Abstract
The brain is a large and complex network of neurons. Specific neuronal connectivity is thought to be based on the combinatorial expression of the 52 protocadherins (Pcdh) membrane adhesion proteins, whereby each neuron expresses only a specific subset. Pcdh genes are arranged in tandem, in a cluster of three families: Pcdhα, Pcdhβ and Pcdhγ. The expression of each Pcdh gene is regulated by a promoter that has a regulatory conserved sequence element (CSE), common to all 52 genes. The mechanism and factors controlling individual Pcdh gene expression are currently unknown. Here we show that the promoter of each Pcdh gene contains a gene-specific conserved control region, termed specific sequence element (SSE), located adjacent and upstream to the CSE and activates transcription together with the CSE. We purified the complex that specifically binds the SSE–CSE region and identified the CCTC binding-factor (CTCF) as a key molecule that binds and activates Pcdh promoters. Our findings point to CTCF as a factor essential for Pcdh expression and probably governing neuronal connectivity.
INTRODUCTION
The brain consists of a large and complex organism-specific network of neurons. A fundamental question in neurobiology is where and how the structure of this network is encoded in each organism's genome, i.e. how individual neurons acquire unique identities that enable them to create highly-specific synaptic connections, leading to the formation of an organism-specific network. There are several well-established examples for such mechanisms: the olfactory system, in which individual olfactory neurons express only 1 out of 1300 olfactory receptor genes, and establish connections based on the receptor expressed (1), and the Down-Syndrome-Cell-Adhesion-Molecules (Dscam) that regulate neural circuit formation in Drosophila (2–4). In mammals, it was hypothesized that the precise patterns of neuronal connectivity are largely determined by neuronal membrane molecules called protocadherins (Pcdh), which promote specific interneuron connections. These molecules are encoded by the clustered Protocadherin (Pcdh) genes (5–11) that represent the largest subgroup in the cadherin superfamily (12,13). The Pcdh genes are present in all known vertebrate genomes, including mammals, chicken, zebrafish, fugu, and coelacanth (13) and are highly conserved in mammals. The Pcdh genes were shown to be highly expressed during neural development, creating Pcdh proteins that are concentrated in the synaptic region. As the brain matures, the expression level of the Pcdh genes decreases (10,12,14–17). Gene-knockout studies have demonstrated that Pcdh gene products play a crucial role in proper axonal projection, synaptic formation and neuronal survival (18,19).
Pcdh genes are located on human chromosome 5 (13) and on mouse chromosome 18 (20,21). In each neuron, Pcdh genes are expressed monoallelically, each allele is independently regulated, i.e. one variable exon is expressed from the paternal chromosome and another variable exon from the maternal chromosome, creating a combinatorial expression at the cell level (22,23). There are 52 tandem-arranged Pcdh genes in human that are divided into three families: Pcdhα (15 genes), Pcdhβ (16 genes), and Pcdhγ (21 genes), which are further subdivided into Pcdhγa and Pcdhγb (21).
In the Pcdhα and Pcdhγ families, each gene has a specific variable exon (24) that is linked to three constant exons (a structure similar to that of immunoglobulins and T-cell-receptors). In the Pcdhβ family, each gene includes a variable exon only. Linking of a specific variable exon with the three constant exons in the Pcdhα and Pcdhγ genes is done by alternative splicing (as opposed to gene recombination used in immunoglobulin genes). Prior to splicing, each precursor mRNA transcribed by the Pcdhα and Pcdhγ genes, is of high molecular weight, since it includes all downstream variable exons (25). During splicing, only the 5′-most variable exon is cis-spliced to the first constant exon to generate functional mRNAs (13,21,26).
Assuming that each cluster expresses, at the most, one gene from each allele, this unusual Pcdh expression may provide sufficient diversity to represent at least 1.5 million unique individual cell labeling (see materials and methods for the calculation). Each Pcdh variable exon is preceded by a distinct promoter, and all promoters contain a similar highly conserved core motif of ∼22 bp, the conserved sequence element (CSE) (26,27). In addition, long-range cis-regulatory DNA elements in the Pcdhα gene cluster, HS5-1 and HS7, were identified and found to possess enhancer activity in reporter assays (28). Interestingly, both elements are conserved among vertebrates and also include a CSE. The Pcdhα genes are likely to be regulated by methylation as the transcription of specific Pcdhα genes was found to be significantly correlated with the methylation state of the first exon. On the other hand, mosaic or mixed methylation states of the CSE in the promoters were associated with both active and inactive transcription (29). Presently, the mechanisms underlying promoter choice and promoter activity are largely unknown.
In the present study, we investigated the mechanism underlying Pcdh gene transcription. Using bioinformatics methods, we have identified a sequence element located near the CSE that is highly conserved among mammals but, unlike the Common Sequence Element, is unique to each of the 52 Pcdh genes. We termed the identified element specific sequence element (SSE). The 20-bp-long SSE is essential for transcription and can activate transcription only in the presence of the CSE. We have purified the complex that binds the SSE–CSE region and identified CCTC-binding factor (CTCF) as a factor that binds the promoters of Pcdh genes through the common CSE and the SSE elements. Remarkably in the context of Pcdh genes CTCF plays an essential positive role in transcription. Our findings point to CTCF as a factor that is essential for Pcdh expression and probably for the control of neuronal connectivity.
MATERIALS AND METHODS
Bioinformatics analysis of the Pcdh promoters
Regulatory elements such as enhancers and locus control regions are highly conserved among different mammalian species. To identify regions containing putative DNA elements that regulate Pcdh expression, we compared the genomic DNA upstream to the first exon of each gene in the three Pcdh families (α, β and γ). Human, chimp, mouse, rat and dog promoter regions from −1000 relative to the TSS were retrieved from UCSC Genome Browser (http://genome.ucsc.edu/). This analysis revealed new conserved sequences located immediately upstream of the previously identified CSE. These regions are more conserved among species (orthologs) than among family members within the same species (paralogs) (Supplementary Figure S1).
For calculation of combinational Pcdh diversity, we used binomial coefficients: the number of ways to choose K elements from N is 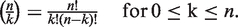
The calculation is as follows: the number of ways to choose two variable exons from 13 (for Pcdhα), 13!/11! × 2!, multiplied by choose 2 from 15 (for Pcdhβ), 15!/13! × 2!, multiplied by choose 2 from 19 (for Pcdh γ), 19!/17! × 2!, is equal to 1.5 million unique labels. This calculation does not take into account that the Pcdhα and Pcdhγ proteins also form oligomers (30), which can further increase the molecular diversity at the cell surface.
Plasmid construction
The promoter regions of the α6 and α3 genes (−217 and −219 relative to translation start site, respectively) were cloned by genomic PCR into pGL3-Basic (Promega) using the HindIII site. The α6, α3 and deletions promoters region was amplified using primers that introduce a HindIII restriction site at the end of the PCR products (Supplementary Data S1, primers 17–20 and 28–30); For the mutatgenesis of the αSSE linker addition, we used oligonucleotides containing the mutated or linker sequences flanked by KpnI in 5′ and NheI in 3′ sites located immediately upstream and downstream, respectively, to the αSSE site (Supplementary Data S1, primers 21–27 and 31–51). All plasmids constructed in this study were verified by DNA sequencing.
Transient transfection assays and RNA analysis
The 293T and SH-SY5Y cells were maintained and transfected as described (31). HEC1-B cells (human endometrial cancer cell line) were maintained in Minimum essential medium (Eagle) with 2 mM l-glutamine and Earle's BSS adjusted to contain 1.5 g/l sodium bicarbonate, 0.1 mM non-essential amino acids, and 1.0 mM sodium pyruvate, 90%; fetal bovine serum, 10%. All transfections were performed by using Lipofectamine 2000 (Invitrogen).
For reporter assays, subconfluent cells were transfected in a 24-well plate using 750 ng luciferase reporter plasmid, 25 ng CMV-GFP, 50 ng Rous sarcoma virus (RSV) promoter-driven Renilla luciferase reporter plasmid, At 24 h after transfection, cells were harvested and their luciferase and Renilla luciferase activities were measured. The mean and the SD values were determined for each construct based on four independent transfections.
For determining the effect of CTCF depletion on the activity of the reporter gene, SH-SY5Y cells were grown on 10-well plates and transfected with CTCF siRNA (Origene, cat # SR307273), or Scrambled siRNA-Scr (Origene, cat # SR30004) as a negative control at a concentration of 10 nM. Twenty-four hours later, the cells were split to 12-well plates. After an additional 72 h (96 h after siRNA transfection) the cells were transfected with 1.5 µg luciferase reporter plasmid, 50 ng CMV-GFP, 100 ng Rous sarcoma virus (RSV) promoter-driven Renilla luciferase reporter plasmid. At 24 h after second transfection, cells were harvested and their luciferase and Renilla luciferase activities were measured. The mean and the SD values were determined for each construct based on four independent transfections.
For determining the effect of CTCF depletion on the activity of the endogenous Pcdh mRNA levels, HEC1-B cells were grown on 6-well plate and tranfected with 10 nM CTCF siRNA (Origene, cat # SR307273), or Scrambled siRNA-Scr (Origene, cat # SR30004) as a negative control. One hundred and twenty hours later total RNA was extracted using Tri-reagent (MRC Inc.). cDNAs were synthesized from 1 µg of total RNA in a 20 µl reaction volume using the SuperScript III Reverse Transcriptase (Invitrogen) and random primers as per the manufacturer's instructions. Quantitative real time PCRs were performed in duplicate using the Applied Biosystems 7300 Real-Time PCR System (Applied Biosystems). Independent PCRs were performed using the same cDNA for genes of interest and the GAPDH gene which serves as an internal control, using the SYBR® Green PCR Master Mix (Applied Biosystems). Gene-specific primers were designed for the genes of interest and for the GAPDH gene using Primer Express® software (Applied Biosystems). The sequences of the primer pairs are listed in Supplementary Data S3, primers 1–5.
Electrophoretic mobility shift assay and ‘super-shift’ assay
DNA oligonucleotides containing the α6SSE–CSE or α3SSE–CSE or α12SSE–CSE or SP1 sequences (Supplementary Data S2, oligonucleotides 1–2, 10 and 18) were fluorescently labeled on the 5′-end with Cy5, HEX, or FAM (Integrated DNA Technologies, Inc). The oligonucleotides were annealed in 20 µl in a concentration of 10 pmol/µl and used as probes to the reaction. The binding reactions containing 2 µg of poly(dI-dC) (Sigma), 2 µg of poly(dA-dT) (Sigma), 10 µM Zn and 10 µg of HEC-1B/SH-SY5Y nuclear extract prepared as described previously (32), with binding buffer consisting of 25 mM HEPES (pH 7.9), 50 mM KCl, 1 mM DTT and 10% glycerol. The reaction mix was incubated on ice for 10 min after which 500 fmol probe was added for an additional 20 min. Competitor doubled stranded DNAs (50×, 25 pmol) were added prior to the addition of the probe (Supplementary Data S2, oligonucleotides 3–9, 11–17 and 19–21). The muted doubled stranded DNA sequences have the same sticky ends. The reactions were separated by native electrophoresis at 4°C in a 6.5% polyacryamide gel with 1× Tris–Glycine buffer at 185 V. The gel visualized with the Typhoon 9400 instrument (Amersham Biosciencs). Supershift assays were carried out with antibodies to CTCF (Abcam ab70303, ab37477, ab37478), BUB3 (Abcam ab4180) and YY1 (Abcam ab12132). Methylation assays were carried out using methylated competitor DNA in 50- to 100-fold excess. The competitor DNA was synthesized with methyl Cs instead of Cs in two places of CpG sequences (Integrated DNA Technologies, Inc).
Fractionation of nuclear extract by Macro-prep high S cation exchange column
SH-SY5Y nuclear extract prepared as described previously was incubated with washed Macro-prep high S cation exchange support beads (Bio-Rad) at 4°C for 30 min. Flow through fractions where collected after separation from the beads using centrifugation at 500 G for 4 min. Elution with 200, 400 and 800 mM KCl buffers containing 25 mM HEPES (pH 7.9), 1 mM EDTA, 1 mM dithiothreitol and 10% glycerol was performed using centrifugation at 500 G for 4 min. Dialysis over night was performed on eluted fractions using 10 KDa dialysis bag (Thermo scientific) in dialysis buffer described previously. A fraction of all the eluted samples was analyzed on SDS–PAGE gradient acryl amid gel 4–12.5% (GEBA) with 1× Tris–Glycine 0.1% SDS buffer at 130 V for 1 h.
Affinity purification and western blot
Affinity magnetic FG plain beads were prepared as follows: oligonucleotides of the α6 and α6_mut sequences, with phosphorylated GGGG or CCCC at the 5′-end of the forward and reverse complement, respectively, were synthesized (Integrated DNA Technologies, Inc) (Supplementary Data S2, oligonucleotides 22–23).
The oligonucleotides were annealed and then ligated using T4 ligase 5 U/µl (Fermentas) using the manufacturer ligation buffer for 16 h in 4°C to generate a concatamer of 5–15 repeats. The muted sequence has the same sticky ends. The ligated DNA was then phenol–chloroform extracted and desalted using Nick column (GE healthcare) equilibrated with water. The ligated samples were run on agarose electrophoresis gel to verify that the ligation process has succeeded (Supplementary Figure S5). The ligated DNA was then immobilized to 10 mg magnetic FG plain beads (Tamagawa seiki Co. Ltd) by incubation at 50°C for 24 h. The immobilized beads where then washed with 2.5 M KCl and incubated with 1 M ethanoamine (pH 8) over night for masking. Prior to purification the immobilized beads were washed three times using a magnetic stand in washing buffer containing 25 mM HEPES (pH 7.9), 100 mM KCl, 1 mM EDTA, 1 mM dithiothreitol and 10% glycerol. Crude nuclear extract or elution fraction collected from cation fractionation was incubated with beads at 4°C for 4 h using rotator in binding buffer containing 2 µg of poly(dI-dC) (Sigma), 1 µg of poly(dA-dT) (Sigma) consisting of 25 mM HEPES (pH 7.9), 50 mM KCl, 1 mM dithiothreitol and 10% glycerol. Beads were then washed four times using a magnetic stand with washing buffer previously described. Bound proteins were then eluted from the beads using a magnetic stand with elution buffer at room temperature for 30 min consisting of, 25 mM HEPES (pH 7.9), 1 M KCl, 1 mM EDTA, 1 mM dithiothreitol and 10% glycerol. Dialysis over night was performed on a fraction of the sample using dialysis bag (Thermo scientific) in dialysis buffer described previously. A fraction of the sample was sent to Mass-spec analysis for protein identification. Tandem mass spectrometry (MS/MS) analysis coupled with liquid chromatography (LC) was carried out by the Smoler Proteomics Center (Faculty of Biology, Technion, Israeli Institute of Technology, Israel). Top candidates are those proteins that were identified by mass spectrometry analysis following DNA affinity purification only from the wild-type SSE–CSE but not detected at the elution of the mutated SSE–CSE sequences in three repeats.
For western blot, a fraction of the sample was analyzed on SDS–PAGE gradient acryl amid gel 4–12.5% (GEBA) with 1× Tris–Glycine 0.1% SDS buffer at 130 V for 1 h, transferred to nitrocellulose membranes blots, and probed with anti-CTCF (Abcam ab70303) and anti-Histone H3 (Abcam ab1791) antibodies. Secondary antibodies consisted of goat anti-rabbit conjugated to IRDye800 or to IRDye680 (LI-COR Biosciences), were used and then the membrane was scanned for infrared signal using the Odyssey Imaging System (LI-COR Biosciences).
Production of CTCF Proteins in vitro
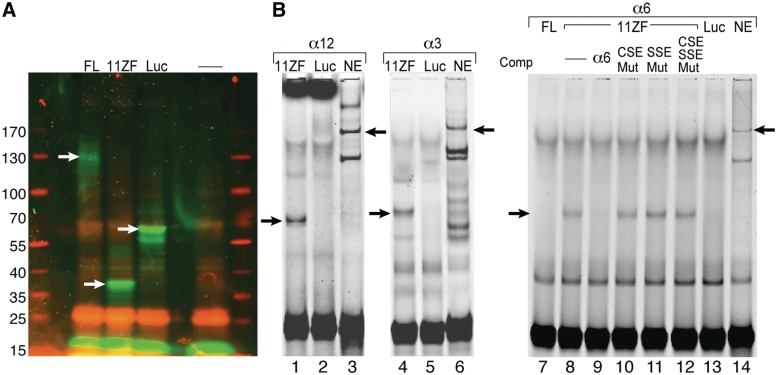
Full-length human CTCF (pET-7.1) and the 11 ZF CTCF-binding domain (pET-11ZF) which were verified in several papers (33,34), were kindly donated by V. Lobanenkov (National Institute of Allergy and Infectious Diseases, National Institutes of Health, Rockville, MD, USA). These plasmids along with Luciferase T7 Control DNA, which serves as a positive control, were synthesized using the TnT® T7 Quick Coupled Transcription/Translation System (Promega). The in vitro synthesized proteins were labeled with a fluorescently labeled lysine amino acid using the FluoroTect GreenLys labeling system according to the manufacturer’s instructions (Promega). We verified the identity of the plasmids using restriction enzyme analysis and SDS–PAGE analysis which verified that the synthesized proteins have the correct molecular mass (Figure 6A).
Figure 6.
Recombinant CTCF binds specifically to several αSSE–CSE DNA sequences. (A) SDS–PAGE analysis of the full-length CTCF and 11ZF CTCF-binding domain proteins, which were synthesized in vitro from the pET-7.1 and pET-11ZF constructs. Luciferase T7 Control DNA no plasmid were used for positive and negative controls, respectively. The in vitro synthesized proteins are fluorescently labeled. Positions of the molecular mass protein markers (on the left) are indicated. The white arrows point to the positions of the in vitro synthesized proteins. (B) EMSAs using in vitro-translated luciferase, human CTCF full-length (FL), 11ZF or SH-SY5Y nuclear extract with αSSE–CSE sequences as probes. Lanes 1–3, α12SSE–CSE probe, lanes 4–6, α3SSE–CSE probe and lanes 7–14, α6SSE–CSE probe. The proteins used for binding and the competitor DNA are indicated on the top. The specific complexes between the probe and the recombinant proteins or the nuclear extract are indicated by arrows.
RESULTS
Identification of a novel cis-element in the promoters of the Pcdh genes
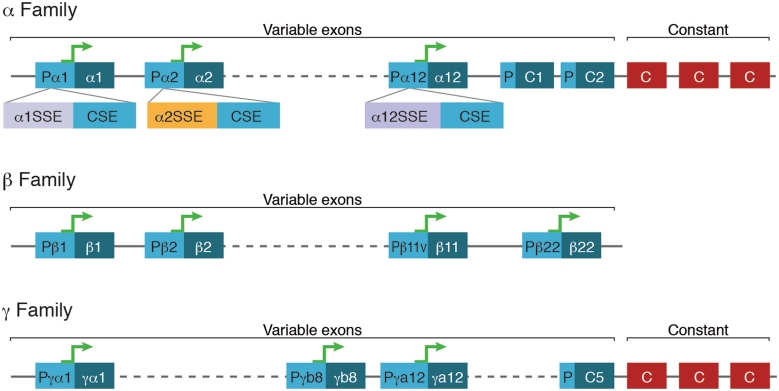
In order to identify the regulatory elements that control Pcdh expression, we first looked for additional segments in the Pcdh cluster that are conserved among mammalian species, namely: chimp, mouse, rat, dog and human. The 1000 bp upstream of each of the 52 Pcdh V-exons from the above species were aligned. This analysis revealed novel highly conserved regions located immediately upstream of the previously defined conserved sequence element, CSE (16,35,36) (Figure 1). Interestingly, unlike the CSE which is common to the promoters of all families, these conserved regions, although they may have common regions, are unique to each gene (Supplementary Figure S1). We therefore termed these regions, Specific Sequence Elements (SSE). All 52 occurrences of the CSE and the 52 unique SSEs are highly conserved in mammals (Supplementary Figure S1). The SSEs we analyze consists of α(1 to 13)SSE, β(1 to 16)SSE, γa(1 to 12)SSE and γb(1 to 7)SSE. The PcdhαC1, αC2, PcdhγC3, γC4, and γC5 were omitted from this analysis.
Figure 1.
Schematic representation of the Pcdh gene cluster promoters. Genomic organization of the Pcdh genes, with tandem variable region exons (blue), promoter regions (turquoise) and constant region exons (red). Each promoter region contains an SSE followed by a CSE, which is common to all genes of the family. V, variable region; C, constant region.
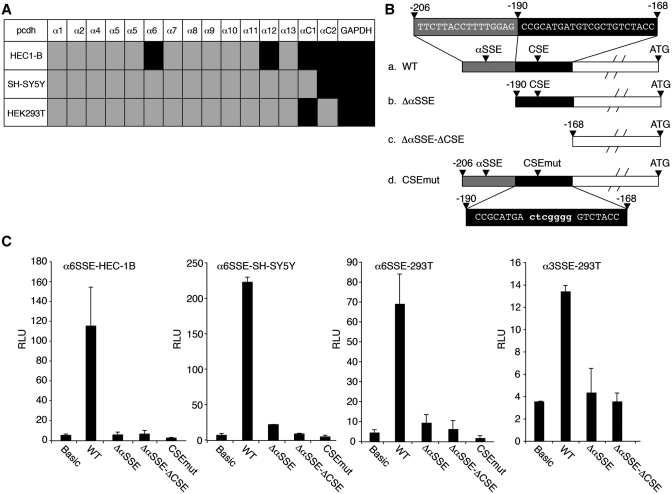
The αSSE is essential for Pcdh transcriptional activity
To investigate the transcriptional control of the Pcdh gene clusters we chose to focus on the Pcdhα family. We first examined the expression of all the Pcdhα genes in HEC-1B, 293T and SH-SY5Y cell lines by RT–PCR and found that each of these cell lines expresses at least one Pcdhα gene (Figure 2A). While the CSE has been previously shown to be important for the transcription of Pcdh genes (16,35,36) we wished to examine the function of the newly discovered αSSE in the Pcdhα family, and for this purpose Pcdhα6 was selected. The α6 promoter containing the α6SSE and the CSE was cloned in front of a luciferase reporter gene (Figure 2B). Upon transfection into HEC-1B, 293T and SH-SY5Y cell lines, this promoter fragment displayed a high transcriptional activity compared to the promoter-less (basic) reporter gene in all these cells (Figure 2C, WT columns). When the α6SSE or α6SSE plus CSE were deleted (Δα6SSE or Δα6SSE-ΔCSE, respectively) the transcriptional activity was severely diminished in all the cell lines (Figure 2C). Likewise, point mutations in the CSE (CSEmut) also diminished transcription (Figure 2C), in agreement with previous reports (16,35,36). We also examined the SSE function of the Pcdhα3 gene promoter (α3SSE) and found it to be essential for α3 promoter activity (Figure 2C, fourth panel). These findings confirm that CSE is crucial but not sufficient for Pcdh expression and also requires in addition the SSE to drive the Pcdh transcription.
Figure 2.
αSSE and CSE activate transcription cooperatively. (A) The expression profile of all the Pcdhα genes in HEC-1B, 293T and SH-SY5Y cell lines by RT–PCR, (Supplementary Figure S2). Gray and black boxes indicate for absence or presence of Pcdhα mRNA, respectively. (B) A scheme depicting the sequences of α6SSE–CSE WT and mutants (b–d). (C) WT (α6SSE–CSE and α3SSE–CSE) and the mutated promoters (fused to firefly luciferase reporter gene) were transfected into HEC-1B, 293T and SH-SY5Ycell lines together with RSV-Renilla that serves as internal control. The parental pGL3-basic (Basic) was also transfected as a control. Twenty-four hours post-transfection firefly and renilla luciferase activities were measured. The normalized results are the mean of at least four independent experiments (±SD).
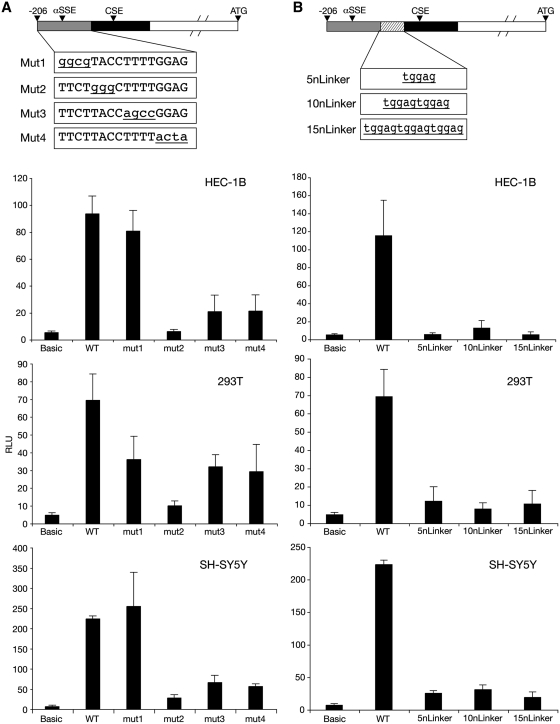
To investigate α6SSE further, α6SSE mutants were created by site-directed mutagenesis (Figure 3A, Mut1–Mut4). The α6SSE Mut2 construct reduced the transcriptional activity to the level of the promoter-less (basic) construct, Mut3–Mut4 promoters caused reduction of 70% while Mut1 did not have a significant effect on the transcription level (Figure 3A).
Figure 3.
Sequence requirements for the function of α6SSE–CSE as transcriptional elements. (A) Successive blocks within αSSE (underlined) in the α6SSE–CSE promoter were mutated (Mut1-Mut4). The wild-type and mutated constructs were transfected into HEC-1B, 293T and SH-SY5Y cell lines together with RSV-Renilla that serves as internal control. Twenty-four hours post-transfection firefly and Renilla luciferase activities were measured. The normalized results are the mean of at least four independent experiments (±SD). (B) Linkers of 5, 10 and 15 bp (5nLinker, 10nLinker and 5nLinker) were introduced between α6SSE and CSE and their effect was analyzed as described earlier.
To determine whether the distance between the SSE and the CSE is important for their function we increased the spacing between the SSE and the CSE sites by 5, 10 and 15 bp, which corresponded to ∼0.5, 1 and 1.5 helical turns (5nLinker, 10nLinker and 5nLinker). The results revealed that the transcriptional activity was reduced to the level of promoter-less construct in all the examined cell lines, regardless of the size of the spacer that was introduced (Figure 3B). These findings suggest that the specific location of the SSE relative to the CSE is crucial for their promoter activity.
Identification of a specific DNA-binding complex that is αSSE and CSE dependent
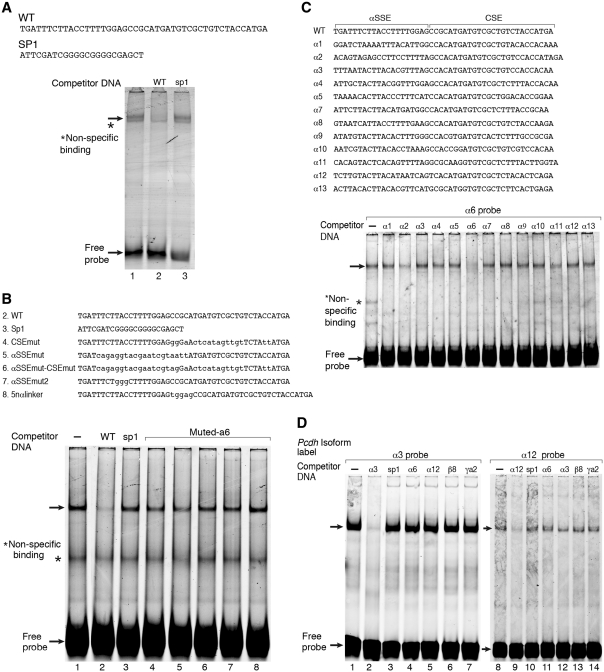
To investigate further the mechanism underlying αSSE–CSE activity we analyzed the proteins that interact with the αSSE–CSE region. A fluorescently labeled DNA fragment comprising the α6SSE–CSE sequence was incubated with a nuclear extract prepared from HEC-1B cells and then subjected to electrophoresis mobility shift assay (EMSA). Two major complexes were formed between the DNA and the extract (Figure 4A). The complexes were competed out by excess (50-fold) of unlabeled α6SSE–CSE DNA (Figure 4A, lane 2) but not by excess of the non-relevant Sp1 sequence DNA (Figure 4A, lane 3).
Figure 4.
Gene-specific complex binds to the αSSE–CSE. (A) EMSA using HEC-1B cell nuclear extract and a fluorescently labeled double stranded oligonucleotide containing αSSE–CSE sequence as a probe. Lane 1, the probe is incubated with HEC-1B nuclear extract. Competitor DNAs were added to the reactions in lanes 2 and 3 as indicated on the top. The sequences of the oligonucleotides used for binding and competition are shown on the top. (B) αSSE–CSE display cooperative DNA binding activity. EMSA using HEC-1B cell nuclear extract and a fluorescently labeled double stranded oligonucleotide containing α6SSE–CSE as a probe. Unlabeled competitor DNAs were added to the reactions as indicated in the top panel. The sequences of the oligonucleotides used for binding and competition are shown earlier. (C) The specific complex is shown to bind with high affinity to α6SSE–CSE. EMSA using SH-SY5Y cell nuclear extract and a fluorescently labeled double stranded oligonucleotide containing α6SSE–CSE as a probe. Unlabeled αSSE–CSE sequences of Pcdhα genes as competitor DNAs were added in excess to the reactions as indicated in the top panel. The sequences of the oligonucleotides used for binding and competition are shown earlier. (D) Competition assay for α3SSE–CSE and to α12SSE–CSE specific complex. EMSA using HEC-1B cell nuclear extract and a fluorescently labeled double stranded oligonucleotide comprising of α3SSE–CSE as a probe in the left panel and α12SSE–CSE as a probe in the right panel. Unlabeled competitor DNAs were added to the reactions as indicated in the top panel. The specific DNA complex and the free probes are indicated by arrows.
To examine the importance of the α6SSE–CSE region, competition assays with excess of various types of unlabeled DNA were performed: (i) DNA with mutations in either α6SSE, CSE or both (ii) DNA with spacing between the α6SSE and CSE regions. We found that the complex was not competed out by excess DNA containing mutations in either the CSE (Figure 4B, lane 4), α6SSE (Figure 4B, lane 5), both (Figure 4B, lane 6) or mut2 α6SSE (Figure 4B, lane 7). In addition, a 5-bp linker between the α6SSE and CSE also failed to compete (Figure 4B, lane 8) indicating that the spacing between the elements is important for binding. These results are consistent with the effect of the mutations and the linker on promoter activity (Figure 3B) and suggest that the two elements are critical for the DNA binding complex.
A recent study reported that the CSE shows mosaic methylation or hypo-methylation, regardless of transcription level (29). Our findings also show that CpG methylation of the αSSE–CSE regions does not affect complex formation (Supplementary Figure S3).
Next, we carried out competition assays with a similar DNA sequence derived from the other promoters of the Pcdhα cluster (with α1SSE–CSE, α2SSE–CSE, … , α13SSE–CSE) and of the Pcdhγ cluster, γA2SSE–CSE. Remarkably, the complex was not competed out by excess of most of the Pcdhα SSE–CSE and not by excess the Pcdh γA2SSE–CSE (Figure 4C). We similarly analyzed the complex that binds to the α3SSE–CSE (Figure 4D, left panel) and the α12SSE–CSE (Figure 4D, right panel). A competition assay was performed, in which the α3SSE–CSE or α12SSE–CSE labeled DNA probes were competed out by excess of unlabelled DNA from parallel regions of other Pcdh genes (Figure 4D). The α3SSE–CSE and α12SSE–CSE complexes failed to be competed out by SSE–CSE regions corresponding to other genes in the cluster. The results suggest the existence of a unique component for each SSE that confers specificity of the complex that binds to each promoter of the Pcdhα.
Purification of the αSSE–CSE complex and its identification by mass spectrometry
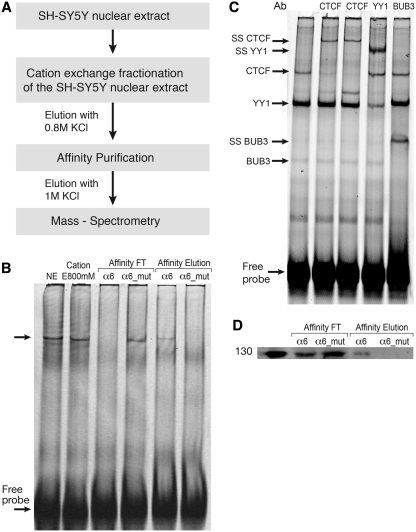
To further characterize αSSE–CSE binding proteins, we developed a purification scheme composed of two chromatographic steps (Figure 5A). First, SY5Y nuclear extract was loaded onto Macro-prep high S cation exchange column and proteins were eluted by increased salt concentrations. The fractions where analyzed by EMSA and the α6SSE–CSE specific complex was found in the 800 mM KCl fraction (Figure 5B, lane 2), resulting in ∼3-fold enrichment. Next, the 800 mM fraction was subjected to DNA affinity chromatography using magnetic nanoparticles (37). The affinity matrix was generated by chemical cross-linking immobilization to the magnetic nanoparticles the α6SSE–CSE or the α6SSE–CSE_mut DNAs that were multimerized. The α6SSE–CSE_mut DNA was mutated in both the α6SSE and the CSE elements and served as negative control. After incubation of the 800 mM enriched fraction with the DNA-beads, unbound and eluted fractions were analyzed by EMSA using α6SSE–CSE DNA (Figure 5B). It was apparent that the α6SSE–CSE was efficiently depleted from the unbound fraction of the α6SSE–CSE DNA containing beads (Figure 5B, lane 3). In contrast, in the unbound fraction of the α6SSE–CSE_mut the specific complex was still present (Figure 5B, lane 4). Consistently, the elution of the α6SSE–CSE DNA affinity contained the α6SSE–CSE complex whereas the complex was completely absent from the elution of the mutated DNA beads (Figure 5B, lanes 5 and 6). This purification step resulted in approximately a 13-fold enrichment. To identify the proteins that specifically bind to α6SSE–CSE, the affinity eluted fractions of both α6SSE–CSE and α6SSE–CSE_mut DNA were subjected to comparative mass-spectrometry. The top significant candidates from mass-spectrometry analysis of proteins that specifically bind to the α6SSE–CSE are depicted in Table 1.
Figure 5.
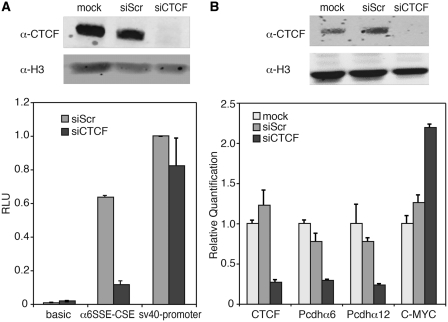
Purification of αSSE–CSE DNA-bound proteins. (A) Schematic diagram of the purification procedure. (B) Binding activity of α6SSE–CSE to the purified fractions was assessed by EMSA using the α6SSE–CSE as a probe. Lane 1, the probe is incubated with SH-SY5Y nuclear extract. Lane 2, with the 800 mM KCl elution fraction of the cation exchange. The flow-through (lanes 3 and 4) and the eluted fractions (lanes 5 and 6) of the affinity purification stage of α6SSE–CSE DNA (α6) and α6SSE–CSE_mut DNA (α6_mut), respectively. (C) Identification of the proteins that bind to the α6SSE–CSE using specific antibodies according to the MS results. Lane 1, the probe is incubated with SH-SY5Y nuclear extract; lanes 2 and 3 EMSAs were carried out in the presence of two different CTCF antibodies, lane 4 YY1 antibody; lane 5 BUB3 antibody. The specific DNA complexes, ‘super-shift’ (SS) complexes and the free probes are indicated by arrows. (D) Western blot analysis of CTCF protein in the flow-through and in the eluted fractions of the affinity purification stage of α6SSE–CSE DNA (α6) and muted DNA (α6_mut).
Table 1.
Significant candidate proteins which specifically bind to the α6SSE–CSE identified by mass-spectrometry analysis
| Accession Number | Protein name | Gene symbol | Function | Number of peptides | |
|---|---|---|---|---|---|
| 1 | P49711 | Transcriptional repressor | CTCF | Transcription regulator and chromatin insulator | 4 |
| 2 | O43684 | Mitotic checkpoint protein | BUB3 | Involved in spindle checkpoint function | 3 |
| 3 | O14776 | Transcription elongation regulator 1 | TCR1 | Regulates transcriptional elongation and pre-mRNA splicing | 3 |
| 4 | Q9P016 | Thymocyte nuclear protein 1 | THYN1 | May be involved in the induction of apoptosis | 3 |
| 5 | P18615 | Negative elongation factor E | NELFE | Part of a complex which represses RNA polymerase II transcript elongation | 2 |
| 6 | A6NFI3 | Zinc finger protein 691 | ZN316 | May be involved in transcriptional regulation | 3 |
| 7 | P51858 | Hepatoma-derived growth factor | HDGF | Acts as a transcriptional repressor | 2 |
| 8 | Q96K17 | Transcription factor BTF3 homolog 4 | BT3L4 | Basic transcription factor—unknown | 2 |
| 9 | P06748 | Nucleophosmin | NPM | Involved in diverse cellular processes such as ribosome biogenesis, centrosome duplication, protein chaperoning, histone assembly, cell proliferation, and regulation of tumor suppressors TP53/p53 and ARF | 2 |
| 10 | Q9H5H4 | Zinc finger protein 768 | ZN768 | May be involved in transcriptional regulation | 2 |
| 11 | O60828 | Polyglutamine-binding protein | PQBP1 | Involved with transcription activation | 2 |
| 12 | Q6DD87 | Zinc finger protein 787 | ZN787 | May be involved in transcriptional regulation | 2 |
| 13 | Q86U70 | LIM domain-binding protein 1 | LDB1 | Binds to the LIM domain of a wide variety of LIM domain-containing transcription factors | 3 |
| 14 | A6NFI3 | Zinc finger protein 316 | ZN316 | May be involved in transcriptional regulation | 3 |
| 15 | Q99417 | C-Myc-binding protein | MYCBP | Stimulates the activation of E box-dependent transcription by MYC | 5 |
| 16 | Q5T6F2 | Ubiquitin-associated protein 2 | UBAP2 | The function of this protein has not been determined | 5 |
| 17 | Q6PJG2 | Uncharacterized protein C14orf43 | CN043 | unknown | 6 |
To test which of the proteins identified by the mass-spectrometry binds specifically to α6SSE–CSE, we examined whether antibodies against these factors interfere or ‘super-shift’ the α6SSE–CSE complex in EMSA. Three different anti-CTCF antibodies were added to the reaction mix. The first anti-CTCF antibody (ab70303) eliminated the specific transcription complex (Supplementary Figure S4) while the two other anti-CTCF antibodies (ab37477, ab37478) retarded the migration of the specific transcription complex and a slower migration was formed instead of the original one (Figure 5C, lanes 2 and 3). Interestingly this was the case also for another, non-specific complex which was super-shifted with anti-BUB3 antibody (Figure 5C, lane 5). In addition we used an antibody for YY1, a transcription factor that appeared in the mass-spectrometry results in both the α6SSE–CSE and in the control α6SSE–CSE_mut DNA. The addition of anti-YY1 to the reaction mix retarded another complex that binds to the α6SSE–CSE fluorescently labeled probe (Figure 5C, lane 4), which was not specific to these elements. Inspection of the sequence α6SSE–CSE DNA that was used as a probe revealed the presence of a perfect YY1 binding site at the 3′-end of the fragment which was unaffected by the mutations. These results imply that there are several protein-DNA complexes formed between the nuclear extract and the α6SSE–CSE labeled DNA probe and provide validation that the purification scheme we utilized led to the identification of relevant proteins. Next, we examined the effect of the CTCF antibody on the complexes formed with a DNA fragment from other members of the Pcdhα family (α12SSE–CSE and α3SSE–CSE) or from another family (γa2SSE–CSE). The same pattern, which eliminated the specific complex (ab70303), was observed with all these probes (Supplementary Figure S4). These findings are consistent with in vivo data as analysis of the recently released ChIP-seq data from UCSC genome-browser and from Handoko et al. (38) which revealed that CTCF was associated with the promoter of each of the Pcdh genes in vivo.
To further validate that CTCF binds specifically to the SSE–CSE we performed western blot following affinity purification using anti-CTCF (Abcam ab70303). The CTCF was depleted from the unbound fraction of the WT but not the mutated DNA beads. Consistently, CTCF was exclusively eluted from the WT and not in the muted affinity column (Figure 5D).
To gain further support that SSE–CSE is directly bound by CTCF, we used in vitro transcribed and translated full-length human CTCF and 11ZFs fragments (Figure 6A) in EMSA. A specific retarded complex was observed using 11ZFs protein for DNA promoters of several members of the Pcdhα family: α6SSE–CSE (Figure 6B, lane 8), α12SSE–CSE (Figure 6B, lane 1) and α3SSE–CSE (Figure 6B, lane 4). Recombinant luciferase protein prepared by the same in vitro translation reaction was used as a negative control for site-specific DNA-binding experiments (Figure 6B, lanes 2, 4, 13). No complex observed, however, with the full-length CTCF (Figure 6B, lane 7) possibly since the DNA binding domain of CTCF in the full-length context is masked or not properly folded. The specificity of the interaction was confirmed by a competition assays with excess of unlabeled DNA sequence of WT (Figure 6B, lane 9) and DNA with mutations in either CSE (Figure 6B, lane 10) α6SSE (Figure 6B, lane 11), or both (Figure 6B, lane 12).
The in vitro data provides strong evidence for the novel interaction of CTCF with highly conserved CSE-SSE sequences present in the Pcdh gene promoters. These sequences were sufficient for function, for binding and for biochemical purification of CTCF.
CTCF is essential for Pcdhα expression
To investigate further the role of CTCF in regulation of Pcdh expression, we tested whether knocking down the endogenous CTCF would affect Pcdh promoter activity. SH-SY5Y cells were transfected with siRNA against CTCF or a scrambled negative control. Seventy-two hours post-transfection Pcdhα6 promoter-luciferase reporter genes were transfected into these cells and luciferase activity was determined 24 h later. Down-regulation of endogenous CTCF by siRNA-CTCF but not scrambled siRNA was confirmed by western blot analysis with anti-CTCF (ab70303) (Figure 7A). The results clearly show that CTCF siRNA but not scrambled siRNA severely diminished the luciferase activity directed by the WT Pcdhα6 promoter by >3-fold (Figure 7A). This reduction in luciferase activity was specific to Pcdh because siRNA-CTCF did not affect the luciferase activity driven by the SV40 promoter (Figure 7A).
Figure 7.
CTCF is essential for Pcdhα gene-expression. (A) Knocking down CTCF down-regulated the expression of α6 promoter-driven reporter gene. Top panel, western blot analysis with either CTCF or H3 antibodies of SH-SY5Y cells transfected with siRNA-CTCF scrambled siRNA or mock. Bottom panel, 72-h post-siRNA transfction, luciferase reporter gene driven by Pcdhα6, basic and SV40 promoters were transfected into SH-SY5Y cell lines together with RSV-Renilla that served as internal control. Twenty-four hours post-transfection firefly and Renilla luciferase activities were measured. The normalized results are the mean of four independent experiments (±SD). (B) Knocking down CTCF down-regulated the expression of endogenous Pcdhα genes. Top panel, western blot analysis with either CTCF or H3 antibodies of HEC-1B cells transfected with siRNA-CTCF scrambled siRNA or mock. Bottom panel, 120-h post-siRNA transfection, mRNA level was measured using relative quantification for endogenous Pcdha6, Pcdha12 genes and c-Myc gene as a positive control.
To gain further support for the involvement of CTCF in the transcriptional activity of endogenous Pcdh mRNA, we measured the mRNA levels of Pcdhα6 and Pcdhα12 in HEC1-B cells that selectively expressed these genes, following knock down of CTCF by siRNA (Figure 7B). In these cells CTCF protein level was decreased by 80% whereas its levels were unchanged by the scrambled siRNA. Down-regulation of CTCF caused similar reduction in the levels of the endogenous Pcdhα6 and Pcdhα12 mRNA levels. As a positive control, we selected the c-Myc gene, for which CTCF was known to act as a repressor (39), and found that c-Myc levels were up-regulated as expected. These findings strongly suggest that CTCF acts as a major activator of Pcdh genes.
DISCUSSION
In the present study we have identified a novel specific element (SSE) found in the promoters of all 52 Pcdh genes, positioned immediately upstream of each CSE. In contrast to the CSE, SSEs are rather unique sequences and much less conserved between paralogs of the same gene family. We have shown that Pcdh promoter activity was governed by the combined activity of the newly identified SSE and the CSE. Thus, while CSE is indeed crucial for transcription, it is definitely not sufficient. Using DNA binding assay we found a nuclear complex that specifically interacts with αSSE–CSE in vitro. Subsequently, we have isolated and identified CCTC-binding-factor (CTCF) as the protein that binds and specifically binds and regulates the SSE–CSE in each promoter region of the Pcdh. First, we found that CTCF binds in a highly specific manner to the promoter region of several representative genes from the Pcdhα and Pcdhγ families in vitro. Second, knocking down CTCF down-regulated the expression of endogenous active Pcdh genes as well as Pcdh promoter-driven reporter gene. These findings suggest that CTCF binds to the promoters of the clustered Pcdh genes and is acting as a master positive regulator of all the Pcdh genes. Our findings are consistent with previous reports that show that binding of CTCF to the target site is methylation-independent (40,41).
CTCF is a diverse protein and has a unique structure of 11 zinc-finger-DNA binding domains which are conserved among vertebrates. This distinct structure gives CTCF an exceptional degree of flexibility for DNA binding site recognition, which led to the description of CTCF as a ‘multivalent’ transcription factor (42). CTCF is known to perform numerous functions including enhancer blocking, X-chromosome inactivation, gene imprinting, monoallelic gene expression and promoter activation or repression (42–46). It has been demonstrated that CTCF can mediate contact between CTCF binding sites, to stabilize intra- and inter chromosomal long range interactions to affect transcription (38,47,48). Conditional CTCF Knock out models have demonstrated that CTCF affects transcription in enhancers of both beta and alpha Globin loci (49,50), the HOX cluster (51), the XITE of the X-chromosome (52) and the EBV (53). Another recent study, which investigates the transcriptional activity in CTCF mutant limbs, has shown that the CTCF sites in RNA splicing may regulate the production of specific alternative transcript variants (51).
CTCF is known to interact with diverse protein partners that determine its specific function; hence, the regulation of CTCF activity might be achieved by neighboring factors bound to DNA. These partners factors include the RNA polymerases I, II and III, another zinc finger factor VEZF1 and the factors YY1, SMAD, TR and Oct4. Each of these seems to influence, modulate or determine the function of CTCF (54). Of particular interest to this study is YY1 which was shown to interact with CTCF and to function together in X-chromosome inactivation (52). YY1 is a ubiquitous four-zinc-finger transcription factor that has been implicated in biological processes such as embryogenesis, differentiation, cell proliferation and tumorigenesis (55). Surprisingly, in the course of our research YY1 were found to bind specifically to the SSE–CSE, most likely through a YY1 binding site that partially overlap the CSE of α6, α3 and α12. Although YY1 and CTCF appeared in our experiments as distinct complexes on EMSA, it is possible that under physiological conditions they bind cooperatively to the promoters of Pcdhα genes.
Considering that the Pcdh gene cluster contains several promoter elements in tandem, with only one or two are active, and the fact that Pcdh expression is monoallelic, it is possible that in addition to its ability to activate Pcdh transcription, it also has a central role in insulating nearby promoters in the monoallelic expression of Pcdh genes. Depending on the promoter context and cell background, CTCF may repress or activate transcription; however, its repression function predominates. We found that CTCF acts primarily as a positive transcription factor for the Pcdh genes. We cannot conclude from our results that the SSE dictates cell type specific expression. However, if we take into account the fact that CTCF is associated with most of the Pcdh promoters in vivo (56,57) (according to the ChIP-seq data found in the UCSC genome browser), as well as in conserved enhancers HS5-1 (28) and HS16–20 (58), it may well be possible that CTCF also serves as a repressor that acts to silence the inactive Pcdh genes. However in these studies the exact site to which CTCF binds and the functional significance of CTCF for Pcdh expression were not addressed. This repression can be achieved through different conformation of DNA/CTCF-complexes (which CTCF binding to itself) that allow it to form chromatin hubs by selective intra- and interchromosomal interactions bridging together specific subsets of genomic CTCF sites. The divergence of the CTCF-binding sequences (as a result of unique SSEs) can serve as ‘CTCF CODE’ (59,60) encrypting interactions with co-partner in a site specific manner (through the SSEs) and establishing structure-functional 3D organization involved in regulating the expression of individual Pcdh genes. The nature and composition of this co-partner is yet to be determined. Profiling the gene expression pattern of several different cell lines following profiling the DNA binding location of the CTCF and other candidate proteins will give us a predictive mechanistic model for the specific regulation of the Pcdh gene expression.
In summary, the expression of distinct Pcdh mRNAs in individual neurons is regulated by the activation or repression of subsets of promoters preceding individual genes. The choice of a gene included in a Pcdh mRNA appears to be a direct consequence of promoter selection. The mechanism of differential Pcdh promoter activation or repression in individual neurons is therefore likely to be a key step in regulating the cell-specific expression of distinct Pcdh genes. Our work is, therefore, another step forward in the effort to understand the mechanism for Pcdh gene expression, which may have significant implications on mammalian brain development. These findings may also have implications to human neuronal diseases linked to aberrant Pcdh function such as autism (61), bipolar disorder, schizophrenia (62), auditory deficiencies (63) and tumors (64).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–5 and Supplementary Data 1–3.
FUNDING
Kahn Family Research Center for System Biology of the Human Cell; John von Neumann Minerva Center for the Development of Reactive Systems; European Union (FP7-ERC-AdG). Funding for open access charge: European Union (FP7-ERC-AdG).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank V. Lobanenkov (National Institute of Allergy and Infectious Diseases, National Institutes of Health, Rockville, MD, USA) for generously providing plasmids pET-7.1 and pET-11ZF. We would like to thank Eran Segal for help in bioinformatics analysis and discussions. We would like to thank Ido Amit for the valuable advice and insightful comments, which were very helpful in improving our manuscript. E.S. is the incumbent of The Harry Weinrebe Professorial Chair of Computer Science and Biology. R.D. is the incumbent of the Ruth and Leonard Simon Chair of Cancer Research.
REFERENCES
- 1.Lomvardas S, Barnea G, Pisapia D, Mendelsohn M, Kirkland J, Axel R. Interchromosomal interactions and olfactory receptor choice. Cell. 2006;126:403–413. doi: 10.1016/j.cell.2006.06.035. [DOI] [PubMed] [Google Scholar]
- 2.Schmucker D, Clemens J, Shu H, Worby C, Xiao J, Muda M, Dixon J, Zipursky S. Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell. 2000;101:671–684. doi: 10.1016/s0092-8674(00)80878-8. [DOI] [PubMed] [Google Scholar]
- 3.Wojtowicz WM, Flanagan JJ, Millard SS, Zipursky SL, Clemens JC. Alternative splicing of Drosophila Dscam generates axon guidance receptors that exhibit isoform-specific homophilic binding. Cell. 2004;118:619–633. doi: 10.1016/j.cell.2004.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hattori D, Demir E, Kim H, Viragh E, Zipursky S, Dickson B. Dscam diversity is essential for neuronal wiring and self-recognition. Nature. 2007;449:223–227. doi: 10.1038/nature06099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hamada S, Yagi T. The cadherin-related neuronal receptor family: a novel diversified cadherin family at the synapse. Neurosci. Res. 2001;41:207–215. doi: 10.1016/s0168-0102(01)00281-4. [DOI] [PubMed] [Google Scholar]
- 6.Shapiro L, Colman DR. The diversity of cadherins and implications for a synaptic adhesive code in the CNS. Neuron. 1999;23:427–430. doi: 10.1016/s0896-6273(00)80796-5. [DOI] [PubMed] [Google Scholar]
- 7.Serafini T. Finding a partner in a crowd: neuronal diversity and synaptogenesis. Cell. 1999;98:133–136. doi: 10.1016/s0092-8674(00)81008-9. [DOI] [PubMed] [Google Scholar]
- 8.Yagi T, Takeichi M. Cadherin superfamily genes: functions, genomic organization, and neurologic diversity. Genes Dev. 2000;14:1169–1180. [PubMed] [Google Scholar]
- 9.Emond M, Jontes J. Inhibition of protocadherin-alpha function results in neuronal death in the developing zebrafish. Dev. Biol. 2008;321:175–187. doi: 10.1016/j.ydbio.2008.06.011. [DOI] [PubMed] [Google Scholar]
- 10.Phillips GR, Tanaka H, Frank M, Elste A, Fidler L, Benson DL, Colman DR. Gamma-protocadherins are targeted to subsets of synapses and intracellular organelles in neurons. J. Neurosci. 2003;23:5096–5104. doi: 10.1523/JNEUROSCI.23-12-05096.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weiner J, Wang X, Tapia J, Sanes J. Gamma protocadherins are required for synaptic development in the spinal cord. Proc. Natl Acad. Sci. USA. 2005;102:8–14. doi: 10.1073/pnas.0407931101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kohmura N, Senzaki K, Hamada S, Kai N, Yasuda R, Watanabe M, Ishii H, Yasuda M, Mishina M, Yagi T. Diversity revealed by a novel family of cadherins expressed in neurons at a synaptic complex. Neuron. 1998;20:1137–1151. doi: 10.1016/s0896-6273(00)80495-x. [DOI] [PubMed] [Google Scholar]
- 13.Wu Q, Maniatis T. A striking organization of a large family of human neural cadherin-like cell adhesion genes. Cell. 1999;97:779–790. doi: 10.1016/s0092-8674(00)80789-8. [DOI] [PubMed] [Google Scholar]
- 14.Wang X, Weiner JA, Levi S, Craig AM, Bradley A, Sanes JR. Gamma protocadherins are required for survival of spinal interneurons. Neuron. 2002;36:843–854. doi: 10.1016/s0896-6273(02)01090-5. [DOI] [PubMed] [Google Scholar]
- 15.Blank M, Triana-Baltzer G, Richards C, Berg D. Alpha-protocadherins are presynaptic and axonal in nicotinic pathways. Mol. Cell. Neurosci. 2004;26:530–543. doi: 10.1016/j.mcn.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 16.Frank M, Ebert M, Shan W, Phillips GR, Arndt K, Colman DR, Kemler R. Differential expression of individual gamma-protocadherins during mouse brain development. Mol. Cell. Neurosci. 2005;29:603–616. doi: 10.1016/j.mcn.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 17.Zou C, Huang W, Ying G, Wu Q. Sequence analysis and expression mapping of the rat clustered protocadherin gene repertoires. Neuroscience. 2007;144:579–603. doi: 10.1016/j.neuroscience.2006.10.011. [DOI] [PubMed] [Google Scholar]
- 18.Fukuda E, Hamada S, Hasegawa S, Katori S, Sanbo M, Miyakawa T, Yamamoto T, Yamamoto H, Hirabayashi T, Yagi T. Down-regulation of protocadherin-alpha A isoforms in mice changes contextual fear conditioning and spatial working memory. Eur. J. Neurosci. 2008;28:1362–1376. doi: 10.1111/j.1460-9568.2008.06428.x. [DOI] [PubMed] [Google Scholar]
- 19.Katori S, Hamada S, Noguchi Y, Fukuda E, Yamamoto T, Yamamoto H, Hasegawa S, Yagi T. Protocadherin-alpha family is required for serotonergic projections to appropriately innervate target brain areas. J. Neurosci. 2009;29:9137–9147. doi: 10.1523/JNEUROSCI.5478-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sugino H, Hamada S, Yasuda R, Tuji A, Matsuda Y, Fujita M, Yagi T. Genomic organization of the family of CNR cadherin genes in mice and humans. Genomics. 2000;63:75–87. doi: 10.1006/geno.1999.6066. [DOI] [PubMed] [Google Scholar]
- 21.Wu Q, Zhang T, Cheng J, Kim Y, Grimwood J, Schmutz J, Dickson M, Noonan J, Zhang M, Myers R, et al. Comparative DNA sequence analysis of mouse and human protocadherin gene clusters. Genome Res. 2001;11:389–404. doi: 10.1101/gr.167301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Esumi S, Kakazu N, Taguchi Y, Hirayama T, Sasaki A, Hirabayashi T, Koide T, Kitsukawa T, Hamada S, Yagi T. Monoallelic yet combinatorial expression of variable exons of the protocadherin-alpha gene cluster in single neurons. Nat. Genet. 2005;37:171–176. doi: 10.1038/ng1500. [DOI] [PubMed] [Google Scholar]
- 23.Kaneko R, Kato H, Kawamura Y, Esumi S, Hirayama T, Hirabayashi T, Yagi T. Allelic gene regulation of Pcdh-alpha and Pcdh-gamma clusters involving both monoallelic and biallelic expression in single Purkinje cells. J. Biol. Chem. 2006;281:30551–30560. doi: 10.1074/jbc.M605677200. [DOI] [PubMed] [Google Scholar]
- 24.Zhang T, Haws P, Wu Q. Multiple variable first exons: a mechanism for cell- and tissue-specific gene regulation. Genome Res. 2004;14:79–89. doi: 10.1101/gr.1225204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu Q, Maniatis T. Large exons encoding multiple ectodomains are a characteristic feature of protocadherin genes. Proc. Natl Acad. Sci. USA. 2000;97:3124–3129. doi: 10.1073/pnas.060027397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tasic B, Nabholz CE, Baldwin KK, Kim Y, Rueckert EH, Ribich SA, Cramer P, Wu Q, Axel R, Maniatis T. Promoter choice determines splice site selection in protocadherin alpha and gamma pre-mRNA splicing. Mol. Cell. 2002;10:21–33. doi: 10.1016/s1097-2765(02)00578-6. [DOI] [PubMed] [Google Scholar]
- 27.Wang X, Su H, Bradley A. Molecular mechanisms governing Pcdh-gamma gene expression: evidence for a multiple promoter and cis-alternative splicing model. Genes Dev. 2002;16:1890–1905. doi: 10.1101/gad.1004802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ribich S, Tasic B, Maniatis T. Identification of long-range regulatory elements in the protocadherin-alpha gene cluster. Proc. Natl Acad. Sci. USA. 2006;103:19719–19724. doi: 10.1073/pnas.0609445104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kawaguchi M, Toyama T, Kaneko R, Hirayama T, Kawamura Y, Yagi T. Relationship between DNA methylation states and transcription of individual isoforms encoded by the protocadherin-alpha gene cluster. J. Biol. Chem. 2008;283:12064–12075. doi: 10.1074/jbc.M709648200. [DOI] [PubMed] [Google Scholar]
- 30.Murata Y, Hamada S, Morishita H, Mutoh T, Yagi T. Interaction with protocadherin-gamma regulates the cell surface expression of protocadherin-alpha. J. Biol. Chem. 2004;279:49508–49516. doi: 10.1074/jbc.M408771200. [DOI] [PubMed] [Google Scholar]
- 31.Amir-Zilberstein L, Ainbinder E, Toube L, Yamaguchi Y, Handa H, Dikstein R. Differential regulation of NF-kappaB by elongation factors is determined by core promoter type. Mol. Cell Biol. 2007;27:5246–5259. doi: 10.1128/MCB.00586-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Amir-Zilberstein L, Dikstein R. Interplay between E-box and NF-kappaB in regulation of A20 gene by DRB sensitivity-inducing factor (DSIF) J. Biol. Chem. 2008;283:1317–1323. doi: 10.1074/jbc.M706767200. [DOI] [PubMed] [Google Scholar]
- 33.Kurukuti S, Tiwari VK, Tavoosidana G, Pugacheva E, Murrell A, Zhao Z, Lobanenkov V, Reik W, Ohlsson R. CTCF binding at the H19 imprinting control region mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to Igf2. Proc. Natl Acad. Sci. USA. 2006;103:10684–10689. doi: 10.1073/pnas.0600326103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hancock AL, Brown KW, Moorwood K, Moon H, Holmgren C, Mardikar SH, Dallosso AR, Klenova E, Loukinov D, Ohlsson R, et al. A CTCF-binding silencer regulates the imprinted genes AWT1 and WT1-AS and exhibits sequential epigenetic defects during Wilms' tumourigenesis. Hum. Mol. Genet. 2007;16:343–354. doi: 10.1093/hmg/ddl478. [DOI] [PubMed] [Google Scholar]
- 35.Noonan JP, Li J, Nguyen L, Caoile C, Dickson M, Grimwood J, Schmutz J, Feldman MW, Myers RM. Extensive linkage disequilibrium, a common 16.7-kilobase deletion, and evidence of balancing selection in the human protocadherin alpha cluster. Am. J. Hum. Genet. 2003;72:621–635. doi: 10.1086/368060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Noonan JP, Grimwood J, Schmutz J, Dickson M, Myers RM. Gene conversion and the evolution of protocadherin gene cluster diversity. Genome Res. 2004;14:354–366. doi: 10.1101/gr.2133704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sakamoto S, Kabe Y, Hatakeyama M, Yamaguchi Y, Handa H. Development and application of high-performance affinity beads: toward chemical biology and drug discovery. Chem. Rec. 2009;9:66–85. doi: 10.1002/tcr.20170. [DOI] [PubMed] [Google Scholar]
- 38.Handoko L, Xu H, Li G, Ngan CY, Chew E, Schnapp M, Lee CW, Ye C, Ping JL, Mulawadi F, et al. CTCF-mediated functional chromatin interactome in pluripotent cells. Nat. Genet. 2011;43:630–638. doi: 10.1038/ng.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Filippova GN, Fagerlie S, Klenova EM, Myers C, Dehner Y, Goodwin G, Neiman PE, Collins SJ, Lobanenkov VV. An exceptionally conserved transcriptional repressor, CTCF, employs different combinations of zinc fingers to bind diverged promoter sequences of avian and mammalian c-myc oncogenes. Mol. Cell. Biol. 1996;16:2802–2813. doi: 10.1128/mcb.16.6.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kosaka-Suzuki N, Suzuki T, Pugacheva EM, Vostrov AA, Morse HC, Loukinov D, Lobanenkov V. Transcription factor BORIS (Brother of the Regulator of Imprinted Sites) directly induces expression of a cancer-testis antigen, TSP50, through regulated binding of BORIS to the promoter. J. Biol. Chem. 2011;286:27378–27388. doi: 10.1074/jbc.M111.243576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Renaud S, Pugacheva EM, Delgado MD, Braunschweig R, Abdullaev Z, Loukinov D, Benhattar J, Lobanenkov V. Expression of the CTCF-paralogous cancer-testis gene, brother of the regulator of imprinted sites (BORIS), is regulated by three alternative promoters modulated by CpG methylation and by CTCF and p53 transcription factors. Nucleic Acids Res. 2007;35:7372–7388. doi: 10.1093/nar/gkm896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ohlsson R, Renkawitz R, Lobanenkov V. CTCF is a uniquely versatile transcription regulator linked to epigenetics and disease. Trends Genet. 2001;17:520–527. doi: 10.1016/s0168-9525(01)02366-6. [DOI] [PubMed] [Google Scholar]
- 43.Gaszner M, Felsenfeld G. Insulators: exploiting transcriptional and epigenetic mechanisms. Nat. Rev. Genet. 2006;7:703–713. doi: 10.1038/nrg1925. [DOI] [PubMed] [Google Scholar]
- 44.Filippova GN. Genetics and epigenetics of the multifunctional protein CTCF. Curr. Top. Dev. Biol. 2008;80:337–360. doi: 10.1016/S0070-2153(07)80009-3. [DOI] [PubMed] [Google Scholar]
- 45.Klenova EM, Morse HC, Ohlsson R, Lobanenkov VV. The novel BORIS + CTCF gene family is uniquely involved in the epigenetics of normal biology and cancer. Semin. Cancer Biol. 2002;12:399–414. doi: 10.1016/s1044-579x(02)00060-3. [DOI] [PubMed] [Google Scholar]
- 46.Shu W, Chen H, Bo X, Wang S. Genome-wide analysis of the relationships between DNaseI HS, histone modifications and gene expression reveals distinct modes of chromatin domains. Nucleic Acids Res. 2011;39:7428–7443. doi: 10.1093/nar/gkr443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bell AC, West AG, Felsenfeld G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 1999;98:387–396. doi: 10.1016/s0092-8674(00)81967-4. [DOI] [PubMed] [Google Scholar]
- 48.Liu Z, Scannell DR, Eisen MB, Tjian R. Control of embryonic stem cell lineage commitment by core promoter factor, TAF3. Cell. 2011;146:720–731. doi: 10.1016/j.cell.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Heath H, Ribeiro de Almeida C, Sleutels F, Dingjan G, van de Nobelen S, Jonkers I, Ling KW, Gribnau J, Renkawitz R, Grosveld F, et al. CTCF regulates cell cycle progression of alphabeta T cells in the thymus. EMBO J. 2008;27:2839–2850. doi: 10.1038/emboj.2008.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Splinter E, Heath H, Kooren J, Palstra RJ, Klous P, Grosveld F, Galjart N, de Laat W. CTCF mediates long-range chromatin looping and local histone modification in the beta-globin locus. Genes Dev. 2006;20:2349–2354. doi: 10.1101/gad.399506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Soshnikova N, Montavon T, Leleu M, Galjart N, Duboule D. Functional analysis of CTCF during mammalian limb development. Dev. Cell. 2010;19:819–830. doi: 10.1016/j.devcel.2010.11.009. [DOI] [PubMed] [Google Scholar]
- 52.Donohoe ME, Zhang LF, Xu N, Shi Y, Lee JT. Identification of a Ctcf cofactor, Yy1, for the X chromosome binary switch. Mol. Cell. 2007;25:43–56. doi: 10.1016/j.molcel.2006.11.017. [DOI] [PubMed] [Google Scholar]
- 53.Tempera I, Wiedmer A, Dheekollu J, Lieberman PM. CTCF prevents the epigenetic drift of EBV latency promoter Qp. PLoS Pathog. 2010;6:e1001048. doi: 10.1371/journal.ppat.1001048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weth O, Renkawitz R. CTCF function is modulated by neighboring DNA binding factors. Biochem Cell Biol. 2011;89:459–468. doi: 10.1139/o11-033. [DOI] [PubMed] [Google Scholar]
- 55.Gordon S, Akopyan G, Garban H, Bonavida B. Transcription factor YY1: structure, function, and therapeutic implications in cancer biology. Oncogene. 2006;25:1125–1142. doi: 10.1038/sj.onc.1209080. [DOI] [PubMed] [Google Scholar]
- 56.Kim TH, Abdullaev ZK, Smith AD, Ching KA, Loukinov DI, Green RD, Zhang MQ, Lobanenkov VV, Ren B. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kehayova P, Monahan K, Chen W, Maniatis T. Regulatory elements required for the activation and repression of the protocadherin-alpha gene cluster. Proc. Natl Acad. Sci. USA. 2011;108:17195–17200. doi: 10.1073/pnas.1114357108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yokota S, Hirayama T, Hirano K, Kaneko R, Toyoda S, Kawamura Y, Hirabayashi M, Hirabayashi T, Yagi T. Identification of the cluster control region for the protocadherin-{beta} genes located beyond the protocadherin-{gamma} cluster. J. Biol. Chem. 2011;286:31885–31895. doi: 10.1074/jbc.M111.245605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ohlsson R, Lobanenkov V, Klenova E. Does CTCF mediate between nuclear organization and gene expression? Bioessays. 2010;32:37–50. doi: 10.1002/bies.200900118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Phillips JE, Corces VG. CTCF: master weaver of the genome. Cell. 2009;137:1194–1211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Morrow E, Yoo S, Flavell S, Kim T, Lin Y, Hill R, Mukaddes N, Balkhy S, Gascon G, Hashmi A, et al. Identifying autism loci and genes by tracing recent shared ancestry. Science. 2008;321:218–223. doi: 10.1126/science.1157657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pedrosa E, Stefanescu R, Margolis B, Petruolo O, Lo Y, Nolan K, Novak T, Stopkova P, Lachman H. Analysis of protocadherin alpha gene enhancer polymorphism in bipolar disorder and schizophrenia. Schizophr. Res. 2008;102:210–219. doi: 10.1016/j.schres.2008.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zheng Q, Yu H, Washington, Kisley L, Kikkawa Y, Pawlowski K, Wright C, Alagramam K. A new spontaneous mutation in the mouse protocadherin 15 gene. Hear Res. 2006;219:110–120. doi: 10.1016/j.heares.2006.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dallosso A, Hancock A, Szemes M, Moorwood K, Chilukamarri L, Tsai H, Sarkar A, Barasch J, Vuononvirta R, Jones C, et al. Frequent long-range epigenetic silencing of protocadherin gene clusters on chromosome 5q31 in Wilms’ tumor. PLoS Genet. 2009;5:e1000745. doi: 10.1371/journal.pgen.1000745. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.