Abstract
The myogenic differentiation 1 (MyoD) gene is a master regulator of myogenesis. We previously reported that the expression of MyoD mRNA oscillates over 24 h in skeletal muscle and that the circadian clock transcription factors, BMAL1 (brain and muscle ARNT-like 1) and CLOCK (circadian locomotor output cycles kaput), were bound to the core enhancer (CE) of the MyoD gene in vivo. In this study, we provide in vivo and in vitro evidence that the CE is necessary for circadian expression of MyoD in adult muscle. Gel shift assays identified a conserved non-canonical E-box within the CE that is bound by CLOCK and BMAL1. Functional analysis revealed that this E-box was required for full activation by BMAL1/CLOCK and for in vitro circadian oscillation. Expression profiling of muscle of CEloxP/loxP mice found approximately 1300 genes mis-expressed relative to wild-type. Based on the informatics results, we analyzed the respiratory function of mitochondria isolated from wild-type and CEloxP/loxP mice. These assays determined that State 5 respiration was significantly reduced in CEloxP/loxP muscle. The results of this work identify a novel element in the MyoD enhancer that confers circadian regulation to MyoD in skeletal muscle and suggest that loss of circadian regulation leads to changes in myogenic expression and downstream mitochondrial function.
INTRODUCTION
The MyoD (myogenic differentiation 1) gene is the founding member of the myogenic regulatory factor family of transcription factors and is known to have a central role in myogenesis (1). In adult skeletal muscle, MyoD is thought to function primarily in the myogenic differentiation of satellite cells during muscle regeneration (2,3). Recently, however, MyoD expression has been found to oscillate over a 24 h period in both mouse and rat skeletal muscle (4,5). Andrews and colleagues (4) also determined that MyoD was a direct transcriptional target of the molecular clock. Both BMAL1 and CLOCK were found to bind to the MyoD core enhancer (CE) 20 kb upstream (6). The molecular link between the core clock and MyoD implicated a role for the circadian expression of MyoD in skeletal muscle homeostasis. This was supported by phenotypic analysis of muscle from two different genetic mouse models in which expression of the core circadian genes, Bmal1 or Clock, were disrupted (4). Andrews and colleagues (4) reported that in skeletal muscle of clock-compromised mice MyoD expression did not oscillate and this was associated with a disruption of muscle structure and diminished mechanical function.
The CE is a relatively small enhancer of 258 bp and resides ∼20 kb upstream of the MyoD transcription start site. Previous studies have shown that loss of the CE affects expression of MyoD in all embryonic skeletal muscle compartments, including somitic myotomes, limb buds and branchial arches (6,7). The CE appears to be primarily regulated by a positive control mechanism operating through multiple, discrete cis-elements that are scattered throughout the enhancer (8,9). Kablar et al. (7) reported that deletion of the CE in the mouse caused a reduction in MyoD mRNA expression in both embryos and neonates and resulted in a delay in muscle differentiation. In contrast to its role during development, the CE was found to be dispensable for expression during late fetal stages and was assumed to be inactive in adult muscle (10). Thus, these studies showed that the CE was critical for initiation of MyoD transcription during development but was argued to be not necessary for expression of MyoD in adult skeletal muscle (8,11).
The molecular clock that governs circadian rhythms can be described as a transcription–translation feedback loop (12). The forward arm of the molecular clock is comprised of CLOCK (circadian locomotor output cycles kaput) and BMAL1 (brain and muscle ARNT-like 1) which are bHLH-PAS transcription factors that bind to an E-box motif (5′-CACGTG-3′) as a heterodimer. As part of the molecular clock, CLOCK and BMAL1 activate transcription of factors that constitute the negative loop of the molecular clock, including the Period (Per) and Cryptochrome (Cry) gene isoforms Per1, Per2, Cry1 and Cry2. In turn, CRY and PER proteins act together to inhibit the trans-activation function of CLOCK and BMAL1 and this mechanism oscillates over an approximate 24 h cycle (12,13). Consistent with the feedback mechanism, the circadian phase of Bmal1 expression was in anti-phase to circadian phase of Per1/2 and Cry1/2 expression (14–17).
The purpose of the current study was to determine whether the CE is the module through which the molecular clock is regulating the circadian expression of MyoD. In addition, we wanted to identify the cis-acting sites [E-box(es)] within the CE that bind CLOCK and BMAL1. In this study, we provide evidence showing that the MyoD CE is necessary for circadian oscillation of MyoD expression in vivo. We also show that the CE is sufficient to confer circadian expression in vitro. We identify a previously unknown non-canonical E-box in the MyoD CE that mediates CLOCK and BMAL1 binding and using an in vitro circadian assay we demonstrate that this non-canonical E-box is required for oscillation of a MyoD reporter. Expression profiling of muscle from adult CEloxP/loxP mice identified mis-expressed genes that overlap with MyoD target genes from ChIP-Seq studies and also disrupted genes identified from analysis of muscle from the ClockΔ19 mouse (18). Further experiments indicated that mitochondrial respiration was impaired in the CEloxP/loxP muscle. These findings support a model in which the molecular clock factors directly regulate MyoD expression in skeletal muscle through regulation of a non-canonical E-box in the CE and that loss of circadian expression of MyoD contributes to downstream changes in the expression of MyoD target genes with effects on mitochondrial function.
MATERIALS AND METHODS
Animals
All animal procedures were conducted in accordance with institutional guidelines for the care and use of laboratory animals as approved by the University of Kentucky Institutional Animal Care and Use Committee. Mice (C57BL/6J strain) were purchased from The Jackson Laboratory. The MyoD core enhancer knockout strain (CEloxP/loxP mice) was originally developed and described by Chen and Goldhamer in 2004 (10). These mice are germline knock-outs for the CE so no further breeding is required. We bred the original CEloxP/loxP mice onto a C57BL/6J background by backcrossing for 10 generations. The CEloxP/loxP mice pups were genotyped by PCR as described by (10). The CEloxP/loxP mouse was also crossed with Per2::Luc knock-in mice (19) to produce homozygous mice harboring both CEloxP/loxP and Per2::Luc.
Circadian tissue collection
We followed the established protocol in the circadian field for collection of tissues for circadian analysis (18,20). Briefly, mice (10–12 weeks of age) were entrained to a 12 h light/12 h dark (12L:12D) cycle for 2 weeks and then placed in total darkness for 30 h prior to the start of the tissue collection to remove any light cues. Tissues were collected in darkness with red light from mice (n = three/genotype) every 4 h for 32 h (nine time points). By circadian convention, the onset of activity for a nocturnal animal is defined as circadian time 12 (CT12) so the first collection was performed at CT18.
Cell culture
The mouse myogenic cell line C2C12 was obtained from ATCC (CRL 1722). Myoblasts were grown in Dulbecco's modified Eagles medium (DMEM; Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum and penicillin–streptomycin in a humidified incubator at 37°C under 5% CO2. To establish stable clones, C2C12 cells (5 × 104) were co-transfected with 1 µg of a particular reporter gene plasmid and 1 µg of pCI-neo plasmid (Promega, Madison, WI, USA). Stable clones (n = 3/construct) were selected following treatment with G418 at 600 µg/ml and maintained in culture with 200 µg/ml.
Transient transfection
C2C12 cells were transiently transfected with expression vectors for Clock and Bmal1 (200 ng/vector) and one of the MyoD reporter genes (50 ng) using Fugene 6 reagent according to the manufacturer's instructions (Roche, Indianapolis, IN, USA) (21). Five ng of pRL Renilla Luciferase vector (Promega) was co-transfected to control for transfection efficiency. When necessary, an empty pGEM plasmid was used to equalize the total amount of DNA transfected. Cells were collected 24–48 h after transfection and luciferase activity was determined using Dual Luciferase Assay System according to the manufacturer's instructions (Promega).
In vitro circadian assay
Serum shock was used to synchronize molecular clock in C2C12 myotubes in vitro using a method previously described (22,23). Briefly, C2C12 myoblasts were grown to confluence and then switched to differentiation medium (DM) consisting of DMEM supplemented with 2% horse serum. DM was changed every 48 h. After 5 days in DM, C2C12 myotubes were treated with 50% horse serum for 2 h, washed with phosphate buffered saline (PBS) and switched to DMEM supplemented with 2% horse serum for the duration of the experiment. At selected time points following serum stimulation, cells were washed with cold PBS and total RNA was isolated from cells using TRIzol reagent (Invitrogen).
MyoD reporter genes
Based on our finding that the CE and not the DRR were bound by CLOCK:BMAL1 we generated a MyoD luciferase reporter gene lacking the DRR (CE-4.7MyoD) using the CE-6.8MyoD:luciferase as a template (4). Approximately 4.7 kb of MyoD upstream 5′-flanking sequence was cloned into the pGL3 basic vector (Promega) to replace the 6.8 kb fragment in the CE-6.8MyoD:luciferase reporter. The CE region was amplified from mouse genomic DNA and was subsequently inserted upstream of 4.7 kb sequence to produce CE-4.7MyoD:luciferase reporter gene. The primers used for amplification of the 4.7 kb flanking sequence (−4566 to +103) were 5′-CACAAGTGGTTTGAAGTGCTTC-3′ (forward) and 5′-AGGTTCTGTGGGTTGGAATG-3′ (reverse). The primers for amplifying the CE were 5′-AATGCCCAAAGAGTGGCAGA-3′ (forward) and 5′-CCAATCTCAAAGCCCTGGA-3′ (reverse). The DNA sequence of the reporter gene was verified by sequencing.
Mutagenesis
To create the mutant CE-4.7MyoD:luciferase reporter gene (CEmt-4.7MyoD), mutagenesis was performed with plasmid CE-4.7MyoD:luciferase using GeneEditor in vitro Site-Directed Mutagenesis System (Promega). The sequence of the oligonucleotide used for mutagenesis of the non-canonical E-box of the MyoD CE was 5′-ATCTCCCAGAGTGCTGCGCTTAAAACCCGTG-3′ with the underlined sequence indicating the position of the E-box and the bold letters indicating the mutated bases. The successful mutation of the non-canonical E-box was verified by sequencing.
Real-time PCR
To analyze gene expression levels, 1 µg of RNA was reverse-transcribed to cDNA using Superscript III First Strand Synthesis system (Invitrogen). Real-time PCR was performed using Power SYBR Green PCR Master mix (Applied Biosystems, Carlsbad, CA, USA). The PCR conditions were as follows: 10 min at 95°C, 15 s at 95°C, 60 s at 60°C and 15 s at 95°C. The mRNA levels of target genes were normalized to the corresponding level of Rpl26 mRNA. The following primers were used: Rpl26, 5′-CGAGTCCAGCGAGAGAAGG-3′ and 5′-GCAGTCTTTAATGAAAGCCGTG-3′; Bmal1, 5′-GCAGTGCCACTGACTACCAAGA-3′ and 5′-TCCTGGACATTGCATTGCAT-3′; Per2, 5′-GGCTTCACCATGCCTGTTGT-3′ and 5′-GGAGTTATTTCGGAGGCAAGTGT-3′; MyoD, 5′-ATGGATTACAGCGGCCCC-3′ and 5′-TGTGGAGATGCGCTCCACTA-3′. The specificity of each PCR product was analyzed by electrophoresis and melting curve.
DNA mobility shift assay
Bmal1 and Clock expression plasmids were transfected into C2C12 cells and nuclear extracts were prepared as previously described (24). DNA duplexes were synthesized for each of the five E-boxes in the MyoD CE. The sequence for each oligonucleotide was as follows: E-box-1, 5′-GAGCCCCACAGCA TTTGGGGGCATTTATG-3′; E-box-2, 5′-GTAT CCTCCTCCAGCAGCTGGTCACAAAGC-3′; E-box-3, 5′-CACAACACAGCCAGTTGGGGGAAGGGG-3′; E-box-4, 5′-GCGCCCAGAGTCAGCT GTTCCTGGGTCTTC-3′; the non-canonical E-box, 5′-CCCAGAGTGCTCAGCTTAAAACCCG TGAC-3′; the non-canonical E-box mutant, 5′-CCCAGAGTGCTGCTAGGAAAACCCGTGAC-3′ (25). The underlined sequences indicate the position of E-boxes. Duplexes of these DNA oligonucleotides were 5′-end labeled with 32P-γ-ATP using T4 polynucleotide kinase (Promega). DNA mobility shift assays were performed as previously described (26). For super-shift experiment, CLOCK (Abcam ab3517) and BMAL1 Ab (Abcam, ab3350) were used. The protein–DNA complexes were resolved on a 5% polyacrylamide gel with 0.4 × Tris/Borate/EDTA. The images were obtained using a PhosphoImager and scanned using Storm Scanner 860.
Bioluminescent analysis of circadian expression in vitro
Stable clones of C2C12 cells (n = 3/construct) were established with luciferase based reporter genes containing either the Bmal1 promoter, CE-4.7MyoD or the CE non-canonical E-box mutant CEmt-4.7MyoD promoter. The mouse Bmal1 promoter–luciferase reporter vector was kindly provided by Dr J. Hogenesch (University of Pennsylvania) (27). These reporter genes were individually co-transfected with pCI-neo plasmid into C2C12 myoblast cells. Three selected stable clones for each reporter gene were grown in 35 mm dishes to confluence and differentiated for 5 days in DM. Serum shock is a standard technique used in the circadian field to synchronize the molecular clocks in vitro (28–31). In this study, myotubes were serum shocked and then incubated in recording medium which contained 0.1 mM luciferin in phenol red free DMEM with 2% horse serum as previously described (32). Bioluminescence was measured at 10 min intervals for 4 days using the Lumicycle (Actimetrics Inc., Evanston IL) (32). Both raw and baseline subtracted data were analyzed for circadian characteristics including period and phase using Lumicycle Analysis software. The period length of luminescence for each reporter gene was calculated based on the distances between peaks or troughs over the 24–60 h post-serum shock from the same three clones. For these studies, the phase of expression is defined as the time of the first peak 24 h after serum shock. For the bioluminescent studies of muscle tissue, mPER2::LUC homozygous mice were sacrificed by cervical dislocation. Intact soleus muscles were dissected out and incubated in recording medium which contained 0.1 mM luciferin in phenol red free DMEM with 5% fetal bovine serum. Results from the luminescence data (period, phase) were analyzed as described above.
Microarray
For the microarray experiments, we collected gastrocnemius muscles from the CE knockout and wild-type mice (n = three/genotype/time point) at four time points (CT26, CT30, CT38 and CT42). Total RNA was isolated from each muscle using the TRIzol reagent (Invitrogen) as previously described (18,20). RNA integrity was analyzed using Agilent 2100 Bioanalyzer. For this study, we wanted to determine differences in gene expression and not circadian expression so samples for the array were generated with equal amounts of total RNA/genotype pooled from the CT26 and CT30 samples, or from the CT38 and CT42 samples with each pooled sample analyzed by a separate Affymetrix chip (430 2.0). For each probe set, we calculated the average expression for the two chips from each genotype. To identify differentially expressed genes we set a cutoff of more than 400 intensity and either a >2-fold increase or >50% decrease in expression between the CEloxP/loxP and wild-type probe sets.
Mitochondrial preparation and measurement of mitochondrial respiration
Skeletal muscle mitochondria were isolated using isolation buffer (215 mM mannitol, 75 mM sucrose, 0.1% BSA, 20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), 1 mM EGTA and pH is adjusted to 7.2 with KOH) at 4°C as describe earlier (33). Mitochondrial respiration was performed using a miniature Clark-type electrode (Hansatech Instruments, Norfolk, UK) in a sealed, thermostatically controlled chamber at 37°C as described previously (34). Briefly, oxymetric traces were taken by addition of mitochondria to yield a final concentration of ∼200–300 µg protein/ml respiration buffer (125 mM KCl, 2 mM MgCl2, 2.5 mM KH2PO4, 20 mM HEPES and 0.1% BSA, pH 7.2). Mitochondrial respiration is initiated by addition of the Complex I, Nicotinamide adenine dinucleotide (NADH)-linked oxidative substrates pyruvate and malate (State 2 respiration). This is followed by addition of ADP which causes flow of protons through the ATP synthase (State 3 respiration) as the added ADP is phosphorylated to ATP. Mitochondria enter State 4 respiration as all the added ADP has been phosphorylated (oligomycin, an inhibitor of the ATP synthase is added to ensure that no protons cross the inner membrane via the ATP synthase). Complex I-State 5 respiration is initiated by the addition of carbonyl cyanide-p-trifluoromethoxyphenylhydrazone, which acts as a protonophore that collapses the proton gradient and result in maximum electron transport and NADH-driven respiration (Complexes I–IV assessed); to assess Complex II, FADH-linked respiration, the Complex I inhibitor rotenone is added to the chamber which completely stops Complex I electron transport and NADH-driven respiration, and the Complex II substrate succinate is added to the chamber to assess Complexes II, III and IV (Succ-State 5). Respiration rate was calculated as nanomoles of oxygen consumed/min/mg of mitochondrial protein. Two independent oxymetric traces for each sample were taken and respiration rates were averaged to yield n = 1/sample.
Statistical analysis
Data are reported as mean ± SEM. To determine if a significant difference existed between treatment and/or groups, one-way ANOVA was performed followed by the Tukey post-hoc test to identify specific significant differences between means. Differences between groups were considered significant if P ≤ 0.05. For circadian analysis of gene expression, we used the JTK_CYCLE program (35). JTK_CYCLE is a non-parametric algorithm developed to determine whether a gene expression pattern can be considered circadian and if so, it will provide an objective analysis of phase and period length.
RESULTS
The MyoD CE is required for circadian expression of MyoD in vivo
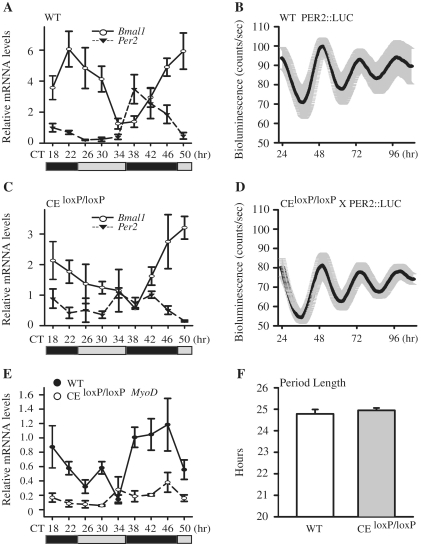
Previously, we showed that BMAL1 and CLOCK bind to the MyoD CE but not the MyoD DRR, suggesting the CE might be necessary for circadian expression of MyoD (4). To test this prediction, we determined the expression of MyoD in adult skeletal muscle of the CEloxP/loxP mouse (10). To allow for comparison with our initial study describing the circadian expression of MyoD, the CEloxP/loxP mice were backcrossed to C57BL/6J for 10 generations to produce a congenic strain that is considered to be 99.9% C57BL/6J background. These backcrosses were necessary to ensure any differences observed in the temporal pattern of MyoD expression could be attributed to the loss of the CE and not genetic background. Since little is known about the behavior of the adult CEloxP/loxP mice, we carried out a series of experiments to characterize the strain before analyzing circadian gene expression. There was no difference in either body weight or muscle weight in comparison to wild-type C57BL/6J mice (data not shown). Analysis of circadian behavior found that adult CEloxP/loxP mice exhibited normal behavior as assessed by wheel running activity (36) under 12L:12D and free running conditions 12D:12D (Supplementary Figure S1). Under free running conditions, CEloxP/loxP mice had an average period length of 23.75 ± 0.28 h compared to 23.57 ± 0.37 h period length for wild-type C57BL/6J mice (Supplementary Figure S1).
Next, we compared the circadian expression of core clock factors, Bmal1 and Per2 in tibialis anterior (TA) muscles of the CEloxP/loxP and wild-type mice. To be consistent with previously published circadian gene expression studies, skeletal muscle collection started 30 h after the lights had been turned off, corresponding to CT18 (18,20). First we examined expression of Bmal1 and Per2 mRNA in the muscles of the wild-type mice every 4 h over 32 h (9 time points). As shown in Figure 1A, Bmal1 mRNA (empty circle) levels oscillated, peaking at CT22 and CT50, and Per2 mRNA (filled triangle) oscillated anti-phase to Bmal1 with peak expression at CT38. Next we determined Bmal1 and Per2 mRNA in the muscle of the CEloxP/loxP mice (Figure 1C). As shown in Figure 1C, expression of Bmal1 (empty circle) and Per2 (filled triangle) mRNAs are oscillating across time points however, the amplitude of expression was slightly lower. To statistically evaluate the circadian pattern of gene expression in the muscle of wild-type and CEloxP/loxP mice, we used the JTK_CYCLE program (35). Using this analysis, we confirmed that Bmal1 and Per2 in both wild-type and the CE KO mice muscle were expressed in a circadian manner with P < 0.05 (Supplementary Table S1).
Figure 1.
Circadian oscillation of MyoD mRNA is dampened in skeletal muscle tissues of the CEloxP/loxP mouse. Adult C57BL/6 J and CEloxP/loxP mice were entrained to 12 h light/12 h dark conditions for 2 weeks. Thirty hours after lights were turned off (CT18), TA muscles were collected every 4 h from CT18 to CT50. Transcript levels were determined by RT–PCR. Bmal1, Per2 and MyoD mRNA levels were normalized to Rpl26 mRNA levels. (A) Bmal1 and Per2 expression in wild-type mice TA muscle; (B) Luminescence recording from Per2::Luc soleus muscles. The data for the graphs represent the mean values ± SEM of raw data for five animals. (C) Bmal1 and Per2 expression in CEloxP/loxP mice TA muscle; (D) Luminescence recordings from Per2::Luc × CEloxP/loxP soleus muscles. The data for the graphs represent the mean values ± SEM of raw data for five animals. (E) MyoD expression in wild-type and CEloxP/loxP mice TA muscle; (F) Average period length data of bioluminescence from Per2::Luc and Per2::Luc × CEloxP/loxP soleus muscles. The graphs in (A, C and E) display the results as the mean value ± SEM for three independent samples for each genotype. The gray bars indicate the subjective circadian light period and the black bars indicate subjective circadian dark period. For each respective gene, one-way ANOVA followed by Tukey's post-hoc test was performed to determine significant differences across all time points. For circadian analysis of gene expression, the JTK_CYCLE program was used.
To more rigorously evaluate molecular clock function in the muscle of CEloxP/loxP mice we employed real-time bioluminescent analysis using the Lumicycle (Actimetrics). Previous studies have shown that the bioluminescence rhythm obtained from tissue explants from the circadian reporter Per2::Luc mouse matches endogenous PER2 protein in vivo (19,32,37). We crossed the CEloxP/loxP mice with Per2::Luc mice and produced homozygous mice harboring both the CEloxP/loxP and Per2::Luc. For these studies both Per2::Luc and the Per2::Luc × CEloxP/loxP mice were entrained for 2 weeks. At that time, the mice were sacrificed and soleus muscles were collected and placed in culture with recording media plus luciferin and real-time bioluminescence was recorded every 10 min for 4 days. The power of this experimental approach for circadian analysis is that it allows us to follow PER2 expression for each sample over a longer period of time with much higher temporal frequency (once every 10 min versus tissue collections every 4 h). The increased sampling frequency and longer duration permits a more robust quantitative analysis of circadian expression with the capacity to more precisely define important characteristics including period length and phase.
Raw data are presented in Figure 1B (Per2::Luc) and Figure 1D (Per2::Luc × CEloxP/loxP) represent the mean ± SEM luminescence recordings averaged for all five explants per genotype. Compared to Per2::Luc, the average level of Per2::Luc × CEloxP/loxP bioluminescence is slightly reduced. However baseline subtracted data are basically the same for Per2::Luc and the Per2::Luc × CEloxP/loxP tissues (Supplementary Figure S2). Analysis of circadian parameters determined that there were no significant differences for either the period length (Figure 1F) or the phase between the muscles from Per2::Luc × CEloxP/loxP and the Per2::Luc.
Once we confirmed the molecular clock functions properly in the muscle of the CEloxP/loxP mice we tested whether loss of the CE affects expression of MyoD mRNA. Consistent with our previous findings in the gastrocnemius muscle, MyoD mRNA levels oscillated in the TA muscle of wild-type mice with peak expression from CT38 to CT46 (Figure 1E, filled circle) (4). We then examined expression of MyoD mRNA in the TA muscle of the CEloxP/loxP mice. As illustrated in Figure 1E (empty circle), the expression pattern of MyoD mRNA was significantly dampened. Analysis of MyoD expression using JTK_CYCLE confirmed that expression was oscillating in a circadian manner in the wild-type mice but was not circadian in the muscle of the CEloxP/loxP mice. These results confirm that the CE is necessary for circadian expression of MyoD in vivo. We also noted that while expression of MyoD in the CEloxP/loxP mice was reduced compared to wild-type across most time points there was no difference in expression between the two genotypes at the trough of MyoD expression, CT34 suggesting that the lower expression of MyoD reported previously maybe more related to time of analysis.
The non-canonical E-box in the MyoD CE is required for BMAL1 and CLOCK Binding and activation
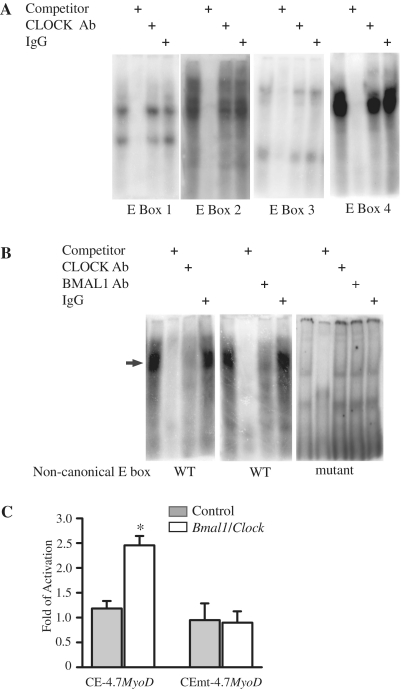
The nucleotide sequence of the CE is highly conserved across mouse, rat and human genomes (∼89–98%). There are four canonical E-boxes (5′-CANNTG-3′) in the MyoD CE as previously described (13,38). The canonical E-box sequences from upstream to downstream are CATTTG (E1), CAGCTG (E2), CAGTTG (E3) and CAGCTG (E4) (Supplementary Figure S3). All four E-boxes are identical in the rat and mouse whereas only E-boxes E2 and E4 are conserved in human. In addition to these four canonical E-boxes, there was one non-canonical E-box (5′-CAGCTT-3′) that is located between E2 and E3 (indicated by hash). The sequence of the non-canonical E-box and its flanking sequences are conserved across mouse, rat and human MyoD CE sequences (Supplementary Figure S3). Interestingly, the non-canonical E-box is similar to the BMAL1:CLOCK binding site (5′-CACGTT-3′) found within the promoter of the core clock gene Per2 (39).
Our previous work demonstrated that BMAL1 and CLOCK can bind to the MyoD CE using chromatin immunoprecipitation assays with nuclear extracts from mouse adult skeletal muscle (4). In order to determine if CLOCK and BMAL1 proteins were capable of binding any of the aforementioned E-boxes we performed electrophoretic mobility shift assay (EMSA) experiments. The binding reaction contained one of four different E-box oligonucleotide probes and nuclear extracts derived from C2C12 myoblasts that were transfected with Bmal1 and Clock expression plasmids. Figure 2A shows representative images from EMSA using each of the canonical E-box probes. For each probe, protein:DNA complexes were detected and shown to be specific as indicated by successful competition with excess cold oligonucleotide competitor (Figure 2A). While the protein:DNA complexes were specific, none of these complexes contained CLOCK as incubation with a CLOCK antibody failed to deplete or super-shift the observed protein:DNA complexes.
Figure 2.
The non-canonical E-box of the MyoD CE is required for CLOCK and BMAL1 binding and activation of the MyoD promoter by BMAL1: CLOCK. EMSA analysis of CLOCK and BMAL1 binding at the E-boxes located within the MyoD CE was performed with nuclear extract derived from C2C12 cells over-expressing Clock and Bmal1 and an oligonucleotide probe containing one of the five CE E-boxes. (A) The DNA:protein complexes formed using a probe containing one of the canonical E-boxes (E1, E2, E3 or E4). (B) The DNA:protein complexes formed using the non-canonical E-box and the mutant of the non-canonical E-box. The arrow indicates the specific DNA:protein complex that contained CLOCK and BMAL1. (C) A wild-type (CE-4.7MyoD:luciferase) or mutant (CEmt-4.7MyoD:luciferase) reporter gene construct was transiently co-transfected with Clock plus Bmal1 expression plasmids into C2C12 myoblasts. Luciferase activity was measured 24–48 h later. The histogram displays the results from three independent experiments with values presented as mean ± SEM. A one-way ANOVA test indicated CE-4.7MyoD:luciferase reporter gene activation by CLOCK plus BMAL1 (empty bar) was significantly greater than control (gray bar) denoted by asterisks.
We next tested whether the non-canonical E-box located between E2 and E3 within the CE was capable of binding CLOCK and/or BMAL1. Figure 2B shows that the non-canonical E-box formed a protein:DNA complex that contained both CLOCK and BMAL1. The addition of either CLOCK (left panel) or BMAL1 (middle panel) antibody to the binding reaction was able to prevent formation of the protein:DNA complex whereas addition of control IgG did not alter binding. The specificity of the E-box for CLOCK and BMAL1 binding was further confirmed by mutagenesis. EMSA performed with a mutated form of the non-canonical E-box resulted in two binding complexes that were not altered by addition of either the CLOCK antibody or the BMAL1 antibody to the binding reaction (Figure 2B, right panel). Taken together, these results demonstrate that CLOCK and BMAL1 bind to the non-canonical E-box in the MyoD CE.
The EMSA results demonstrated that both CLOCK and BMAL1 bind to the non-canonical E-box (5′-CAGCTT-3′) so our next set of experiments tested whether this E-box was functional in vitro. We generated the reporter construct, CEmt-4.7MyoD:luciferase and performed transient co-transfection experiments in C2C12 cells. We found that transfection of the CE-4.7MyoD:luciferase reporter gene with Clock plus Bmal1 expression vectors resulted in a 2.5-fold increase of CE-4.7MyoD:luciferase expression (Figure 2C). In contrast, over-expression of both Clock and Bmal1 did not result in an activation of the CEmt-4.7MyoD:luciferase reporter gene. We concluded from these transfection experiments that the non-canonical E-box within the CE is functional and required for full activation of MyoD reporter by BMAL1 and CLOCK.
The MyoD CE is required for circadian expression of the MyoD reporter gene in C2C12 myotubes
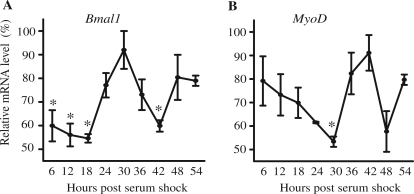
Having established that the CE is required for MyoD oscillation in vivo, and that the non-canonical Ebox binds CLOCK and BMAL1, we next wanted to determine whether this E-box could confer circadian oscillation using an in vitro assay. This required the validation of a serum shock cell culture model for evaluating molecular clock factor expression in C2C12 muscle cells. Serum shock is commonly used to synchronize molecular clocks across cells in vitro. This is necessary to allow for objective analysis of circadian gene expression taking into account the proper pattern (cosinar) and temporal oscillations (∼24 h). To determine whether serum stimulation is sufficient to synchronize circadian gene expression in C2C12 myotubes we evaluated expression of Bmal1 mRNA using real-time PCR. Myotube cultures were serum shocked and samples were collected every 6 h for 54 h. As shown in Figure 3A, Bmal1 mRNA expression varies over time following serum stimulation. We found that the peak and trough expression of Bmal1 mRNA was separated by 12 h (18 versus 30 h, 30 versus 42 h) and the difference in expression between peak and trough was statistically significant (P < 0.05). Likewise, endogenous MyoD mRNA expression showed an oscillation in expression with the peak and trough levels occurring 12 h apart (Figure 3B). In addition, the pattern of Bmal1 and MyoD oscillations were anti-phase and this is similar to the pattern observed in vivo and consistent with MyoD expression being regulated by the clock factors, BMAL1 and CLOCK (4). Use of JTK-CYCLE software also confirmed these observations that expression of endogenous Bmal1 and MyoD mRNAs in myotubes following serum shock were circadian (Supplementary Table S2).
Figure 3.
Serum shock is sufficient to synchronize circadian gene expression in C2C12 myotubes. Stable clones of C2C12 cells (CE-4.7MyoD) were differentiated for 5 days prior to serum shock. RNA was isolated at the indicated time post-serum shock and used to determine the expression level of Bmal1 and MyoD by RT-PCR. The target transcript levels were normalized to Rpl26 transcript levels and then normalized to the highest point across all time points. The data for the graphs represent the mean values ± SEM for three independent clones for the reporter gene. The expression of endogenous Bmal1 (A), endogenous MyoD (B) all displayed a circadian oscillation as indicated by significant differences in peak and trough expression. For each respective gene, one-way ANOVA followed by Tukey post-hoc test identified significant difference (P < 0.05) between peak expression and other time points as indicated by an asterisk. For circadian analysis of gene expression the JTK_CYCLE program was used.
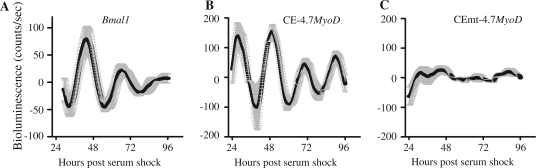
To more quantitatively evaluate the necessity of the non-canonical E-box for the circadian expression of MyoD, we employed real-time bioluminescent analysis using the Lumicycle. For this set of experiments, we generated stable C2C12 cells lines containing either the Bmal1:luciferase reporter gene, CE-4.7MyoD:luciferase reporter gene or CEmt-4.7MyoD:luciferase reporter gene. C2C12 stable clones (n = three/reporter) were differentiated into myotubes for 5 days and serum shocked as previously described. The medium was then replaced with non-phenol red DMEM containing luciferin plus 2% horse serum. Light emission was captured at 10 min intervals in real time using the Lumicycle for 4 days. Raw data for each clone was baseline subtracted and the data for the graphs in Figure 4 represent the mean values ± SEM for three independent clones for the reporter gene.
Figure 4.
Mutation of the MyoD CE non-canonical E-box disrupts circadian oscillation in bioluminescent assay. Stable clones of C2C12 cells (carrying the Bmal1 promoter:luciferase, CE-4.7MyoD:luciferase or CEmt-4.7MyoD:luciferase reporter) were differentiated for 5 days prior to serum shock. Then the cultures were incubated in Lumicycle at 37°C and bioluminescence was recorded. The graphs shown here are baseline subtracted results as the mean value ± SEM from three independent clones for the reporter gene. (A) Bmal1 promoter:luciferase reporter. (B) CE-4.7MyoD:luciferase reporter. (C) CEmt-4.7MyoD:luciferase reporter. The graphs display the results as the mean value ± SEM from three independent clones for each reporter. For circadian analysis of gene expression, the JTK_CYCLE program was used.
We first evaluated expression of the Bmal1 reporter gene in C2C12 myotubes. Consistent with our real-time PCR analysis (Figure 3A), the results of these experiments showed that the Bmal1 promoter directs circadian expression of luciferase (Figure 4A). As seen in Figure 4, the circadian amplitude damped gradually with time resulting from the depletion of nutrients and luciferin as replacement with fresh medium was able to restore the amplitude (data not shown).
Next, we tested whether the CE-4.7MyoD:luciferase reporter gene directed circadian expression. As shown in Figure 4B, there was a robust oscillation. In contrast, the CEmt-4.7MyoD:luciferase reporter gene expression (Figure 4C) was greatly dampened. JTK analysis determined that Bmal1:luciferase and CE-4.7MyoD:luciferase were circadian, but CEmt-4.7MyoD:luciferase was not expressed in a circadian manner (Supplementary Table S2). These results provide further evidence that the non-canonical E-box is the cis-regulatory element within the CE that confers circadian rhythmicity to the MyoD promoter.
Differentially expressed genes in skeletal muscle of the CEloxP/loxP mice are also mis-expressed in the ClockΔ19 mutant muscle
Similar to the CEloxP/loxP mice, we previously reported a loss of MyoD circadian expression in the ClockΔ19 mutant strain and the Bmal1 knockout strain (4,18). The mis-expression of MyoD in skeletal muscle of ClockΔ19 mutant was associated with the down-regulation of muscle-specific genes, many of which are known MyoD target genes. To determine if a comparable response in gene expression occurred in the muscle of CEloxP/loxP mice, we performed a microarray analysis of RNA from gastrocnemius muscles of adult mice. Using the criteria (either 2-fold increased or 50% decreased) outlined in ‘Materials and Methods’ section, we identified 1271 genes that were differentially expressed between wild-type and CEloxP/loxP muscle with 84% of the genes showing decreased expression in the CEloxP/loxP (Supplementary Table S3, in black). Gene ontology classification using DAVID (http://david.abcc.ncifcrf.gov/) revealed enrichment for genes related to contractile fiber (30 genes), endoplasmic reticulum (105 genes), cytosol (70 genes) and mitochondrion (134 genes) (Supplementary Table S6A). The expression of a number of skeletal muscle-specific genes were down-regulated in the muscle of the CEloxP/loxP mice including alpha actinin 3 (Actn3), titin (Ttn), dihydropyridine receptor, calcium channel, voltage-dependent calcium channel (Cacnb1) and calpain 3 (Capn3).
We next interrogated a ChIP-Seq derived MyoD target database to determine whether any of the mis-expressed genes were MyoD target genes (40). This analysis identified 461 genes (36.3%) that were scored as MyoD target genes according to the ChIP-Seq database (Supplementary Table S4). Among the 461 genes, Gene ontology classification using DAVID revealed enrichment for genes related to sarcomere (13 genes), cytosol (35 genes) and mitochondrion (59 genes) (Supplementary Table S6B).
We also interrogated the differentially expressed genes from the CEloxP/loxP data set versus the data set reported for the ClockΔ19 skeletal muscle (18). This analysis revealed that 207 genes altered in the muscle of the CEloxP/loxP mice are also altered in ClockΔ19 mice (Supplementary Table S5). Among the 207 genes, we found 177 (86%) genes (Supplementary Table S5, in black) are down-regulated. Gene ontology classification using DAVID revealed enrichment for genes related to sarcomere (seven genes), cytosol (17 genes) and mitochondrion (26 genes) (Supplementary Table S6C). The overlapping genes include several muscle specific genes, for example, Myl3, Myo18a, Ttn, Tpm1, Tpm2, Myoz2 and Myom2 (Supplementary Table S5). These results suggest that genes differentially expressed in the muscle of the CEloxP/loxP mice could be downstream of the BMAL1:CLOCK to MyoD circadian transcriptional loop.
Respiratory function is reduced in the mitochondria isolated from muscle of CEloxP/loxP mice
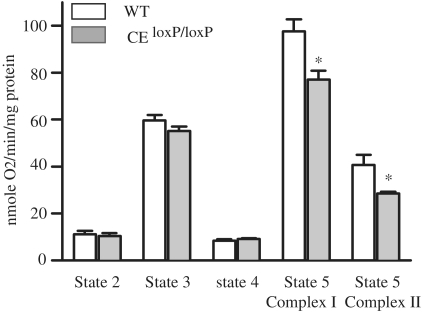
The gene ontology analysis indicated that many genes related to mitochondria are altered in the CEloxP/loxP mouse muscle. We analyzed the respiratory function of mitochondria isolated from 5- to 6-month-old wild-type versus CEloxP/loxP mice to determine whether the changes in gene expression were associated with any functional effect. As shown in Figure 5, we found no significant differences in States 2, 3 and 4 respiration rates between wild-type and CEloxP/loxP muscle. However, we found that there was a significant difference in Complexes I and II driven maximum electron transport (Complex I-State 5 and succinate State 5 respiration rates). Specifically, we found that both Complexes I and II of State 5 respiration rates were reduced. In Complex I, NADH dehydrogenase catalyzes the electron transfer from NADH to Ubiquinone (Q) and in Complex II succinate dehydrogenase catalyzes electron transfer from succinate to Ubiquinone (Q) via FAD. The changes in respiration were consistent with our data from expression analysis as level of Sdhc mRNA, which encodes a subunit of succinate dehydrogenase is down-regulated by 67% in the CEloxP/loxP muscle. These findings demonstrate that loss of the CE of MyoD is associated with a disruption in mitochondrial function.
Figure 5.
Respiration rates are lower in the mitochondria of the CEloxP/loxP than in wild-type. Gastrocnemius muscles from 5- to 6-month-old mice were isolated from wild-type (empty bars) or the CEloxP/loxP (gray bars) and used for the determination of respiration rates. The data for the graphs represent the mean values ± SEM for four animals.
DISCUSSION
The main finding from this study was the identification of a novel non-canonical E-box in the CE of the MyoD promoter that is necessary for CLOCK:BMAL1 binding and confers circadian expression. The deletion of the CE abolished circadian expression of MyoD mRNA in vivo and transfection assays demonstrated that mutation of the E-box abolished the circadian expression in vitro. Of the E-boxes located within the CE, only the non-canonical E-box was capable of binding the core clock factors CLOCK and BMAL1. However, previous studies have shown that the canonical E-boxes in the CE are required for MyoD expression in developing skeletal muscle (9). Conclusions from this work were that the E-box 1 region is essential for basal MyoD expression while the E-box 4 region defines the CE as a target for Myf5-dependent activation of MyoD in myotomal muscle. Thus, our current findings expand the role of the CE in the regulation of MyoD expression to include BMAL1 and CLOCK binding and activation in adult skeletal muscle.
The finding that CLOCK and BMAL1 are capable of binding to a non-canonical E-box (5′-CAGCTT-3′) within the CE is not without precedent as these factors have been shown to bind a range of different E-box sequences (41,42). For instance, the CLOCK:BMAL1 heterodimer has been shown to regulate the circadian expression of Per1 via a canonical E-box (5′-CACGTG-3′) while analysis of the promoter of the clock-controlled gene, Dbp, demonstrated BMAL1 and CLOCK binding at multiple E-boxes, including one non-canonical E-box important for circadian expression (26). In Per2, the BMAL1: CLOCK heterodimer binds to an E-box-like sequence (5′-CACGTT-3′) that was shown to be sufficient and indispensable for the circadian oscillation of Per2 expression (39,43). The same non-canonical E-box was also shown to have circadian function in another clock-controlled gene, bHLH family member e41 (Bhlhe41) (44). In cryptochrome 1 (Cry1), a 47 bp DNA fragment containing a canonical E-box and a non-canonical E-box, both E-boxes were required and sufficient to drive Cry1 circadian expression with the appropriate phase (38,44). Collectively, these findings clearly show that both canonical and non-canonical like E-boxes we identified in the MyoD CE are commonly found as targets for BMAL1 and CLOCK binding.
The loss of circadian MyoD expression seen in the muscle of the CEloxP/loxP mouse clearly demonstrated that this regulatory region was necessary for conferring a circadian pattern of expression in adult muscle. This finding was somewhat surprising given that previous studies had shown that the DRR (distal regulatory region) was the enhancer responsible for regulating MyoD expression in adult skeletal muscle (8,11). The necessity of the CE for the circadian expression of MyoD, however, was consistent with our previous ChIP results showing that CLOCK and BMAL1 bound only the CE, not the DRR, in a diurnal manner (4). However this does not exclude the possibility that DRR and other elements within the MyoD promoter, which may also contribute to proper circadian MyoD expression. The CE and the DRR enhancers have been shown to exhibit complementary activities and together are able to recapitulate MyoD expression in developing and mature skeletal muscle (8,11,45,46,). The molecular mechanisms are not well defined at this time and future studies will be required to delineate all the necessary elements that cooperate to direct the proper MyoD circadian expression.
Since circadian MyoD mRNA expression is disrupted in the CEloxP/loxP mice, we used expression profiling to ask whether there was an alteration in muscle gene expression and if so, whether any of the changes were in common with those defined for the muscle of the ClockΔ19 mouse. This concept emerged from our earlier work using the ClockΔ19 mouse in which the loss of MyoD cycling, but not loss of MyoD expression, was associated with mis-expression of many muscle-specific genes. Furthermore, in agreement with the ClockΔ19 findings, the expression level of some of the same muscle-specific genes (Ttn, Tpm1, Tpm2, Myom2) were found to be mis-expressed in the CEloxP/loxP mice. While the downstream consequences of this alteration in MyoD target gene expression remains to be fully explored, preliminary studies indicate the muscle from the CEloxP/loxP mice does not phenocopy the ClockΔ19 in terms of mechanical function (Zhang, X., Wolff, G. and Esser, K.A., unpublished data). The reason for a lack of a muscle phenotype in the CEloxP/loxP mice at 4 months of age remains to be determined but may be an issue of time and/or age—it may take longer for the muscle phenotype to emerge in the CEloxP/loxP mice than in ClockΔ19 mouse. This is a reasonable hypothesis given that the muscle pathologies seen in the ClockΔ19 mouse occur in a system in which all cells/tissues exhibit circadian misalignment due to the mutation in Clock. Future studies will test whether long-term loss of MyoD oscillation in the CEloxP/loxP mice leads to downstream alterations associated with changes in the myogenic expression program.
Previously, we reported decreased mitochondrial volume and depressed respiratory function of muscle from circadian mutant Bmal1 KO and ClockΔ19 mice. However in MyoD KO mouse muscle, we did not detect any decrease in mitochondrial volume based on analysis of electron micrographs (4). In this study, we evaluated mitochondrial respiration in the muscle of the CEloxP/loxP mice based on our expression profiling data. We found a significant reduction in State 5 which is consistent with the array results but was not what we expected based on our morphological analysis of mitochondria from the MyoD KO mouse. One explanation for this difference is that we did not measure mitochondrial respiration in the muscle of the MyoD KO mouse, thus while the volume was unchanged the biochemical capacity was not assessed. Additionally, previous studies have reported that the loss of MyoD is compensated by other functionally redundant genes, such as Myf5, and this could act to preserve mitochondrial volume. In the muscle of the CEloxP/loxP mice we do not see any changes in expression of the other myogenic regulatory factor family. This leads us to suggest that circadian expression of MyoD in the adult mouse is important for the regulation of mitochondrial function.
In conclusion, we have identified a new function for the CE of MyoD in adult skeletal muscle. We determined that the CE was required for MyoD circadian expression in vivo. We also found that a novel non-canonical E-box located within the CE was required for the full activation of MyoD expression as well as its circadian expression. The loss of the MyoD circadian expression is associated with significant changes in expression of many MyoD target genes and disrupted mitochondrial function. These results provide evidence that the molecular clock in skeletal muscle is important for daily regulation of myogenic gene expression and contributes to the maintenance of mitochondrial/metabolic function of adult skeletal muscle.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–6 and Supplementary Figures 1–3.
FUNDING
The National Institutes of Health (R01AR055246 to K.A.E., R01AR44878 to D.J.G.). Funding for open access charge: NIH (R01AR055246).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Mitsunori Miyazaki, Gretchen Wolff and Mellani Lefta for their help with the circadian collections, Drs Elizabeth Schroder and Michael Hughes for help with the JTK analysis and Dr Ok-Kyong Park-Sarge for helpful discussions with this article.
REFERENCES
- 1.Tapscott SJ, Davis RL, Thayer MJ, Cheng PF, Weintraub H, Lassar AB. MyoD1: a nuclear phosphoprotein requiring a Myc homology region to convert fibroblasts to myoblasts. Science. 1988;242:405–411. doi: 10.1126/science.3175662. [DOI] [PubMed] [Google Scholar]
- 2.Megeney LA, Kablar B, Garrett K, Anderson JE, Rudnicki MA. MyoD is required for myogenic stem cell function in adult skeletal muscle. Genes Dev. 1996;10:1173–1183. doi: 10.1101/gad.10.10.1173. [DOI] [PubMed] [Google Scholar]
- 3.Rudnicki MA, Schnegelsberg PN, Stead RH, Braun T, Arnold HH, Jaenisch R. MyoD or Myf-5 is required for the formation of skeletal muscle. Cell. 1993;75:1351–1359. doi: 10.1016/0092-8674(93)90621-v. [DOI] [PubMed] [Google Scholar]
- 4.Andrews JL, Zhang X, McCarthy JJ, McDearmon EL, Hornberger TA, Russell B, Campbell KS, Arbogast S, Reid MB, Walker JR, et al. CLOCK and BMAL1 regulate MyoD and are necessary for maintenance of skeletal muscle phenotype and function. Proc. Natl Acad. Sci USA. 2010;107:19090–19095. doi: 10.1073/pnas.1014523107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miyazaki M, Schroder E, Edelmann SE, Hughes ME, Kornacker K, Balke CW, Esser KA. Age-associated disruption of molecular clock expression in skeletal muscle of the spontaneously hypertensive rat. PloS ONE. 2011;6:e27168. doi: 10.1371/journal.pone.0027168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goldhamer DJ, Brunk BP, Faerman A, King A, Shani M, Emerson CP., Jr Embryonic activation of the myoD gene is regulated by a highly conserved distal control element. Development. 1995;121:637–649. doi: 10.1242/dev.121.3.637. [DOI] [PubMed] [Google Scholar]
- 7.Kablar B, Krastel K, Ying C, Tapscott SJ, Goldhamer DJ, Rudnicki MA. Myogenic determination occurs independently in somites and limb buds. Dev. Biol. 1999;206:219–231. doi: 10.1006/dbio.1998.9126. [DOI] [PubMed] [Google Scholar]
- 8.Chen JC, Love CM, Goldhamer DJ. Two upstream enhancers collaborate to regulate the spatial patterning and timing of MyoD transcription during mouse development. Dev. Dyn. 2001;221:274–288. doi: 10.1002/dvdy.1138. [DOI] [PubMed] [Google Scholar]
- 9.Kucharczuk KL, Love CM, Dougherty NM, Goldhamer DJ. Fine-scale transgenic mapping of the MyoD core enhancer: MyoD is regulated by distinct but overlapping mechanisms in myotomal and non-myotomal muscle lineages. Development. 1999;126:1957–1965. doi: 10.1242/dev.126.9.1957. [DOI] [PubMed] [Google Scholar]
- 10.Chen JC, Goldhamer DJ. The core enhancer is essential for proper timing of MyoD activation in limb buds and branchial arches. Dev. Biol. 2004;265:502–512. doi: 10.1016/j.ydbio.2003.09.018. [DOI] [PubMed] [Google Scholar]
- 11.Gerber AN, Klesert TR, Bergstrom DA, Tapscott SJ. Two domains of MyoD mediate transcriptional activation of genes in repressive chromatin: a mechanism for lineage determination in myogenesis. Genes Dev. 1997;11:436–450. doi: 10.1101/gad.11.4.436. [DOI] [PubMed] [Google Scholar]
- 12.Ko CH, Takahashi JS. Molecular components of the mammalian circadian clock. Hum Mol Genet. 2006;15(Spec. No. 2):R271–R277. doi: 10.1093/hmg/ddl207. [DOI] [PubMed] [Google Scholar]
- 13.Hastings MH, Herzog ED. Clock genes, oscillators, and cellular networks in the suprachiasmatic nuclei. J. Biol. Rhythms. 2004;19:400–413. doi: 10.1177/0748730404268786. [DOI] [PubMed] [Google Scholar]
- 14.Yu W, Nomura M, Ikeda M. Interactivating feedback loops within the mammalian clock: BMAL1 is negatively autoregulated and upregulated by CRY1, CRY2, and PER2. Biochem. Biophys. Res. Commun. 2002;290:933–941. doi: 10.1006/bbrc.2001.6300. [DOI] [PubMed] [Google Scholar]
- 15.Shearman LP, Sriram S, Weaver DR, Maywood ES, Chaves I, Zheng B, Kume K, Lee CC, van der Horst GT, Hastings MH, et al. Interacting molecular loops in the mammalian circadian clock. Science. 2000;288:1013–1019. doi: 10.1126/science.288.5468.1013. [DOI] [PubMed] [Google Scholar]
- 16.Lefta M, Wolff G, Esser KA. Circadian rhythms, the molecular clock, and skeletal muscle. Curr. Top. Dev. Biol. 2011;96:231–271. doi: 10.1016/B978-0-12-385940-2.00009-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang X, Dube TJ, Esser KA. Working around the clock: circadian rhythms and skeletal muscle. J. Appl. Physiol. 2009;107:1647–1654. doi: 10.1152/japplphysiol.00725.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCarthy JJ, Andrews JL, McDearmon EL, Campbell KS, Barber BK, Miller BH, Walker JR, Hogenesch JB, Takahashi JS, Esser KA. Identification of the circadian transcriptome in adult mouse skeletal muscle. Physiol. Genomics. 2007;31:86–95. doi: 10.1152/physiolgenomics.00066.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoo SH, Yamazaki S, Lowrey PL, Shimomura K, Ko CH, Buhr ED, Siepka SM, Hong HK, Oh WJ, Yoo OJ, et al. PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc. Natl Acad. Sci. USA. 2004;101:5339–5346. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller BH, McDearmon EL, Panda S, Hayes KR, Zhang J, Andrews JL, Antoch MP, Walker JR, Esser KA, Hogenesch JB, et al. Circadian and CLOCK-controlled regulation of the mouse transcriptome and cell proliferation. Proc. Natl Acad. Sci. USA. 2007;104:3342–3347. doi: 10.1073/pnas.0611724104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Escobedo J, Koh TJ. Improved transfection technique for adherent cells using a commercial lipid reagent. Biotechniques. 2003;35:936–938, 940. doi: 10.2144/03355bm06. [DOI] [PubMed] [Google Scholar]
- 22.Balsalobre A, Damiola F, Schibler U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell. 1998;93:929–937. doi: 10.1016/s0092-8674(00)81199-x. [DOI] [PubMed] [Google Scholar]
- 23.Grechez-Cassiau A, Rayet B, Guillaumond F, Teboul M, Delaunay F. The circadian clock component BMAL1 is a critical regulator of p21WAF1/CIP1 expression and hepatocyte proliferation. J. Biol. Chem. 2008;283:4535–4542. doi: 10.1074/jbc.M705576200. [DOI] [PubMed] [Google Scholar]
- 24.Schreiber E, Matthias P, Muller MM, Schaffner W. Rapid detection of octamer binding proteins with ‘mini-extracts’, prepared from a small number of cells. Nucleic Acids Res. 1989;17:6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakahata Y, Yoshida M, Takano A, Soma H, Yamamoto T, Yasuda A, Nakatsu T, Takumi T. A direct repeat of E-box-like elements is required for cell-autonomous circadian rhythm of clock genes. BMC Mol. Biol. 2008;9:1. doi: 10.1186/1471-2199-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ripperger JA, Schibler U. Rhythmic CLOCK-BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions. Nat. Genet. 2006;38:369–374. doi: 10.1038/ng1738. [DOI] [PubMed] [Google Scholar]
- 27.Sato TK, Panda S, Miraglia LJ, Reyes TM, Rudic RD, McNamara P, Naik KA, FitzGerald GA, Kay SA, Hogenesch JB. A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron. 2004;43:527–537. doi: 10.1016/j.neuron.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 28.Ballesta A, Dulong S, Abbara C, Cohen B, Okyar A, Clairambault J, Levi F. A combined experimental and mathematical approach for molecular-based optimization of irinotecan circadian delivery. PLoS Comput. Biol. 2011;7:e1002143. doi: 10.1371/journal.pcbi.1002143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pan X, Zhang Y, Wang L, Hussain MM. Diurnal regulation of MTP and plasma triglyceride by CLOCK is mediated by SHP. Cell Metab. 2010;12:174–186. doi: 10.1016/j.cmet.2010.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yin L, Joshi S, Wu N, Tong X, Lazar MA. E3 ligases Arf-bp1 and Pam mediate lithium-stimulated degradation of the circadian heme receptor Rev-erb alpha. Proc. Natl Acad. Sci. USA. 2010;107:11614–11619. doi: 10.1073/pnas.1000438107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tamanini F. Manipulation of mammalian cell lines for circadian studies. Methods Mol. Biol. 2007;362:443–453. doi: 10.1007/978-1-59745-257-1_36. [DOI] [PubMed] [Google Scholar]
- 32.Yamazaki S, Takahashi JS. Real-time luminescence reporting of circadian gene expression in mammals. Methods Enzymol. 2005;393:288–301. doi: 10.1016/S0076-6879(05)93012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patel SP, Gamboa JL, McMullen CA, Rabchevsky A, Andrade FH. Lower respiratory capacity in extraocular muscle mitochondria: evidence for intrinsic differences in mitochondrial composition and function. Invest. Ophthalmol. Vis. Sci. 2009;50:180–186. doi: 10.1167/iovs.08-1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sullivan PG, Krishnamurthy S, Patel SP, Pandya JD, Rabchevsky AG. Temporal characterization of mitochondrial bioenergetics after spinal cord injury. J. Neurotrauma. 2007;24:991–999. doi: 10.1089/neu.2006.0242. [DOI] [PubMed] [Google Scholar]
- 35.Hughes ME, Hogenesch JB, Kornacker K. JTK_CYCLE: an efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. J. Biol. Rhythms. 2010;25:372–380. doi: 10.1177/0748730410379711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Antoch MP, Song EJ, Chang AM, Vitaterna MH, Zhao Y, Wilsbacher LD, Sangoram AM, King DP, Pinto LH, Takahashi JS. Functional identification of the mouse circadian Clock gene by transgenic BAC rescue. Cell. 1997;89:655–667. doi: 10.1016/s0092-8674(00)80246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kornmann B, Schaad O, Bujard H, Takahashi JS, Schibler U. System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol. 2007;5:e34. doi: 10.1371/journal.pbio.0050034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fustin JM, O'Neill JS, Hastings MH, Hazlerigg DG, Dardente H. Cry1 circadian phase in vitro: wrapped up with an E-box. J. Biol. Rhythms. 2009;24:16–24. doi: 10.1177/0748730408329267. [DOI] [PubMed] [Google Scholar]
- 39.Yoo SH, Ko CH, Lowrey PL, Buhr ED, Song EJ, Chang S, Yoo OJ, Yamazaki S, Lee C, Takahashi JS. A noncanonical E-box enhancer drives mouse Period2 circadian oscillations in vivo. Proc. Natl Acad. Sci. USA. 2005;102:2608–2613. doi: 10.1073/pnas.0409763102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cao Y, Yao Z, Sarkar D, Lawrence M, Sanchez GJ, Parker MH, MacQuarrie KL, Davison J, Morgan MT, Ruzzo WL, et al. Genome-wide MyoD binding in skeletal muscle cells: a potential for broad cellular reprogramming. Dev. Cell. 18:662–674. doi: 10.1016/j.devcel.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gekakis N, Staknis D, Nguyen HB, Davis FC, Wilsbacher LD, King DP, Takahashi JS, Weitz CJ. Role of the CLOCK protein in the mammalian circadian mechanism. Science. 1998;280:1564–1569. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- 42.Jin X, Shearman LP, Weaver DR, Zylka MJ, de Vries GJ, Reppert SM. A molecular mechanism regulating rhythmic output from the suprachiasmatic circadian clock. Cell. 1999;96:57–68. doi: 10.1016/s0092-8674(00)80959-9. [DOI] [PubMed] [Google Scholar]
- 43.Akashi M, Ichise T, Mamine T, Takumi T. Molecular mechanism of cell-autonomous circadian gene expression of Period2, a crucial regulator of the mammalian circadian clock. Mol. Biol. Cell. 2006;17:555–565. doi: 10.1091/mbc.E05-05-0396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ueda HR, Hayashi S, Chen W, Sano M, Machida M, Shigeyoshi Y, Iino M, Hashimoto S. System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nat. Genet. 2005;37:187–192. doi: 10.1038/ng1504. [DOI] [PubMed] [Google Scholar]
- 45.Chen JC, Ramachandran R, Goldhamer DJ. Essential and redundant functions of the MyoD distal regulatory region revealed by targeted mutagenesis. Dev. Biol. 2002;245:213–223. doi: 10.1006/dbio.2002.0638. [DOI] [PubMed] [Google Scholar]
- 46.Asakura A, Lyons GE, Tapscott SJ. The regulation of MyoD gene expression: conserved elements mediate expression in embryonic axial muscle. Dev. Biol. 1995;171:386–398. doi: 10.1006/dbio.1995.1290. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.