Abstract
Class II transfer RNAs (tRNAs), including tRNALeu and tRNASer, have an additional stem and loop structure, the long variable arm (V-arm). Here, we describe Class II tRNAs with a unique anticodon corresponding to neither leucine nor serine. Because these tRNAs are specifically conserved among the nematodes, we have called them ‘nematode-specific V-arm-containing tRNAs’ (nev-tRNAs). The expression of nev-tRNA genes in Caenorhabditis elegans was confirmed experimentally. A comparative sequence analysis suggested that the nev-tRNAs derived phylogenetically from tRNALeu. In vitro aminoacylation assays showed that nev-tRNAGly and nev-tRNAIle are only charged with leucine, which is inconsistent with their anticodons. Furthermore, the deletion and mutation of crucial determinants for leucylation in nev-tRNA led to a marked loss of activity. An in vitro translation analysis showed that nev-tRNAGly decodes GGG as leucine instead of the universal glycine code, indicating that nev-tRNAs can be incorporated into ribosomes and participate in protein biosynthesis. Our findings provide the first example of unexpected tRNAs that do not consistently obey the general translation rules for higher eukaryotes.
INTRODUCTION
Transfer RNAs (tRNAs) are small RNA molecules, of ∼70–85 nt, that play a critical role in protein biosynthesis as the links between the codons of messenger RNAs (mRNAs) and the amino acids in the growing polypeptide chains. tRNAs can be classified into two groups based on structural differences in their variable regions: Class I tRNAs have a short variable region of 4–5 nt, whereas Class II tRNAs (e.g. tRNALeu, tRNASer and bacterial tRNATyr) have a long variable arm (V-arm) structure containing ≥10 nt (1). The attachment of the correct amino acid to the 3′ end of each tRNA is catalyzed by an aminoacyl-tRNA synthetase (aaRS), which collectively comprise a protein family, with very high accuracy (2). The recognition elements for Class I tRNAs are mainly found in their acceptor stems and anticodon domains (3). In contrast, the recognition elements for Class II tRNAs occur in the acceptor stem, D-stem and long V-arm (4–7), with the anticodons generally less important for their aminoacylation (8). The long V-arms of tRNALeu and tRNASer are conserved in all organisms and organelles, except animal mitochondria, whereas the long V-arms of archaeal and eukaryotic tRNATyr were probably lost soon after the separation of the domain Bacteria (9). Hence, the evolutionary divergence of Class II tRNAs occurred after the differentiation of the three domains of life.
To understand the diversity of tRNA genes in living organisms, we analyzed the genome sequences of Archaea and primitive eukaryotes, and found that various types of tRNAs are encoded in a unique manner. These include: (i) intron-containing tRNAs, which contain a maximum of three introns located at various nucleotide positions, and are found predominantly in crenarchaeal species (10–13); (ii) tri-split tRNAs, comprising three transcripts of Caldivirga maquilingensis (14); and (iii) permuted tRNAs, in which the 5′ and 3′ halves are encoded in a permuted orientation. These permuted tRNAs have been found in Cyanidioschyzon merolae and other unicellular red/green algae (15,16), and more recently, in the crenarchaeon Thermofilum pendens by Todd M. Lowe's group (17). The tRNA genes in higher eukaryotes were thought to contain a single intron at most, located in the canonical position (37/38), 1 nt downstream from the anticodon (13,18), but a comprehensive analysis of eukaryotic tRNA genes has revealed two putative tRNAGly and tRNAIle genes (with Class I anticodons) in the Caenorhabditis elegans genome that harbor 15–16 nt V-arm structures (18). These tRNA-like sequences have features found only in typical Class II tRNAs, and their evolutionary origins and cellular functions remain unclear.
To assess the evolutionary origins, diversity and functions of these putative tRNA genes, we comprehensively reanalyzed the eukaryotic tRNA genes and identified over 100 Class II tRNA candidates with anticodons corresponding to neither leucine nor serine, but possessing the long V-arm. These sequences are specifically conserved among the nematodes and contain recognition elements for aaRSs similar to those of tRNALeu. Surprisingly, we found that these tRNAs in C. elegans can only be aminoacylated with leucine, but recognize codons corresponding to other amino acids. Our findings identify a new type of tRNA that translates nucleotides in a manner that transgresses the genetic code, emphasizing the importance of considering the evolutionary processes underlying the general translation rules.
MATERIALS AND METHODS
Phylogenetic analysis of the tRNA genes of three nematode species
All mature tRNA sequences, including nematode-specific V-arm-containing tRNAs (nev-tRNAs) in C. elegans and Caenorhabditis brenneri, were collected from the Genomic tRNA database (19), and the Caenorhabditis japonica and Pristionchus pacificus tRNA sequences were predicted using tRNAscan-SE (20). Any redundant tRNA sequences (multicopy tRNAs with identical sequences) were discarded. A neighbor-joining tree of non-redundant mature tRNA sequences was constructed using ClustalX (21) and visualized with iTOL (22).
Preparation of total RNA
The N2 nematode strain and Escherichia coli strain OP50 used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the NIH National Center for Research Resources. Total RNA was isolated from mixed-stage C. elegans, including eggs, larvae 1–4 and adults, using TRIzol Reagent (Invitrogen, Carlsbad, CA, USA), according to the manufacturer's instructions. For acid–urea polyacrylamide gel electrophoresis (PAGE)/northern blot analysis, the isolated RNA was incubated with 0.2 M Tris–HCl (pH 9.5) at 37°C for 2 h to deacylate the charged tRNAs.
Northern blot analysis
Total RNA (10 μg per lane) was separated on a denaturing 6% polyacrylamide gel containing 8 M urea and transferred onto Hybond-N+ membranes (GE Healthcare, Buckinghamshire, UK) by electroblotting. The membranes were hybridized with specific oligonucleotide probes (Supplementary Table S1A), labeled with a Biotin 3′-End DNA Labeling Kit (Pierce Biotechnology, Rockford, IL, USA) in ULTRAhyb-Oligo Hybridization Buffer (Ambion, Austiin, TX, USA) at room temperature. The hybridized membranes were treated with 5× Wash Buffer (Ambion) at room temperature. The non-isotopic blots were detected with the BrightStar BioDetect Kit (Ambion) using the Enhanced ChemiFluorescence (ECF) substrate (GE Healthcare). The images were visualized with a Molecular Imager FX Pro (Bio-Rad Laboratories, Hercules, CA, USA).
RT–PCR analysis and nucleotide sequencing
Reverse transcription (RT)–PCR was performed with the enzymes, ReverTra Dash and KOD FX (Toyobo Biochemicals, Osaka, Japan). All amplification reactions, except those amplifying nev-tRNAGly (CCC), consisted of 30 cycles of denaturation at 98°C for 10 s, annealing at 55°C for 3 s and extension at 74°C for 6 s. PCR of nev-tRNAGly was performed with stepdown amplification, consisting of 15 cycles of denaturation at 98°C for 10 s, annealing at 74°C (5 cycles), 72°C (5 cycles), 70°C (5 cycles) and 60°C (15 cycles) for 6 s, and extension at 74°C for 6 s. The PCR products were separated by electrophoresis on 3% NuSieve 3 : 1 agarose gels (Cambrex Bio Science, Rockland, ME), which were stained with ethidium bromide. The bands were visualized with a Molecular Imager FX Pro (Bio-Rad Laboratories). The primers used in this analysis are summarized in Supplementary Table S1B. The RT–PCR products were further purified with an Illustra GFX PCR DNA and Gel Band Purification Kit (GE Healthcare), and subcloned into the pCR–Blunt II–TOPO vector (Invitrogen). The nucleotide sequences of the inserted DNAs were determined on an ABI PRISM 3100 DNA Sequencer (Applied Biosystems, Foster City, CA, USA).
Preparation of recombinant aminoacyl-tRNA synthetases from C. elegans
Caenorhabditis elegans total RNA was used as the template for RT–PCR of four genes encoding aminoacyl-tRNA synthetases (aaRSs): glycyl-tRNA synthetase (GlyRS, GenBank accession no. NP_871640.1), isoleucyl-tRNA synthetase (IleRS, GenBank accession no. NP_501914.1), leucyl-tRNA synthetase (LeuRS, GenBank accession no. NP_497837.1) and seryl-tRNA synthetase (SerRS, GenBank accession no. NP_501804.1). RT–PCR was performed using the ReverTra Dash Kit and KOD FX (Toyobo Biochemicals), and the amplified products were cloned into the pET–23 b expression vector (Novagen, Madison, WI, USA). Each resulting vector encoded a full-length aaRS with a six-histidine tag at its C-terminal end. The nucleotide sequences of the inserted DNA fragments were determined on an ABI PRISM 3100 DNA Sequencer (Applied Biosystems) and confirmed to be identical to those in the database. The primers and restriction enzymes used for cloning are summarized in Supplementary Table S1C.
The resulting vectors were used to transform E. coli strain BL21(DE3)pLysS (IleRS and LeuRS) or HMS174(DE3)pLysS (GlyRS and SerRS). The transformants, growing logarithmically at 37°C in Luria–Bertani medium containing 50 μg/ml ampicillin and 35 μg/ml chloramphenicol, were supplemented with 0.1–0.4 mM isopropyl-β-d-thiogalactopyranoside. After 14–16 h of further growth at 16°C, the cells were pelleted by centrifugation (5000 g for 5 min at 4°C) and sonicated (3 min) in 1× phosphate buffered saline (PBS) with 10 mM imidazole. The extracts were centrifuged at 12 000 g for 10 min at 4°C to remove any debris, and the recombinant proteins were purified with a Proteus IMAC Protein Purification Kit (Pro-Chem, Littleton, MA, USA). The eluted protein solutions were pooled and gel filtered to remove any salt on a HiTrap column (GE Healthcare) with buffer A (50 mM Tris–HCl [pH 8.0], 1 mM EDTA, 0.02% Tween 20, 7 mM 2-mercaptoethanol and 10% glycerol).
In vitro aminoacylation assay
The tRNAs synthesized in vitro with T7 RNA polymerase were incubated at room temperature with purified recombinant aaRSs from C. elegans for 0, 2.5, 5, 10, 20, 40 and 60 min in a 25 μl reaction mixture containing 50 mM Hepes (pH 7.2), 50 mM KCl, 4 mM ATP, 15 mM MgCl2, 3 mM dithiothreitol, 15 mM amino acid (glycine, isoleucine, leucine, or serine), 3 μg of tRNA and 1 μg of aaRS (except IleRS, for which we used 5 μg to accommodate its lower aminoacylation efficiency). The reaction was stopped by the addition of an equal volume of acid–urea stop buffer (23). The charged tRNAs were separated on acid–urea 6.5% polyacrylamide gels, which were stained with SYBR Green II (Lonza, Rockland, ME, USA). The bands were detected with a Molecular Imager FX Pro (Bio-Rad Laboratories).
Acid–urea PAGE/northern blot analysis
Alkali-treated total RNA (100 μg; see ‘Preparation of total RNA’ section) was used in the in vitro aminoacylation assay, with purified recombinant LeuRS and GlyRS titrated from 0.01 to 1 μg. The assay products were separated on acid–urea 6.5% polyacrylamide gels, followed by northern blot hybridization to detect the individual tRNAs.
Construction of expression vectors for cell-free protein synthesis
For in vitro transcription/translation, we prepared two expression vectors encoding PF1549 protein (RNA 3′-terminal-phosphate cyclase of Pyrococcus furiosus (24), GenBank accession no. NP_579278) and firefly luciferase (GenBank accession no. ACF93193.1). Pyrococcus furiosus genomic DNA and luciferase ICE T7 Control DNA (Promega, Madison, WI, USA) were used as the templates for the PCR of PF1549 (24) and luciferase, respectively. PCR was performed with KOD FX (Toyobo Biochemicals), and the amplified products were cloned into the pF25A ICE T7 Flexi Vector (Promega) containing the T7 promoter. The resulting expression vectors encoded full-length PF1549 or luciferase, with six-histidine tags at their C-terminal ends. The nucleotide sequences of the inserted DNA fragments were determined on an ABI PRISM 3100 DNA Sequencer (Applied Biosystems) and confirmed to be identical to those in the database. The primers and restriction enzymes used for cloning are summarized in Supplementary Table S1C.
In vitro translation analysis
Each 46 μl cell-free protein expression reaction mixture, with coupled transcription (driven by the T7 promoter) and translation, contained 40 μl of cell extract from the Spodoptera frugiperda Sf21 cell line (Promega), 4 μg of the T7 expression vector encoding PF1549 protein or firefly luciferase and 4 μg of in vitro-transcribed tRNA [tRNALeu (AAG) or nev-tRNAGly (CCC)]. After incubation at 29°C for 4 h, the cell lysates were sonicated (1 min) with a Polytron sonicator (Kinematica, Bohemia, NY, USA). The samples containing the in vitro-translated PF1549 proteins were incubated at 80°C for 15 min to destroy any endogenous insect proteins. The homogenates were centrifuged at 13 000 g for 10 min at 4°C to remove any debris, and the His-tagged recombinant proteins were purified with a Talon Magnetic Beads Buffer Kit (Clontech Laboratories, Mountain View, CA, USA).
Nano liquid chromatography–tandem mass spectrometry (NanoLC–MS/MS) analysis
The purified proteins were extracted with 100 mM Tris–HCl (pH 9.0) containing 12 mM sodium deoxycholate (SDC) and 12 mM sodium lauroyl sarcosinate (SLS), then reduced with 10 mM dithiothreitol at room temperature for 30 min and alkylated with 55 mM iodoacetamide in the dark at room temperature for 30 min. The samples were diluted 5-fold with 50 mM ammonium bicarbonate and digested with MS-grade lysyl endoprotease, followed by overnight trypsin digestion. The SDC and SLS were removed from the sample solutions (25,26) by the addition of ethyl acetate, acidification with 0.5% trifluoroacetic acid, shaking for 1 min, and centrifugation at 15 700 g for 2 min. The aqueous phase was desalted with StageTips on a C18 Empore Disk (27).
An LTQ-Orbitrap (Thermo Fisher Scientific, Bremen, Germany) equipped with a nanoLC interface (Nikkyo Technos, Tokyo, Japan) and a Dionex Ultimate3000 pump with a FLM-3000 flow manager (Dionex, Germering, Germany) and an HTC-PAL autosampler (CTC Analytics, Zwingen, Switzerland) were used for nanoLC–MS/MS measurements. Reprosil C18 materials (3 μm; Dr Maisch, Ammerbuch, Germany) were packed into a self-pulled needle (150 mm length, 375 μm O.D. × 100 μm I.D.) with a nitrogen-pressurized column loader cell (Nikkyo Technos, Tokyo, Japan) to prepare an analytical column needle with a ‘stone-arch’ frit (28). The flow rate was 500 nl/min. The mobile phases consisted of (A) 0.5% acetic acid and (B) 0.5% acetic acid and 80% acetonitrile. A three-step linear gradient was used: 5–10% B in 5 min, 10–40% B in 60 min, 40–100% B in 5 min and 100% B for 10 min. A spray voltage of 2400 V was applied. The MS scan range was m/z 300–1500 in the Orbitrap analyzer and the top 10 precursor ions were selected for subsequent MS/MS scans in the ion trap.
Data analysis for protein identification
The raw data files were analyzed with Mass Navigator v1.2 (Mitsui Knowledge Industry, Tokyo, Japan) to create peak lists based on the recorded fragmentation spectra. The peptides were identified with Mascot v2.2 (Matrix Science, London, UK) in a comparison with an in-house protein database containing PF1549, luciferase and keratin proteins and trypsin sequences. A precursor mass tolerance of 3 ppm and a fragment ion mass tolerance of 0.8 Da were used with strict trypsin specificity, allowing up to two missed cleavages (29). Carbamidomethylation of cysteine was set as a fixed modification, and methionine oxidation was allowed as a variable modification. Peptides were rejected if the Mascot score was below the 99% confidence limit based on the ‘identity’ score of each peptide. False-positive rates were estimated by comparison with a randomized decoy database created with the Mascot Perl program and supplied by Matrix Science.
RESULTS AND DISCUSSION
Nematode-specific Class II tRNAs
Although a set of unique Class II tRNA-like sequences with Class I tRNA anticodons has been identified in the genome of C. elegans, the evolutionary and functional implications of these tRNA genes remain unclear. To assess whether these tRNAs are conserved in the eukaryotes and to determine their possible functions, we collected the sequences of 49 872 tRNAs from 44 eukaryote species registered in the Genomic tRNA Database (19). We also obtained the genomic sequences of C. japonica and P. pacificus from the University of California Santa Cruz (UCSC) Genome Bioinformatics database (30). Application of tRNAscan-SE (20) with default parameters predicted an additional 2138 tRNA genes. We screened these 52 010 tRNA sequences for candidate Class II tRNAs that contain the conserved basic tRNA features (1,18), including a 7-bp acceptor stem, a 3–4-bp D-stem, a 5-bp anticodon stem, a 5-bp T-stem, a 7-nt T-loop, a 7-nt anticodon loop and a Class I tRNA anticodon specifying an amino acid other than leucine or serine. We identified a total of 115 candidate tRNAs from six nematode species: C. brenneri, C. briggsae, C. elegans, C. japonica, C. remanei and P. pacificus (summarized in Supplementary Data set S1). All the candidate tRNAs were observed in nematodes, with none in any other eukaryotic genome. Therefore, we designated these unusual Class II tRNAs ‘nematode-specific V-arm-containing tRNAs’ (nev-tRNAs).
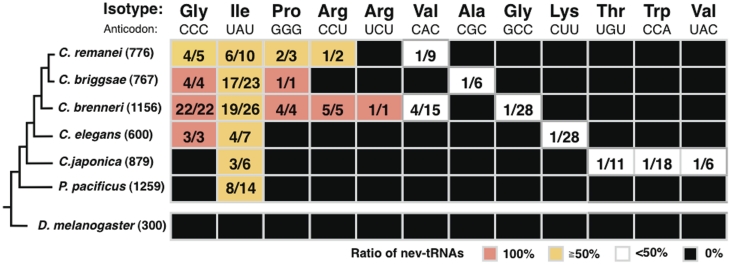
When we assessed the distribution of the predicted nev-tRNA genes among the six nematode species (Figure 1), we found that three species, C. remanei, C. briggsae and C. brenneri, encoded an average of 31 nev-tRNAs each, with C. brenneri encoding 56 nev-tRNAs, corresponding to seven different anticodons. This indicates the rapid development of many nev-tRNAs. In contrast, C. elegans, C. japonica and P. pacificus each encoded only 6–8 nev-tRNA genes. These three species diverged earlier within the nematode lineage than the former three species, suggesting that the number of nev-tRNAs and their anticodon variations have increased during the evolution of the nematode taxon, and are especially prominent in C. brenneri. These findings also indicate that nev-tRNAGly (CCC) and nev-tRNAIle (UAU) are the two major types of nev-tRNAs and are widely conserved among the nematode genomes. Intriguingly, nev-tRNAGlys (CCC), found only in C. briggsae, C. brenneri and C. elegans, are the only tRNAs that correspond to the codon GGG (shown as red boxes in Figure 1). Although all nematode genomes encode a Class I tRNAIle (UAU) containing a single intron, most tRNAs corresponding to the UAU anticodon are nev-tRNAIles (UAU) (shown as yellow boxes in Figure 1). Furthermore, three nev-tRNAs, Pro(GGG), Arg(CCU) and Arg(UCU), in C. brenneri constitute the entire population of tRNAs that possess these cognate anticodons. Therefore, the abundance and evolutionary conservation of nev-tRNAs suggest that they play important roles in cellular processes.
Figure 1.
Evolutionary conservation of the nev-tRNAs in nematodes. For each nematode species, the ratios of nev-tRNA genes to tRNA genes corresponding to each specific anticodon are shown (red, 100%; yellow, >50% but <100%; white, <50%). The total number of tRNA genes in each species is indicated in brackets.
Sequence characteristics of nev-tRNAs and their evolutionary background
We selected three nematode species to examine the origins and evolution of the nev-tRNA genes: C. brenneri (which has more nev-tRNAs than any other species), C. elegans (used for experimental verification, as described below) and P. pacificus (representing the earliest branch within the nematode lineage). Using 151 tRNA exons, including the exons of 13 nev-tRNAs from these three nematode species, we performed a phylogenetic analysis of the non-redundant tRNA sequences (Supplementary Figure S1). We found that four types of nev-tRNAs, Ile(UAU), Gly(CCC), Arg(CCU) and Lys(CUU), clustered into a Class II tRNA clade, especially close to the tRNALeu (UAA) family. Moreover, the Class I tRNAs with these same anticodons clustered separately from these nev-tRNAs on the phylogenetic tree of nematode tRNAs. Because nev-tRNAIle (UAU) and nev-tRNAGly (CCC) are widely conserved in the nematode linage (Figure 1), these results suggest that at least the major nev-tRNAs probably evolved from Class II tRNAs, rather than by gene duplication of synonymous tRNAs. In contrast, six types of nev-tRNAs in C. brenneri clustered within the Class I tRNA family, suggesting that nev-tRNAs emerged on two independent occasions during tRNA evolution. We also found that a single C. brennneri tRNALeu (UAA) occurred within the nev-tRNA clade. Its sequence was almost identical (91.6%) to that of C. brennneri nev-tRNAArg (CCU), but was only 55.9% similar to the synonymous tRNALeu (UAA), indicating that it originated from the nev-tRNA gene (shown as an asterisk in Supplementary Figure S1).
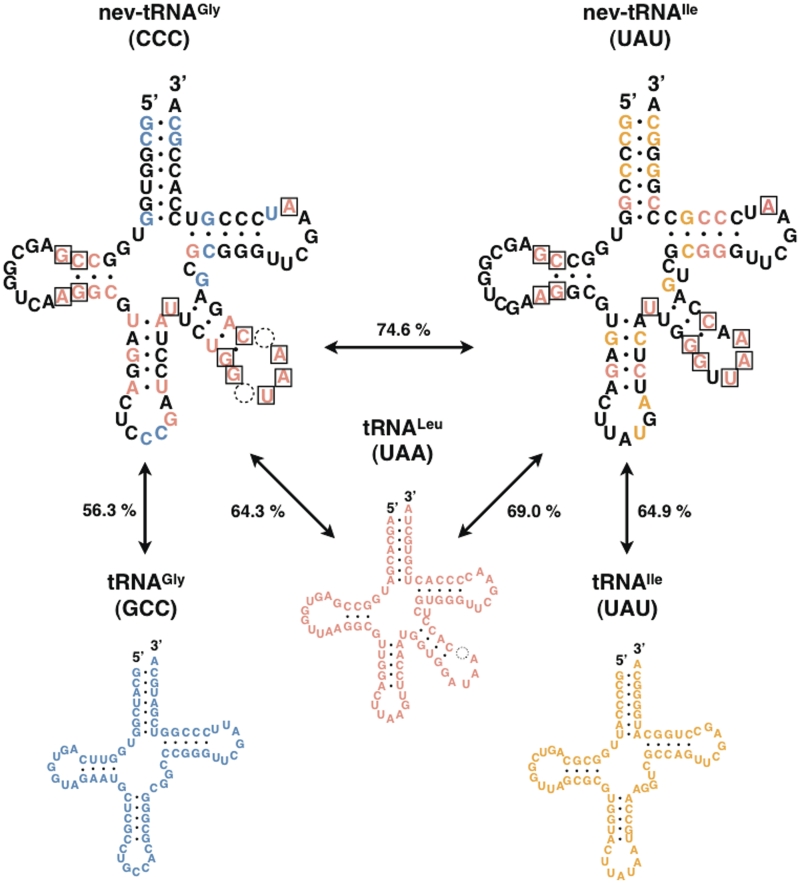
To analyze the characteristics of the nev-tRNA sequences relative to their nucleotides at each position, we compared C. elegans nev-tRNAGly (CCC) and nev-tRNAIle (UAU) with their synonymous Class I tRNAGly (GCC), tRNAIle (UAU) and tRNALeu (UAA) sequences (Figure 2). Each of these nev-tRNAs forms a cloverleaf secondary structure, has endogenous A-box (5′-TGGCNNAGTGG-3′) and B-box (5′-GGTCGANNCC-3′) promoters (31), and contains a V-arm of 15–16 nucleotides that forms a stem–loop structure. We found that nev-tRNAGly (CCC) showed greater sequence similarity to tRNALeu (UAA) (64.3%) than to Class I tRNAGly (GCC) (56.3%). The results for the nev-tRNAIle (UAU) sequence are similar. The nucleotides conserved among these three Class II tRNAs were concentrated in the D-arm and V-arm regions, which are both recognition elements for LeuRS (4,5) (shown as boxed nucleotides in Figure 2). These results indicate that the nev-tRNAs may be recognized by LeuRS and charged with leucine.
Figure 2.
Comparison of nucleotide sequences and secondary structures of two nev-tRNAs and their related tRNAs in C. elegans. Nucleotides conserved between nev-tRNAGly (CCC) and the synonymous Class I tRNAGly (GCC) are shown in blue; those between nev-tRNAIle (UAU) and the synonymous Class I tRNAIle (UAU) are shown in yellow; and those between the two nev-tRNAs and the Class II tRNALeu (UAA) are shown in red. Nucleotides conserved among the three V-arm-containing tRNAs are boxed. Sequence similarities are also shown for each pair of tRNAs.
nev-tRNAs are weakly expressed in C. elegans
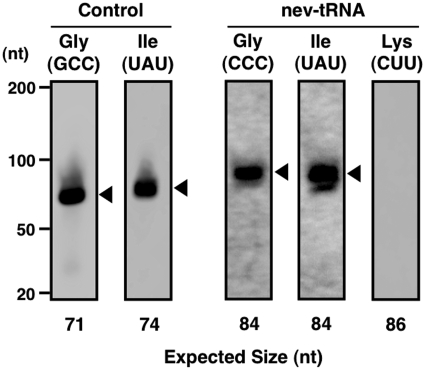
Although our bioinformatics approach indicated that the nev-tRNAs probably originated from tRNALeu and are expanding among the nematode genomes, it was unclear whether they are expressed in cells. Therefore, we assayed the expression of the nev-tRNA genes by northern blot analysis of the total RNA extracted from mixed stages of C. elegans, including eggs, L1–4 larvae and adults, using specific oligonucleotide probes (Supplementary Table S1A). We detected the expression of nev-tRNAGly (CCC) and nev-tRNAIle (UAU) at their predicted lengths, but no expression of nev-tRNALys (CUU) (Figure 3). RT–PCR analysis confirmed the expression of all three nev-tRNAs at their expected lengths (Supplementary Figure S2), with the nucleotide sequences of the amplified products identical to those of the nev-tRNAs. Moreover, none of these nev-tRNAs contained a V-arm spliced tRNA band of ∼70 bp, indicating that the V-arm domain is part of the mature tRNA, not an intronic sequence that is spliced out during tRNA processing. These results demonstrate that although expressed, the levels of nev-tRNAs are lower than those of general tRNAs.
Figure 3.
Expression of nev-tRNAs in C. elegans. The results of the northern blot analysis of the three nev-tRNA genes found in C. elegans, nev-tRNAGly (CCC), nev-tRNAIle (UAU) and nev-tRNALys (CUU), and their synonymous Class I tRNA genes, tRNAGly (GCC) and tRNAIle (UAU) (control), are shown. Black triangles indicate that the band sizes were approximately consistent with the predicted lengths of the nev-tRNAs based on the bioinformatics approach used in this study. nev-tRNALys was not detected, even when the amount of total RNA was increased to 30 μg per lane. Contrast levels were adjusted for the nev-tRNA images.
nev-tRNAGly and nev-tRNAIle are specifically aminoacylated with leucine
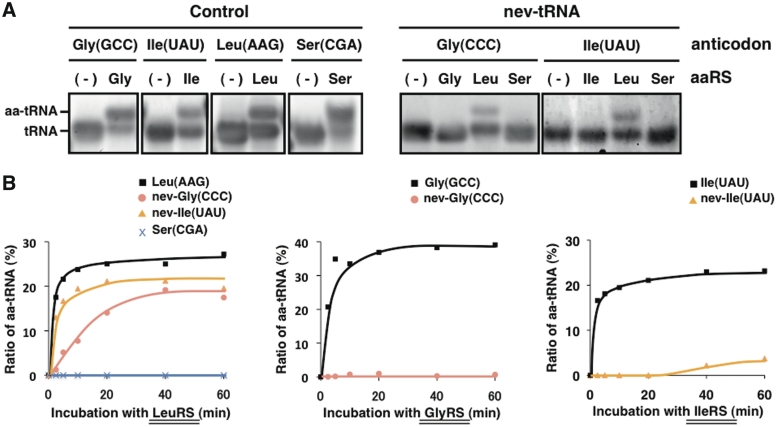
The V-arm domain and the other determinant sites for LeuRS that are conserved in the nev-tRNAs suggest that they are leucylated, whereas their anticodons correspond to other amino acids. To understand the functions of the cellular nev-tRNAs, we determined the amino acids with which they are charged. We attempted to aminoacylate C. elegans nev-tRNAGly (CCC) and nev-tRNAIle (UAU) in vitro with four purified recombinant C. elegans aaRSs: GlyRS, IleRS, SerRS and LeuRS (Supplementary Figure S3). Each control tRNA was successfully aminoacylated with the aaRS and the amino acid corresponding to its anticodon (Figure 4A, left). Under the same conditions, nev-tRNAGly (CCC) and nev-tRNAIle (UAU) were not aminoacylated with the amino acids corresponding to their respective anticodons, nor with serine, but with leucine in the presence of LeuRS (Figure 4A, right). When we assessed the aminoacylation efficiency and specificity of each nev-tRNA at different time points, we found that both nev-tRNAs had a lower leucylation efficiency than tRNALeu (AAG) and that, conversely, almost no tRNA was aminoacylated by GlyRS or IleRS (Figure 4B).
Figure 4.
Aminoacylation assay of nev-tRNAs. (A) In vitro-transcribed tRNAs [four types of control tRNAs, nev-tRNAGly (CCC) and nev-tRNAIle (UAU)] were aminoacylated using four recombinant C. elegans aaRSs: GlyRS, IleRS, SerRS and LeuRS (Supplementary Figure S3). The aminoacylated tRNAs (aa-tRNAs) were separated on an acid–urea polyacrylamide gel and stained with SYBR Green II. (B) Measurement of the aminoacylation efficiencies of the nev-tRNAs at 0, 2.5, 5, 10, 20, 40 and 60 min. (Left panel) Leucylation of tRNALeu (AAG), tRNASer (CGA) and two nev-tRNAs by LeuRS. (Middle panel) Glycylation of tRNAGly (GCC) and nev-tRNAGly (CCC) by GlyRS. (Right panel) Isoleucylation of tRNAIle (UAU) and nev-tRNAIle (UAU) by IleRS.
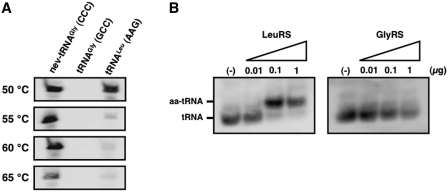
Because many examples have so far been reported of tRNA modifications that affect the recognition by aaRSs (32,33), we conducted further aminoacylation assays using total RNA fractions, followed by the identification of the mature nev-tRNAs by northern blot analysis, to validate the leucylation of native nev-tRNAs in vivo. Because the expression levels of native nev-tRNAs are low, we designed an oligonucleotide complementary to full-length nev-tRNAGly as the northern hybridization probe (Supplementary Table S1A) to improve the signal strength. We used the most stringent hybridization conditions to detect nev-tRNAGly with the full-length probe and found that almost all the cross-hybridization signals could be reduced by increasing the hybridization and wash temperatures above 55°C (Figure 5A). Accordingly, we performed the hybridization reactions for in vivo nev-tRNAs at 60°C and successfully demonstrated that native nev-tRNAGly is aminoacylated with leucine but not glycine (Figure 5B). These results suggest that nev-tRNAs are only ever charged with leucine both in vitro and in vivo.
Figure 5.
In vivo-modified nev-tRNAGly (CCC) is specifically charged with leucine. (A) Validation of the hybridization stringency conditions for detecting nev-tRNAGly (CCC) using the full-length probe. In vitro-transcribed nev-tRNAGly (CCC), tRNAGly (GCC) and tRNALeu (AAG) were detected using an oligonucleotide fully complementary to nev-tRNAGly. Northern hybridization and washing treatments were performed at 50, 55, 60 and 65°C. (B) The aminoacylation of native nev-tRNAGly (CCC) determined by acid–urea PAGE/northern hybridization. An aminoacylation assay was performed at room temperature for 60 min with purified recombinant LeuRS or GlyRS titrated from 0.01 to 1 μg.
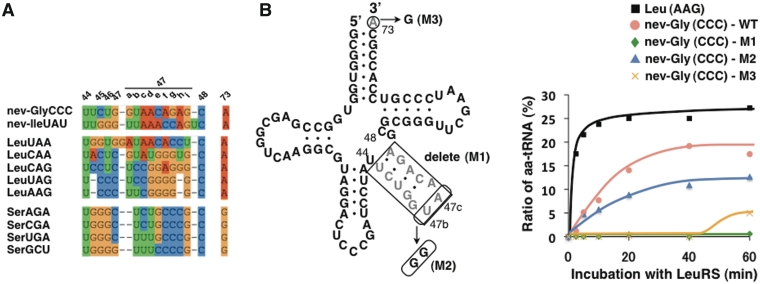
To determine the nucleotides in the nev-tRNAs recognized by LeuRS, we tested the ability of LeuRS to aminoacylate nev-tRNAGly (CCC) variants with mutations at several recognition sites. The V-arm domains are major determinants of the recognition of Class II tRNAs (4–7), and X-ray structural analysis of the Pyrococcus horikoshii LeuRS-tRNALeu complex showed that the protruding C-terminal domain of LeuRS specifically recognizes the bases at the tip of the V-arm (34). Caenorhabditis elegans encodes five synonymous tRNALeu genes, and the V-arm sequences of its nev-tRNAs are most similar to that of tRNALeu (UAA), with the bases at the tip of the nev-tRNA V-arms (U47b and A47c) conserved in both tRNALeus (UAA/CAA) (Figure 6A). Complete deletion of the nev-tRNAGly (CCC) V-arm domain led to a severe loss of leucylation (Figure 6B, M1), and replacement of the 2 nt at the tip of the V-arm reduced the leucylation efficiency ∼2-fold (Figure 6B, M2). This suggests that the V-arm domains of the nev-tRNAs, primarily the 2 nt at their tips, are necessary for leucylation by C. elegans LeuRS, as are the determinants in tRNALeu. In contrast, the discriminator base A73 of tRNALeu is highly conserved throughout all living organisms and is important for leucylation in E. coli (35), yeast (36) and humans (4). A73 is also conserved in all tRNALeu genes in the six nematode species examined, as well as in all 115 nev-tRNAs, whereas the Class II tRNASer genes contain a conserved G73 (Figure 6A). We found that the leucylation efficiency of nev-tRNAGly (CCC) was strongly influenced by base substitutions at A73 (Figure 6B, M3). The strong evolutionary conservation of A73 and the V-arm suggest that nev-tRNAs have been under evolutionary pressure to be specifically recognized by LeuRS.
Figure 6.
Mutagenesis of nev-tRNAGly (CCC) sites recognized by LeuRS. (A) Sequence alignments of the V-arm domains of Class II tRNAs, including nev-tRNAs, in C. elegans. The nucleotide positions are numbered as described previously (1). (B) (Left panel) The nev-tRNAGly (CCC) variants used in this study. (Right panel) Leucylation efficiencies of tRNALeu (AAG), wild-type nev-tRNAGly (WT) and the three nev-tRNAGly variants (M1–3). Each experiment was performed in duplicate to confirm the reproducibility of the enzymatic reactions.
nev-tRNAGly is incorporated into eukaryotic ribosomes during translation and decodes the GGG codon as leucine in vitro
Finally, to demonstrate whether nev-tRNAs are incorporated into ribosomes and function in translation, we used these nev-tRNAs in an in vitro translation assay and determined the sequences of the resultant peptides with MS. In these experiments, we attempted to synthesize two proteins, PF1549 (24) and firefly luciferase, because (i) their mRNAs contain 6–8 GGG codons, corresponding to glycine; and (ii) PF1549, an RNA 3′-terminal-phosphate cyclase in the hyperthermophilic archaeon Pyrococcus furiosus, can be purified easily by heat treatment. The in vitro translation reactions were performed in an insect cell-free protein expression system using in vitro-transcribed tRNAs, either tRNALeu (AAG) (control) or nev-tRNAGly (CCC). After partial purification with a magnetic separator, SDS/PAGE analysis showed that the PF1549 protein and luciferase were successfully synthesized at their expected sizes (Supplementary Figure S4). To examine whether specific leucines, derived from the nev-tRNA decoding of GGG codons, were contained in the proteins synthesized in the presence of nev-tRNAGly, we analyzed the purified proteins with nanoLC–MS/MS (summarized in Supplementary Data set S2). Surprisingly, proteins containing leucine residues translated from GGG codons were only observed in the presence of nev-tRNAGly, in both PF1549 (Table 1) and luciferase (Supplementary Table S2 and Supplementary Figure S5). Although leucine is indistinguishable from isoleucine on MS, these amino acid residues were almost certainly leucines because in vitro aminoacylation assays showed that nev-tRNAGly is not charged with isoleucine. Peptides containing glycine translated from GGG codons were also observed in the presence of nev-tRNAGly, suggesting that the GGG codon is also decoded by endogenous insect tRNAGly (UCC) via G–U wobble base pairing (37). These results strongly suggest that in insect cells, (i) nev-tRNAGly is aminoacylated with leucine; (ii) leucylated nev-tRNAGly is incorporated into ribosomes during translation; and (iii) leucylated nev-tRNAGly recognizes GGG codons, resulting in the incorporation of leucine, which does not correspond to their anticodons.
Table 1.
In vitro translation analysis using nev-tRNAGly
| Sequence (PF1549) | Mascot score (n = 4) |
|
|---|---|---|
| tRNALeu (control) | nev-tRNAGly | |
| GGGIVAGYVKPWIER | 45.0 | 37.4 |
| GGGIVAGYVKPWIERK | 57.6 | 44.5 |
| GKPAEEVGREAAQELLSQVK | 69.1 | 63.3 |
| KGKPAEEVGREAAQELLSQVK | 74.9 | 73.6 |
| KLANAKVEGAEVGSR | 75.1 | 83.4 |
| LANAKVEGAEVGSR | 49.2 | 61.8 |
| GGLIVAGYVKPWIER | – | 46.1 |
| GGLIVAGYVKPWIERK | – | 20.1 |
| KLKPAEEVGR | – | 42.8 |
| KLKPAEEVGREAAQELLSQVK | – | 20.3 |
| LKPAEEVGR | – | 30.5 |
| LKPAEEVGREAAQELLSQVK | – | 42.1 |
| VEGAEVLSR | – | 63.0 |
The PF1549 protein was translated in vitro in the presence of tRNALeu (AAG) or nev-tRNAGly (CCC). Peptide sequences with the amino acid residues (underlined) arising from the decoding of the GGG codon are listed. Mascot scores were determined from four independent experiments.
nev-tRNAs tend to correspond to rare codons
In this work, we have identified a new group of Class II tRNA genes, which probably originated and diversified from the tRNALeu gene. These unique tRNAs are found only in several nematode species, and their numbers and the types of anticodons involved have tended to increase during nematode evolution. On the basis of the codon usage of C. elegans, we found that the major nev-tRNAs correspond to the three rarest codons in the organism, Gly(GGG), Pro(CCC) and Arg(AGG), which comprise 0.44, 0.44 and 0.38%, respectively, of the codons in C. elegans (Supplementary Figure S6). The usage of codon Ile(ATA) is lowest among the three synonymous isoleucine codons (0.94%), but is higher than that of the other rare codons. This may be attributable to the coexistence of both Class I tRNAIle (UAU) and nev-tRNAIle (UAU) in the nematode genomes (Figure 1). Because isoleucine and leucine are chiral amino acids with highly similar chemical properties, the penetration of nev-tRNA against the Ile(ATA) codon would have the least effect on the proteome. This hypothesis is also supported by the findings that other major nev-tRNAs tend to selectively penetrate the rare codons and are weakly expressed in the cell.
Recent studies have indicated that tRNAs have alternative functions associated with the stress responses (38,39). In Gram-negative bacteria, such as E. coli, uncharged tRNAs block protein synthesis by penetrating the A site of the ribosome in response to amino acid starvation (38). In mammalian cells, the response to oxidative stress involves the selective mismethionylation of non-methionyl tRNAs, with the resulting Met-misacylated tRNAs used in translation (39). These data suggest that nev-tRNAs have non-canonical roles under certain conditions, indicating the need for additional research into their expression patterns under nutritional, heat and oxidative stress, and at different developmental stages. In conclusion, we have shown here that: (i) nev-tRNAs are present only in the nematode lineage and tend to correspond to rare codons; (ii) nev-tRNAs are weakly expressed in C. elegans under laboratory breeding conditions; (iii) nev-tRNAs are only charged with leucine in vitro; and (iv) nev-tRNAs are incorporated into eukaryotic ribosomes during translation and decode rare codons to leucine in vitro, contrary to the genetic code. These results may provide further insight into the evolution of alternative genetic codes in eukaryotes. Because the precise biological roles of nev-tRNAs in nematode cells remain unclear, future studies are required to identify their functions in vivo.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1 and 2, Supplementary Figures 1–6, Supplementary Data sets 1 and 2.
FUNDING
Funding for open access charge: Grant-in-Aid for Scientific Research (B) #22370066 from the Ministry of Education, Culture, Sports, Science and Technology of Japan; research fund of the Institute for Fermentation, Osaka, Japan; research funds of the Yamagata Prefectural Government and Tsuruoka City, Japan.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr Takashi Yokogawa (Gifu University, Japan) for his technical comments on the aminoacylation assay and Dr Yoshiki Ikeda (Keio University, Japan) for his useful comments on the article. They also thank all the members of the RNA Group at the Institute for Advanced Biosciences of Keio University, Japan, for their insightful discussions.
REFERENCES
- 1.Sprinzl M, Horn C, Brown M, Ioudovitch A, Steinberg S. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1998;26:148–153. doi: 10.1093/nar/26.1.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Freist W. Mechanisms of aminoacyl-tRNA synthetases: a critical consideration of recent results. Biochemistry. 1989;28:6787–6795. doi: 10.1021/bi00443a001. [DOI] [PubMed] [Google Scholar]
- 3.McClain WH. Rules that govern tRNA identity in protein synthesis. J. Mol. Biol. 1993;234:257–280. doi: 10.1006/jmbi.1993.1582. [DOI] [PubMed] [Google Scholar]
- 4.Breitschopf K, Achsel T, Busch K, Gross HJ. Identity elements of human tRNA(Leu): structural requirements for converting human tRNA(Ser) into a leucine acceptor in vitro. Nucleic Acids Res. 1995;23:3633–3637. doi: 10.1093/nar/23.18.3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Soma A, Uchiyama K, Sakamoto T, Maeda M, Himeno H. Unique recognition style of tRNA(Leu) by Haloferax volcanii leucyl-tRNA synthetase. J. Mol. Biol. 1999;293:1029–1038. doi: 10.1006/jmbi.1999.3219. [DOI] [PubMed] [Google Scholar]
- 6.Biou V, Yaremchuk A, Tukalo M, Cusack S. The 2.9 A crystal structure of T. thermophilus seryl-tRNA synthetase complexed with tRNA(Ser) Science. 1994;263:1404–1410. doi: 10.1126/science.8128220. [DOI] [PubMed] [Google Scholar]
- 7.Yaremchuk A, Kriklivyi I, Tukalo M, Cusack S. Class I tyrosyl-tRNA synthetase has a class II mode of cognate tRNA recognition. EMBO J. 2002;21:3829–3840. doi: 10.1093/emboj/cdf373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saks ME, Sampson JR, Abelson JN. The transfer RNA identity problem: a search for rules. Science. 1994;263:191–197. doi: 10.1126/science.7506844. [DOI] [PubMed] [Google Scholar]
- 9.Achsel T, Gross HJ. Identity determinants of human tRNA(Ser): sequence elements necessary for serylation and maturation of a tRNA with a long extra arm. EMBO J. 1993;12:3333–3338. doi: 10.1002/j.1460-2075.1993.tb06003.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sugahara J, Yachie N, Sekine Y, Soma A, Matsui M, Tomita M, Kanai A. SPLITS: a new program for predicting split and intron-containing tRNA genes at the genome level. In Silico Biol. 2006;6:411–418. [PubMed] [Google Scholar]
- 11.Sugahara J, Yachie N, Arakawa K, Tomita M. In silico screening of archaeal tRNA-encoding genes having multiple introns with bulge-helix-bulge splicing motifs. RNA. 2007;13:671–681. doi: 10.1261/rna.309507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sugahara J, Kikuta K, Fujishima K, Yachie N, Tomita M, Kanai A. Comprehensive analysis of archaeal tRNA genes reveals rapid increase of tRNA introns in the order thermoproteales. Mol. Biol. Evol. 2008;25:2709–2716. doi: 10.1093/molbev/msn216. [DOI] [PubMed] [Google Scholar]
- 13.Sugahara J, Fujishima K, Morita K, Tomita M, Kanai A. Disrupted tRNA gene diversity and possible evolutionary scenarios. J. Mol. Evol. 2009;69:497–504. doi: 10.1007/s00239-009-9294-6. [DOI] [PubMed] [Google Scholar]
- 14.Fujishima K, Sugahara J, Kikuta K, Hirano R, Sato A, Tomita M, Kanai A. Tri-split tRNA is a transfer RNA made from 3 transcripts that provides insight into the evolution of fragmented tRNAs in archaea. Proc. Natl Acad. Sci. USA. 2009;106:2683–2687. doi: 10.1073/pnas.0808246106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Soma A, Onodera A, Sugahara J, Kanai A, Yachie N, Tomita M, Kawamura F, Sekine Y. Permuted tRNA genes expressed via a circular RNA intermediate in Cyanidioschyzon merolae. Science. 2007;318:450–453. doi: 10.1126/science.1145718. [DOI] [PubMed] [Google Scholar]
- 16.Maruyama S, Sugahara J, Kanai A, Nozaki H. Permuted tRNA genes in the nuclear and nucleomorph genomes of photosynthetic eukaryotes. Mol. Biol. Evol. 2009;27:1070–1076. doi: 10.1093/molbev/msp313. [DOI] [PubMed] [Google Scholar]
- 17.Chan PP, Cozen AE, Lowe TM. Discovery of permuted and recently split transfer RNAs in Archaea. Genome Biol. 2011;12:R38. doi: 10.1186/gb-2011-12-4-r38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marck C, Grosjean H. tRNomics: analysis of tRNA genes from 50 genomes of Eukarya, Archaea, and Bacteria reveals anticodon-sparing strategies and domain-specific features. RNA. 2002;8:1189–1232. doi: 10.1017/s1355838202022021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chan PP, Lowe TM. GtRNAdb: a database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res. 2009;37:D93–D97. doi: 10.1093/nar/gkn787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 22.Letunic I, Bork P. Interactive tree of life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics. 2007;23:127–128. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]
- 23.Kohrer C, Rajbhandary UL. The many applications of acid urea polyacrylamide gel electrophoresis to studies of tRNAs and aminoacyl-tRNA synthetases. Methods. 2008;44:129–138. doi: 10.1016/j.ymeth.2007.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sato A, Soga T, Igarashi K, Takesue K, Tomita M, Kanai A. GTP-dependent RNA 3′-terminal phosphate cyclase from the hyperthermophilic archaeon Pyrococcus furiosus. Genes Cells. 2011;16:1190–1199. doi: 10.1111/j.1365-2443.2011.01561.x. [DOI] [PubMed] [Google Scholar]
- 25.Masuda T, Tomita M, Ishihama Y. Phase transfer surfactant-aided trypsin digestion for membrane proteome analysis. J Proteome Res. 2008;7:731–740. doi: 10.1021/pr700658q. [DOI] [PubMed] [Google Scholar]
- 26.Masuda T, Saito N, Tomita M, Ishihama Y. Unbiased quantitation of Escherichia coli membrane proteome using phase transfer surfactants. Mol. Cell Proteomics. 2009;8:2770–2777. doi: 10.1074/mcp.M900240-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rappsilber J, Ishihama Y, Mann M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 2003;75:663–670. doi: 10.1021/ac026117i. [DOI] [PubMed] [Google Scholar]
- 28.Ishihama Y, Rappsilber J, Andersen JS, Mann M. Microcolumns with self-assembled particle frits for proteomics. J. Chromatogr. A. 2002;979:233–239. doi: 10.1016/s0021-9673(02)01402-4. [DOI] [PubMed] [Google Scholar]
- 29.Olsen JV, Ong S-E, Mann M. Trypsin cleaves exclusively C-terminal to arginine and lysine residues. Mol. Cell Proteomics. 2004;3:608–614. doi: 10.1074/mcp.T400003-MCP200. [DOI] [PubMed] [Google Scholar]
- 30.Fujita PA, Rhead B, Zweig AS, Hinrichs AS, Karolchik D, Cline MS, Goldman M, Barber GP, Clawson H, Coelho A, et al. The UCSC Genome Browser database: update 2011. Nucleic Acids Res. 2010;39:D876–D882. doi: 10.1093/nar/gkq963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Galli G, Hofstetter H, Birnstiel M. Two conserved sequence blocks within eukaryotic tRNA genes are major promoter elements. Nature. 1981;294:626–631. doi: 10.1038/294626a0. [DOI] [PubMed] [Google Scholar]
- 32.Stern L, Schulman LH. The role of the minor base N4-acetylcytidine in the function of the Escherichia coli noninitiator methionine transfer RNA. J. Biol. Chem. 1978;253:6132–6139. [PubMed] [Google Scholar]
- 33.Szweykowska-Kulinska Z, Senger B, Keith G, Fasiolo F, Grosjean H. Intron-dependent formation of pseudouridines in the anticodon of Saccharomyces cerevisiae minor tRNA(Ile) EMBO J. 1994;13:4636–4644. doi: 10.1002/j.1460-2075.1994.tb06786.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fukunaga R, Yokoyama S. Aminoacylation complex structures of leucyl-tRNA synthetase and tRNALeu reveal two modes of discriminator-base recognition. Nat. Struct. Mol. Biol. 2005;12:915–922. doi: 10.1038/nsmb985. [DOI] [PubMed] [Google Scholar]
- 35.Asahara H, Himeno H, Tamura K, Hasegawa T, Watanabe K, Shimizu M. Recognition nucleotides of Escherichia coli tRNA(Leu) and its elements facilitating discrimination from tRNASer and tRNA(Tyr) J. Mol. Biol. 1993;231:219–229. doi: 10.1006/jmbi.1993.1277. [DOI] [PubMed] [Google Scholar]
- 36.Soma A, Kumagai R, Nishikawa K, Himeno H. The anticodon loop is a major identity determinant of Saccharomyces cerevisiae tRNA(Leu) J. Mol. Biol. 1996;263:707–714. doi: 10.1006/jmbi.1996.0610. [DOI] [PubMed] [Google Scholar]
- 37.Varani G, McClain WH. The G x U wobble base pair. A fundamental building block of RNA structure crucial to RNA function in diverse biological systems. EMBO Rep. 2000;1:18–23. doi: 10.1093/embo-reports/kvd001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wendrich TM, Blaha G, Wilson DN, Marahiel MA, Nierhaus KH. Dissection of the mechanism for the stringent factor RelA. Mol. Cell. 2002;10:779–788. doi: 10.1016/s1097-2765(02)00656-1. [DOI] [PubMed] [Google Scholar]
- 39.Netzer N, Goodenbour JM, David A, Dittmar KA, Jones RB, Schneider JR, Boone D, Eves EM, Rosner MR, Gibbs JS, et al. Innate immune and chemically triggered oxidative stress modifies translational fidelity. Nature. 2009;462:522–526. doi: 10.1038/nature08576. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.