Abstract
Cdx2, a gene of the paraHox cluster, encodes a homeodomain transcription factor that plays numerous roles in embryonic development and in homeostasis of the adult intestine. Whereas Cdx2 exerts a tumor suppressor function in the gut, its abnormal ectopic expression in acute leukemia is associated to a pro-oncogenic function. To try to understand this duality, we have hypothesized that Cdx2 may interact with different protein partners in the two tissues and set up experiments to identify them by tandem affinity purification. We show here that Cdx2 interacts with the Ku heterodimer specifically in intestinal cells, but not in leukemia cells, via its homeodomain. Ku proteins do not affect Cdx2 transcriptional activity. However, Cdx2 inhibits in vivo and in vitro the DNA repair activity mediated by Ku proteins in intestinal cells. Whereas Cdx2 does not affect the recruitment of Ku proteins and DNA-PKcs into the DNA repair complex, it inhibits DNA-PKcs activity. Thus, we report here a new function of Cdx2, acting as an inhibitor of the DNA repair machinery, that may contribute to its tumor suppressor function specifically in the gut.
INTRODUCTION
The Cdx2 gene, a member of the paraHox cluster, plays numerous roles in embryos including trophectoderm differentiation, posterior elongation and patterning, and intestinal determination (1–3) By midgestation onwards, it becomes restricted to the intestinal epithelium and is later involved in the homeostasis of the adult gut (4–6). The primary function of Cdx2 depends on its activity of DNA-binding transcription factor (7), but recent data suggest that it can also act via non-conventional mechanisms. For instance, it interacts with Smad3 to activate TGFβ reporter systems in the absence of TGFβ (8). Cdx2 is also able to interact with the p65 subunit of NFκB and therefore prevent the NFκB binding and activation of the Cox-2 promoter (9,10). Similarly, by interacting with β-catenin, and consequently preventing the recruitment of β-catenin on Tcf4, Cdx2 interferes with Wnt signalling (11). Recently, Cdx2 has also been reported to inhibit cell proliferation by stabilizing the cell cycle regulator p27Kip1 independently of its DNA-binding activity (12).
Cdx2 deregulation has several outcomes in pathological situations. In the normal site of expression, the gut, it becomes reduced and heterogeneous in human colorectal cancers. Experimentally, the reduced expression of Cdx2 in Cdx2+/− mice facilitates tumor progression in models of genetically and chemically induced intestinal cancers (13–14), and it also increases the migration and dissemination of colon cancer cells (15). Together, these data led to attribute a tumor suppressor role to Cdx2 in the gut. However, beside the gut, Cdx2 is ectopically expressed in a number of pathologies including acute leukemia, where it has been described as an oncogene since forced expression in haematopoietic progenitors induces leukemia (16,17). Thus, Cdx2 has opposite effects depending on the tissue context in sites of normal or ectopic expression.
Interestingly, the susceptibility of Cdx2+/− mice to intestinal cancer has been correlated to a higher resistance of the colon epithelium to γ-irradiation-induced apoptosis and to an increased chromosomal instability (13,14). In line with this, Cdx2 suppresses cell proliferation by blocking G0/G1-S progression at the first DNA checkpoint (12). These observations open the possibility that Cdx2 may have a role in genome maintenance and DNA repair. Double-strand DNA breaks (DSBs) induced by many pro-oncogenic stresses, like oxidative stress or radiations, play a major role in cancer. Indeed, massive unrepaired DSBs lead to cell cycle arrest or apoptosis whereas misrepaired DSBs induce neoplastic transformations due to chromosomal translocation or mutations (18). In higher eukaryotes, the non-homologous DNA end-joining (NHEJ) repair mechanism prevails when DSBs occur, notably during G1 phase of the cell cycle (19). During this process, the Ku antigen heterodimer (Ku70/Ku80) recognizes broken DNA ends and allows the recruitment of the repair machinery, including DNA-PK and ligase IV. The lack of either Ku antigen or DNA-PK large catalytic subunit (DNA-PKcs) contributes to high radiosensitivity, immunodeficiency and premature aging (20–22).
The reason why Cdx2 has opposite effects in various pathological settings, especially in colon cancers and leukemia, remains elusive. One possibility is that the function of this homeoprotein depends on the cellular context, or in other words that Cdx2 interacts with different partners in different tissues, leading to different effects. In the present work, we challenged this hypothesis, which allowed us to identify the Ku heterodimer as a partner of Cdx2 specifically in intestinal cells but not in leukemia cells. Functional studies provided evidence for a new and unexpected role of the Cdx2 protein, as a regulator of the DNA repair machinery.
MATERIELS AND METHODS
Cell lines, plasmids and cell transfections
HCT116, SW480, K562 and Nalm6 were grown as recommended (40–43). HT29-TW6, -TW10 or -TG8 cells have been described previously (15). pFlag-Cdx2 and pFlag-miniCdx2 have been described (26) pFOXO4 and 6DBE-luc plasmids have been described (27). Mutant plasmids were prepared in pFlag2 vector (Sigma Aldrich, St. Louis, MO, USA). DNA fragments were amplified from pFlag-Cdx2 using the 5′ primer AAAAAGCTTTCCCTCGGCAGCCAAGTGAAA and the 3′ primers AAAAGATCTAGTTCCCGAGACCCTGTGAAG and AAAAGATCTACGGCGACGTCTTGGGCCA for Cdx2[175-250] and Cdx2[175-313], respectively, and the 5′ primer AAAAAAGCTTTACGTGAGCTACCTCCTGGACAAGG and 3′ primer AAAAGATCTCGTTGAAGAAGAACAACTAAAAGG for Cdx2[1-220]. pCTAP-Cdx2 was prepared by cloning the human Cdx2 cDNA amplified with the primers 5′AAAAAGCTTGCCACCATGTACGTGAGCTACTCCTGGACAAGGACG 3′, 5′AAAGTCGACCTGGGTGACGGTGGGGTTTAGCA 3′ into HindIII/SalI digested pCTAP-A vector (Agilent Technologies, Santa Clara, CA). For SI-luc and CDH17-luc plasmids construction, the two promoter regions were amplified from human DNA using primers AAAGAGCTCCACAGCTTTGAGAAATCAAAGA and AAAAAGCTTGCCTGTTCTCTTTGCTATGTTGT for SI promoter and AAAGAGCTCTTTCACTTGCAGGGTCCTCAGG and AAAAAGCTTCGAGACTCTTGCTACGACTGGA for CDH17 promoter. DNA were then cloned into SacI and HindIII sites of pGl3-basic vector (Promega, Madison, WI) Underlined sequences indicate restriction sites inserted and used for cloning. Colon cancer cells transfections were performed using JetPEI (Polyplus Transfection, Illkirch, France) or Lipofectamin2000 (Invitrogen, Carlsbad, CA, USA) and leukemia cells were nucleofected using the Amaxa system (program X1; Lonza, Basel, Switzerland), as recommended by the suppliers in Viaspan buffer (Bristol-Myers Squibb Pharmaceuticals, Dublin, Ireland).
Luciferase activity measurement
For luciferase activity measurement, 1.5 × 105 cells were co-transfected with 100 ng of reporter plasmid (SI-luc and CDH17-luc) together with 10 ng of Cdx2 and Ku70, Ku80 encoding plasmids when indicated 200 ng of reporter 6DBE-luc together with 200 ng of FOXO4 and Ku70 encoding plasmids when indicated. pRL-null was used as internal control (Promega, Madison, WI, USA). Firefly and Renilla luciferase activities were measured using a dual reporter luciferase assay (Promega, Madison, WI) with a Lumistar luminometer (BMG, Offenburg, Germany) 48 h after DNA transfection. Each transfection experiment was repeated at least three times in triplicate. Firefly luciferase activity was normalized with Renilla luciferase activity.
Antibodies and western blot
Proteins separated on SDS–PAGE were analyzed by western blot using different primary antibodies: mouse monoclonal anti-Cdx2 (CDX2-88, BioGenex, San Ramon, CA, USA; 1/2000), goat polyclonal anti-Ku70 (Santa Cruz Biotechnology, Santa Cruz, CA, USA; 1/2000), rabbit polyclonal anti-Ku70 (Millipore, Billerica, MA, USA; 1/5000), mouse monoclonal anti-Ku70 (S5C11, Abcam, Cambridge, MA, USA; 1/10 000), rabbit monoclonal anti-Ku80 (EPR3468, Abcam, Cambridge, MA, USA; 1/10 000), mouse monoclonal anti-Flag (M2 affinity purified, Sigma Aldrich, St Louis, MO, USA; 1/4000), rabbit polyclonal anti-DNA-PKcs (Santa Cruz Biotechnology, Santa Cruz, CA, USA; 1/200), rabbit polyclonal anti S2056-phosphorylated DNA-PKcs (Abcam, Cambridge, MA, USA; 1/200), mouse monoclonal anti-β-actin (C4, Millipore, Billerica, MA, USA; 1/25 000). HRP-conjugated anti-mouse or anti-rabbit secondary antibodies (GE company, Fairfield, CT, USA) were used before ECL detection (GE company, Fairfield, CT, USA).
Cell survival assay of colon cancer cells
HCT116 cells (3 × 106) were transfected for 24 h or HT29-TW6, -TW10 or –TG8 cells were treated with vehicle or doxycyclin (1 µg µl−1; Sigma Aldrich, St Louis, MO, USA) for 24 h. Cells were washed in PBS and treated by 20 µM etoposide (Sigma Aldrich, St Louis, MO, USA) containing medium for 24 h (with 1 µg µl−1 of doxycyclin for HT29 cells). Cells were trypsinized and 1 × 106 cells were seeded in culture medium (with 1 µg µl−1 doxycyclin for HT29), grown in 25 cm2 for 2 weeks. Adherent cells were fixed with 70% ethanol, stained with 0.1% crystal violet and colonies were estimated by ImageJ software (W Rasband, Bethesda, MA, USA).
Cell survival analysis in leukemia cancer cells
Nalm6 or K562 cancer cells (5 × 106 cells) were nucleofected with 5 µg plasmid and cultured for 24 h for recovering. Nucleofected cells were treated with 60 µM of etoposide for 24 h. Apoptotic cell number was estimated at indicated time point on the FACScalibur machine (BD Biosciences, Franklin Lakes, NJ, USA) using a green-versus-red fluorescent plot after staining cells 10 min with 2.5 µg ml−1 FITC-labeled annexin V (Roche Diagnostics, Mannheim, Germany) in buffer AP (10 mM HEPES/NaOH, pH 7,4, 150 mM NaCl, 5 mM Kcl, 1 mM MgCl2, 1,8 mM CaCl2) supplemented with 10 µg ml−1 propidium iodide (Sigma Aldrich, St Louis, MO, USA). Annexin V positive cells (PI negative and positive) were considered as dead cells.
In situ protein–protein interaction assay (DuoLink®)
SW480 cells seeded in eight-well CultureSlides (BD Biosciences, Mountain View, CA, USA) were fixed (PFA 4%, 15 min) and permeabilized (Triton X-100 0.5% in PBS, 30 min). Slides were processed for in situ Proximity Ligation Assay following manufacturer's instructions using the Duolink® II Detection Reagents Green, Duolink® II PLA probe anti-Mouse Plus, Duolink® II PLA probe anti-Rabbit Minus (Olink Bioscience, Uppsala, Sweden), and mouse anti-Cdx2, and rabbit anti-Ku70 or rabbit anti-Ku80 as primary antibodies. To check the specificity of the PLA signal, control experiments omitting one of the primary antibodies were performed. Imaging was performed using an Imager.Z2 microscope and Axiovision software (Zeiss, Jena, Germany). Nuclei were labeled with DAPI 1 µg ml−1.
Tandem affinity purification (TAP-Tag)
For tandem affinity purification of Cdx2 containing complex, we used the InterPlay® mammalian TAP system (Agilent Technologies, Santa Clara, CA, USA). For purification, 12 × 106 HCT116 were transfected with 72 µg of pCTAP or pCTAP-Cdx2, 24 × 106 SW480 with 96 µg of pCTAP or pCTAP-Cdx2 and 100 × 106 NALM6 or K562 cells were nucleofected with 5 µg of pCTAP or pCTAP-Cdx2 for 5 × 106 cells. Double-tagged protein was recovered according to manufacturer's protocol. Proteins were eluted in 125 μl of 2× laemmli buffer and boiled. The recovered proteins were separated on a 14-cm long 7.5–15% gradient gel together with a prestained protein ladder (Euromedex, Strasbourg, France), silver stained (PlusOne™ Silver Staining Kit, GE company, Fairfield, CT, USA) and analyzed by mass spectrometry. Gel slices containing protein bands of interest were excised and processed as described previously (44). After digestion, the resulting peptide extracts were directly analyzed by nanoLC-MS/MS using an Agilent 1100 series nanoHPLC-Chip/MS system (Agilent Technologies, Palo Alto, USA) coupled to a HCT Plus ion trap (Bruker Daltonics, Bremen, Germany). Mass data collected during the nanoLC-MS/MS analysis were processed and converted into *.mgf files using the DataAnalysis 3.3 Build 146 software (Bruker Daltonics, Bremen, Germany). The MS and the MS/MS data were searched using a local Mascot server (MASCOT 2. 2.0, MatrixScience, UK) against a composite target-decoy database including the SwissProt 55.5 protein sequences. Mascot results were loaded in MuDPIT mode into the Scaffold 2.00.03 software (Proteome Software, Portland, USA).
Immunoprecipitation
Cells were rinsed with PBS and lysed for 30 min in lysis buffer [10 mM Tris–HCl pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 1% NP-40, 1 mM EGTA and protease inhibitor cocktail (Roche Diagnostics, Mannheim, Germany)]. The lysate was centrifuged at 20 000 g for 20 min at 4°C. One mg of protein was incubated with 2 µg of primary antibodies in 1 ml of lysis buffer overnight at 4°C with gentle rocking. Immunoprecipations were performed with 30 μl of 50% G- or A-agarose beads (Roche Applied Science) for 2 h at 4°C. Immunoprecipitates were washed twice with lysis buffer, twice with lysis buffer containing 500 mM NaCl and eluted using SDS–PAGE sample loading buffer (Biorad, Hercules, CA, USA). When precised, HCT116 cell extracts (1 mg ml−1) were treated for 30 min with 400 µg ml−1 Ethidium Bromide (EtBr) prior to addition of antibodies.
In vivo reparation assay
pEGFP-Pem1-Ad2 plasmid was kindly provided by G. Iliakis (Essen, germany). HindIII restricted pEGFP-Pem1-Ad2 was purified using Qiagen PCR purification kit (Qiagen, Hilden, Germany), ethanol precipitated and resuspended in water. One day after splitting, exponentially growing cells were transfected with 10 ng of digested plasmid pEGFP-Pem1-Ad2, 10 ng of pCherry (Clontech, Mountain View, CA, USA) as an internal control and 0.8 µg of pFlag or wt/mutant Cdx2 encoding plasmid. Cells were analyzed on the FACScalibur machine (BD Biosciences, Mountain View, CA) using a green-versus-red fluorescent plot. Cells with repaired pEGFP-Pem1-Ad2 DNA are identified in the up-right panel. Results are represented as the percentage of green and red cells in the red cells population.
In vitro DNA repair assay
Cell free extract was prepared as described (45). Briefly, 48 h after transfection, 3 × 107 cells were washed in cold PBS, resuspended in four packed cell volumes of cold hypotonic lysis buffer A (10 mM Tris–HCl pH 7.6, 1 mM DTT, 5 mM MgCl2, 1 mM EDTA and protease inhibitors), incubated for 40 min on ice and then disrupted using a Dounce homogenizer (40 strokes). Subsequently, sucrose was added to reach a final concentration of 250 mM. The extract was homogenized and centrifuged 10 min at 1000 g to discard cell debris, and then washed again in the hypotonic lysis buffer containing 250 mM sucrose. Nuclear pellet was then resuspended in two volumes of nuclear extraction buffer B (20 mM Tris–Hcl pH 7.6, 1 mM DTT, 2 mM EDTA, 20% glycerol, 500 mM NaCl and protease inhibitors) and incubated for 30 min on ice. Nuclear extracts were then clarified 30 min at 21 000 g and dialysed overnight against dialysis buffer D (20 mM Tris–HCl pH 7.6, 1 mM EDTA, 1 mM DTT, 20% glycerol, 25 mM NaCl and 0.2 mM PMSF).
End-joining reaction was performed in a final volume of 30 µl by incubating 50 ng of BamHI digested pcDNA3 plasmid with 5 µg of nuclear extract for 1 h at 37°C in buffer E (50 mM Tris–HCl pH 7.6, 5 mM MgCl2, 1 mM ATP, 1 mM DTT, 50 µM dNTP, 80 mM NaCl and protease inhibitors). Reaction was stopped by RNAse treatment (0.25 µg µl−1 of RNAse for 10 min at 37°C) followed by a proteinase K treatment (0.5% SDS, 50 mM EDTA and 1 µg µl−1 of proteinase K at 37°C for 1 h). DNA was purified by phenol and chloroform extraction and recovered by ethanol precipitation, separated on a 0.85% agarose gel at 2 V cm−1 and visualized by UV light.
Wortmanin (Sigma Aldrich, St Louis, MO, USA) was prepared in DMSO at 10 mM and diluted in 10% DMSO to 20 µM immediately before use. Diluted wortmanin (1 µl) was added to the reaction and incubated for 10 min at 4°C prior the addition of ATP and DNA.
DNA ends-binding activity of Ku70
Preparation of nuclear extracts and assessment of DNA ends-binding activity of Ku70 were carried out using a Nuclear Extract kit and a Ku70/Ku80 DNA Repair Protein ELISA kit (Active Motif, Carlsbad, CA, USA). In brief, 3 × 106 HCT116 cells were transfected with 8 µg of pFlag or pFlag-Cdx2 plasmid and nuclear proteins were extracted following the kit instructions. Equivalent amounts of proteins (5 μg) from each lysate were loaded into wells coated with blunt ends linear oligonucleotides that can be specifically bound by Ku70 protein. Amount of bound Ku70 was quantified by using the provided anti-Ku70 primary antibody and the secondary HRP-conjugated antibody in a sensitive colorimetric assay using the Universal Microplate Reader EL800UV (Biotek Instruments, Winooski, VT, USA).
DNA-PKcs activity assay
At 48 h after Cdx2 induction by Doxycyclin, cells were treated with etoposide (50 µM), neocarzinostatin (200 ngml−1; Sigma Aldrich, St Louis, MO, USA) or DMSO for 1 h. The amount of p53 phosphorylated on serine 15 is measured following the PathScan® Sandwich ELISA Kit instructions (Cell Signaling Technology, Danvers, MA, USA). Briefly, an aliquot of 10 ng ml−1 of cell extract is added to a microwell coated with p53 antibodies and incubated overnight at 4°C. The amount of phospho-p53 is quantified by spectrophotometry by using a phosphoSer15-p53 specific antibody revealed by the provided HRP-conjugated secondary antibody. The absorbance is read at 450 nm with the Universal Microplate Reader EL800UV (Biotek Instruments, Winooski, VT, USA).
RT-qPCR
Total RNA was isolated using TRI-Reagent (Ambion Applied Biosystem, Houston, TX, USA). For reverse transcription, we used the high capacity reverse transcription kit (Applied Biosystems, Carlsbad, CA, USA) and 2 µg of RNA. For qPCR, we used TaqMan Universal PCR Master Mix (Applied Biosystems, Carlsbad, CA, USA), TaqMan human TBP, CDH17 and SI probes (TBP, Hs99999910_m1; CDH17, Hs00184865_m1; SI, Hs00356112_m1; Applied Biosystems, Carlsbad, CA), and 7500 Real-Time PCR system (Applied Biosystems, Carlsbad, CA, USA). Results analysis was performed using the Applied 7500 Software v2.0.1.
Statistics
The results were analyzed using GraphPad Prism version 5.0. Mean of results of at least three independent experiments with SEM is generally represented. The statistical difference of the mean was analyzed using Student's paired two-tailed t-test.
RESULTS
Identification of Ku70/Ku80 proteins as specific partners of Cdx2 in intestinal cells
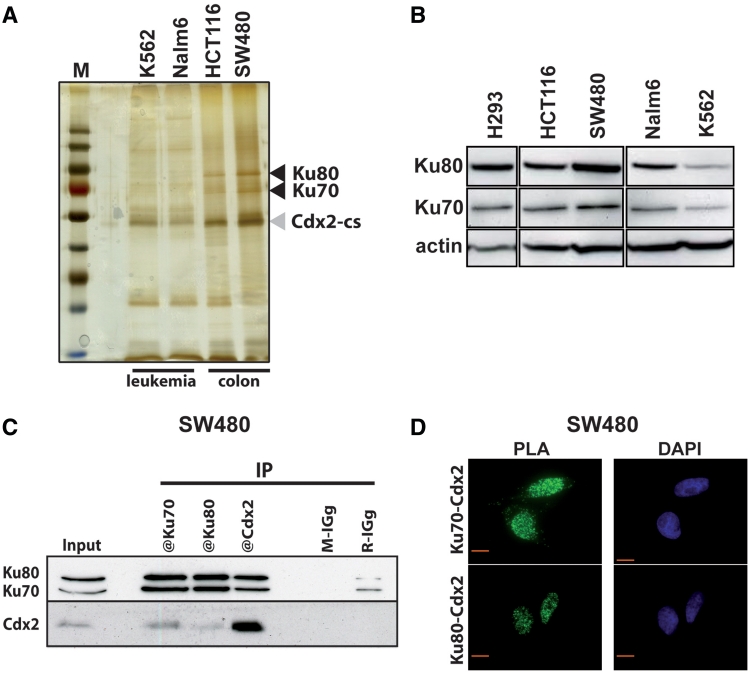
In an attempt to identify Cdx2 interacting proteins, we used the TAP-Tag technology (Tandem Affinity Purification). For this purpose, we expressed a double tagged Cdx2 protein, containing at its C-terminus end CBP (Calmodulin Binding Protein) and SBP (Streptavidin Binding Protein) motives, in two human colon cancer cell lines that express endogenous Cdx2 either at high (SW480) or low levels (HCT116), and in two leukemia cell lines that also express Cdx2 at intermediate (Nalm6) or very low levels (K562). The purified proteins were separated on SDS–polyacrylamide gels and silver stained. This revealed distinct patterns in intestinal and leukemia cells, mainly in the region corresponding to proteins of 70–90 kDa. Indeed, two bands appeared in this region in both intestinal cell lines but were virtually absent in the two leukemia cell lines (Figure 1A). Mass spectrometry analysis of the proteins present in this region identified the Ku70 and Ku80 proteins, specifically in the intestinal cells as opposed to leukemia cells. This result combining TAP-Tag purification and mass spectrometry analysis was repeated in three separate experiments.
Figure 1.
Cdx2 interacts with Ku70/Ku80 proteins. (A) Silver-stained protein gel obtained after tandem affinity purification from K562, Nalm6, HCT116 and SW480 cells transfected with pCTAP-Cdx2. M: molecular weight protein ladder. Cdx2-CBP-SBP (Cdx2-cs) and Ku70/Ku80 proteins are indicated with gray or black arrowheads, respectively. (B) Western blot revealing the presence of Ku70 and Ku80 proteins in all studied cell lines. Nearly 20 µg of total proteins were separated on a gradient SDS–PAGE. Actin was used as loading control. (C) Coimmunoprecipitation of endogenous Cdx2 and Ku proteins. SW480 extracts were prepared and Cdx2, Ku70 or Ku80 were immunoprecipitated using corresponding antibodies. Mouse IGg (M-IGg) or rabbit IGg (R-IGg) were used as negative control. Western blot was revealed using rabbit anti-Ku70 and rabbit anti-Ku80 in the upper panel and using mouse anti-Cdx2 in the lower panel. About 20 µg of protein extracts were loaded in the input line. (D) In situ proximity ligation assay. Mouse anti-Cdx2 and rabbit anti-Ku70 (left panel) or mouse anti-Cdx2 and rabbit anti-Ku80 antibodies (right panel) were used to reveal endogenous proteins in SW480 cells. Green fluorescence corresponds to the PLA positive signal and indicates that the two molecules belong to the same protein complex; blue fluorescence corresponds to nuclei (DAPI staining). Red bar is 10 µm.
We wondered whether the different Cdx2 interaction patterns observed in intestinal and leukemia cells resulted from a different level of expression of Ku70 and Ku80 in these cells. Western blot analysis revealed that both proteins were expressed at nearly similar levels in intestinal and leukemia cells (Figure 1B), indicating that the absence of interaction with Cdx2-CBP-SBP in leukemia cells did not reflect a loss of Ku70 or Ku80 expression, but instead a differential interaction capacity.
The interaction of Cdx2 with Ku70/80 uncovered by TAP-Tag purification in intestinal cells used transfected cells to produce high levels of Cdx2-CBP-SBP. Two series of experiments were performed to rule out any technical artifact and to actually demonstrate the interaction at the level of the endogenous proteins. For this purpose, we used the intestinal SW480 cells that express both Ku70/80 and Cdx2 proteins. First, coimmunoprecipitations of endogenous proteins were performed. As illustrated in Figure 1C, antibodies raised against Ku70 or Ku80 were able to coimmunoprecipitate Cdx2, and reciprocally, anti-Cdx2 antibody coimmunoprecipitated the Ku70 and Ku80 proteins. These results were confirmed in DLD1, an intestinal cell line that also expresses endogenous Cdx2 at a relatively high level, and in HT29 and HCT116 cells after overexpression of Cdx2 since these two intestinal cell lines express only barely detectable levels of endogenous Cdx2 (Supplementary Figure S1 and Figure 2). Second, we used the Proximity Ligation Assay (PLA) technology that is appropriate to reveal the proximity of two proteins in situ in the cells (23). The Figure 1D shows that both anti-Ku70 and anti-Ku80 antibodies produced a positive fluorescent signal in the nuclei of SW480 cells when combined with anti-Cdx2 antibody, indicating the interaction of the two proteins in this cell compartment. Altogether, these results demonstrate that the Ku70, Ku80 and Cdx2 proteins can associate within a same nuclear multiprotein complex in intestinal cells.
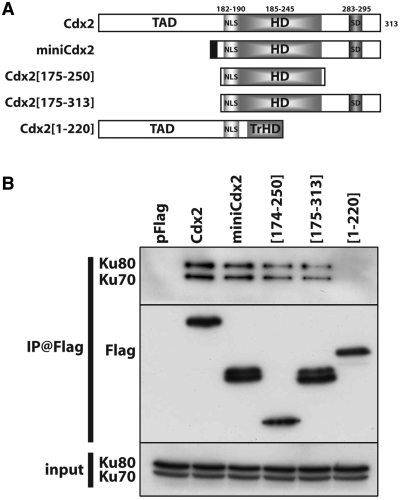
Figure 2.
The homeodomain of Cdx2 is necessary for its interaction with Ku70/Ku80. (A) Schematic representation of the different Cdx2 deletion mutants tested. TAD: transactivator domain; NLS: nuclear localization signal; HD: homeodomain; SD: stabilization domain. The upper numbers represents the position of the different domains in the 313aa long Cdx2 protein. (B) Coimmunoprecipitations of the different mutant proteins with Ku proteins. Co-IP was performed in HCT116 transfected with the indicated mutant plasmids using anti-Flag antibodies. Immunoprecipitates were separated on a SDS–PAGE and analyzed by western blot using anti-Ku70 and anti-Ku80 antibodies (upper panel), anti-Flag antibodies (middle panel). Ku70 and Ku80 proteins present in the protein extracts before immunoprecipitation were revealed by western blot (input, lower panel).
Coimmunoprecipitations performed in Nalm6 and K562 leukemia cells failed to reveal any interaction between Cdx2 and Ku proteins, consistent with the absence of the Ku protein in the TAP-Tag experiments (Supplementary Figure S1).
Cdx2 interacts with Ku70/Ku80 via the third α-helix of the homeodomain
Two homeoproteins of the Hox clusters, HoxB7 and HoxC4, have been shown to interact with Ku70, and the homeodomain was demonstrated to be essential for this interaction (24,25). To delineate the domain of interaction with Ku proteins within the Cdx2 homeoprotein, we constructed plasmids encoding truncated forms of Cdx2, which were all Flag-tagged at their N-terminus: the miniCdx2 form corresponds to a splicing variant of Cdx2 in which the transactivation domain is replaced by a 13 amino acids extension at its N-part (26); the mutant Cdx2[175–313] overlaps the homeodomain and the C-terminal regulatory region but lacks the transactivation domain; the mutant Cdx2[1–220] contains the transactivation domain and the first two α-helixes of the homeodomain but has lost the third α-helix and the downstream regulatory region; the mutant Cdx2[175–250] contains the full-length homeodomain without the N-ter transactivation domain and the C-ter regulatory domain (Figure 2A). After transfection of these plasmids into HCT116 colon cancer cells, coimmunoprecipitation experiments were performed using anti-Flag antibody. The full-length Cdx2 protein, the miniCdx2 variant, and the mutants containing an intact homeodomain, Cdx2[175–250] and Cdx2[175–313], co-immunoprecipitated Ku70 and Ku80; however, the mutant lacking the third α-helix of the homeodomain, Cdx2[1–220], did not interact with Ku proteins (Figure 2B). These data indicate that the homeodomain is involved in the interaction of Cdx2 with the Ku70/80 complex and that the third α-helix is required for this interaction.
The Ku70/Ku80 complex does not alter Cdx2 transcription activity
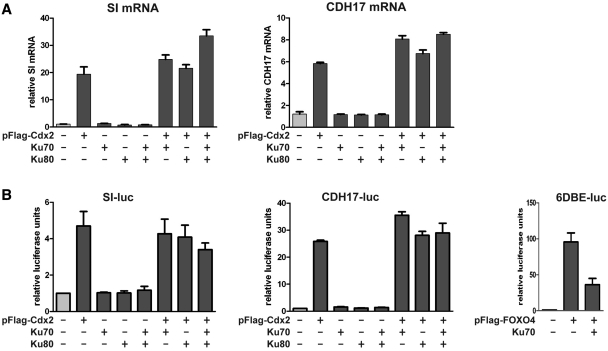
To take up the biological relevance of the interaction between Cdx2 and Ku70/80, we first addressed the issue of the impact of Ku proteins on the transcriptional activity of Cdx2. For this goal, HCT116 cells were transfected with the plasmid encoding Cdx2 together with combinations of plasmids encoding Ku70 and Ku80. Then, the expression of two well-established targets of Cdx2, the liver–intestine cadherin (CDH17) and the sucrase-isomaltase (SI) genes was analyzed by RT-qPCR. Cdx2 alone strongly stimulated the expression of both genes, and the level of stimulation was not significantly modified in the presence of Ku70 and/or Ku80 (Figure 3A). To corroborate these results, HCT116 cells were cotransfected with reporter luciferase plasmids containing the promoter of the CDH17 or SI genes with their Cdx2-responding elements. Again, Cdx2 stimulated the basal activity of these promoters, but Ku70 and/or Ku80 did not significantly change the stimulatory effect exerted by Cdx2 (Figure 3B, left and middle panels). In the same conditions, Ku70 inhibited the transcriptional activity of the FOXO4 transcription factor (Figure 3B, right panel), as previously described (27). We conclude from these data, that, albeit Ku70/80 interacts with Cdx2, it does neither stimulate nor inhibit its transcriptional activity.
Figure 3.
Ku proteins do not affect significantly Cdx2 transcriptional activity. (A) Stimulation of endogenous Cdx2 target genes. HCT116 were transfected with 1 µg of each plasmid in a combined manner as indicated and RNA were extracted and analyzed by RT-qPCR using CDH17, SI or TBP TaqMan probes. Results are represented as fold induction relative to pFlag (normalized with TBP expression). Data correspond to the mean of several experiments and error bars represent SEM (n = 2). (B) Stimulation of Cdx2 reporter plasmids. HCT116 were transfected with CDH17-luc or SI-luc reporter plasmids and with indicated expression plasmids. Protein extracts were analyzed for dual luciferase activity. Relative promoter activity (normalized with Renilla values) of reporter plasmid cotransfected with empty plasmids was set at 1. Data correspond to the mean of several experiments and error bars represent SEM (n = 3). Right panel: FOXO4 transcription factor was used as positive control for Ku70 inhibition on the 6DBE-luc reporter plasmid as described in Brenkman et al. (27).
Cdx2 inhibits in vitro DNA repair specifically in intestinal cell lines
We then asked whether Cdx2 may affect the function of Ku70/80 proteins. The Ku70/80 heterodimer is largely involved in DNA repair. Indeed, the mechanism of DNA repair primarily activated by DSBs, namely C-NHEJ, is dependent on Ku70/80, on the wortmanin-sensitive DNA-PK and on ATP. Ku protein heterodimers bind and encircle broken DNA, allowing the recruitment of DNA-PKcs and DNA-ligase IV. Under conditions preventing the activation of C-NHEJ, for example when Ku70/80 heterodimers fail, an alternative pathway, named A-NHEJ, can be turned on to compensate the missing activity of C-NHEJ. Whereas C-NHEJ is primed by the binding of Ku70/80 on the broken DNA ends, A-NHEJ is activated in the absence of Ku70/80 by the recognition of the DNA ends by PARP-1 (28,29).
DNA repair can be assayed in vitro by incubating linear DNA with nuclear extracts and measuring the appearance of slow-migrating electrophoretic DNA fragments corresponding to circularized and multimeric forms (30). We set up this assay and confirmed previous data by Fattah et al. (28) showing that the C-NHEJ pathway is predominant in intestinal HCT116 cells since DNA repair is wortmanin-sensitive and insensitive to PARP1 antibody (Figure 4A, left). In addition, when cells were transfected with siRNA@Ku70 to deplete the nuclear extracts in Ku70, the DNA repair activity became wortmanin-insensitive, but significantly decreased in the presence of anti-PARP-1 antibody (Figure 4A, right), indicating that, in these conditions, DNA repair involves the A-NHEJ instead of the C-NHEJ pathway.
Figure 4.
Cdx2 inhibits in vitro DNA repair in intestinal cells but not in leukemia cells. (A) NHEJ-C predominates in HCT116 cells. Linear DNA was incubated with nuclear extracts of HCT116 cells transfected with the siRNA against Ku70 or control siRNA. Processed plasmids were separated from the linear DNA by gel electrophoresis. When indicated, wortmanin or anti-PARP-1 antibodies were added to the reaction. (B) Cdx2 inhibits in vitro DNA repair in HCT116 cells. HCT116 cells were transfected with empty vector or pFlag-Cdx2 plasmids and nuclear extracts were assayed for in vitro DNA repair. When indicated (-ATP), ATP was omitted in the reaction. (C) Cdx2[1–220] mutant does not inhibits DNA repair. HCT116 cells were transfected with empty vector or Cdx2 mutant plasmids as indicated and nuclear extracts were assayed for in vitro DNA repair. When indicated (-ATP), ATP was omitted in the reaction. Above, expression of Cdx2 and its mutants was checked by western blot on 10 µg of whole-protein extracts using anti-Flag antibody. Actin was used as loading control. (D) Cdx2 does not alter in vitro DNA repair in Nalm6 cells. Nalm6 cells were transfected with empty vector or pFlag-Cdx2 plasmids and nuclear extracts were assayed for in vitro DNA repair. Gray arrows indicate linear plasmid and black arrows multimeric forms of the plasmid.
We used this in vitro DNA repair assay to investigate if Cdx2 has any effect on the NHEJ activity in intestinal cells. For this purpose, nuclear extracts were prepared from HCT116 cells transfected either with the pFlag2 vector as control or with expression plasmids encoding Cdx2 or truncated forms of the protein. As shown in Figure 4B, the DNA repair activity observed by incubating linear DNA with control nuclear extracts was inhibited when Cdx2 was present in the extracts. Omitting ATP in the reaction mix also strongly compromised DNA repair activity. Consequently, since Cdx2 inhibits the overall DNA repair activity while Ku70/80 is present in the nuclear extracts, we concluded that Cdx2 compromises C-NHEJ activity without turning on the A-NHEJ pathway.
To strengthen this conclusion, we analyzed the dependency of the inhibitory effect exerted by Cdx2 on the interaction between Cdx2 and Ku70/80. DNA repair activity determined using the in vitro assay was inhibited by Cdx2 as well as miniCdx2 (Figure 4C) which both interacted with Ku70/80 (Figure 2B). Inversely, the truncated Cdx2[1–220] mutant, that failed to interact with Ku70/80, did not inhibit DNA repair, demonstrating that the physical interaction between Cdx2 and the Ku70/80 complex is required for Cdx2 to exert its inhibitory effect of C-NHEJ activity.
Having demonstrated on the one hand that Cdx2 interacts with Ku70/80 in intestinal cells to inhibit the Ku70/80-dependent DNA repair activity via the C-NHEJ pathway, and on the other hand that, unlike colon cancer cells, Cdx2 does not interact with Ku70/80 in leukemia cells, we checked whether Cdx2 has any effect on the DNA repair activity measured in blood cells using the in vitro assay. To this end, linear DNA was incubated with nuclear extracts of Nalm6 cells transfected with the Cdx2 expressing or with the control pFlag2 plasmids and the resulting DNA fragments were analyzed by gel electrophoresis. The Figure 4D shows that Cdx2 failed to inhibit DNA repair activity in leukemia cells, despite its high level of expression, which clearly contrasted with the results obtained in colon cancer cells.
Cdx2 inhibits in vivo DNA repair in intestinal cell lines
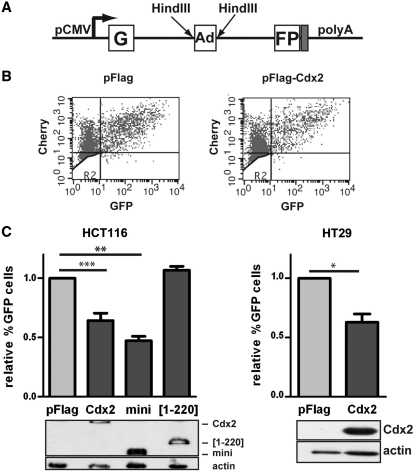
Next, we analyzed DNA repair in vivo, using the model developed by Wang et al. (29). This reporter system is based on the plasmid pEGFP-Pem1-Ad2, consisting in the GFP-encoding cDNA sequence interrupted by an intron containing itself an adenovirus exon flanked by two HindIII restriction sites (Figure 5A). When this plasmid is cut with HindIII and transfected into cells, it presents cohesive ends mimicking DSBs, which are then processed by the NHEJ machinery to circularize the plasmid and subsequently allow the expression of the reporter GFP after splicing of the intron.
Figure 5.
Cdx2 inhibits DNA repair in vivo in intestinal cells. (A) Schematic representation of the pEGFP-Pem1-Ad2 plasmid. G and FP corresponds to the 5′ and 3′ part of the GFP encoding cDNA. Ad corresponds to the adenoviral intron flanked by two HindIII sites. pCMV indicates the CMV promoter. (B) Example of green-versus-red fluorescent plots. The experiment was performed in HCT116 transfected with pFlag (left panel) or pFlag-Cdx2 (right panel). X axis: green fluorescence; Y axis: red fluorescence. (C) Cdx2 alters the in vivo DNA repair. Results represent the mean of at least four experiments performed in HCT116 (left panel) or HT29 (right panel) transfected with either pFlag, pFlag-Cdx2, pFlag-miniCdx2 or Cdx2[1-220] plasmids as indicated. The percentage of GFP and Cherry double positive cells in the Cherry positive cell population was set at 1 for pFlag and error bars represent SEM. Asterisk indicate a significant difference (**P < 0.01; *P < 0.05). Above, expression of Cdx2 and its mutants was checked by western blot on 10 µg of whole protein extracts using anti-Flag antibody. Actin was used as loading control.
To evaluate DNA repair activity in vivo, intestinal cells were cotransfected with the HindIII-linearized plasmid pEGFP-Pem1-Ad2 and the plasmid encoding the red fluorescent protein Cherry to normalize the results. The number of GFP and Cherry positive cells were quantified by FACS 72 h after transfection and an example of the green-versus-red fluorescent plot is illustrated on Figure 5B. When HCT116 cells were cotransfected with the Cdx2-encoding plasmid, the number of GFP-positive cells was significantly reduced when compared with cells cotransfected with the control vector pFlag2, indicating that the presence of Cdx2 alters the activity of DSB repair in vivo (Figure 5C, left panel). This result was confirmed in HT29 cells (Figure 5C, right panel). Moreover, miniCdx2, shown above to interact with Ku70/80 like Cdx2, had the same inhibitory effect as Cdx2, whereas the truncated form Cdx2[1–220] that lacks the ability to interact with Ku70/80 proteins, failed to inhibit DNA repair activity in vivo (Figure 5C, left panel).
Neither Ku proteins and DNA-PKcs recruitment nor DNA-PKcs autophosphorylation are affected by Cdx2, but Cdx2 inhibits DNA-PK activity
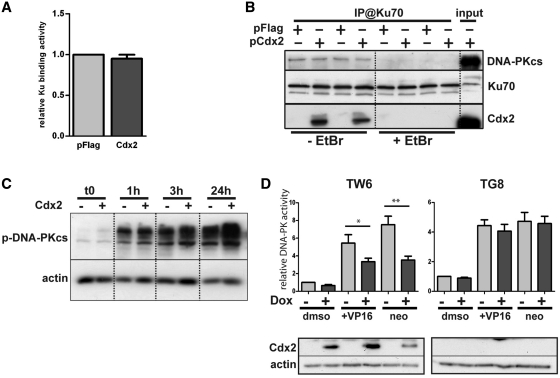
DNA repair by the C-NHEJ machinery involves sequential steps of recruitment and activation of several proteins. The first one consists in the recruitment of Ku70/80 heterodimers on the DNA breaks. Because Cdx2 interacts with Ku70/80 and inhibits DNA repair, we wondered whether this interaction perturbed Ku70/80 recruitment on the broken DNA ends. To address this issue, we used an ELISA method to quantify the amount of Ku70 protein bound on immobilized linear double-strand DNA molecules mimicking broken DNA ends. Nuclear extracts were prepared from HCT116 cells transfected with the Cdx2 encoding or pFlag2 control plasmid and incubated with the immobilized DNA molecules. As illustrated in Figure 6A, the presence of Cdx2 did not change the ability of Ku70 to bind DNA ends, suggesting that Cdx2 does not alter the recruitment of Ku proteins onto broken DNA.
Figure 6.
Cdx2 does alter neither Ku proteins and DNA-PKcs recruitment nor DNA-PK autophosphorylation but inhibits DNA-PKcs activity. (A) Cdx2 does not interfere with DSB Ku70 binding. HCT116 cells were transfected with pFlag (light gray) or Cdx2 (dark gray) expressing plasmid and Ku70 binding activity was assessed. Results represent the mean of at least four independent experiments. pFlag values were set at 1. (B) Cdx2 does not alter DNA-PKcs recruitment. HCT116 were transfected with either pFlag or pFlag-Cdx2 as indicated and immunoprecipitation was performed using anti-Ku70 antibodies in the presence or absence of Ethidium Bromide (EtBr) as indicated at the bottom of the figure. DNA-PKcs, Ku70 or Cdx2 were revealed by western blot. Twenty micrograms of whole-protein extract were loaded on the right line (input). (C) Cdx2 does not modify DNA-PKcs autophosphorylation. Time course analysis of the phosphorylation of the DNA-PKcs after etoposide treatment 100 μM for 1 hour of HCT116 cells transfected with pFlag or pFlag-Cdx2. Phospho-DNA-PKcs was revealed by western blot. β-actin was used to normalized the amount of loaded proteins. (D) Cdx2 inhibits DNA-PKcs activity. DNA-PKcs activity was assessed in HT29-TW6 or -TG8 (control) cells treated or not with etoposide (VP16) 100 μM or neocarzinostatin 200 ng/μL for 1 hour. Cdx2 expression was induced with doxycyclin when indicated (dark gray bars). Experiments without doxycyclin and etoposide treatment were considered as references and results were set at 1. Results illustrate the mean of at least six experiments and error bars represent SEM. Asterisk indicates a significant difference (**P < 0.01; *P < 0.05). Above, Cdx2 expression was checked by western blot using anti-Cdx2 antibody on 10 µg of whole-protein extracts. Actin was used as loading control.
Once Ku70/80 proteins have recognized the broken DNA ends, DNA-PK is recruited into the repair complex and activated by autophosphorylation (31,32). Therefore, we followed the recruitment of DNA-PKcs by coimmunoprecipitation with Ku70 antibody in the presence or absence of Cdx2. As illustrated on Figure 6B, the presence of Cdx2 in HCT116 cells transfected with the Cdx2-encoding plasmid did not change the level of DNA-PKcs coimmunoprecipitated with Ku70 antibody, as compared to cells transfected with the control plasmid pFlag2. As expected, adding ethidium bromide (EtBr) in the protein extracts disrupted Ku70/DNA-PKcs interaction; EtBr also compromised Ku70/Cdx2 interaction, suggesting that it is DNA dependant (Figure 6B).
We then explored the capacity of the DNA-PKcs to autophosphorylate. We compared the Thr2609 phosphorylation of DNA-PKcs after etoposide (VP16) treatment of HCT116 cells transfected with the Cdx2 encoding or control pFlag2 plasmids. As expected, DSBs induced by etoposide rapidly enhanced Thr2609 phosphorylation of DNA-PK which persisted over 24 h; the presence of Cdx2 did neither significantly modify the level nor the time-course of phosphorylation (Figure 6C).
DNA-PK phosphorylates several intracellular proteins, including proteins of the NHEJ pathway as Artemis, Ku70 and Ku80, H2AX and numerous nuclear DNA-binding proteins such as tumor suppressor protein p53 (33). We investigated the phosphorylating activity of DNA-PKcs in an assay based on in vitro solid phase sandwich ELISA that captures total endogenous p53 protein and detects the level of p53 phosphorylated on Ser15. Inducible HT29-TW6 and HT29-TG8 cell lines producing either Cdx2 or GFP as control upon induction with doxycyclin (15) were either uninduced or treated with doxycyclin and then treated for one hour with etoposide or neocarzinostatin to induce DSB or with the vehicle as control. Nuclear extracts were prepared and tested for their DNA-PK phosphorylation activity. As presented on Figure 6D, the level of phosphorylation of the DNA-PKcs target was significantly reduced when Cdx2 expression was induced by doxycyclin, suggesting that Cdx2 inhibits the DNA-PK phosphorylation activity.
Cdx2 inhibits cells survival after etoposide treatment in colon cancer cells but not in leukemia cells
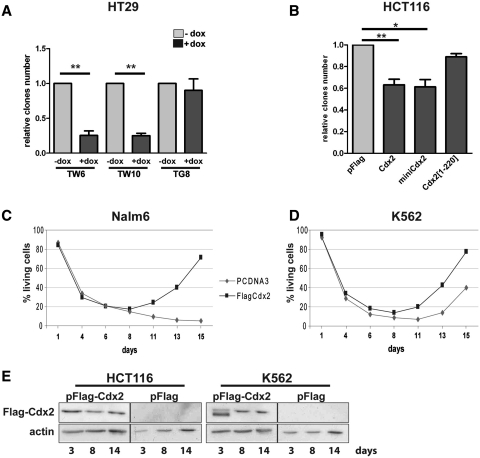
Alteration of DBS repair compromises cell survival following a genotoxic stress, since the accumulation of broken DNA ends induces apoptosis (34). Because Cdx2 inhibits DNA repair in colon cancer cells but not in leukemia cells, we tested the consequence of its expression on cell survival after a genotoxic stress in both types of cells. For this purpose, we used on the one hand the human colon cancer cell lines HT29-TW6 and -TW10 in which Cdx2 expression can be switched on by Doxycyclin (15), and on the other hand, HCT116 cells transiently transfected with the Cdx2-encoding plasmid. Cells were treated for 24 h with the topoisomerase II inhibitor etoposide so as to induce DSBs and allowed to recover for 2 weeks. Cdx2 expression was followed by western blot and was confirmed even at 14-days post-transfection (Figure 7E, left panel). Cell survival was quantified by measuring the number of remaining colonies. As illustrated in Figure 7A, the induction of Cdx2 expression by Doxycyclin strongly reduced cell survival in the HT29-TW6 and -TW10 lines as compared to cells treated with the vehicle, whereas the survival of the control line HT29-TG8 expressing GFP instead of Cdx2 was unchanged upon Doxycyclin treatment. Similarly, the transient expression of Cdx2 reduced the survival of etoposide-treated HCT116 cells (Figure 7B). The effect was less pronounced in HCT116 than in HT29 cells likely because the efficacy of Cdx2 expression was lower by transient cell transfection than in stable cell lines homogeneously treated with Doxycyclin. MiniCdx2, which interacts with Ku70/80 too but lacks the transactivating domain, also compromised HCT116 cell survival after etoposide treatment, thus indicating that this effect is not dependent on the transcriptional activity of Cdx2. Moreover, the truncated mutant Cdx2[1–220] that does not interact with Ku proteins, failed to reduce cell survival, in contrast to Cdx2 and miniCdx2.
Figure 7.
Cdx2 inhibits cell survival in intestinal cells but rather enhances cell recovery in leukemia cells. (A) Cdx2 inhibits cell survival in HT29 cells. HT29-TW6, -TW10 or -TG8 were treated with etoposide in the presence (dark gray) or not (light gray) of doxycyclin and cultured for 2 weeks. (B) Cdx2 inhibits cell survival in HCT116 cells. HCT116 cells were transfected with either pFlag, pFlag-Cdx2, pFlag-miniCdx2 or pFlag-Cdx2[1-220] plasmids as indicated, treated with etoposide and cultured for 2 weeks. For A and B, the numbers of clones are represented as the mean of four independent experiments. Values corresponding to -dox for HT29 or pFlag for HCT116 were set at 1 and error bars represent SEM. Asterisks indicate a significant difference (**P < 0.01; *P < 0.05). (C) and (D) Cdx2 does not alter cell survival in leukemia cells. Nalm6 or K562 cells were transfected with vehicle or pFlag-Cdx2 and cell death number was quantified using annexinV-FITC/propidium iodide at different times after etoposide treatment as indicated. Results are represented as percentage of living cells. For C and D, results of a representative experiment performed in triplicates are shown. (E) Control of Cdx2 protein expression. At 3, 8, or 14 days after transfection with indicated plasmids, HCT116 or K562 cells were lysed in laemmli buffer and Cdx2 expression was tested by western blot on 10 µg of whole protein extracts using anti-Flag antibody. Actin was used as loading control.
Next, these experiments were repeated in leukemia cells instead of colon cancer cells and cell apoptosis was followed by FACS after AnnexinV labeling. Results present the percentage of living cells. In both K562 and Nalm6 cell lines, etoposide induced a strong decline of living cells during the first week, whether transfected with the pFlag2 control vector or with the Cdx2-encoding plasmid (Figure 7C and D). Protein expression in K562 was confirmed by western blot at 14 days after transfection (Figure 7E, right panel). Interestingly, from 1 week after treatment onwards, cells transfected with Cdx2 entered again a growing phase, in contrast to the cells transfected with the control plasmid. These results indicate that, unlike colon cancer cells, Cdx2 does not compromise cell survival after genotoxic stress in leukemia cells, and that it even facilitates cell recovery and/or growth at long term. Hence the effect of Cdx2 on the response to a genotoxic stress is different in colon cancer cells compared to leukemia cells.
DISCUSSION
The homeoprotein Cdx2 is recognized as a classical transcription factor that acts via DNA binding to a consensus AT-rich sequence and activation of transcription by the transactivation domain (7,35,36). In the present study, we uncovered a new role for Cdx2 in regulating DNA repair and we provide evidence that this function does not require its transactivation domain. This new role depends on the interaction of Cdx2 with the Ku70/80 proteins and subsequently the inhibition of the NHEJ DNA repair machinery, independently of the transcriptional activity of Cdx2. Moreover, this new function is cell type specific since it is active in intestinal cells where the gene is normally expressed, but inactive in leukemia cells where Cdx2 becomes ectopically turned on only during the transformation process of hematopoietic cells. This differential function in intestinal and leukemia cells is correlated to the cell specific ability of Cdx2 to interact with Ku70/80. The reason why Cdx2 is unable to interact with Ku70/80 in leukemia cells remains to be elucidated. One possibility is that Cdx2 and/or Ku70/80 undergo different post-translational modifications that differentially control their interaction in intestinal compared to leukemia cells. Another possibility is that the interaction between Cdx2 and Ku70/80 is indirect and depends on an intermediary factor present in intestinal but not in leukemia cells.
The binding of Ku70/80 to broken DNA ends is the first step that initiates DNA repair by the conventional C-NHEJ machinery. In the presence of Cdx2, neither the binding of Ku70/80 to the DNA nor the recruitment of DNA-PKcs is altered despite the interaction of Cdx2 with the Ku70/80-DNA-PKcs complex; however, the kinase activity of the DNA-PK, a major component of C-NHEJ recruited by Ku70/80, is reduced. This ultimately results in a decreased DNA repair demonstrated here using in vivo and in vitro assays. Because Cdx2 does not compromise DNA recognition by Ku70/80, the Ku70/80 occupancy of the broken DNA ends prevents the recruitment of PARP-1 and thus the activation of the alternative A-NHEJ pathway normally activated in case of the low activity of C-NHEJ. Therefore, in the presence of Cdx2, the reduced activity of C-NHEJ is not compensated by A-NHEJ.
An increasing number of studies documents new functions for homeoproteins besides their primary role of transcription factors. Especially, several classical or distantly-related homeodomain-containing proteins interact with Ku proteins, such as HoxC4, HoxD4, Oct-1, Oct-2 or Dlx2, and are phosphorylated by the DNA-PK. Yet, the functional significance of this interaction remains elusive (25,37). The case of the homeoprotein HoxB7, which is also a partner of Ku70/80 (24), is of particular interest regarding the results we have obtained for Cdx2. Indeed, HoxB7 elicits a transformed phenotype in mammary cancer cells, increases the cell resistance to a genotoxic stress, and interacts with the Ku70/80-dependent NHEJ machinery to stimulate DNA repair activity (24); this signifies that the stimulation of DNA repair by NHEJ is linked to tumor progression, likely because of the genetic alterations caused by this error-prone DNA repair pathway. Inversely, the interaction of Cdx2 with Ku70/80 in intestinal cells inhibits the NHEJ DNA repair activity and hampers cell recovery after a genotoxic stress, associated to a tumor suppressor function of Cdx2 in the gut. Despite their opposite effect on NHEJ activity, HoxB7 and Cdx2 both interact with Ku70/80 through the third α-helix of the homeodomain. Further studies will allow determining the mechanisms by which the homeodomains of HoxB7 and Cdx2 act, respectively, to stimulate or inhibit the NHEJ activity.This study supports the hypothesis that the opposite effects reported for Cdx2 in colon cancers and leukemia, respectively, as tumor suppressor and oncogene, relies at least in part on the fact that this homeoprotein interacts with a different set of partners depending on the cellular context. Indeed, in leukemia cells the ectopically expressed Cdx2 protein is not able to interact with Ku proteins and to inhibit the activity of the error-prone pro-oncogenic NHEJ pathway, whereas in colon cancer cells the interaction of Cdx2 with the NHEJ machinery reduces the DNA repair activity. In the normal gut, Cdx2 is expressed in most of the epithelial cells including the stem cells and transit amplifying cells. On the basis of the results shown here, we propose that in this rapidly renewing system, there is a benefit to prevent DNA repair by NHEJ because of the risk of fixing oncogenic mutations, whereas when cells keep the DNA breaks unrepaired, they are eliminated by gatekeeper pathways. Indeed, several lines of evidence suggest that aberrant repair of DSBs by error-prone NHEJ could contribute to the accumulation of genomic alterations, as in the case of Bloom syndrome for example, and lead to chromosomal instability and cancer (38). This is corroborated by our finding that a decreased expression of Cdx2 in Cdx2+/− heterozygote mice lessens apoptosis in the colon epithelium after γ-irradiation (14), thus rendering the gut epithelium more resistant to a genotoxic stress. This is also consistent with the fact that Cdx2 overexpression, together with Cdx1, in human colon cancer cells leads to reduced malignancy associated with higher sensitivity to apoptosis (39). Therefore, the effect of Cdx2 on the NHEJ DNA repair activity may contribute to the tumor suppressor function attributed to this gene in the gut.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figure 1.
FUNDING
Institut National du Cancer (INCa, ACM 07), the Ligue du Haut-Rhin contre le Cancer and the Canceropôle Grand-Est. B.R. was funded by the INCa (ACM 07), and C.S. by the Ministère de l'Enseignement Supérieur et de la Recherche. Funding for open access charge: INSERM academic institution.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr Iliakis for providing the plasmid pEGFP-Pem1-Ad2, Dr van den Broek for FOXO4 and Ku protein expression plasmids and 6DBE-luc construct, Dr Hendrickson for technical advises and H. Domon and M. Maarouf for comments on the manuscript. B.R., C.D.D. and J.N.F. designed the experiments; B.R., C.S., M.V., F.D., E.M., T.S. and C.D.D. performed the experiments; B.R., C.D.D., I.D., I.G. and J.N.F. analyzed the data; I.G. provided essential reagents; C.D.D. and J.N.F. provided funding; C.D.D. and J.N.F. wrote the article.
REFERENCES
- 1.Chawengsaksophak K, James R, Hammond VE, Kontgen F, Beck F. Homeosis and intestinal tumours in Cdx2 mutant mice. Nature. 1997;386:84–87. doi: 10.1038/386084a0. [DOI] [PubMed] [Google Scholar]
- 2.Chawengsaksophak K, De Graaff W, Rossant J, Deschamps J, Beck F. Cdx2 is essential for axial elongation in mouse development. Proc. Natl Acad. Sci. USA. 2004;101:7641–7645. doi: 10.1073/pnas.0401654101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van Den Akker E, Forlani S, Chawengsaksophak K, De Graaff W, Beck F, Meyer BI, Deschamps J. Cdx1 and Cdx2 have overlapping functions in anteroposterior patterning and posterior axis elongation. Development. 2002;129:2181–2193. doi: 10.1242/dev.129.9.2181. [DOI] [PubMed] [Google Scholar]
- 4.Beck F, Chawengsaksophak K, Waring P, Playford RJ, Furness JB. Reprogramming of intestinal differentiation and intercalary regeneration in Cdx2 mutant mice. Proc. Natl Acad. Sci. USA. 1999;96:7318–7323. doi: 10.1073/pnas.96.13.7318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gao N, White P, Kaestner KH. Establishment of intestinal identity and epithelial-mesenchymal signaling by Cdx2. Dev. Cell. 2009;16:588–599. doi: 10.1016/j.devcel.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crissey MA, Guo R, Funakoshi S, Kong J, Liu J, Lynch JP. Cdx2 levels modulate intestinal epithelium maturity and paneth cell development. Gastroenterology. 2011;140:517–528. doi: 10.1053/j.gastro.2010.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Verzi MP, Shin H, He HH, Sulahian R, Meyer CA, Montgomery RK, Fleet JC, Brown M, Liu XS, Shivdasani RA. Differentiation-specific histone modifications reveal dynamic chromatin interactions and partners for the intestinal transcription factor CDX2. Dev. Cell. 2010;19:713–726. doi: 10.1016/j.devcel.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calon A, Gross I, Lhermitte B, Martin E, Beck F, Duclos B, Kedinger M, Duluc I, Domon-Dell C, Freund JN. Different effects of the Cdx1 and Cdx2 homeobox genes in a murine model of intestinal inflammation. Gut. 2007;56:1688–1695. doi: 10.1136/gut.2007.125542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim SP, Park JW, Lee SH, Lim JH, Jang BC, Lee SH, Jang IH, Freund JN, Suh SI, Mun KC, et al. Homeodomain protein CDX2 regulates COX-2 expression in colorectal cancer. Biochem. Biophys. Res. Commun. 2004;315:93–99. doi: 10.1016/j.bbrc.2004.01.020. [DOI] [PubMed] [Google Scholar]
- 10.Mutoh H, Hayakawa H, Sakamoto H, Sugano K. Homeobox protein CDX2 reduces Cox-2 transcription by inactivating the DNA-binding capacity of nuclear factor-κB. J. Gastroenterol. 2007;42:719–729. doi: 10.1007/s00535-007-2088-y. [DOI] [PubMed] [Google Scholar]
- 11.Guo RJ, Funakoshi S, Lee HH, Kong J, Lynch JP. The intestine-specific transcription factor Cdx2 inhibits beta-catenin/TCF transcriptional activity by disrupting the beta-catenin-TCF protein complex. Carcinogenesis. 2010;31:159–166. doi: 10.1093/carcin/bgp213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aoki K, Kakizaki F, Sakashita H, Manabe T, Aoki M, Taketo MM. Suppression of colonic polyposis by homeoprotein CDX2 through its nontranscriptional function that stabilizes p27Kip1. Cancer Res. 2011;71:593–602. doi: 10.1158/0008-5472.CAN-10-2842. [DOI] [PubMed] [Google Scholar]
- 13.Aoki K, Tamai Y, Horiike S, Oshima M, Taketo MM. Colonic polyposis caused by Mtor-mediated chromosomal instability in Apc+/delta716 Cdx2+/- compound mutant mice. Nat. Genet. 2003;35:323–330. doi: 10.1038/ng1265. [DOI] [PubMed] [Google Scholar]
- 14.Bonhomme C, Duluc I, Martin E, Chawengsaksophak K, Chenard MP, Kedinger M, Beck F, Freund JN, Domon-Dell C. The Cdx2 homeobox gene has a tumour suppressor function in the distal colon in addition to a homeotic role during gut development. Gut. 2003;52:1465–1471. doi: 10.1136/gut.52.10.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gross I, Duluc I, Benameur T, Calon A, Martin E, Brabletz T, Kedinger M, Domon-Dell C, Freund JN. The intestine-specific homeobox gene Cdx2 decreases mobility and antagonizes dissemination of colon cancer cells. Oncogene. 2008;27:107–115. doi: 10.1038/sj.onc.1210601. [DOI] [PubMed] [Google Scholar]
- 16.Scholl C, Bansal D, Dohner K, Eiwen K, Huntly BJ, Lee BH, Rucker FG, Schlenk RF, Bullinger L, Dohner H, et al. The homeobox gene CDX2 is aberrantly expressed in most cases of acute myeloid leukemia and promotes leukemogenesis. J. Clin. Invest. 2007;117:1037–1048. doi: 10.1172/JCI30182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thoene S, Rawat VP, Heilmeier B, Hoster E, Metzeler KH, Herold T, Hiddemann W, Gokbuget N, Hoelzer D, Bohlander SK, et al. The homeobox gene CDX2 is aberrantly expressed and associated with an inferior prognosis in patients with acute lymphoblastic leukemia. Leukemia. 2009;23:649–655. doi: 10.1038/leu.2008.355. [DOI] [PubMed] [Google Scholar]
- 18.Rothkamm K, Lobrich M. Misrepair of radiation-induced DNA double-strand breaks and its relevance for tumorigenesis and cancer treatment (Review) Int J Oncol. 2002;21:433–440. [PubMed] [Google Scholar]
- 19.Delacote F, Lopez BS. Importance of the cell cycle phase for the choice of the appropriate DSB repair pathway, for genome stability maintenance: the trans-S double-strand break repair model. Cell Cycle. 2008;7:33–38. doi: 10.4161/cc.7.1.5149. [DOI] [PubMed] [Google Scholar]
- 20.Downs JA, Jackson SP. A means to a DNA end: the many roles of Ku. Nat. Rev. Mol. Cell Biol. 2004;5:367–378. doi: 10.1038/nrm1367. [DOI] [PubMed] [Google Scholar]
- 21.Lieber MR, Ma Y, Pannicke U, Schwarz K. The mechanism of vertebrate nonhomologous DNA end joining and its role in V(D)J recombination. DNA Repair. 2008;3:817–826. doi: 10.1016/j.dnarep.2004.03.015. [DOI] [PubMed] [Google Scholar]
- 22.Van Gent DC, Van Der Burg M. Non-homologous end-joining, a sticky affair. Oncogene. 2007;26:7731–7740. doi: 10.1038/sj.onc.1210871. [DOI] [PubMed] [Google Scholar]
- 23.Soderberg O, Leuchowius KJ, Gullberg M, Jarvius M, Weibrecht I, Larsson LG, Landegren U. Characterizing proteins and their interactions in cells and tissues using the in situ proximity ligation assay. Methods. 2008;45:227–232. doi: 10.1016/j.ymeth.2008.06.014. [DOI] [PubMed] [Google Scholar]
- 24.Rubin E, Wu X, Zhu T, Cheung JC, Chen H, Lorincz A, Pandita RK, Sharma GG, Ha HC, Gasson J, et al. A role for the HOXB7 homeodomain protein in DNA repair. Cancer Res. 2007;67:1527–1535. doi: 10.1158/0008-5472.CAN-06-4283. [DOI] [PubMed] [Google Scholar]
- 25.Schild-Poulter C, Pope L, Giffin W, Kochan JC, Ngsee JK, Traykova-Andonova M, Haché RJG. The binding of Ku antigen to homeodomain proteins promotes their phosphorylation by DNA-dependent protein kinase. J. Biol. Chem. 2001;276:16848–16856. doi: 10.1074/jbc.M100768200. [DOI] [PubMed] [Google Scholar]
- 26.Modica S, Morgano A, Salvatore L, Petruzzelli M, Vanier MT, Valanzano R, Esposito DL, Palasciano G, Duluc I, Freund JN, et al. Expression and localisation of insulin receptor substrate 2 in normal intestine and colorectal tumours. Regulation by intestine-specific transcription factor CDX2. Gut. 2009;58:1250–1259. doi: 10.1136/gut.2008.158386. [DOI] [PubMed] [Google Scholar]
- 27.Brenkman AB, van den Broek NJF, de Keizer PLJ, van gent DC, Burgering BMT. The DNA damage repair protein Ku70 interacts with FOXO4 to coordinate a conserved cellular stress response. FASEB J. 2010;24:4271–4280. doi: 10.1096/fj.10-158717. [DOI] [PubMed] [Google Scholar]
- 28.Fattah F, Lee EH, Weisensel N, Wang Y, Lichter N, Hendrickson EA. Ku regulates the non-homologous end joining pathway choice of DNA double-strand break repair in human somatic cells. Plos Genet. 2010;6:E1000855. doi: 10.1371/journal.pgen.1000855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang M, Wu W, Wu W, Rosidi B, Zhang L, Wang H, Iliakis G. PARP-1 And Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways. Nucleic Acids Res. 2006;34:6170–6182. doi: 10.1093/nar/gkl840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iliakis G, Rosidi B, Wang M, Wang H. Plasmid-based assays for DNA end-joining in vitro. Methods Mol. Biol. 2006;314:123–131. doi: 10.1385/1-59259-973-7:123. [DOI] [PubMed] [Google Scholar]
- 31.Lieber MR. The mechanism of human nonhomologous DNA end joining. J. Biol. Chem. 2008;283:1–5. doi: 10.1074/jbc.R700039200. [DOI] [PubMed] [Google Scholar]
- 32.Lieber MR, Lu H, Gu J, Schwarz K. Flexibility in the order of action and in the enzymology of the nuclease, polymerases, and ligase of vertebrate non-homologous DNA end joining: relevance to cancer, aging, and the immune system. Cell Res. 2008;18:125–133. doi: 10.1038/cr.2007.108. [DOI] [PubMed] [Google Scholar]
- 33.Collis SJ, Deweese TL, Jeggo PA, Parker AR. The life and death of DNA-PK. Oncogene. 2004;24:949–961. doi: 10.1038/sj.onc.1208332. [DOI] [PubMed] [Google Scholar]
- 34.Roos WP, Kaina B. DNA damage-induced cell death by apoptosis. Trends Mol. Med. 2006;12:440–450. doi: 10.1016/j.molmed.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 35.Rings EH, Boudreau F, Taylor JK, Moffett J, Suh ER, Traber PG. Phosphorylation of the Serine 60 residue within the Cdx2 activation domain mediates its transactivation capacity. Gastroenterology. 2001;121:1437–1450. doi: 10.1053/gast.2001.29618. [DOI] [PubMed] [Google Scholar]
- 36.Verzi MP, Hatzis P, Sulahian R, Philips J, Schuijers J, Shin H, Freed E, Lynch JP, Dang DT, Brown M, et al. TCF4 and CDX2, major transcription factors for intestinal function, converge on the same Cis-regulatory regions. Proc. Natl Acad. Sci. USA. 2010;107:15157–15162. doi: 10.1073/pnas.1003822107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schild-Poulter C, Shih A, Tantin D, Yarymowich NC, Soubeyrand S, Sharp PA, Hache RJG. DNA-PK phosphorylation sites on Oct-1 promote cell survival following DNA damage. Oncogene. 2007;26:3980–3988. doi: 10.1038/sj.onc.1210165. [DOI] [PubMed] [Google Scholar]
- 38.Gaymes TJ, North PS, Brady N, Hickson ID, Mufti GJ, Rassool FV. Increased error-prone non homologous DNA end-joining—a proposed mechanism of chromosomal instability in Bloom's syndrome. Oncogene. 2002;21:2525–2533. doi: 10.1038/sj.onc.1205331. [DOI] [PubMed] [Google Scholar]
- 39.Mallo GV, Soubeyran P, Lissitzky JC, Andre F, Farnarier C, Marvaldi J, Dagorn JC, Iovanna JL. Expression of the Cdx1 and Cdx2 homeotic genes leads to reduced malignancy in colon cancer-derived cells. J. Biol. Chem. 1998;273:14030–14036. doi: 10.1074/jbc.273.22.14030. [DOI] [PubMed] [Google Scholar]
- 40.Brattain MG, Fine WD, Khaled FM, Thompson J, Brattain DE. Heterogeneity of malignant cells from a human colonic carcinoma. Cancer Res. 1981;41:1751–1756. [PubMed] [Google Scholar]
- 41.Leibovitz A, Stinson JC, Mccombs WB, Mccoy CE, Mazur KC, Mabry ND. Classification of human colorectal adenocarcinoma cell lines. Cancer Res. 1976;36:4562–4569. [PubMed] [Google Scholar]
- 42.Lozzio CB, Lozzio BB. Human chronic myelogenous leukemia cell-line with positive philadelphia chromosome. Blood. 1975;45:321–334. [PubMed] [Google Scholar]
- 43.Han T, Dadey B, Minowada J. Unique leukemic non-T/Non-B lymphoid cell lines (REH And KM-3): absence of MLR-S and presence of suppressor cell activity for normal T-cell response. J. Clin. Lab. Immunol. 1978;1:237–243. [PubMed] [Google Scholar]
- 44.Delalande F, Carapito C, Brizard JP, Brugidou C, Van Dorsselaer A. Multigenic families and proteomics: extended protein characterization as a tool for paralog gene identification. Proteomics. 2005;5:450–460. doi: 10.1002/pmic.200400954. [DOI] [PubMed] [Google Scholar]
- 45.Pastwa E, Neumann RD, Winters TA. In vitro repair of complex unligatable oxidatively induced DNA double-strand breaks by human cell extracts. Nucleic Acids Res. 2001;29:E78. doi: 10.1093/nar/29.16.e78. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.