orientations of the PCP and C-terminal subdomains differ, and that they do not behave as a single rigid body in the states captured crystallographically in the SrfA-C and PA1221 structures. Therefore, there are two components of the rotation needed to align both the C-terminal subdomain and the PCP (Figure 7C).
The structures of SrfA-C (14), PA1221, and EntE-B (11) provide exquisite insights into the molecular choreography of modular NRPSs. To catalyze the first partial reaction, the adenylation of the amino acid and displacement of pyrophosphate, the C-terminal subdomain and PCP will be positioned away from the adenylation N-terminal subdomain in a conformation similar to that of SrfA-C. Upon completing the adenylation reaction, the C-terminal domain rotates into the thioester-forming conformation as seen in EntE-B and PA1221. This domain alternation in the adenylation subdomain does not, on its own, bring the PCP domain directly into the active site. Rather, the rotation brings the PCP close to the newly formed binding interface, composed of regions from both the N- and C-terminal subdomains, and flexibility in the linker joining the adenylation and PCP domains allows the PCP to rotate into the proper position to enable the delivery of the pantetheine to the phosphopantetheine tunnel. Upon thioester formation, the loaded cofactor exits the adenylation domain and the C-terminal subdomain returns to a conformation that approximates the adenylate-forming conformation. This rotation allows delivery of the loaded PCP domain to the upstream condensation domain in the conformation adopted by SrfA-C.
From here, upon peptide bond formation, the PCP must be delivered to a downstream catalytic domain, either a condensation domain of the next module or a thioesterase domain. Structures of PCP thioesterase domain complexes derived from NMR and crystallography exist (12, 13) and further analysis of these structures in the context of the new insights into adenylation-PCP domain interactions will ultimately provide a better understanding of all of the steps in the choreography of the NRPS catalytic cycle.
Supplementary Material
Acknowledgments
This work is supported in part by NIH Grant GM-068440 (to A.M.G.). Diffraction data were collected at the Stanford Synchrotron Radiation Laboratory, a national user facility operated by Stanford University on behalf of the U.S. Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and by the National Institutes of Health National Center for Research Resources, Biomedical Technology Program, and the National Institute of General Medical Sciences.
Footnotes
Supporting Information Available
Supporting documentation is provided for general methods, materials, and experimental procedures for synthesis of the Val-AVS inhibitor. Additionally, a illustration of the amino acid binding pocket is included in Figure S1. This material is available free of charge via the internet at http://pubs.acs.org.
The genome of Pseudomonas aeruginosa strain PAO1 is predicted to encode 5530 open reading frames (56). These have been designated using PA#### as a template for each gene and protein.
References
- 1.Fischbach MA, Walsh CT. Assembly-line enzymology for polyketide and nonribosomal Peptide antibiotics: logic, machinery, and mechanisms. Chem Rev. 2006;106:3468–3496. doi: 10.1021/cr0503097. [DOI] [PubMed] [Google Scholar]
- 2.Flugel RS, Hwangbo Y, Lambalot RH, Cronan JE, Jr., Walsh CT. Holo-(acyl carrier protein) synthase and phosphopantetheinyl transfer in Escherichia coli. J Biol Chem. 2000;275:959–968. doi: 10.1074/jbc.275.2.959. [DOI] [PubMed] [Google Scholar]
- 3.Koglin A, Walsh CT. Structural insights into nonribosomal peptide enzymatic assembly lines. Nat Prod Rep. 2009;26:987–1000. doi: 10.1039/b904543k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yonus H, Neumann P, Zimmermann S, May JJ, Marahiel MA, Stubbs MT. Crystal structure of DltA. Implications for the reaction mechanism of non-ribosomal peptide synthetase adenylation domains. J Biol Chem. 2008;283:32484–32491. doi: 10.1074/jbc.M800557200. [DOI] [PubMed] [Google Scholar]
- 5.Conti E, Stachelhaus T, Marahiel MA, Brick P. Structural basis for the activation of phenylalanine in the non-ribosomal biosynthesis of gramicidin S. EMBO J. 1997;16:4174–4183. doi: 10.1093/emboj/16.14.4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drake EJ, Duckworth BP, Neres J, Aldrich CC, Gulick AM. Biochemical and structural characterization of bisubstrate inhibitors of BasE, the self-standing nonribosomal peptide synthetase adenylate-forming enzyme of acinetobactin synthesis. Biochemistry. 2010;49:9292–9305. doi: 10.1021/bi101226n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.May JJ, Kessler N, Marahiel MA, Stubbs MT. Crystal structure of DhbE, an archetype for aryl acid activating domains of modular nonribosomal peptide synthetases. Proc Natl Acad Sci U S A. 2002;99:12120–12125. doi: 10.1073/pnas.182156699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keating TA, Marshall CG, Walsh CT, Keating AE. The structure of VibH represents nonribosomal peptide synthetase condensation, cyclization and epimerization domains. Nat Struct Biol. 2002;9:522–526. doi: 10.1038/nsb810. [DOI] [PubMed] [Google Scholar]
- 9.Koglin A, Mofid MR, Lohr F, Schafer B, Rogov VV, Blum MM, Mittag T, Marahiel MA, Bernhard F, Dotsch V. Conformational switches modulate protein interactions in peptide antibiotic synthetases. Science. 2006;312:273–276. doi: 10.1126/science.1122928. [DOI] [PubMed] [Google Scholar]
- 10.Claxton HB, Akey DL, Silver MK, Admiraal SJ, Smith JL. Structure and functional analysis of RifR, the type II thioesterase from the rifamycin biosynthetic pathway. J Biol Chem. 2009;284:5021–5029. doi: 10.1074/jbc.M808604200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sundlov JA, Shi C, Wilson DJ, Aldrich CC, Gulick AM. Structural and functional investigation of intermolecular interaction between NRPS adenylation and carrier protein domains. Chem. Biol. 2012;19:188–198. doi: 10.1016/j.chembiol.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frueh DP, Arthanari H, Koglin A, Vosburg DA, Bennett AE, Walsh CT, Wagner G. Dynamic thiolation-thioesterase structure of a non-ribosomal peptide synthetase. Nature. 2008;454:903–906. doi: 10.1038/nature07162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Y, Zheng T, Bruner SD. Structural basis for phosphopantetheinyl carrier domain interactions in the terminal module of nonribosomal Peptide synthetases. Chem Biol. 2011;18:1482–1488. doi: 10.1016/j.chembiol.2011.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tanovic A, Samel SA, Essen LO, Marahiel MA. Crystal structure of the termination module of a nonribosomal peptide synthetase. Science. 2008;321:659–663. doi: 10.1126/science.1159850. [DOI] [PubMed] [Google Scholar]
- 15.Gulick AM. Conformational dynamics in the Acyl-CoA synthetases, adenylation domains of non-ribosomal peptide synthetases, and firefly luciferase. ACS Chem Biol. 2009;4:811–827. doi: 10.1021/cb900156h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reger AS, Wu R, Dunaway-Mariano D, Gulick AM. Structural characterization of a 140 degrees domain movement in the two-step reaction catalyzed by 4-chlorobenzoate:CoA ligase. Biochemistry. 2008;47:8016–8025. doi: 10.1021/bi800696y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu R, Cao J, Lu X, Reger AS, Gulick AM, Dunaway-Mariano D. Mechanism of 4-chlorobenzoate:coenzyme a ligase catalysis. Biochemistry. 2008;47:8026–8039. doi: 10.1021/bi800698m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu R, Reger AS, Lu X, Gulick AM, Dunaway-Mariano D. The mechanism of domain alternation in the acyl-adenylate forming ligase superfamily member 4-chlorobenzoate: coenzyme A ligase. Biochemistry. 2009;48:4115–4125. doi: 10.1021/bi9002327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reger AS, Carney JM, Gulick AM. Biochemical and crystallographic analysis of substrate binding and conformational changes in acetyl-CoA synthetase. Biochemistry. 2007;46:6536–6546. doi: 10.1021/bi6026506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rusnak F, Faraci WS, Walsh CT. Subcloning, expression, and purification of the enterobactin biosynthetic enzyme 2,3-dihydroxybenzoate-AMP ligase: demonstration of enzyme-bound (2,3-dihydroxybenzoyl)adenylate product. Biochemistry. 1989;28:6827–6835. doi: 10.1021/bi00443a008. [DOI] [PubMed] [Google Scholar]
- 21.Drake EJ, Nicolai DA, Gulick AM. Structure of the EntB multidomain nonribosomal peptide synthetase and functional analysis of its interaction with the EntE adenylation domain. Chem Biol. 2006;13:409–419. doi: 10.1016/j.chembiol.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 22.Olsen SK, Capili AD, Lu X, Tan DS, Lima CD. Active site remodelling accompanies thioester bond formation in the SUMO E1. Nature. 2010;463:906–912. doi: 10.1038/nature08765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu X, Zhou R, Sharma I, Li X, Kumar G, Swaminathan S, Tonge PJ, Tan DS. Stable analogues of OSB-AMP: potent inhibitors of MenE, the o-succinylbenzoate-CoA synthetase from bacterial menaquinone biosynthesis. Chembiochem. 2012;13:129–136. doi: 10.1002/cbic.201100585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Law CJ, Maloney PC, Wang DN. Ins and outs of major facilitator superfamily antiporters. Annu Rev Microbiol. 2008;62:289–305. doi: 10.1146/annurev.micro.61.080706.093329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kapust RB, Tozser J, Fox JD, Anderson DE, Cherry S, Copeland TD, Waugh DS. Tobacco etch virus protease: mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Protein Eng. 2001;14:993–1000. doi: 10.1093/protein/14.12.993. [DOI] [PubMed] [Google Scholar]
- 26.Deng J, Davies DR, Wisedchaisri G, Wu M, Hol WG, Mehlin C. An improved protocol for rapid freezing of protein samples for long-term storage. Acta Crystallogr D Biol Crystallogr. 2004;60:203–204. doi: 10.1107/s0907444903024491. [DOI] [PubMed] [Google Scholar]
- 27.Lambalot RH, Gehring AM, Flugel RS, Zuber P, LaCelle M, Marahiel MA, Reid R, Khosla C, Walsh CT. A new enzyme superfamily - the phosphopantetheinyl transferases. Chem Biol. 1996;3:923–936. doi: 10.1016/s1074-5521(96)90181-7. [DOI] [PubMed] [Google Scholar]
- 28.Mootz HD, Finking R, Marahiel MA. 4'-phosphopantetheine transfer in primary and secondary metabolism of Bacillus subtilis. J Biol Chem. 2001;276:37289–37298. doi: 10.1074/jbc.M103556200. [DOI] [PubMed] [Google Scholar]
- 29.Babbitt PC, Kenyon GL, Martin BM, Charest H, Slyvestre M, Scholten JD, Chang KH, Liang PH, Dunaway-Mariano D. Ancestry of the 4-chlorobenzoate dehalogenase: analysis of amino acid sequence identities among families of acyl:adenyl ligases, enoyl-CoA hydratases/isomerases, and acyl-CoA thioesterases. Biochemistry. 1992;31:5594–5604. doi: 10.1021/bi00139a024. [DOI] [PubMed] [Google Scholar]
- 30.Drake EJ, Cao J, Qu J, Shah MB, Straubinger RM, Gulick AM. The 1.8 A crystal structure of PA2412, an MbtH-like protein from the pyoverdine cluster of Pseudomonas aeruginosa. J Biol Chem. 2007;282:20425–20434. doi: 10.1074/jbc.M611833200. [DOI] [PubMed] [Google Scholar]
- 31.Weinreb PH, Quadri LE, Walsh CT, Zuber P. Stoichiometry and specificity of in vitro phosphopantetheinylation and aminoacylation of the valine-activating module of surfactin synthetase. Biochemistry. 1998;37:1575–1584. doi: 10.1021/bi9719859. [DOI] [PubMed] [Google Scholar]
- 32.Soltis SM, Cohen AE, Deacon A, Eriksson T, Gonzalez A, McPhillips S, Chui H, Dunten P, Hollenbeck M, Mathews I, Miller M, Moorhead P, Phizackerley RP, Smith C, Song J, van dem Bedem H, Ellis P, Kuhn P, McPhillips T, Sauter N, Sharp K, Tsyba I, Wolf G. New paradigm for macromolecular crystallography experiments at SSRL: automated crystal screening and remote data collection. Acta Crystallogr D Biol Crystallogr. 2008;64:1210–1221. doi: 10.1107/S0907444908030564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 34.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vagin AA, Steiner RA, Lebedev AA, Potterton L, McNicholas S, Long F, Murshudov GN. REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr D Biol Crystallogr. 2004;60:2184–2195. doi: 10.1107/S0907444904023510. [DOI] [PubMed] [Google Scholar]
- 37.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Battye TG, Kontogiannis L, Johnson O, Powell HR, Leslie AG. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr D Biol Crystallogr. 2011;67:271–281. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Diederichs K, Karplus PA. Improved R-factors for diffraction data analysis in macromolecular crystallography. Nat Struct Biol. 1997;4:269–275. doi: 10.1038/nsb0497-269. [DOI] [PubMed] [Google Scholar]
- 41.Stachelhaus T, Mootz HD, Marahiel MA. The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chem Biol. 1999;6:493–505. doi: 10.1016/S1074-5521(99)80082-9. [DOI] [PubMed] [Google Scholar]
- 42.Rausch C, Weber T, Kohlbacher O, Wohlleben W, Huson DH. Specificity prediction of adenylation domains in nonribosomal peptide synthetases (NRPS) using transductive support vector machines (TSVMs) Nucleic Acids Res. 2005;33:5799–5808. doi: 10.1093/nar/gki885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Horswill AR, Escalante-Semerena JC. Characterization of the propionyl-CoA synthetase (PrpE) enzyme of Salmonella enterica: residue Lys592 is required for propionyl-AMP synthesis. Biochemistry. 2002;41:2379–2387. doi: 10.1021/bi015647q. [DOI] [PubMed] [Google Scholar]
- 44.Baltz RH. Function of MbtH homologs in nonribosomal peptide biosynthesis and applications in secondary metabolite discovery. J Ind Microbiol Biotechnol. 2011;38:1747–1760. doi: 10.1007/s10295-011-1022-8. [DOI] [PubMed] [Google Scholar]
- 45.Felnagle EA, Barkei JJ, Park H, Podevels AM, McMahon MD, Drott DW, Thomas MG. MbtH-like proteins as integral components of bacterial nonribosomal peptide synthetases. Biochemistry. 2010;49:8815–8817. doi: 10.1021/bi1012854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang W, Heemstra JR, Jr., Walsh CT, Imker HJ. Activation of the pacidamycin PacL adenylation domain by MbtH-like proteins. Biochemistry. 2010;49:9946–9947. doi: 10.1021/bi101539b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lautru S, Oves-Costales D, Pernodet JL, Challis GL. MbtH-like protein-mediated cross-talk between non-ribosomal peptide antibiotic and siderophore biosynthetic pathways in Streptomyces coelicolor M145. Microbiology. 2007;153:1405–1412. doi: 10.1099/mic.0.2006/003145-0. [DOI] [PubMed] [Google Scholar]
- 48.Wolpert M, Gust B, Kammerer B, Heide L. Effects of deletions of mbtH-like genes on clorobiocin biosynthesis in Streptomyces coelicolor. Microbiology. 2007;153:1413–1423. doi: 10.1099/mic.0.2006/002998-0. [DOI] [PubMed] [Google Scholar]
- 49.Qiao C, Wilson DJ, Bennett EM, Aldrich CC. A mechanism-based aryl carrier protein/thiolation domain affinity probe. J Am Chem Soc. 2007;129:6350–6351. doi: 10.1021/ja069201e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gulick AM, Starai VJ, Horswill AR, Homick KM, Escalante-Semerena JC. The 1.75 A crystal structure of acetyl-CoA synthetase bound to adenosine-5'-propylphosphate and coenzyme A. Biochemistry. 2003;42:2866–2873. doi: 10.1021/bi0271603. [DOI] [PubMed] [Google Scholar]
- 51.Kochan G, Pilka ES, von Delft F, Oppermann U, Yue WW. Structural snapshots for the conformation-dependent catalysis by human medium-chain acyl-coenzyme A synthetase ACSM2A. J Mol Biol. 2009;388:997–1008. doi: 10.1016/j.jmb.2009.03.064. [DOI] [PubMed] [Google Scholar]
- 52.Challis GL, Ravel J, Townsend CA. Predictive, structure-based model of amino acid recognition by nonribosomal peptide synthetase adenylation domains. Chem Biol. 2000;7:211–224. doi: 10.1016/s1074-5521(00)00091-0. [DOI] [PubMed] [Google Scholar]
- 53.Strieker M, Tanovic A, Marahiel MA. Nonribosomal peptide synthetases: structures and dynamics. Curr Opin Struct Biol. 2010;20:234–240. doi: 10.1016/j.sbi.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 54.Branchini BR, Rosenberg JC, Ablamsky DM, Taylor KP, Southworth TL, Linder SJ. Sequential bioluminescence resonance energy transfer-fluorescence resonance energy transfer-based ratiometric protease assays with fusion proteins of firefly luciferase and red fluorescent protein. Anal Biochem. 2011;414:239–245. doi: 10.1016/j.ab.2011.03.031. [DOI] [PubMed] [Google Scholar]
- 55.Du L, He Y, Luo Y. Crystal structure and enantiomer selection by D-alanyl carrier protein ligase DltA from Bacillus cereus. Biochemistry. 2008;47:11473–11480. doi: 10.1021/bi801363b. [DOI] [PubMed] [Google Scholar]
- 56.Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, Hickey MJ, Brinkman FS, Hufnagle WO, Kowalik DJ, Lagrou M, Garber RL, Goltry L, Tolentino E, Westbrock-Wadman S, Yuan Y, Brody LL, Coulter SN, Folger KR, Kas A, Larbig K, Lim R, Smith K, Spencer D, Wong GK, Wu Z, Paulsen IT, Reizer J, Saier MH, Hancock RE, Lory S, Olson MV. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature. 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.