T cells typically recognize peptide fragments derived from antigenic proteins when these fragments are bound to a groove in the major histocompatibility complex (MHC)-encoded class I and II molecules. This phenomenon is known as antigen presentation (1). CD1 proteins constitute a third family of antigen-presenting molecules with a specialized function: the presentation of (glyco)lipid antigens (2). Although several reports have described T lymphocytes that are reactive to such CD1-bound (glyco)lipids, overall there is only a limited knowledge of their biology. Two papers in this issue of PNAS now describe an elegant method that allows the efficient refolding of recombinant CD1 proteins (3, 4). Moreover, they show that this method is useful for the construction of tetramers, reagents that can be used to identify and purify CD1-reactive cells with particular specificities. Reagents of this type should enhance our understanding of the function of CD1-reactive T lymphocytes greatly.
CD1 molecules are expressed predominantly by professional antigen-presenting cells (APCs) such as dendritic cells, macrophages, and B cells (2, 5). Four CD1 molecules (CD1a, CD1b, CD1c, and CD1d) are expressed as β2-microglobulin (β2m)-associated cell-surface proteins in humans (6), but mice have an ortholog of CD1d only. As outlined in Table 1, T cells reactive with CD1-bound glycolipids can be divided into three broad categories in terms of their specificity and T cell receptor usage (Table 1). First, human T cell clones reactive to lipids from mycobacterial cell walls that are presented by either CD1a, CD1b, or CD1c have been described (2, 7, 8). These clones express highly diverse T cell antigen receptors (TCRs) of the αβ type, which is typical of the majority of T lymphocytes, and there is evidence indicating that these cells are important for host defense against mycobacteria (2, 9). Most of the data in support of this hypothesis are derived from T cell clones obtained from long-term cultures, however, and such clones may not be representative of immune responses in vivo. It is highly significant, therefore, that the CD1c-mediated T cell response to a hexosyl-1-phosphoisoprenoid from mycobacteria is increased in short-term cultures of cells from tuberculosis patients when compared with normal controls (9).
Table 1.
Lipid antigens presented by CD1 molecules
| Antigen | T cell | CD1 specificity |
|---|---|---|
| Microbial glycolipids | ||
| Undefined microbial lipid | αβ T cells | CD1a |
| Glucose monomycolate | αβ T cells | CD1b |
| Lipoarabinomannan | αβ T cells | CD1b |
| Phosphatidyl inositols | αβ T cells | CD1b |
| Hexosyl-1-phosphoisoprenoids | αβ T cells | CD1c |
| Autologous glycolipids | ||
| Undefined autologous | αβ T cells | CD1a |
| Brain lipids/gangliosides/sulfatide | αβ T cells | CD1b |
| Undefined autologous | αβ and γδ T cells | CD1c |
| Undefined autologous | NK (?) T cells* | CD1d |
| Synthetic | ||
| αGalCer | NK T cells† | CD1d |
Have diverse TCRs, may or may not express NK1.1.
Mostly NK1.1-positive cells with an invariant TCR α-chain.
A second category of CD1-reactive T lymphocytes seems to recognize autologous glycolipids. Most of these also express αβ TCRs, although some can express the alternative γδ form of the antigen receptor. There is evidence suggesting that these cells are important for immune regulation, the prevention of autoimmune disease, and the surveillance for tumors (10, 11). For example, human T cell clones reactive with brain-derived glycolipids, including gangliosides, have been isolated from the blood of patients with multiple sclerosis (12, 13).
The third category of glycolipid-reactive lymphocytes is the natural killer (NK) T cells, and these cells represent a distinct lymphocyte sublineage (11, 14). NK T cells are conserved between mice and humans, and they constitute 1–2% of the total T cells in mice. They have a highly restricted diversity of their TCRs with a nearly identical or invariant TCR α chain. NK T cells are thought to be involved in immune regulation. These cells are strongly reactive to a marine-sponge-derived glycosphingolipid, α-galactosyl ceramide (αGalCer), when presented by CD1d (15). This αGalCer stimulation results in the production of copious quantities of cytokines. αGalCer, however, is not distributed widely in nature and therefore is unlikely to be the natural ligand recognized by NK T cells. In addition, these NK T cells display some level of autoreactivity for CD1d even in the absence of αGalCer (16), and there is evidence indicating that NK T cells can respond to an autologous glycolipid (17, 18). The structure of this self-ligand, however, is not known. Strikingly, evidence for T cells reactive to microbial antigens presented by CD1d is lacking, and CD1d−/− mice are not more susceptible to Mycobacterium tuberculosis infection (19). CD1d-reactive T cells, therefore, seem not to be involved in the defense against mycobacteria.
Together, these results indicate that CD1-restricted T cells have multiple functions in the immune system. To understand fully their biological roles, however, it would be very helpful to have tools to determine their number, antigen specificity, and distributions. A recent development is the generation of MHC tetramers that are loaded with a defined peptide and that can be used to detect T cells with a particular specificity (20). The use of such reagents has revolutionized the study of T lymphocyte biology, yielding a wealth of data on specific immune responses and providing insights into T cell homeostasis, trafficking, and memory. Typically, MHC class I tetramers are constructed by refolding the soluble recombinant forms of the heavy chain and β2m obtained from bacterial inclusion bodies in the presence of a single peptide, yielding homogeneously loaded class I molecules (21). By engineering the soluble proteins with a BirA enzymatic biotinylation site, tetramers can be formed with streptavidin and biotinylated forms of the peptide-loaded class I molecule. Multimerization is required because of the relatively modest affinity of the TCR for peptide–class I or peptide–class II complexes (22, 23), although dimers also have proven to be useful (24).
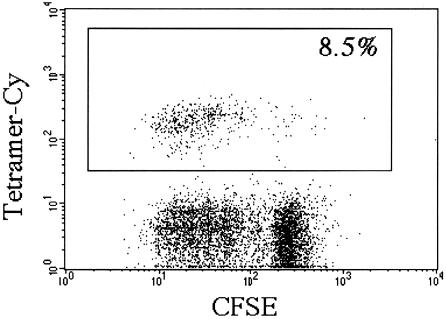
The method outlined above has not been successful generally for the construction of tetramers of peptide-loaded MHC class II molecules or CD1 loaded with glycolipid antigens. In the case of CD1, this failure may be due to the hydrophobic nature of the glycolipid antigens that bind to CD1, or because CD1 heavy chains might have special requirements for refolding with β2m. Therefore, another strategy has been used to produce CD1 tetramers successfully. Soluble mouse CD1d molecules have been produced in native form by using insect tissue cells. These CD1d molecules can be biotinylated, complexed with streptavidin, and loaded in vitro with αGalCer. The resulting αGalCer–CD1d tetramers have been used to detect and purify NK T cells that express the invariant TCR α chain (25, 26). As shown in Fig. 1, a similarly produced human CD1d tetramer loaded with αGalCer specifically stains human NK T cells proliferating in response to this antigen.
Figure 1.
Tetramer staining of human NK T cells. Human peripheral blood mononuclear cells were labeled with 5,6-carboxyfluorescein diacetate succinimidyl ester (CFSE) and cultured subsequently in RPMI medium 1640 supplemented with 10% (vol/vol) pooled human serum/100 units rIL-2/100 ng/ml αGalCer. After 4 days, cells were harvested and stained with a phycoerythrin-labeled Vβ11-specific antibody and a tricolor-labeled αGalCer–CD1 tetramer and analyzed on a FACScan (fluorescence-activated cell sorter). Both the Vβ11-specific antibody (data not shown) and the αGalCer–CD1 tetramer stained a similar population of cells, the majority of which had divided. The percentage of Vβ11/tetramer-positive cells at the start of the culture was 0.6%; after 7 days, it was 20% (data not shown).
A potential disadvantage to this strategy is that the insect cell-derived CD1 molecules might have a lipid loaded into their antigen-binding groove already, and therefore the CD1 molecules in the tetramer might not be loaded uniformly with the lipid antigen of interest. The αGalCer–CD1d tetramers work quite well despite this potential problem, possibly because of relatively high affinity of the invariant NK T cell receptor for the αGalCer–CD1d ligand. The replacement of a ligand(s) bound to native CD1 molecules, however, could be a serious issue when other CD1 molecules and antigens are studied. To circumvent this problem, two groups have now used a method they call oxidative refolding chromatography to generate refolded human CD1a, CD1b, and CD1d molecules (3, 4). In this method, refolding of the CD1 heavy chain with β2m from inclusion bodies is facilitated greatly by the addition of three refolding “helpers” (chaperones) immobilized on agarose beads. These helpers include the apical domain of the bacterial chaperone GroEL (which prevents protein aggregation), a protein disulfide isomerase that promotes correct disulfide bond formation, and a peptidyl-prolyl cis-trans isomerase. The use of immobilized chaperones offers the great advantage of easy separation of the refolded protein from the refolding matrix, yielding pure refolded protein. Moreover, the refolding reaction goes to completion in an astonishing 6 min at 4°C. Altamirano et al. (4) have carried out a rigorous biochemical characterization to demonstrate that the CD1a and CD1b molecules have been refolded to generate native molecules. Importantly, successful refolding occurs in the presence of glycosphingolipid ligands that bind to CD1a and CD1b but also in the absence of such ligands. Thus, properly folded CD1 molecules can be loaded homogeneously with a ligand of choice.
Although the method of oxidative refolding chromatography has been used previously to refold a relatively small toxin (27), the present papers are the first to describe its application to the refolding of much larger and multi-subunit proteins. The apparent success of the method suggests that it could have wide applications in the refolding of Ig-gene superfamily members and other relatively complex proteins. This hypothesis is supported by the work described in the second manuscript, in which Karadimitris et al. (3) have used oxidative refolding chromatography to produce tetramers of human CD1d refolded with αGalCer or control glycolipids. The αGalCer–human CD1d tetramers show the expected reactivity for human T cells with the invariant α chain TCR (AV24) coexpressed with a particular TCR β chain—BV11. They also show human-mouse interspecies cross reactivity, consistent with the results from several previous studies (25, 26, 28, 29). It is still early, but several conclusions can be drawn from the analysis of NK T cells with αGalCer–CD1d tetramers. First, in the mouse, NK T cells undergo a rapid cell death in vivo in response to antigenic stimulation with αGalCer, with no evidence for either antigen-driven clonal expansion or memory (26). These findings are consistent with the hypothesis that these cells function essentially as part of the innate immune system. The situation in humans seems quite different. First, the analysis with the tetramers shows up to 30-fold differences between individuals in the frequency of αGalCer-reactive T cells in the blood and liver (ref. 3; unpublished results). Second, the frequency of αGalCer-reactive NK T cells in humans is at least 10-fold lower than it is in mice. Finally, human NK T cells vigorously proliferate in response to αGalCer in the presence of growth factors (Fig. 1). Although previously existing methods permitted the detection of NK T cells in mice and humans, and as a consequence they permitted conclusions similar to some of those outlined above (30), these methods were not based directly on detecting TCR specificity, and therefore the data obtained were more difficult to interpret. CD1 tetramers now can be used to delineate further the similarities and difference between the human and mouse NK T cell populations. Furthermore, they will be used to study different ligands for CD1d molecules, to detect CD1d-reactive T cells without the invariant TCR α chain. Finally, they are certain to be used with different CD1 molecules. As these experiments are about to be carried out, one cannot help but think that we are about to cross a threshhold leading to a greatly expanded knowledge of the functions of lipid-reactive T cells. Because these cells seem to function in areas as diverse as tumor eradication and resistance to pathogens, this knowledge should prove to be very important.
Acknowledgments
We thank P. Rogers for help with the tetramer staining. This work was supported by National Institutes of Health Grant RO1 CA52511 (M.K.). F.K. is on leave from the Department of Immunohematology and Blood Transfusion, (Leiden University Medical Center, Leiden, The Netherlands) and supported by a Fulbright grant and stipends from the Dutch Organization for Scientific Research and the Child Wellbeing fund. This is manuscript 414 from the La Jolla Institute for Allergy and Immunology.
Footnotes
References
- 1.Pamer E, Cresswell P. Annu Rev Immunol. 1998;16:323–358. doi: 10.1146/annurev.immunol.16.1.323. [DOI] [PubMed] [Google Scholar]
- 2.Porcelli S A, Modlin R L. Annu Rev Immunol. 1999;17:297–329. doi: 10.1146/annurev.immunol.17.1.297. [DOI] [PubMed] [Google Scholar]
- 3.Karadimitris A, Gadola S, Altamirano M, Brown D, Woolfson A, Klenerman P, Chen J-L, Koezuka Y, Roberts I A G, Dusheiko G, et al. Proc Natl Acad Sci USA. 2001;98:3294–3298. doi: 10.1073/pnas.051604498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Altamirano M M, Woolfson A, Donda A, Shamshiev A, Briseño-Roa L, Foster N W, Veprintsev D B, De Libero G, Fersht A R, Milstein C. Proc Natl Acad Sci USA. 2001;98:3288–3293. doi: 10.1073/pnas.041596598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burdin N, Kronenberg M. Curr Opin Immunol. 1999;11:326–331. doi: 10.1016/s0952-7915(99)80052-1. [DOI] [PubMed] [Google Scholar]
- 6.Calabi F, Jarvis J M, Martin L, Milstein C. Eur J Immunol. 1989;19:285–292. doi: 10.1002/eji.1830190211. [DOI] [PubMed] [Google Scholar]
- 7.Beckman E M, Porcelli S A, Morita C T, Behar S M, Furlong S T, Brenner M B. Nature (London) 1994;372:691–694. doi: 10.1038/372691a0. [DOI] [PubMed] [Google Scholar]
- 8.Matsuda J L, Kronenberg M. Curr Opin Immunol. 2001;13:19–25. doi: 10.1016/s0952-7915(00)00176-x. [DOI] [PubMed] [Google Scholar]
- 9.Moody D B, Ulrichs T, Muhlecker W, Young D C, Gurcha S S, Grant E, Rosat J P, Brenner M B, Costello C E, Besra G S, Porcelli S A. Nature (London) 2000;404:884–888. doi: 10.1038/35009119. [DOI] [PubMed] [Google Scholar]
- 10.Godfrey D I, Hammond K J, Poulton L D, Smyth M J, Baxter A G. Immunol Today. 2000;21:573–583. doi: 10.1016/s0167-5699(00)01735-7. [DOI] [PubMed] [Google Scholar]
- 11.Bendelac A, Rivera M N, Park S H, Roark J H. Annu Rev Immunol. 1997;15:535–562. doi: 10.1146/annurev.immunol.15.1.535. [DOI] [PubMed] [Google Scholar]
- 12.Shamshiev A, Donda A, Carena I, Mori L, Kappos L, De Libero G. Eur J Immunol. 1999;29:1667–1675. doi: 10.1002/(SICI)1521-4141(199905)29:05<1667::AID-IMMU1667>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 13.Shamshiev A, Donda A, Prigozy T I, Mori L, Chigorno V, Benedict C A, Kappos L, Sonnino S, Kronenberg M, De Libero G. Immunity. 2000;13:255–264. doi: 10.1016/s1074-7613(00)00025-x. [DOI] [PubMed] [Google Scholar]
- 14.Elewaut D, Kronenberg M. Semin Immunol. 2000;12:561–568. doi: 10.1006/smim.2000.0275. [DOI] [PubMed] [Google Scholar]
- 15.Kawano T, Cui J, Koezuka Y, Toura I, Kaneko Y, Motoki K, Ueno H, Nakagawa R, Sato H, Kondo E, et al. Science. 1997;278:1626–1629. doi: 10.1126/science.278.5343.1626. [DOI] [PubMed] [Google Scholar]
- 16.Bendelac A, Lantz O, Quimby M E, Yewdell J W, Bennink J R, Brutkiewicz R R. Science. 1995;268:863–865. doi: 10.1126/science.7538697. [DOI] [PubMed] [Google Scholar]
- 17.Cardell S, Tangri S, Chan S, Kronenberg M, Benoist C, Mathis D. J Exp Med. 1995;182:993–1004. doi: 10.1084/jem.182.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burdin N, Brossay N, Koezuka Y, Smiley S T, Grusby M J, Gui M, Taniguchi M, Hayakawa K, Kronenberg M. J Immunol. 1998;161:3271–3281. [PubMed] [Google Scholar]
- 19.Behar S M, Dascher C C, Grusby M J, Wang C R, Brenner M B. J Exp Med. 1999;189:1973–1980. doi: 10.1084/jem.189.12.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McMichael A J, O'Callaghan C A. J Exp Med. 1998;187:1367–1371. doi: 10.1084/jem.187.9.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Altman J D, Moss P A H, Goulder P J R, Barouch D H, McHeyzer-Williams M G, Bell J I, McMichael A J, Davis M M. Science. 1996;274:94–96. [PubMed] [Google Scholar]
- 22.Davis M M, Boniface J J, Reich Z, Lyons D, Hampl J, Arden B, Chien Y. Annu Rev Immunol. 1998;16:523–544. doi: 10.1146/annurev.immunol.16.1.523. [DOI] [PubMed] [Google Scholar]
- 23.Garcia K C, Teyton L, Wilson I A. Annu Rev Immunol. 1999;17:369–397. doi: 10.1146/annurev.immunol.17.1.369. [DOI] [PubMed] [Google Scholar]
- 24.Schneck J P. Immunol Invest. 2000;29:163–169. doi: 10.3109/08820130009062300. [DOI] [PubMed] [Google Scholar]
- 25.Benlagha K, Weiss A, Beavis A, Teyton L, Bendelac A. J Exp Med. 2000;191:1895–1903. doi: 10.1084/jem.191.11.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsuda J L, Naidenko O V, Gapin L, Nakayama T, Taniguchi M, Wang C R, Koezuka Y, Kronenberg M. J Exp Med. 2000;192:741–754. doi: 10.1084/jem.192.5.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Altamirano M M, Garcia C, Possani L D, Fersht A R. Nat Biotechnol. 1999;17:187–191. doi: 10.1038/6192. [DOI] [PubMed] [Google Scholar]
- 28.Spada F M, Koezuka Y, Porcelli S A. J Exp Med. 1998;188:1529–1534. doi: 10.1084/jem.188.8.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brossay L, Chioda M, Burdin N, Koezuka Y, Casorati G, Dellabona P, Kronenberg M. J Exp Med. 1998;188:1521–1528. doi: 10.1084/jem.188.8.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eberl G, MacDonald HR. Immunity. 1998;9:345–353. doi: 10.1016/s1074-7613(00)80617-2. [DOI] [PubMed] [Google Scholar]