Abstract
Yeast snR30 is an essential box H/ACA small nucleolar RNA (snoRNA) that promotes 18S rRNA processing through forming transient base-pairing interactions with the newly synthesized 35S pre-rRNA. By using a novel tandem RNA affinity selection approach, followed by coimmunoprecipitation and in vivo cross-linking experiments, we demonstrate that in addition to the four H/ACA core proteins, Cbf5p, Nhp2p, Nop10p and Gar1p, a fraction of snR30 specifically associates with the Utp23p and Kri1p nucleolar proteins. Depletion of Utp23p and Kri1p has no effect on the accumulation and recruitment of snR30 to the nascent pre-ribosomes. However, in the absence of Utp23p, the majority of snR30 accumulates in large pre-ribosomal particles. The retained snR30 is not base-paired with the 35S pre-rRNA, indicating that its aberrant tethering to nascent preribosomes is likely mediated by pre-ribosomal protein(s). Thus, Utp23p may promote conformational changes of the pre-ribosome, essential for snR30 release. Neither Utp23p nor Kri1p is required for recruitment of snR30 to the nascent pre-ribosome. On the contrary, depletion of snR30 prevents proper incorporation of both Utp23p and Kri1p into the 90S pre-ribosome containing the 35S pre-rRNA, indicating that snR30 plays a central role in the assembly of functionally active small subunit processome.
INTRODUCTION
In eukaryotic cells, the nucleolar biogenesis of cytoplasmic ribosomes is a highly complex process that includes the synthesis, modification and processing of precursor rRNAs (pre-rRNAs), the assembly of rRNAs with ribosomal proteins and the transport of the newly assembled ribosomal subunits to the cytoplasm (for recent reviews, see refs 1–3). In the yeast Saccharomyces cerevisiae, the 18S, 5.8S and 25S rRNAs are synthesized within the 35S pre-rRNA that also contains long external and internal transcribed spacer sequences (Figure 1A). Maturation of the 18S, 5.8S and 25S rRNAs and their assembly with ribosomal proteins into functional ribosomes require the coordinated activity of more than 200 protein and small nucleolar ribonucleoprotein (snoRNP) factors which transiently interact with the nucleolar preribosomal particles, but are missing from the mature cytoplasmic ribosomes.
Figure 1.
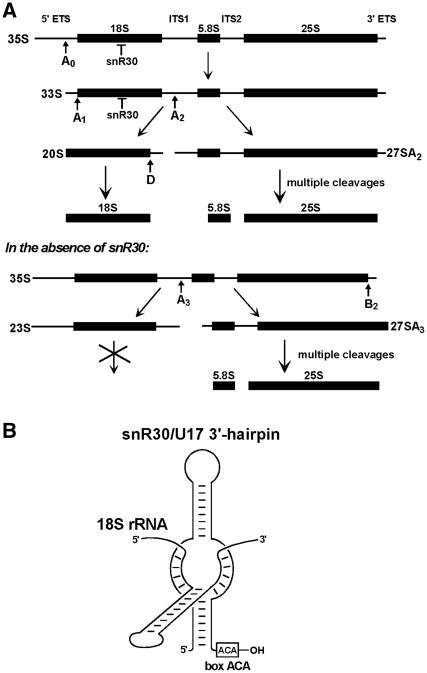
The snR30/U17 snoRNA functions in the nucleolytic processing of 18S rRNA. (A) Processing of yeast 35S pre-rRNA in the presence and absence of snR30. The 18S, 5.8S and 25S rRNAs and the external (5′ETS and 3′ETS) and internal (ITS1 and ITS2) transcribed spacer sequences, the major processing sites (arrows A0, A1, A2, A3 and D) and the resulting pre-rRNA intermediates (35S, 33S, 23S, 20S, 27SA2 and 27SA3) are indicated. Interaction of 18S rRNA with snR30 is needed for cleavages at the A0, A1 and A2 sites. (B) Schematic presentation of the evolutionarily conserved interaction of snR30 and 18S rRNA (23).
During transcription, the nascent pre-rRNA undergoes extensive covalent modification that requires formation of transient base-pairing interactions with 43 box C/D 2′-O-ribose methylation and 28 box H/ACA pseudouridylation guide snoRNPs (4). The newly synthesized 35S pre-RNA also associates with many non-ribosomal processing factors and some ribosomal proteins to form the early 90S preribosomal particle (2,5,6). Since the 90S particle predominantly lacks large (60S) subunit processing factors, it is considered as a molecular machine (processome) supporting the assembly of the small (40S) ribosomal subunit. Within the nascent 90S complex, early cleavages of the 35S pre-rRNA at the A0, A1 and A2 processing sites separate the biogenesis of the small and large ribosomal subunits (Figure 1A).
Efficient nucleolytic processing of the 35S pre-rRNA at the A0, A1 and A2 sites leading to 18S rRNA production also requires the U3, U14, snR10 and snR30 snoRNPs (7–11). These, so-called processing snoRNPs apparently lack nucleolytic activity; they likely promote pre-rRNA processing through safeguarding its correct folding via formation of dynamic base-pairing interactions with 35S sequences. The snR30 snoRNP belongs to the family of box H/ACA snoRNPs which function predominantly in rRNA pseudouridylation (12–15). Eukaryotic H/ACA snoRNAs are minimally composed of two hairpins connected and followed by short single-stranded hinge and tail sequences carrying the conserved H (ANANNA) and ACA box motifs (16–18). All H/ACA RNAs are associated with four conserved core proteins, the pseudouridine synthase Cbf5p, Nhp2p, Nop10p and Gar1p (16,17,19,20). The 5′- and/or 3′-terminal hairpins of H/ACA pseudouridylation guide snoRNAs contain large internal target recognition loops which form bipartite helices with rRNA sequences flanking the selected uridine (21). The 3′-terminal hairpin of snR30 also contains an evolutionarily conserved rRNA recognition loop (22). However, in contrast to the canonical pseudouridylation guide sequences which occupy the distal (upper) part of the internal loop, the 18S rRNA recognition elements of snR30 are located in the proximal (lower) part of the loop (Figure 1B) (22,23). Disruption of the base-pairing capacity of snR30 with 18S rRNA abolishes pre-rRNA processing at the A0, A1 and A2 sites and induces 35S processing at the A3 site to produce the 23S and 27SA3 pre-rRNAs (Figure 1A). While the 27SA3 pre-rRNA is efficiently processed into 5.8S and 25S rRNAs, the 23S pre-RNA encompassing 18S rRNA is rapidly degraded.
Previous studies proposed several snR30-associated proteins (20,24–26). However, these putative snR30 proteins failed to reproducibly copurify with snR30 and/or turned out to be non-specific for snR30, since they also interacted with many other snoRNAs, including both C/D and H/ACA classes. In this study, to identify proteins which specifically interact with snR30, we purified the yeast snR30 snoRNP by using a newly developed tandem RNA affinity purification technique. We demonstrated that besides the four H/ACA core proteins, Cbf5p, Gar1p, Nhp2p and Nop10p, a fraction of snR30 specifically associates with Utp23p and Kri1p, two conserved nucleolar proteins required for the biogenesis of the 40S ribosomal subunit (27,28). We found that expression of snR30 is essential for pre-ribosomal recruitment of both Kri1p and Utp23p, whereas accumulation of Utp23p is required for the release of snR30 from nascent 90S pre-ribosomal particles.
MATERIALS AND METHODS
General procedures and strains
Unless stated otherwise, standard laboratory procedures were used to manipulate DNA, RNA and oligodeoxynucleotides. Likewise, yeast S. cerevisiae cells were cultured according to standard protocols (29). Strains used in this study are described in Supplemental Table S1. HeLa cells were grown in Dulbecco's modified Eagle medium supplemented with antibiotics, 1% sodium pyruvate and 10% fetal calf serum (Invitrogen). Transfection of plasmids was performed by using the JET PRIME transfection reagent according to the instructions of the manufacturer (Ozyme).
Plasmid construction
Construction of the pR30 expression plasmid has been described (22). To generate pTOB-MS2-R30, 5′-terminally phosphorylated synthetic oligodeoxynucleotides were annealed and inserted into the KpnI site of pR30. To obtain the pMAL-MS2 bacterial expression vector, the open reading frame of the MS2 coat protein gene was PCR-amplified and fused in frame to the C-terminus of the maltose-binding protein gene in the pMAL-c2X vector (New England BioLabs). The pCMV-Flag-UTP23 expression plasmid was purchased from Origene. All constructs were verified by sequence analysis.
RNA affinity selection
Tobramycin-coupled sepharose beads were prepared by rotating NHS-activated Sepharose 4 Fast Flow beads (GE Healthcare) in 90% dimethylformamide, 75 mM Hepes-KOH, pH 7.9, 5 µM tobramycin overnight at 4°C. The beads were collected and incubated in blocking buffer (0.2 M Tris-HCl, pH 8.4) for 2 h at RT and in 20 mM Tris–HCl, pH 8.1, 300 mM KCl, 1 mM CaCl2, 1 mM MgCl2, 0.2 mM dithiothreitol (DTT), 0.5 mg/ml bovine serum albumin, 0.01% Nonidet P-40 overnight at 4°C. Recombinant MBP-MS2 protein was expressed and purified from E. coli TB1 cells carrying the pMAL-MS2 expression plasmid as described by the supplier (New England BioLabs).
Yeast GAL::SNR30-pTOB-MS2-R30 cells collected from 2 l of culture (OD600 ∼1) were washed twice with cold water and frozen in liquid nitrogen. Cells were broken in 12 ml of tobramycin binding buffer (20 mM Tris–HCl, pH 8.1, 145 mM KCl, 1 mM CaCl2, 1 mM MgCl2, 0.2 mM DTT) at 500 rpm for 10 min in Fritsch Pulverisette 6. The crude cell extract was clarified by centrifugation twice at 10 000 g for 10 min and filtered through a 250 -µm nylon filter. After ultracentrifugation at 100 000g for 1 h, the extract was filtered again through a 0.45 -µm cellulose-acetate filter. The clear extract was incubated with 100 µg MBP-MS2 protein and 200 µl of tobramycin beads with gentle agitation for 1 h at 4°C. Beads were collected and washed three times with tobramycin wash buffer (20 mM Tris–HCl pH, 8.1, 75 mM KCl, 1 mM CaCl2, 1 mM MgCl2, 0.2 mM DTT). To elute tobramycin-bound snR30, the beads were washed twice for 5 min with 0.5 ml of tobramycin elution buffer (20 mM Tris–HCl pH, 8.1, 145 mM KCl, 1 mM CaCl2, 3 mM MgCl2, 0.2 mM DTT, 5 mM tobramycin). The supernatants were combined and diluted with 10 ml of amylose column buffer (20 mM Tris–HCl, pH 7.4, 200 mM NaCl, 1 mM EDTA, 2 mM DTT). About 200 µg of amylose resin (New England BioLabs) was added and the mixture was incubated with gentle agitation for 1 h at 4°C. The amylose resin was washed three times with both amylose column buffer and low salt amylose column buffer (30 mM Tris-HCl, pH 8.1, 10 mM NaCl, 2 mM DTT). To recover MBP-MS2-bound snR30, the resin was incubated twice with 0.5 ml low salt amylose column buffer containing 20 µM maltose for 5 min. The supernatants were combined and the eluted proteins were collected by 2,2,2-trichloroacetic acid-precipitation. The protein mixture was analyzed by mass spectrometry using Velos LTQ linear ion traps and Orbitraps from Thermo Electron (Taplin Mass Spectrometry Facility, Harvard Medical School).
Protein analysis and sucrose gradient sedimentation
IP of TAP-tagged yeast proteins was performed as described (30), except that we used IP buffer containing 20 mM Tris–Cl, pH 8.0, 5 mM Mg-acetate, 200 mM NaCl, 0.2% Triton X-100, supplemented with 1 mM DTT, 1× EDTA-free protease inhibitor cocktail (Roche) and 0.5 U/µl RNasin (Promega). To obtain high speed extracts devoid of large RNPs, the original (low speed) extracts were subjected to further centrifugation at 180 000g for 45 min. IP of human proteins has been described (31). Proteins separated by electrophoresis on 10% or 12% SDS polyacrylamide gels and electroblotted onto Hybond-C extra membranes (Amersham Pharmacia Biotech) were detected using rabbit peroxidase antiperoxidase (PAP; Dako) diluted to 1:10 000 in PBST buffer containing 5% milk. The Nhp2p was detected with a rabbit anti-serum kindly provided by Dr Y. Henry (LBME, Toulouse). Sucrose gradient velocity sedimentation was described (23). To deproteinize yeast preribosomal complexes, yeast low speed extracts were treated with proteinase K at 2 mg/ml final concentration in the presence of 1% SDS for 2 h at 16°C.
In vivo cross-linking
Yeast cells were cross-linked in YPD medium supplemented with 1% of formaldehyde with continuous shaking for 30 min at 30°C. The reaction was terminated by adding glycine to 125 mM final concentration. After incubation for 5 min, the cross-linked cells were washed twice with Tris-buffered saline and frozen in liquid nitrogen. Cells were broken in RNA IP buffer (50 mM Tris–Cl, pH 7.4, 1 mM EDTA, 1% triton X-100, 1% Na-deoxycholate, 0,1% SDS) containing 200 mM NaCl by vigorous shaking with glass beads for 45 min. The extract was clarified by ultracentrifugation at 180 000g for 45 min. ZZ-tagged proteins were collected by incubation with 20 μl of IgG beads for 2 h and the beads were washed with RNA IP buffer containing 500 mM NaCl. The reverse cross-linking reaction was carried out at 70°C for 1 h with continuous rotation at 1200 rpm in 50 mM Tris–Cl, pH 6.8, 5 mM EDTA, 10 mM DTT and 1% SDS.
RNA analysis
RNA extraction, northern blot and primer extension analysis were preformed as described (22,30). To detect pre-rRNAs (32), the snR30 (5′-AGATGTCTGCAGTATGGTTT-3′), U17 (5′-TTCCTGCATGGTTTGTCTCC-3′), U3 (5′-ATGGGGCTCATCAACCAACTTGG-3′), 7SK (5′-GTGTCTGGAGTCTTGGAAGC-3′), U14 (5′-CTCAGACATCCTAGGAAGG-3′) and snR10 (5′-CATGGGTCAAGAACGCCCCGGAGGGG-3′) snoRNAs, terminally labeled specific oligodeoxynucleotide probes were used.
RESULTS
Affinity purification of yeast snR30
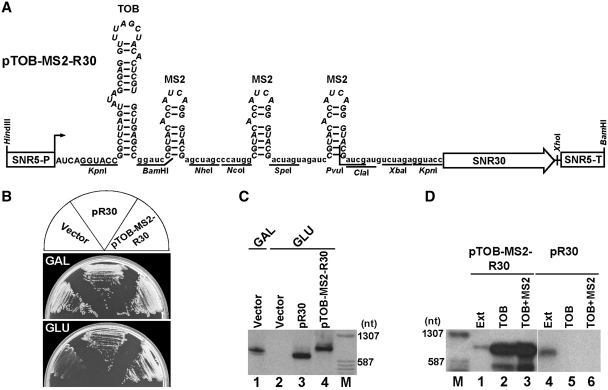
To identify yeast proteins interacting with the snR30 snoRNA, a yeast expression plasmid was constructed to express an extended version of snR30 suitable for RNA affinity selection (Figure 2A). A synthetic DNA fragment encoding one tobramycin- and three tandemly arranged bacteriophage MS2 coat protein-binding motifs was fused to the coding region of the SNR30 gene. The 5′-terminally tagged SNR30 DNA was placed under the control of the promoter and terminator of the constitutively expressed yeast SNR5 gene in the pFL45/R5P expression vector (21). The resulting pTOB-MS2-R30 expression plasmid was transformed into the GAL::SNR30 yeast strain in which the endogenous SNR30 gene is transcribed from the galactose-inducible GAL10 promoter (22). As controls, the empty pFL45/R5P vector and the pR30 expression plasmid encoding wild-type snR30 were transformed into GAL::SNR30 cells. In contrast to cells carrying the empty vector, cells transformed with pTOB-MS2-R30 or pR30 showed efficient growth on glucose-containing medium and, as demonstrated by northern blot analysis, efficiently accumulated the plasmid-encoded wild-type and 5′-terminally extended snR30 RNAs (Figure 2B and 2C, lanes 3 and 4). This demonstrated that the TOB-MS2-tagged snR30, similarly to wild-type snR30, supports ribosome biogenesis and cell growth.
Figure 2.
Affinity purification of yeast snR30 snoRNP. (A) Schematic structure of the pTOB-MS2-R30 expression construct. The promoter (SNR5-P) and terminator (SNR5-T) of the SNR5 and the coding region of the SNR30 gene (open arrow) are indicated. Sequences and predicted structures of the tobramycin (TOB) and MS2 coat protein-binding motifs (italics) and the spacer regions (lower case letters) are shown. The 5′-terminal nucleotides of the predicted snR30 transcript represent authentic snR30 sequences (capitals). Restriction sites are indicated. (B) Growth properties of yeast GAL::SNR30 cells carrying the pTOB-MS2-R30 and pR30 expression plasmids or the pFL45/R5P empty vector on galactose- (GAL) and glucose-containing (GLU) solid medium. (C) Expression of wild type and TOB-MS2-tagged snR30 RNAs. About 5 μg of RNAs extracted from yeast GAL::SNR30 cells transformed by the indicated plasmids and grown in galactose- or glucose-containing liquid medium were fractionated on a 6% sequencing gel and analyzed by northern blotting with a snR30-specific oligonucleotide probe. Lane M, relevant size markers in nucleotides. (D) Purification of TOB-MS2-R30 RNP. Extracts (Ext) prepared from GAL::SNR30 cells expressing TOB-MS2-R30 or wild-type snR30 were used as starting materials for tandem RNA affinity-selection. RNAs isolated from cell extracts (5 μg) or from the pellets of affinity-selection reactions (0.1 μg) were analyzed by northern blotting.
The accumulating TOB-MS2-R30 snoRNA was purified by two subsequent steps of affinity selection. An extract prepared from the GAL::SNR30-pTOB-MS2-R30 cells was incubated with tobramycin immobilized on sepharose matrix and a recombinant fusion protein composed of the maltose-binding protein and the MS2 coat protein (MBP-MS2) (33). The selected TOB-MS2-R30/MBP-MS2 complex was eluted from the matrix with tobramycin and affinity-selected again through incubation with amylose resin. Finally, the immobilized TOB-MS2-R30/MS2-MBP complex was recovered from the amylose resin with maltose solution. Purification of TOB-MS2-R30 was monitored by northern blot analysis (Figure 2D). In the TOB-MS2-R30 snoRNA preparation obtained by two subsequent affinity purification reactions the tagged TOB-MS2-R30 RNA showed about 600 - to 800-fold enrichment compared to the cell extract that was used as starting material (lanes 1 and 3). Mock affinity-purification performed from control cells expressing the untagged wild-type snR30 failed to recover detectable amount of snR30 (lanes 5 and 6).
Proteins co-purified with TOB-MS2-R30 were identified by mass spectrometric analysis (Table 1). After removal of common contaminants present in the side-by-side control mock purification as well as those proteins which were represented by three or less peptides (predominantly ribosomal and C/D snoRNP proteins), we provisionally defined six snR30-specific proteins. As it was highly predictable, on the top of proteins coselected with snR30 were the Cbf5p, Nhp2p, Nop10p and Gar1p H/ACA core proteins which were represented by 27, 9, 4 and 7 oligopeptides covering 41, 45, 50 and 22% of their sequences, respectively. Besides the four H/ACA proteins, we detected two additional putative snR30-associated proteins, Utp23p (U three-associated protein 23, YOR004w) (four peptides, 17% coverage) and Kri1p (Krr1p interacting protein 1, YNL308C) (eight peptides, 16% coverage). Importantly, both Utp23p and Kri1p had been earlier reported to function in 18S rRNA processing, pointing to the relevance of their co-purification with snR30 (27,28).
Table 1.
Mass spectrometric identification of yeast proteins specifically copurified with snR30
| No | Position | Oligopeptide |
|---|---|---|
| Cbf5 (protein coverage by a.a. count: 40.6%) | ||
| 1 | 4–27 | EDFVIKPEAAGASTDTSEWPLLLK; |
| 2 | 36–50 | SGHYTPIPAGSSPLK; |
| 3 | 36–51 | SGHYTPIPAGSSPLKR |
| 4 | 142–161 | SLENLTGALFQRPPLISAVK |
| 5 | 129–141 | LHDALKDEKDLGR |
| 6 | 142–162 | SLENLTGALFQRPPLISAVKR |
| 7 | 168–180 | TIYESNLIEFDNK |
| 8 | 168–181 | TIYESNLIEFDNKR |
| 9 | 221–247 | SGALSENDNMVTLHDVMDAQWVYDNTR |
| 10 | 254–267 | SIIQPLETLLVGYK |
| 11 | 254–268 | SIIQPLETLLVGYKR |
| 12 | 349–358 | RWGLGPVAQK |
| 13 | 350–358 | WGLGPVAQK |
| 14 | 374–383 | VNENTPEQWK |
| 15 | 374–384 | VNENTPEQWKK |
| 16 | 384–403 | KEYVPLDNAEQSTSSSQETK |
| 17 | 385–403 | EYVPLDNAEQSTSSSQETK |
| 18 | 412–419 | AKEDSLIK |
| 19 | 412–435 | AKEDSLIKEVETEKEEVKEDDSKK |
| 20 | 414–425 | EDSLIKEVETEK |
| 21 | 414–429 | EDSLIKEVETEKEEVK |
| 22 | 414–434 | EDSLIKEVETEKEEVKEDDSK |
| 23 | 414–435 | EDSLIKEVETEKEEVKEDDSKK |
| 24 | 414–419 | EDSLIK |
| 26 | 420–434 | EVETEKEEVKEDDSK |
| 26 | 420–435 | EVETEKEEVKEDDSKK |
| 27 | 420–437 | EVETEKEEVKEDDSKKEK |
| Nhp2 (protein coverage by a.a. count: 44.9%) | ||
| 1 | 16–23 | TVDNYEAR |
| 2 | 24–37 | MPAVLPFAKPLASK |
| 3 | 108–117 | QDLGAAGATK |
| 4 | 118–131 | RPTSVVFIVPGSNK |
| 5 | 118–132 | RPTSVVFIVPGSNKK |
| 6 | 134–151 | DGKNKEEEYKESFNEVVK |
| 7 | 137–151 | NKEEEYKESFNEVVK |
| 8 | 137–156 | NKEEEYKESFNEVVKEVQAL |
| 9 | 144–156 | ESFNEVVKEVQAL |
| Nop10 (protein coverage by a.a. count: 50%) | ||
| 1 | 1–12 | MHLMYTLGPDGK |
| 2 | 1–13 | MHLMYTLGPDGKR |
| 3 | 20–28 | VTESGEITK |
| 4 | 52–58 | FGLVPGQ |
| Gar1 (protein coverage by a.a. count: 22.4%) | ||
| 1 | 73–93 | TQVGKVDEILGPLNEVFFTIK |
| 2 | 78–93 | VDEILGPLNEVFFTIK |
| 3 | 105–115 | EGDKFYIAADK |
| 4 | 105–121 | EGDKFYIAADKLLPIER |
| 5 | 109–115 | FYIAADK |
| 6 | 109–121 | FYIAADKLLPIER |
| 7 | 128–135 | VVGPPKPK |
| Utp23 (protein coverage by a.a. count: 16.5%) | ||
| 1 | 74–82 | NDGAINLAK |
| 2 | 117–127 | YVVASQDIDLR |
| 3 | 144–157 | SVMVMEPLSTASAK |
| 4 | 201–208 | APNPLSVK |
| Kri1 (protein coverage by a.a. count: 15.9%) | ||
| 1 | 102–112 | FFEDPESAAAK |
| 2 | 155–162 | QSFVSQQR |
| 3 | 265–278 | YEDPNAAEIISYAR |
| 4 | 324–333 | LTDILEQLTK |
| 5 | 334–345 | EYGAEINADMVK |
| 6 | 440–452 | VNELVENALEQNK |
| 7 | 453–467 | LALIEEVEKEEEERK |
| 8 | 561–571 | DEDTILIPVEK |
Utp23p specifically associates with snR30
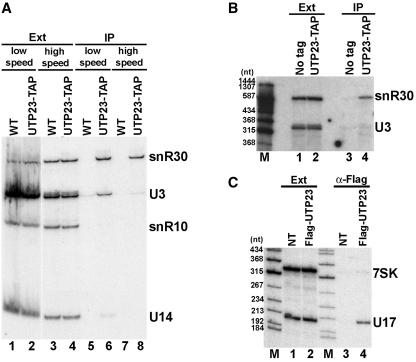
To confirm the in vivo interaction of yeast snR30 with Utp23p, we performed coimmunoprecipitation (co-IP) experiments. When a TAP-tagged version of Utp23p expressed in the yeast UTP23-TAP strain was immunoprecipitated from a cell extract prepared under standard conditions, snR30 was readily detected in the pellet by northern blot analyses (Figure 3A, lane 6). However, probing the same blot with a U3-specific oligonucleotide probe revealed a significant, albeit less efficient interaction of Utp23p-TAP also with the U3 box C/D snoRNA (compare lanes 2 and 6). Other rRNA processing snoRNAs, such as snR10 (H/ACA) and U14 (C/D), showed no significant co-purification with Utp23p-TAP. These observations suggest that Utp23p either interacts with both snR30 and U3 or it is a component of a large processing complex containing these snoRNPs. To distinguish between these possibilities, before IP of Utp23p-TAP, the cell extract was clarified by ultracentrifugation to remove large RNP complexes. Under these conditions, IP of Utp23p-TAP pulled down snR30, but failed to efficiently recover U3, indicating that Utp23p specifically associates with snR30 (lane 8).
Figure 3.
In vivo association of snR30 and Utp23p. (A) Yeast snR30 coprecipitates with TAP-tagged Utp23p. Extracts (Ext) prepared from control cells (WT) or from cells expressing Utp23p-TAP were cleared by centrifugation at 10 000 g for 10 min (low speed) or at 180 000 g for 45 min (high speed) before IP of Utp23-TAP with IgG-sepharose. RNAs from the extracts or from the pellets of IP reactions were analyzed by northern blotting with oligonucleotide probes specific for snR30, U3, snR10 and U14. (B) In vivo cross-linking of snR30 and Utp23p. Yeast cells expressing wild-type (no tag) or TAP-tagged Utp23p were treated with formaldehyde and after extract preparation, Utp23p-TAP was immunoprecipitated under stringent conditions. Coprecipitation of snR30 and U3 was monitored by northern blot analysis. Lane M, size markers. (C) Human Utp23p interacts with the U17 snoRNA. HeLa cells transfected or not transfected (NT) with the pCMV-Flag-UTP23 expression vector were used for preparation of high speed extracts. RNAs prepared from the extracts or from the pellets of anti-Flag IPs were analyzed by northern blotting with U17- and 7SK-specific probes.
To exclude the formal possibility that Utp23p binds to snR30 during extract preparation, we performed in vivo RNA-protein cross-linking experiments (Figure 3B). Yeast cells expressing Utp23p-TAP were treated with formaldehyde before extract preparation and Utp23p-TAP was immunoprecipitated under highly stringent conditions which prevented formation of non-covalent RNA-protein interactions (34,35). Northern blot analysis of RNAs covalently cross-linked to and co-purified with UTP23p-TAP revealed that snR30, but not U3, was present in the pellet (lane 4), confirming that Utp23p specifically interacts with snR30 in living cells.
Both Utp23p and snR30 show strong evolutionarily conservation, suggesting that they play a conserved role in eukaryotic pre-rRNA processing (22). To corroborate this notion, we tested whether human Utp23p can interact in vivo with the human homologue of snR30, called U17 (Figure 3C). A Flag-tagged version of human Utp23p was transiently expressed in HeLa cells. After preparation of a high speed extract devoid of large RNPs, Utp23p was immunoprecipitated with an anti-Flag antibody. Co-precipitation of the endogenous U17 snoRNA and as a negative control, the 7SK snRNA was tested by northern blot analysis. Human U17 efficiently co-purified with Flag-Utp23p (lane 4), but it was not detectable in the pellet of a control IP from non-transfected cells (lane 3). We concluded that Utp23p forms a specific and evolutionarily conserved interaction with the U17/snR30 snoRNP.
Utp23p and Kri1p associate with snR30 in a mutually exclusive manner
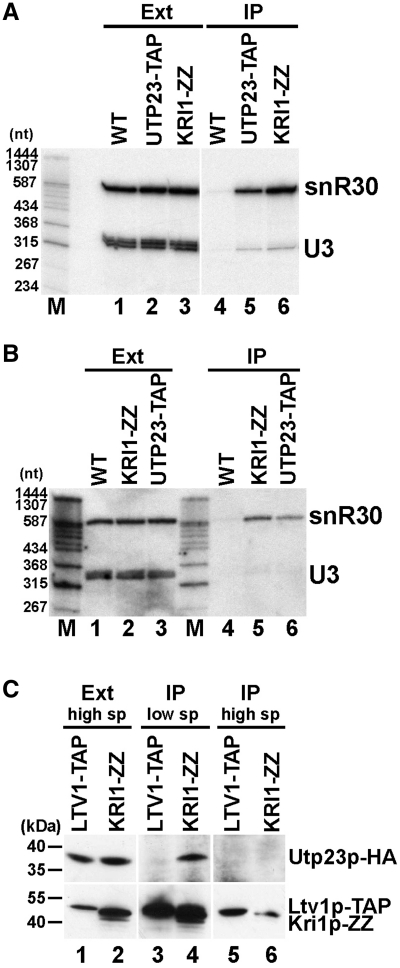
To confirm the in vivo interaction of yeast Kri1p and snR30, an epitope-tagged Kri1p carrying two copies of protein A (Kri1p-ZZ) was expressed and immunoprecipitated with IgG-sepharose from a cell extract that had been depleted of large RNPs by high speed centrifugation (36). Northern blot analysis of RNAs co-purified with Kri1p-ZZ demonstrated that similarly to Utp23p, Kri1p-ZZ specifically associated with snR30 and failed to efficiently interact with U3 (Figure 4A, lanes 5 and 6). We also tested the co-IP of snR30 with Kri1p-ZZ from an extract prepared from formaldehyde cross-linked yeast cells (Figure 4B). In spite of the harsh wash conditions predicted to interrupt non-covalent RNA-protein interactions (34), we detected significant amount of snR30, but not U3 snoRNA, co-purifying with Kri1p-ZZ. We concluded that Kri1p specifically interacts with snR30.
Figure 4.
In vivo association of snR30 and Kri1p. (A) Kri1p interacts with snR30. Extracts were prepared by high speed centrifugation from control cells (WT) or from cells expressing protein A-tagged Kri1p (Kri1p-ZZ) and TAP-tagged Utp23p (Utp23p-TAP). The tagged proteins were immunoprecipitated with IgG-sepharose and the coprecipitated RNAs were analyzed by northern blotting with snR30- and U3-specific probes. Lane M, size markers. (B) In vivo cross-linking of snR30 and Kri1p. Yeast control cells (WT) and cells expressing Utp23p-TAP or Kri1p-ZZ were treated with formaldehyde. After extract preparation, Utp23p-TAP and Kri1p-ZZ were immunoprecipitated under stringent conditions and coprecipitation of snR30 and U3 was assayed by northern blot analysis. (C) Mutually exclusive association of Utp23p and Kri1p with snR30. Extracts were prepared from yeast cells expressing HA-tagged Utp23p (Utp23p-HA) together with Kri1p-ZZ or Ltv1p-TAP following the low speed (low sp) or high speed (high sp) centrifugation protocol. After IP of Kri1p-ZZ and Ltv1p-TAP (lower panel), co-precipitation of Utp23p-HA was tested by western blotting. Size markers are indicated in kDa.
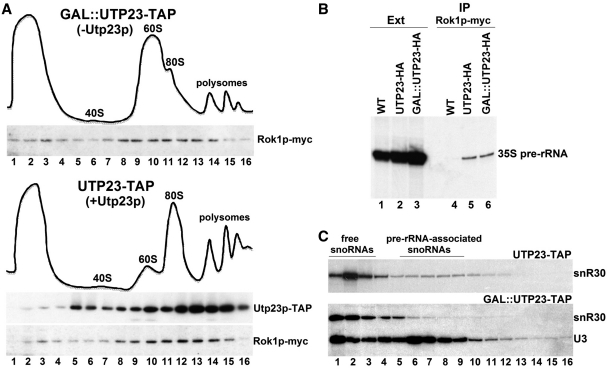
During the course of our co-IP experiments, we noticed that in contrast to the Cbf5p and Nhp2p H/ACA core proteins which efficiently interacted with snR30, IP of Utp23p-TAP and Kri1p-ZZ recovered only small amounts of snR30 (data not shown). This might indicate that Utp23p and Kri1p either weakly associate with snR30 or they interact with only a fraction of yeast snR30. To clarify whether Utp23p and Kri1p bind together or independently to snR30, we tested the co-IP of Kri1p and Utp23p using an extract prepared from yeast cells expressing both Kri1p-ZZ and Utp23p-HA. As western blot analysis demonstrated, IP of Kri1-ZZ from a low speed extract also pulled down Utp23p-HA (Figure 4C, lane 4), but under the same conditions, control IP of Ltv1p-TAP, a late processing factor involved in the nuclear export of the small subunit, failed to recover Utp23p-HA (lane 3). When large RNP complexes were removed from the extract before IP, association of Kri1p-ZZ and Utp23p-HA was no longer detected (lane 6). These results confirmed that Utp23p and Kri1p bind to a fraction of yeast snR30 in a mutually exclusive manner.
Utp23p is required for dissociation of snR30 from large pre-ribosomal particles
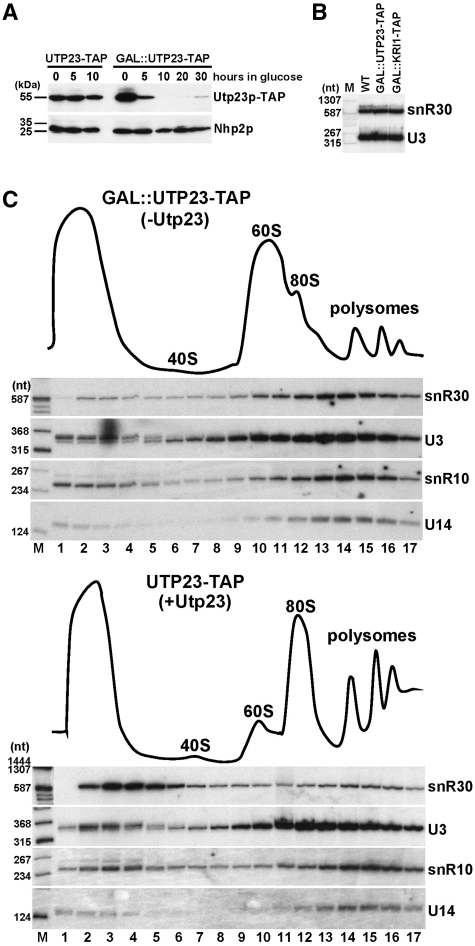
Next, we tested whether Utp23p and Kri1p function in the biogenesis or contribute to the metabolic stability of yeast snR30. We used yeast strains with TAP-tagged chromosomal UTP23 and KRI1 genes under the control of their own promoters or under the GAL10 promoter that is inactive in cells grown on glucose. Therefore, shifting the GAL::UTP23-TAP and GAL::KRI1-TAP cells from galactose- to glucose-containing medium abolished accumulation of Utp23p and Kri1p (Figure 5A and data not shown). Depletion of Utp23p and Kri1p had no effect on the accumulation of the snR30 and the control U3 snoRNAs (Figure 5B). These results suggested that instead of participating in the biogenesis of snR30 snoRNP, Utp23p and Kri1p work in collaboration with snR30 in 18S processing.
Figure 5.
Depletion of Utp23p increases snR30 accumulation in large pre-ribosomal complexes. (A) Expression of Utp23p-TAP in yeast UTP23-TAP and GAL::UTP23-TAP cells grown in glucose medium. Western blot analysis was used to monitor the accumulation of Utp23p-TAP and Nhp2p. Size markers are shown. (B) Utp23p and Kri1p are not required for accumulation of snR30. GAL::UTP23-TAP, GAL::KRI1-TAP and control Y0341 (WT) cells were grown on glucose for 10 h. Accumulation of snR30 and U3 snoRNAs was measured by northern blotting. (C) Sedimentation behavior of yeast snR30 in the absence of Utp23p. Extracts prepared from yeast UTP23-TAP and GAL::UTP23-TAP cells grown on glucose for 10 h were fractionated on 5–45% linear sucrose gradients. Distribution of snR30, U3, snR10 and U14 was detected by northern blotting. Ribosomal sedimentation profiles were obtained by recording absorbance at OD254. Lane M, size markers.
Yeast snR30 base pairs with internal 18S sequences imbedded in the newly synthesized 35S pre-rRNA and packaged into the 90S pre-ribosome (23) (Figure 1). To test whether Utp23p and Kri1p are important for association of snR30 with nascent pre-ribosomes, we compared the sedimentation behaviors of snR30 in the presence and absence of Utp23p and Kri1p. Extracts prepared from UTP23-TAP (+Utp23) and GAL::UTP23-TAP (-Utp23) cells grown on glucose were fractionated by velocity sedimentation through sucrose density gradients. Distribution of the snR30, snR10, U3 and U14 snoRNAs involved in rRNA processing was monitored by northern blot analyses. In the presence of Utp23p, a small fraction of snR30 co-sedimentated with large 90S structures and the majority of snR30 migrated in the upper part of the gradient probably in the form of free snoRNP (23) (Figure 5C, lower panel). In the absence of Utp23p, however, the equilibrium of free and 90S-associated snR30 was robustly shifted towards the fast-migrating large complexes (upper panel), suggesting that Utp23p is required for dissociation of snR30 from large pre-ribosomal complexes. The sedimentation profiles of other rRNA processing snoRNAs, U3, U14 and snR10, showed no apparent alterations in cell extracts containing or lacking Utp23p. Thus, we concluded that Utp23p is specifically required for the release of snR30 from maturing pre-ribosomes. In contrast to Utp23p, depletion of Kri1p had no major effect on the sedimentation behavior of snR30, indicating that Kri1p does not contribute to the dynamic association snR30 with maturing pre-ribosomes (data not shown).
Protein factor(s) tether snR30 to pre-ribosomes in the absence of Utp23p
The release of snR30 from pre-rRNAs and pre-ribosomal particles specifically requires the DEAD box RNA helicase Rok1p (37). To test whether Utp23p works together with Rok1p in snR30 release, we investigated the association of Rok1p with pre-ribosomal particles in cells expressing or lacking Utp23p. UTP23-TAP and GAL::UTP23-TAP cells expressing c-Myc-tagged Rok1p (Rok1p-myc) were grown in glucose medium for 10 h before extract preparation. The extracts were fractionated by sucrose-gradient centrifugation and distribution of Rok1p-myc was measured by western blot analysis (Figure 6A). Both in the presence (UTP23-TAP) and absence (GAL::UTP23-TAP) of Utp23p-TAP, Rok1p-myc efficiently associated with pre-ribosomal particles, indicating that Utp23p is not required for Rok1p recruitment. Further corroborating this notion, primer extension analysis demonstrated that IP of Rok1p-myc from cell extracts accumulating (UTP23-HA) or lacking (GAL::UTP23-HA) Utp23p-HA pulled down the 35S pre-rRNA with comparable efficiency (Figure 6B, lanes 5 and 6). Control mock IP performed from wild-type cells expressing untagged Rok1p failed to recover 35S pre-rRNA (lane 4).
Figure 6.
Protein-mediated tethering of snR30 to pre-ribosomes in the absence of Utp23p. (A) Sedimentation behavior of Rok1p in the absence and presence of Utp23p. Extracts prepared from UTP23-TAP and GAL::UTP23-TAP cells expressing Rok1p-myc and grown on glucose were fractionated on 5–45% sucrose gradients. Distribution of Rok1p-myc and Utp23p-TAP was monitored by western blot analyses. The ribosomal sedimentation profiles are shown. (B) Rok1p binds to the 35S pre-rRNA independently of Utp23p. Rok1p-myc was expressed in control (WT), UTP23-HA and GAL::UTP23-HA cells. After 10 h of growth in glucose medium, extracts were prepared and Rok1p-myc was immunoprecipitated. Coprecipitation of the 35S pre-rRNA was assayed by primer extension analyses using a terminally labeled deoxyoligonucleotide primer. (C) Yeast snR30 is not base-paired with pre-rRNAs in Utp23p-depleted cells. Cell extracts prepared from GAL::UTP23-TAP and UTP23-TAP cells grown on glucose was treated with proteinase K before sucrose gradient fractionation. Distribution of snR30 and U3 was monitored by northern blotting.
While Rok1p is a canonical RNA helicase, Utp23p lacks recognizable helicase domains, suggesting that it promotes snR30 dissociation in a different way. Although the experiments presented above demonstrated that Rok1p can bind to the 35S pre-rRNA-containing particles independently of Utp23p, we tested whether in the absence of Utp23p, snR30 is tethered to the large preribosomes through base-pairing with the 35S pre-rRNA. Extracts prepared from GAL::UTP23-TAP cells grown on glucose was subjected to extensive proteinase treatment to remove pre-rRNA-associated proteins before gradient sedimentation analysis (24,37,38) (Figure 6C). Northern blot analysis revealed that, in contrast to the U3 snoRNA that remained associated with large fast-migrating pre-rRNAs, the majority of snR30 failed to enter the gradient, indicating that it was in the form of free snoRNA. As expected, a significant fraction of snR30 remained associated with large pre-rRNAs when a deproteinized extract of the UTP23-TAP control cells was analyzed by gradient sedimentation (upper panel, lanes 7 to 10). We concluded that in the absence of Utp23p, the snR30 snoRNP remains tethered to the maturing preribosomes through protein–protein or protein–RNA, rather than RNA–RNA interactions. This may indicate that Utp23p is not required for Rok1p-mediated snR30-rRNA unwinding or according to another option, Utp23p is essential for establishment of the base-pairing interaction of snR30 and the 35S pre-rRNA.
snR30 is required for association of Utp23p with 35S pre-rRNA
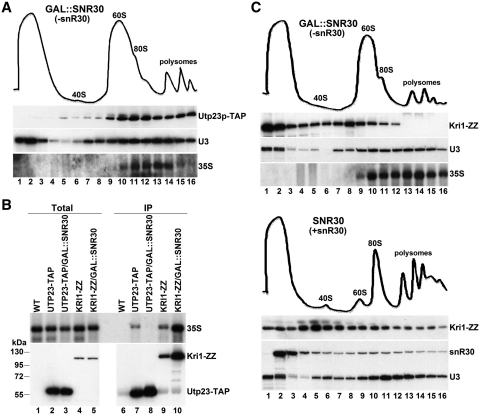
The observation that snR30 efficiently bound to pre-ribosomes in the absence of Utp23p suggested that snR30 and Utp23p are independently recruited to pre-ribosomes. To test whether Utp23p can associate with nascent pre-ribosomes in the absence of snR30, an extract prepared from GAL::SNR30 cells expressing Utp23p-TAP and grown in glucose medium was fractionated on a sucrose gradient (Figure 7A). Northern and western blot analyses revealed that Utp23p-TAP migrated mostly in large complexes together with the 35S pre-rRNA and the U3 snoRNA. A similar gradient distribution was observed for Utp23-TAP in an extract prepared from controls cell expressing snR30 (see Figure 6A, lower panel). These results seemed to support the idea that pre-ribosomal recruitment of Utp23 is independent of snR30. However, IP of Utp23-TAP from the GAL::SNR30 cell extract failed to coprecipitate detectable amount of 35S rRNA, while in control cells accumulating snR30 Utp23-TAP showed an association with 35S (Figure 7A, lanes 7 and 8). Even lowering the stringency of the IP conditions by reducing the ionic strength of the extraction and IP reaction buffers failed to detect association of Utp23p and 35S pre-rRNA in cell extracts lacking snR30 (data not shown). A possible explanation of these observations could be that in the absence of snR30, Utp23p is recruited to large, most probably very loosely assembled pre-ribosomal complexes which are apparently functionally inactive. The snR30 snoRNP is required for proper incorporation of Utp23p into the nascent preribosomal particle. This idea is supported by the recent observation that assembly of functional small subunit processome is abolished in the absence of snR30 (26).
Figure 7.
Association of Utp23p and Kri1p with preribosomes depends on snR30. (A) Utp23p comigrates with the 35S pre-rRNA in the absence of snR30. Yeast GAL::SNR30 cells were grown on glucose to deplete endogenous snR30. After extract preparation, the sedimentation behaviors of Utp23p-TAP, the U3 snoRNA and the 35S pre-rRNA were analyzed. For gradient migration of Utp23p-TAP in the presence of snR30, see Figure 6A. (B) Association of Utp23p and Kri1p with 35S pre-rRNA. Yeast GAL::SNR30/UTP23-TAP, GAL::SNR30/KRI1-ZZ, UTP23-TAP, KRI1-ZZ and control (WT) cells were grown on glucose before extract preparation and IP of with IgG Sepharose. Precipitation of Utp23p-TAP and Kri1p-ZZ and coprecipitation of 35S pre-rRNA were monitored by western and northern blot analyses. (C) Incorporation of Kri1p into large preribosomal particles depends on snR30 expression. Sucrose gradient sedimentation of ectopically expressed Kri1-ZZ and snR30, together with endogenous U3 and 35S RNAs was analyzed in extracts prepared from yeast GAL::SNR30 and SNR30 strains grown on glucose.
Recruitment of Kri1p to the 90S pre-ribosome requires snR30
We found that depletion of Kri1p has no detectable effect on the dynamic interaction of snR30 with nascent preribosomes (data not shown). On the other hand, it still remained possible that snR30 may promote the preribosomal recruitment of Kri1p. To test whether Kri1p can bind to preribosomal particles in the absence of snR30, the yeast GAL::SNR30 and the control SNR30 strains, both expressing protein A-tagged Kri1p (Kri1p-ZZ), were grown on glucose. After extract preparation and glucose gradient fractionation, western blot analysis detected Kri1p-ZZ predominantly in the upper part of the gradient in the absence of snR30, indicating that snR30 is required for incorporation of Kri1p into the 90S small subunit processome (Figure 7C, upper panel). In control cells accumulating snR30, Kri1p showed strong association with pre-ribosomal complexes migrating throughout the gradient (lower panel). The sedimentation profile of the U3 snoRNA showed no alteration in the absence of snR30.
Unpredicted by its gradient distribution, co-IP experiments demonstrated that Kri1-ZZ efficiently associated with the 35S pre-rRNA both in the presence and absence of snR30 (Figure 7B, lanes 9 and 10). These results suggest that the snR30 snoRNP, although dispensable for 35S binding, is required for efficient recruitment of Kri1p to the fully assembled 90S preribosomal particle. These observations further confirmed the idea that snR30 plays a central role in the correct assembly of yeast small subunit processome competent in pre-rRNA processing (26).
DISCUSSION
Protein-based tandem affinity purification of yeast pre-ribosomal particles at different stages of maturation identified a tremendous number of novel biogenesis factors and revealed valuable details of the yeast ribosomal assembly pathway (2). In the hope of identifying ribosomal processing factors which directly interact with the yeast snR30 snoRNP, we established an entirely RNA tag-based tandem affinity purification procedure to purify snR30 under natural conditions (Figure 2). The principle of our approach was based on a previously reported RNA affinity purification protocol developed for isolation of mammalian 7SK snRNP (39). However, instead of applying one PP7 coat protein- and a short tobramycin-binding RNA motif (39), we fused three MS2 coat protein-binding sites and an extended version of the tobramycin-binding motif to the 5′ end of snR30 to increase the specificity and efficiency of the affinity selection reactions (33). Upon expression in yeast cells, the inserted tag sequences folded correctly and independently from snR30, because they did not interfere with the accumulation and function of the tagged snR30 and more tellingly, they proved to be fully active in affinity selection. We propose that upon adopting appropriate RNA expression strategies, the RNA-based tandem affinity purification technique described here may be a powerful tool to define the protein composition of in vivo assembled RNPs in various organisms.
Affinity purification of yeast snR30 followed by co-IP and cross-linking experiments demonstrated that besides the four H/ACA core proteins, a fraction of snR30 specifically associates with the Utp23p and Kri1p nucleolar proteins. It remains unclear whether Utp23p and Kri1p directly recognize the snR30 snoRNA or they interact with the snR30-associated snoRNP proteins. Genetic depletion of Utp23p and Kri1p had no effect on snR30 accumulation, excluding the possibility that these proteins function in the biogenesis of the snR30 snoRNP. Both Utp23p and Kri1p had been reported to be integral components of the small subunit processome and demonstrated to be essential for 35S pre-rRNA processing at the A0, A1 and A2 sites (27,28). Since Utp23p and Kri1p interact with snR30 in a mutually exclusive manner, they likely participate in distinct steps of snR30 function (Figure 4C). Consistent with this idea, depletion of Utp23p, but not Kri1p, dramatically increased the accumulation of snR30 in large pre-ribosomal complexes, indicating that Utp23p promotes the pre-ribosomal release of snR30 (Figure 5C). Utp23p features a putative PIN (PilT N terminus) domain that is presumed to confer nuclease activity to a group of proteins participating in RNA metabolism (40,41). However, alteration of the PIN-like domain of Utp23p had no effect on cell viability, making it unlikely that Utp23p functions as a ribonuclease in 18S rRNA production (27). Depletion of Utp23p had no influence on the preribosomal association of other processing snoRNAs, underlying its specific functional interaction with snR30. It remains unclear how Utp23p can promote the pre-ribosomal release of snR30. It is unlikely that Utp23p works together with the Rok1p RNA helicase that seems to be responsible for specific unwinding of snR30 and 35S pre-rRNA (37,42), since Rok1p associates with the 35S pre-rRNA and seems to disrupt its base-pairing interaction with snR30 even in the absence of Utp23p (Figure 6). On the other hand, it is also conceivable that snR30, although recruited to pre-ribosomal particles, is unable to base-pair with 35S sequences without Utp23p. Nevertheless, the lack of snR30-35S base-pairing interaction in preribosomes devoid of Utp23p indicates that the anomalous retention of snR30 is mediated by stable association with pre-ribosomal protein(s) which probably interact transiently with snR30 under normal conditions. Therefore, affinity purification of snR30 from Utp23p-depleted cells may identify the putative snR30 retention factors which likely represent ribosomal processing factors directly collaborating with snR30.
Co-transcriptional assembly of the small subunit processome on the nascent 35S pre-rRNA is believed to commence with recruitment of the preassembled UTP-A processing complex (43–46). Early binding of UTP-A is essential for stepwise incorporation of other modules of the 90S processome, including the UTP-B and UTP-C protein complexes and the U3 snoRNP. Proper incorporation of Utp23p and Kri1p into pre-ribosomes or in other words, assembly of functionally active small subunit processome depends on the presence of snR30 (Figure 7) (26). Given that snR30 can bind to pre-ribosomes independently of Kri1p and Utp23p, preribosomal docking of these proteins is likely preceded by recruitment of snR30. Of course, we cannot exclude the possibility that one of these proteins, either Utp23p or Kri1p, is recruited to the small subunit processome together with snR30 as a preassembled snoRNP complex. Nevertheless, we favour the idea that Utp23p and Kri1p interact with snR30 only within the newly assembled pre-ribosomal particle. Interestingly, recruitment of snR30 and Utp23p to the nascent 35S pre-rRNA does not depend on the pre-incorporation of the UTP-A early processing complex (Supplementary Figure S1). This might indicate that snR30 and Utp23p play a very important role in the assembly, dynamics or function of the small subunit processome.
During preparation of this article, yeast snR30 has been reported to associate with the nucleolar protein Nop6, the small ribosomal subunit proteins S9 and S18 and histones H2B and H4 (26). However, as acknowledged by the authors, none of these proteins are specific for snR30, since they also interact with other partially overlapping sets of both H/ACA and C/D snoRNAs (26). Thus, similarly to the previously proposed putative snR30-associated proteins (20,24,25), these new snR30 proteins likely bind to snR30 indirectly, as common components of large complexes. Importantly, none of the earlier detected snR30 proteins were present in our snR30 preparation, most probably because in our purification procedure, but not in the protocols used by others, large complexes were eliminated from the extract by extensive ultracentrifugation prior to snR30 selection.
In summary, by employing a novel RNA affinity purification protocol, we demonstrated that the Utp23p and Kri1p small subunit processing factors specifically and transiently interact with the yeast snR30 snoRNP. Utp23p may promote structural rearrangements of the pre-ribosome, essential for snR30 release. We also showed that Utp23p binds to the nascent pre-ribosome independently of the early-binding UTP-A complex, raising the possibility that it plays a more central role in ribosome biogenesis than anticipated before.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table S1 and Supplementary Figure S1.
FUNDING
La Fondation pour la Recherche Médicale (Équipes FRM to T.K.). Carrier Development Award from the Human Frontier Science Program (HFSP to A.K.H.). Funding for open access charge: Grant from FRM.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to Dr Yves Henry for critical reading of the article and for providing with strains and antibodies.
REFERENCES
- 1.Henras AK, Soudet J, Gerus M, Lebaron S, Caizergues-Ferrer M, Mougin A, Henry Y. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell. Mol. Life Sci. 2008;65:2334–2359. doi: 10.1007/s00018-008-8027-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Phipps KR, Charette JM, Baserga SJ. The SSU processome in ribosome biogenesis—progress and prospects. WIREs RNA. 2010;2:1–21. doi: 10.1002/wrna.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kressler D, Hurt E, Bassler J. Driving ribosome assembly. Biochim. Biophys. Acta. 2010;1803:673–683. doi: 10.1016/j.bbamcr.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 4.Kos M, Tollervey D. Yeast pre-rRNA processing and modification occur cotranscriptionally. Mol. Cell. 2010;37:809–820. doi: 10.1016/j.molcel.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grandi P, Rybin V, Bassler J, Petfalski E, Strauss D, Marzioch M, Schafer T, Kuster B, Tschochner H, Tollervey D, et al. 90S pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol. Cell. 2002;10:105–115. doi: 10.1016/s1097-2765(02)00579-8. [DOI] [PubMed] [Google Scholar]
- 6.Dragon F, Gallagher JE, Compagnone-Post PA, Mitchell BM, Porwancher KA, Wehner KA, Wormsley S, Settlage RE, Shabanowitz J, Osheim Y, et al. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature. 2002;417:967–970. doi: 10.1038/nature00769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hughes JM, Ares M., Jr Depletion of U3 small nucleolar RNA inhibits cleavage in the 5′ external transcribed spacer of yeast pre-ribosomal RNA and impairs formation of 18S ribosomal RNA. EMBO J. 1991;10:4231–4239. doi: 10.1002/j.1460-2075.1991.tb05001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li HD, Zagorski J, Fournier MJ. Depletion of U14 small nuclear RNA (snR128) disrupts production of 18S rRNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 1990;10:1145–1152. doi: 10.1128/mcb.10.3.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bally M, Hughes J, Cesareni G. SnR30: a new, essential small nuclear RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 1988;16:5291–5303. doi: 10.1093/nar/16.12.5291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morrissey JP, Tollervey D. Yeast snR30 is a small nucleolar RNA required for 18S rRNA synthesis. Mol. Cell Biol. 1993;13:2469–2477. doi: 10.1128/mcb.13.4.2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tollervey D, Mattaj IW. Fungal small nuclear ribonucleoproteins share properties with plant and vertebrate U-snRNPs. EMBO J. 1987;6:469–476. doi: 10.1002/j.1460-2075.1987.tb04777.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kiss T, Fayet-Lebaron E, Jády BE. Box H/ACA small ribonucleoproteins. Mol. Cell. 2010;37:597–606. doi: 10.1016/j.molcel.2010.01.032. [DOI] [PubMed] [Google Scholar]
- 13.Matera AG, Terns RM, Terns MP. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat. Rev. Mol. Cell Biol. 2007;8:209–220. doi: 10.1038/nrm2124. [DOI] [PubMed] [Google Scholar]
- 14.Meier UT. The many facets of H/ACA ribonucleoproteins. Chromosoma. 2005;114:1–14. doi: 10.1007/s00412-005-0333-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Terns MP, Terns R. Noncoding RNAs of the H/ACA family. Cold Spring Harb Symp Quant Biol. 2006;LXXI:395–405. doi: 10.1101/sqb.2006.71.034. [DOI] [PubMed] [Google Scholar]
- 16.Balakin AG, Smith L, Fournier MJ. The RNA world of the nucleolus: two major families of small RNAs defined by different box elements with related functions. Cell. 1996;86:823–834. doi: 10.1016/s0092-8674(00)80156-7. [DOI] [PubMed] [Google Scholar]
- 17.Ganot P, Caizergues-Ferrer M, Kiss T. The family of box ACA small nucleolar RNAs is defined by an evolutionarily conserved secondary structure and ubiquitous sequence elements essential for RNA accumulation. Genes Dev. 1997;11:941–956. doi: 10.1101/gad.11.7.941. [DOI] [PubMed] [Google Scholar]
- 18.Kiss T, Bortolin ML, Filipowicz W. Characterization of the intron-encoded U19 RNA, a new mammalian small nucleolar RNA that is not associated with fibrillarin. Mol. Cell Biol. 1996;16:1391–1400. doi: 10.1128/mcb.16.4.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henras A, Henry Y, Bousquet-Antonelli C, Noaillac-Depeyre J, Gelugne JP, Caizergues-Ferrer M. Nhp2p and Nop10p are essential for the function of H/ACA snoRNPs. EMBO J. 1998;17:7078–7090. doi: 10.1093/emboj/17.23.7078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Watkins NJ, Gottschalk A, Neubauer G, Kastner B, Fabrizio P, Mann M, Lührmann R. Cbf5p, a potential pseudouridine synthase, and Nhp2p, a putative RNA-binding protein, are present together with Gar1p in all H BOX/ACA-motif snoRNPs and constitute a common bipartite structure. RNA. 1998;4:1549–1568. doi: 10.1017/s1355838298980761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ganot P, Bortolin ML, Kiss T. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell. 1997;89:799–809. doi: 10.1016/s0092-8674(00)80263-9. [DOI] [PubMed] [Google Scholar]
- 22.Atzorn V, Fragapane P, Kiss T. U17/snR30 is a ubiquitous snoRNA with two conserved sequence motifs essential for 18S rRNA production. Mol. Cell. Biol. 2004;24:1769–1778. doi: 10.1128/MCB.24.4.1769-1778.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fayet-Lebaron E, Atzorn V, Henry Y, Kiss T. 18S rRNA processing requires base pairings of snR30 H/ACA snoRNA to eukaryote-specific 18S sequences. EMBO J. 2009;28:1260–1270. doi: 10.1038/emboj.2009.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liang XH, Fournier MJ. The helicase Has1p is required for snoRNA release from pre-rRNA. Mol. Cell. Biol. 2006;26:7437–7450. doi: 10.1128/MCB.00664-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lübben B, Fabrizio P, Kastner B, Lührmann R. Isolation and characterization of the small nucleolar ribonucleoprotein particle snR30 from Saccharomyces cerevisiae. J. Biol. Chem. 1995;270:11549–11554. doi: 10.1074/jbc.270.19.11549. [DOI] [PubMed] [Google Scholar]
- 26.Lemay V, Hossain A, Osheim YN, Beyer AL, Dragon F. Identification of novel proteins associated with yeast snR30 small nucleolar RNA. Nucleic Acids Res. 2011;39:9659–9670. doi: 10.1093/nar/gkr659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bleichert F, Granneman S, Osheim YN, Beyer AL, Baserga SJ. The PINc domain protein Utp24, a putative nuclease, is required for the early cleavage steps in 18S rRNA maturation. Proc. Natl Acad. Sci. USA. 2006;103:9464–9469. doi: 10.1073/pnas.0603673103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sasaki T, Toh EA, Kikuchi Y. Yeast Krr1p physically and functionally interacts with a novel essential Kri1p, and both proteins are required for 40S ribosome biogenesis in the nucleolus. Mol. Cell Biol. 2000;20:7971–7979. doi: 10.1128/mcb.20.21.7971-7979.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–23. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- 30.Gerus M, Bonnart C, Caizergues-Ferrer M, Henry Y, Henras AK. Evolutionarily conserved function of RRP36 in early cleavages of the pre-rRNA and production of the 40S ribosomal subunit. Mol. Cell Biol. 2010;30:1130–1144. doi: 10.1128/MCB.00999-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoareau-Aveilla C, Bonoli M, Caizergues-Ferrer M, Henry Y. hNaf1 is required for accumulation of human H/ACA snoRNPs, scaRNPs and telomerase. RNA. 2006;12:832–840. doi: 10.1261/rna.2344106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beltrame M, Tollervey D. Identification and functional analysis of two U3 binding sites on yeast pre-ribosomal RNA. EMBO J. 1992;11:1531–1542. doi: 10.1002/j.1460-2075.1992.tb05198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hartmuth K, Urlaub H, Vornlocher HP, Will CL, Gentzel M, Wilm M, Lührmann R. Protein composition of human prespliceosomes isolated by a tobramycin affinity-selection method. Proc. Natl Acad. Sci. USA. 2002;99:16719–16724. doi: 10.1073/pnas.262483899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Niranjanakumari S, Lasda E, Brazas R, Garcia-Blanco MA. Reversible cross-linking combined with immunoprecipitation to study RNA-protein interactions in vivo. Methods (San Diego, Calif) 2002;26:182–190. doi: 10.1016/S1046-2023(02)00021-X. [DOI] [PubMed] [Google Scholar]
- 35.Van Herreweghe E, Egloff S, Goiffon I, Jády BE, Froment C, Monsarrat B, Kiss T. Dynamic remodelling of human 7SK snRNP controls the nuclear level of active P-TEFb. EMBO J. 2007;26:3570–3580. doi: 10.1038/sj.emboj.7601783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dez C, Froment C, Noaillac-Depeyre J, Monsarrat B, Caizergues-Ferrer M, Henry Y. Npa1p, a component of very early pre-60S ribosomal particles, associates with a subset of small nucleolar RNPs required for peptidyl transferase center modification. Mol. Cell Biol. 2004;24:6324–6337. doi: 10.1128/MCB.24.14.6324-6337.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bohnsack MT, Kos M, Tollervey D. Quantitative analysis of snoRNA association with preribosomes and release of snR30 by Rok1 helicase. EMBO Rep. 2008;9:1230–1236. doi: 10.1038/embor.2008.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kos M, Tollervey D. The putative RNA helicase Dbp4p is required for release of the U14 snoRNA from preribosomes in Saccharomyces cerevisiae. Mol. Cell. 2005;20:53–64. doi: 10.1016/j.molcel.2005.08.022. [DOI] [PubMed] [Google Scholar]
- 39.Hogg JR, Collins K. RNA-based affinity purification reveals 7SK RNPs with distinct composition and regulation. RNA. 2007;13:868–880. doi: 10.1261/rna.565207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arcus VL, Backbro K, Roos A, Daniel EL, Baker EN. Distant structural homology leads to the functional characterization of an archaeal PIN domain as an exonuclease. J. Biol. Chem. 2004;279:16471–16478. doi: 10.1074/jbc.M313833200. [DOI] [PubMed] [Google Scholar]
- 41.Clissold PM, Ponting CP. PIN domains in nonsense-mediated mRNA decay and RNAi. Curr. Biol. 2000;10:R888–R890. doi: 10.1016/s0960-9822(00)00858-7. [DOI] [PubMed] [Google Scholar]
- 42.Vos HR, Bax R, Faber AW, Vos JC, Raue HA. U3 snoRNP and Rrp5p associate independently with Saccharomyces cerevisiae 35S pre-rRNA, but Rrp5p is essential for association of Rok1p. Nucleic Acids Res. 2004;32:5827–5833. doi: 10.1093/nar/gkh904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gallagher JE, Dunbar DA, Granneman S, Mitchell BM, Osheim Y, Beyer AL, Baserga SJ. RNA polymerase I transcription and pre-rRNA processing are linked by specific SSU processome components. Genes Dev. 2004;18:2506–2517. doi: 10.1101/gad.1226604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krogan NJ, Peng WT, Cagney G, Robinson MD, Haw R, Zhong G, Guo X, Zhang X, Canadien V, Richards DP, et al. High-definition macromolecular composition of yeast RNA-processing complexes. Mol. Cell. 2004;13:225–239. doi: 10.1016/s1097-2765(04)00003-6. [DOI] [PubMed] [Google Scholar]
- 45.Osheim YN, French SL, Keck KM, Champion EA, Spasov K, Dragon F, Baserga SJ, Beyer AL. Pre-18S ribosomal RNA is structurally compacted into the SSU processome prior to being cleaved from nascent transcripts in Saccharomyces cerevisiae. Mol. Cell. 2004;16:943–954. doi: 10.1016/j.molcel.2004.11.031. [DOI] [PubMed] [Google Scholar]
- 46.Perez-Fernandez J, Roman A, De Las Rivas J, Bustelo XR, Dosil M. The 90S preribosome is a multimodular structure that is assembled through a hierarchical mechanism. Mol. Cell Biol. 2007;27:5414–5429. doi: 10.1128/MCB.00380-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.