Abstract
Targeted gene addition to mammalian genomes is central to biotechnology, basic research and gene therapy. For example, gene targeting to the ROSA26 locus by homologous recombination in embryonic stem cells is commonly used for mouse transgenesis to achieve ubiquitous and persistent transgene expression. However, conventional methods are not readily adaptable to gene targeting in other cell types. The emerging zinc finger nuclease (ZFN) technology facilitates gene targeting in diverse species and cell types, but an optimal strategy for engineering highly active ZFNs is still unclear. We used a modular assembly approach to build ZFNs that target the ROSA26 locus. ZFN activity was dependent on the number of modules in each zinc finger array. The ZFNs were active in a variety of cell types in a time- and dose-dependent manner. The ZFNs directed gene addition to the ROSA26 locus, which enhanced the level of sustained gene expression, the uniformity of gene expression within clonal cell populations and the reproducibility of gene expression between clones. These ZFNs are a promising resource for cell engineering, mouse transgenesis and pre-clinical gene therapy studies. Furthermore, this characterization of the modular assembly method provides general insights into the implementation of the ZFN technology.
INTRODUCTION
New genes are routinely introduced to mammalian cells for studies related to functional genomics, cell biology, proteomics, cell-based drug discovery and many other applications in biotechnology and basic science. Most methods for the stable introduction of genes into mammalian cells rely on random integration of the transgene into the genome followed by drug selection and laborious screening to identify cell populations expressing the transgene at desirable levels. This uncontrolled nature of random transgene integration can lead to several confounding effects on gene expression, including multiple integrations per cell, the activation or disruption of endogenous genes at or near the site of integration, and unstable expression of the transgene due to epigenetic regulation. These characteristics of stable cell line engineering often lead to unpredictable cell behavior, irreproducible results and possible erroneous data interpretation. Technologies that facilitate the precise addition of transgenes to specific locations of mammalian genomes have the potential to address these limitations.
Homologous recombination (HR) is a mechanism by which precise changes to defined genomic sequences can be introduced (1,2). Gene targeting by HR, combined with stringent selection methods, is commonly used in mouse embryonic stem cells to generate transgenic mice (1,2). Gene targeting to the mouse ROSA26 locus by HR is the preferred method for mouse transgenesis as this site provides improved targeting efficiency and ubiquitous transgene expression (3,4). Furthermore, gene addition to this locus does not have any adverse consequences on mouse viability or cell phenotype. The mouse ROSA26 locus has a long history of development and application in mouse genetics. Mice modified at the ROSA26 locus were initially derived from pools of embryonic stem cells infected with a retroviral gene trap (5). The ROSA26 locus was cloned and shown to encode a nuclear RNA expressed in a broad variety of tissues (3). The generalized expression at this site suggested that gene targeting to the ROSA26 locus would be a desirable method to achieve ubiquitous transgene expression. From the time of its discovery, hundreds of transgenic animals and cell lines expressing a variety of transgenes including reporters, site-specific recombinases, and non-coding RNAs have been successfully created using the ROSA26 locus.
However, conventional HR methods are not readily transferable to gene targeting in non-embryonic stem cells from mice or in cells from other species. Consequently, the creation of gene-targeted lineage-committed cell types has traditionally required targeting mouse embryonic stem cells followed by directed differentiation into the cell type of interest or generating transgenic mice from which the cell type of interest could be harvested. Technologies for gene targeting in somatic cells and established cell lines would circumvent these costly, inefficient and laborious steps.
Two discoveries have opened new venues toward achieving high rates of HR in mammalian somatic cells. The first was the discovery that the introduction of double-strand breaks at genomic target sites increases the rate of HR at that site by several orders of magnitude (6). The second was the development of engineered zinc finger nucleases (ZFNs), which are able to generate site-specific double-strand breaks at targeted genome sequences (7–9). Synthetic custom-designed ZFNs are a fusion of a DNA-binding domain consisting of an array of individual zinc finger motifs and the non-specific catalytic domain of the FokI restriction endonuclease. Each zinc finger recognizes ∼3 consecutive base pairs of DNA (10). The specificity of particular zinc fingers has been examined extensively through site-directed mutagenesis and rational design (11,12) or the selection of large combinatorial libraries (13–18). Through this work, synthetic zinc finger domains have been established that bind to almost any nucleotide triplet (19–21). Significantly, the modular structure of the zinc finger motif permits the conjunction of several domains in series, allowing for the recognition and targeting of extended sequences in multiples of 3 nt (22).
The cleavage of DNA by ZFNs can theoretically be targeted to nearly any given sequence in the genome based on the DNA-binding specificity of the zinc finger domains (23–25). These targeted double-strand breaks can be repaired by the error-prone, non-homologous end-joining (NHEJ) pathway or HR. DNA breaks repaired by NHEJ frequently exhibit nucleotide deletions and insertions at the cleavage site. This approach can be used to knockout genes in many model organisms, including mice, by expression of ZFNs in early embryos (26–30). However, when a donor repair template is provided with the ZFNs, the rate of HR at the break site is dramatically enhanced (9,23,25). Following this principle, ZFNs have been used to achieve efficient gene targeting at various endogenous genomic loci in mammalian cells (23,31–35).
Several approaches have been described for designing, selecting, and assembling the zinc finger domains that confer targeting specificity to ZFNs (18,36–43). One of the easiest and most straightforward design protocols involves the modular assembly of zinc finger arrays based on specific zinc fingers that have been previously identified to bind certain 3 bp sequences (39,40). Alternatively, selection-based methods have been described in which novel zinc finger arrays are isolated from large libraries (18). Although this approach is significantly more complex and labor-intensive, it has been reported to generate active ZFNs at a much higher frequency than the modular assembly strategy (44). The recently described context-dependent modular assembly (CoDA) method provides an appealing balance of these two approaches (41). In contrast to these ‘open-source’ methods, the most prevalent approach to generating highly active ZFNs targeted to endogenous mammalian genes is a proprietary method developed by scientists at Sangamo Biosciences (25). In fact, ZFNs targeted to the ROSA26 locus and designed by these proprietary methods have recently been reported (29). Although all of these approaches have been used to assemble active ZFNs, there are still many uncertainties regarding the optimal criteria for ZFN design and assembly.
To further evaluate the efficacy of publicly available ZFN assembly methods, we generated ZFNs by modular assembly (39,40) that target the identical sequence of the ROSA26 locus previously reported to be targeted by proprietary ZFNs (29,45). Importantly, although the target site of this ZFN was described, the sequence of the ZFN was not reported. Our novel ZFN pairs created by modular assembly efficiently induce double-strand breaks at the ROSA26 locus in a variety of murine cell lines. ZFN activity was dependent on the number of zinc fingers on the ZFN proteins, time, the ZFN dose, and cell type. A donor vector was constructed that contains a transgene expression cassette flanked by two 800-bp homology arms. When delivered alone, this donor vector integrated randomly into the genome. However, when delivered in conjunction with the ZFN pair targeting the ROSA26 locus, transgene integration at the desired target site was readily detected in selected and unselected cell populations. Targeted transgene expression was relatively uniform amongst clonal cell populations, whereas random plasmid integration led to highly variable levels of expression. Collectively, these results provide insights into strategies and criteria for ZFN design and assembly and characterize a method for gene targeting in murine cell lines that will enable diverse areas of research.
MATERIALS AND METHODS
Cell culture and electroporation
NIH3T3, 293T, C2C12 and Neuro2A cells were obtained from the American Tissue Collection Center (ATCC) through the Duke University Cancer Center Facilities. NIH3T3 cells were maintained in DMEM supplemented with 10% bovine calf serum and 1% penicillin/streptomycin. 293T and Neuro2A cells were maintained in DMEM supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin. C2C12 cells were maintained in DMEM supplemented with 20% fetal bovine serum and 1% penicillin/streptomycin. These cell lines were maintained at 37°C and 5% CO2. Primary skeletal myoblasts, provided by Dr. Terence A. Partridge, were isolated from H-2Kb-tsA58 transgenic mice harboring a thermolabile mutant of the immortalizing SV40 large T antigen under the control of an interferon-gamma inducible promoter (46). These cells were grown in DMEM supplemented with 20% fetal bovine serum, 2% chick embryo extract (Accurate Chemical), 2% l-glutamine, 20 U/ml γ-interferon and 1% penicillin/streptomycin on tissue culture dishes coated with 0.01% porcine gelatin and maintained at 33°C and 5% CO2. Cell culture reagents were obtained from Invitrogen unless stated otherwise.
The 293T cells were transfected with Lipofectamine 2000 (Invitrogen) according to manufacturer’s instructions in 6-well plates with 2 ml of OPTI-MEM. Plasmid DNA was delivered to all other cell lines by electroporation with a Gene Pulser Xcell System (BioRad). Two million cells were electroporated in 400 µl of OPTI-MEM, using conditions optimized for each cell line. Electroporation efficiencies were routinely >90% in Neuro2A cells and primary myoblasts, 80% in C2C12 cells, and 70% in NIH3T3 cells, as determined by flow cytometry following delivery of an EGFP expression plasmid. Unless indicated otherwise, the amount of DNA used for electroporation was 10 µg of each ZFN or empty control expression vector and 40 µg of donor vector.
Plasmids and ZFN assembly
The ZFN expression plasmid (pCMV) was provided by David J. Segal. FokI domains containing both the ELD/KKR obligate heterodimer mutations (47) and the Sharkey mutations (48) were synthesized and codon optimized by Bio Basic. The zinc finger modules were provided by Carlos F. Barbas, III and ZFNs were designed and assembled as previously described (39,40). Full ZFN sequences are provided in the online Supplementary Information. The homology arms in the ROSA26 donor vector were generated by PCR of mouse genomic DNA and insertion of the PCR-amplified right homology arm (left primer: 5′-TTATTGCGGCCGCAGATGGGCGGGAGTCTTCT-3′, right primer: 5′-AACACCGCGGCAGTTTATAAATGGAGAAAAAGGAGA-3′) between the NotI and SacII sites and the left homology arm (left primer: 5′-TCTATGGTACCAGTTAACGGCAGCCGGAGT-3′, right primer: 5′-CGGACGAATTCTCTAGAAAGACTGGAGTTGCAGA-3′) between the KpnI and EcoRI sites in the pZDonor AAVS1 vector obtained from Sigma–Aldrich.
Single-strand annealing recombination assay
Single-strand annealing (SSA) plasmids were provided by David J. Segal (49). A gene encoding EGFP was divided into two segments, which were separated by a stop codon and a ROSA26 ZFN target site. Both EGFP segments contained a 275 bp homology region. Upon cleavage at the ZFN target site, the cellular SSA repair mechanisms remove the stop codon and restore the wt EGFP cDNA (49). Therefore EGFP expression provides a quantitative measurement of ZFN activity. For episomal SSA assays, ZFN expression plasmids and SSA reporters were co-transfected into 293T cells. For genomic SSA assays, the SSA-EGFP open reading frame was cloned into the pcDNA5-FRT vector, which was integrated in a single site of the genome of 293 Flp-In cells (Invitrogen) according to the manufacturer's instructions. The number of EGFP-positive cells was measured by flow cytometry.
Quantification of DNA repair
ZFN-mediated DNA cleavage was measured by quantifying the mutations created at the target site that result from double-strand break repair via NHEJ (50). Following electroporation, the cells were maintained at 32°C for 3 days, which has been shown to enhance the activity of the ZFNs (51), and the genomic DNA was isolated from ZFN-treated cells using the DNeasy kit (Qiagen). The region surrounding the ZFN target site was amplified by PCR with the AccuPrime PCR kit (Invitrogen) and 50–200 ng of genomic DNA as template with primers 5′-AAGGGAGCTGCAGTGGAGTA-3′ and 5′-GCGGGAGAAATGGATATGAA-3′. For NIH3T3 and C2C12 cells the primers were 5′-GCACGTTTCCGACTTGAGTT-3′ and 5′-GGCGGATCACAAGCAATAAT-3′. The PCR products were melted and reannealed using the temperature program: 95°C for 180 s, 85°C for 20 s, 75°C for 20 s, 65°C for 20 s, 55°C for 20 s, 45°C for 20 s, 35°C for 20 s and 25°C for 20 s with a 0.1°C/s decrease rate in between steps. This allows the formation of mutant and wild-type DNA strands with the consequent formation of distorted duplex DNA. Eighteen microliters of the reannealed duplex was combined with 2 µl of the Surveyor nuclease (Transgenomic), which cleaves DNA duplexes at the sites of distortions created by either bulges or mismatches (52), and 1 µl of enhancer solution. This reaction was incubated at 42°C for 60 min and then separated on a 10% TBE polyacrylamide gel. The gels were stained with ethidium bromide and quantified using ImageLab (BioRad) according to the procedure described by Guschin et al. (50). The limit of detection of this assay has been estimated to be ∼3% modified alleles (52).
PCR-based detection of HR events
Neuro2A cells were electroporated with 10 µg of each ZFN and 40 µg of the donor vector. After 4 days genomic DNA was isolated using the DNeasy kit (Qiagen). The targeted region was amplified by PCR with the AccuPrime PCR kit (Invitrogen) and 50–200 ng of genomic DNA as template. For the detection of Zeo-EGFP integration, the primers used were: 5′-CTTCTAGTTGCCAGCCATCTG-3′ and 5′-TCAGGTAATATTGGGGGAGGAGAC-3′ and the cycling program was 95.5°C for 8 min followed by 40 cycles of 95.5°C for 20 s, 55°C for 30 s, and 68°C for 100 s. For detection of MCS integration, the primers used were 5′-GAAATATTCTCGAGGTTTAAACGTC-3′ and 5′-GGTAATATTGGGGGAGGAGACATC-3′ and the cycling program was 95.5°C for 8 min followed by 40 cycles of 95.5°C for 20 s, 53°C for 30 s, and 68°C for 70 s.
Clonal analysis
Four days after electroporation with the ZeocinR-EGFP donor alone or in combination with ZFNs, Neuro2A cells were selected with 1 µg ml−1 Zeocin™ for 14 days. Colonies were isolated and expanded for 7 additional days. Targeted integration events in the established cell lines were identified by PCR as described above and levels of ZeocinR-EGFP expression were measured by flow cytometry. Data was analyzed with the FlowJo software package.
Statistics
SSA assays were performed in duplicate in three independent experiments (n = 3). Data shown are the average of all data points ± standard deviation. Other figures show representative data of experiments performed at least three times. Statistical analysis for Figures 2B, C and 6B was performed using one-way analysis of variance (ANOVA) with α = 0.05 and two-sample Student's t test assuming equal variances. Statistical analyses were performed with Microsoft Office Excel.
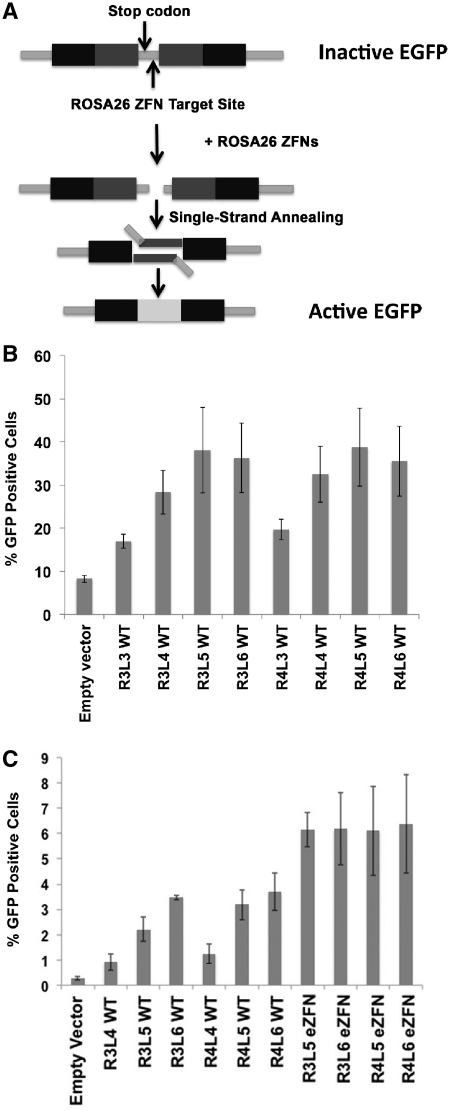
Figure 2.
Activity of the ROSA26 ZFNs in SSA assays. (A) Schematic representation of the vector used in Single Strand Annealing (SSA) assays. A central region in the EGFP cDNA is duplicated and both copies are separated by a sequence that introduces an early stop codon to render a truncated not-functional EGFP. This spacer sequence also contains the ROSA26 ZFN target site. Upon cleavage the endogenous SSA repair mechanisms are expected to remove the stop codon and restore the EGFP cDNA. (B) ROSA26 ZFNs efficiently induce SSA-mediated gene repair in an episomal reporter plasmid. Expression plasmids for ROSA26 ZFNs containing 3, 4, 5, or 6 modules in the left and right array were cotransfected with the SSA reporter vector into 293T cells and the levels of EGFP expression were measured at 48 h by flow cytometry (n = 3, mean ± stdev). ANOVA P = 2E-8. R3L4 versus R3L3 P = 0.001; R3L5 versus R3L4 P = 0.05; R4L3 versus R4L4 P = 0.002; R4L3 versus R4L5 P = 0.001. (C) ROSA26 ZFNs generate targeted genomic double-strand breaks. The SSA reporter construct was stably integrated into the genome of HEK293 cell lines. In the absence of ZFNs, spontaneous EGFP gene reconstitution and protein expression was detected in 0.3% of cells. Transfection of these cells with wild-type (WT) ZFNs or ZFNs containing the obligate heterodimer and Sharkey mutations (eZFNs) led to a significant increase in the number of EGFP-positive cells (n = 3, mean ± stdev). ANOVA P = 7E-19. R3L5 versus R3L4 P = 1E-4; R3L6 versus R3L5 P = 5E-5; R4L5 versus R4L4 P = 2E-5; R4L6 versus R4L4 P = 6E-5.
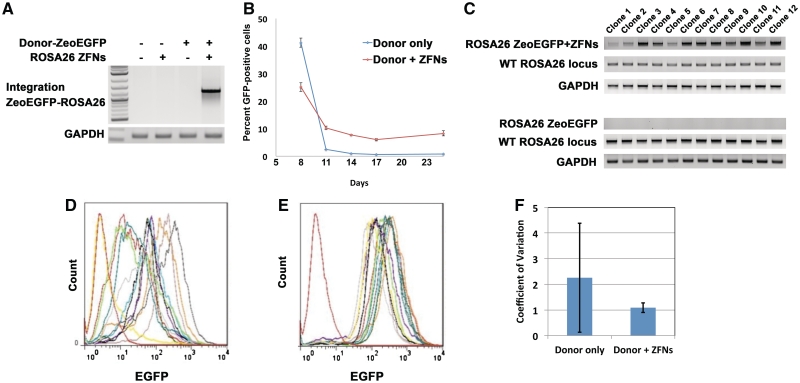
Figure 6.
Targeted integration of a ZeocinR-EGFP expression cassette into the ROSA26 locus. (A) Integration of ZeoEGFP in the ROSA26 locus using ROSA26 R4L6 eZFN in unselected populations. To determine whether the ROSA26 ZFNs facilitate targeted addition of larger expression cassettes at this locus, a ∼1.6 kb sequence consisting of a ZeocinR-EGFP gene (ZeoEGFP) driven by a CMV promoter was inserted between the homology arms of the donor vector. Control plasmid, ROSA26 R4L6 eZFN alone, donor vector alone, or donor vector together with the ROSA26 R4L6 eZFN were electroporated into Neuro2A cells. Targeted gene addition was tested using PCR of the genomic DNA with primers that bind in the Zeo-EGFP expression cassette and genomic DNA in the ROSA26 locus beyond the homology arms. The transgene was inserted correctly in the target site, as confirmed using sequencing of the PCR product, only when the donor vector was delivered with the ZFNs. (B) Efficiency of ZFN-mediated targeted gene addition. In cells transfected with only the donor vector, ZeoEGFP expression was detectable in less than 1% of transfected cells by 14 days post-transfection. However, ∼10% of the cell population expressed EGFP after 25 days in cells transfected with donor vector and the ROSA26 R4L6 eZFNs, representing sustained expression from integrated expression cassettes (ANOVA P = 2E-4; Student's t test P = 1E-4). (C) Clonal analysis of gene targeting by ROSA26 ZFNs. Neuro2A cells were transfected with the ZeoEGFP donor vector alone or cotransfected with the donor vector and the ROSA26 R4L6 eZFNs. The cells were selected with Zeocin and clonal populations were isolated and analyzed for presence of the ZeoEGFP transgene in the ROSA26 locus. All clones contained the desired integration event when the donor vector and the ZFNs were cotransfected but not when the donor vector was delivered alone. Further analysis using PCR with primers that detect the wild-type ROSA26 locus determined that the clones containing targeted ROSA26-ZeoEGFP integration also contained at least one wild-type allele. (D and E) Expression of ZeoEGFP from the ROSA26 locus. To determine the levels of EGFP expressed from the ZeoEGFP transgene stably integrated into the genome, the (D) 12 untargeted and (E) 12 targeted clonal cell lines were analyzed by flow cytometry. Clones derived from transfections with the donor vector alone expressed highly variable levels of EGFP. In contrast, the clones from cells treated with ZFNs and expression vector showed relatively homogenous levels of EGFP expression. (F) Quantification of variability of gene expression in untargeted and targeted clones. The reduced variability of gene expression achieved by ZFN-mediated gene targeting was quantified as a decrease in the mean of the coefficient of variation of cellular fluorescence intensity of each clonal population and a dramatic decrease in the standard deviation of the coefficient of variation for each clone (n = 12, mean ± SD).
RESULTS
Design and construction of ZFNs targeted to ROSA26
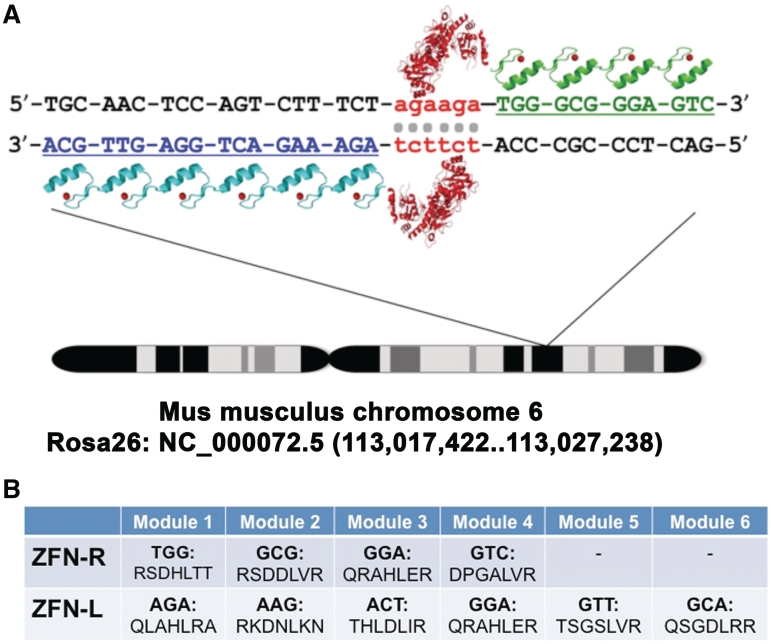
We designed novel ZFNs targeted to a previously reported ZFN target site in the ROSA26 locus (Figure 1A) (29). Our ZFNs were generated following a well-characterized modular assembly approach (39,40). Because ZFNs act as dimers (Figure 1A), two zinc finger protein arrays were assembled using an archive of zinc finger DNA-binding modules (39,40). One array consists of six zinc finger domains (designated the left, or L6 array) and the other array consisted of four zinc finger domains (designated the right, or R4 array) (Figure 1B). These arrays were coupled to either the wild-type DNA cleavage domain of the type IIS restriction enzyme FokI or a codon-optimized FokI domain containing mutations shown to prevent homodimer formation (Q486E, I499L, N496D in L6 and E490K, I538K and H537R in R4) (47). The ‘Sharkey’ mutations S418P and K441E were also added to both heterodimer FokI domains to enhance cleavage activity (48,53). The complete sequence of both ZFNs is provided in Supplementary Figures S1 and S2.
Figure 1.
Schematic representation of the genomic target sequence of the ROSA26 ZFNs. (A) Target sequence of the ROSA26 ZFNs. Located on mouse chromosome 6, the Gt(ROSA)26Sor (ROSA26) locus spans 9 kb at position 113,026,025 of NCBI Reference Sequence NC_000072.5. The ROSA26 ZFN pair was generated by a modular assembly approach and consists of six zinc finger domains in the left array and four zinc finger domains in the right array that target the first intron of the ROSA26 locus. (B) Amino acid sequences of the alpha helices within the zinc finger domains that recognize the indicated triplets of DNA.
Activity of ROSA26 ZFNs in an episomal single-strand annealing assay
Since the modular assembly strategy used here adds individual zinc fingers sequentially (40), we generated ZFNs containing variable numbers of zinc finger modules designated RXLY, where X refers to the number of zinc fingers on the right ZFN and Y refers to the number of zinc fingers on the left ZFN. We then tested whether ROSA26 ZFNs R3L3, R3L4, R3L5, R3L6, R4L3, R4L4, R4L5 and R4L6 were able to induce double strand breaks at their target site in a mammalian cell-based single-strand annealing (SSA) assay. In this assay, a reporter vector containing two partially duplicated sequences of EGFP separated by a DNA sequence that contains a stop codon and the ZFN target site is cotransfected into 293 cells with vectors encoding the ZFNs of interest. Double-strand breaks introduced in the reporter vector by the ZFNs are repaired by the endogenous cell repair machinery via the SSA mechanism, thus eliminating the stop codon and restoring the EGFP open reading frame (Figure 2A) (49,54).
The activity of different ZFN pairs containing the wild-type FokI domain was measured by flow cytometry to determine the percentage of EGFP-positive cells compared to cells transfected with an SSA reporter plasmid and control expression vector (Figure 2B). At 48 h after transfection 17% of the cells transfected with the R3L3 pair and 19% of cells transfected with R4L3 expressed EGFP. This represents a ∼2-fold increase over background levels (8%). When the ZFN R3 was used, the addition of extra modules to the left ZFN resulted in notable increases in the number of cells expressing EGFP: 28% for L4 (P = 0.001 vs. L3), 38% for L5 (P = 0.05 vs. L4) and 36% for L6. When ZFN R4 was used, an increase in the number of EGFP-positive cells was observed between L3 (19%) and L4 (32%) (P = 0.002) and L3 and L5 (38%) (P = 0.001). Addition of module L6 (36%) did not enhance ZFN activity in this assay relative to L5.
Activity of ROSA26 ZFNs in a genomic SSA assay
The episomal SSA assay evaluates ZFN activity in the context of many copies of the reporter plasmid within each cell. To assess ZFN activity at a single and accessible chromosomal locus, we created HEK293 human cell lines in which the SSA reporter construct was stably integrated into a defined chromosomal locus using the Flp-In system from Invitrogen. In the absence of ZFNs, spontaneous EGFP gene reconstitution and protein expression was detected in 0.3% of cells in two independent clonal populations. These cells were transfected with plasmids encoding the ZFN pairs that were most active in the episomal SSA assay, including R3L4, R3L5, R3L6, R4L4, R4L5 and R4L6 (Figure 2C). The percentage of cells expressing EGFP was determined by flow cytometry. The results show that only 0.9% of the cells transfected with ZFN R3L4 expressed EGFP. Addition of the L5 and L6 ZFN modules increased the percent of EGFP-positive cells to 2.2% (P = 1E-4 versus L4) and 3.4% (P = 5E-5 versus L5), respectively. When the R4 ZFN module was added to the R3L4 ZFN the number of GFP-positive cells increased from 0.9% to 1.2%. Addition of the L5 and L6 modules increased the expression of EGFP to 3.2% (P = 2E-5 versus L4) and 3.7% (P = 6E-5 versus L4), respectively. These results suggest that the addition of modules to the left ZFN generates greater ZFN activity. Similarly, addition of the R4 module leads to enhanced activity in the presence of the L4 (P = 0.07) and L5 (P = 0.006) modules. However, no significant differences were found between R3 and R4 when the L6 module was used in this assay.
Off-target DNA cleavage by ZFNs can be caused by the formation of homodimers of the left and right ZFN (53,55). These effects can be significantly reduced by using FokI nuclease variants that are restricted to forming heterodimers (47,53). Furthermore, it has been shown that the ‘Sharkey’ mutations S418P and K441E enhance the endonuclease activity further (47,48). For this reason we introduced both sets of these mutations into the FokI domains of R3, R4, L5 and L6 ZFNs (designated enhanced ZFN, or eZFN) that were most active in the episomal and genomic SSA assays. These modifications significantly increased the percentage of cells that expressed EGFP in the genomic SSA assay when compared to the same R3L5 (P = 2.2E-7), R3L6 (P = 5E-4), R4L5 (P = 0.002), R4L6 (P = 0.02) ZFNs containing a wild-type FokI domain (Figure 2C). We did not observe a significant difference between cells transfected with eZFN R3L5, R3L6, R4L5 or R4L6 in which the percentage of cells that expressed GFP varied between 6.2% and 6.4%.
ZFN activity at the ROSA26 locus
The ZFNs that were active in the SSA assays were further evaluated for activity at the endogenous target sequence by the Surveyor assay (50). A double-strand break induced by ZFNs is typically repaired by error-prone NHEJ. The repaired DNA often contains small insertions or deletions, designated ‘indels’, at the break site. These indel mutations were detected by PCR amplification of the target site from genomic DNA and incubation of the resulting product with the mismatch-sensitive Cel-I nuclease (Figure 3A). The percentage of endogenous gene modification was calculated using image analysis of polyacrylamide gels as previously described (50).
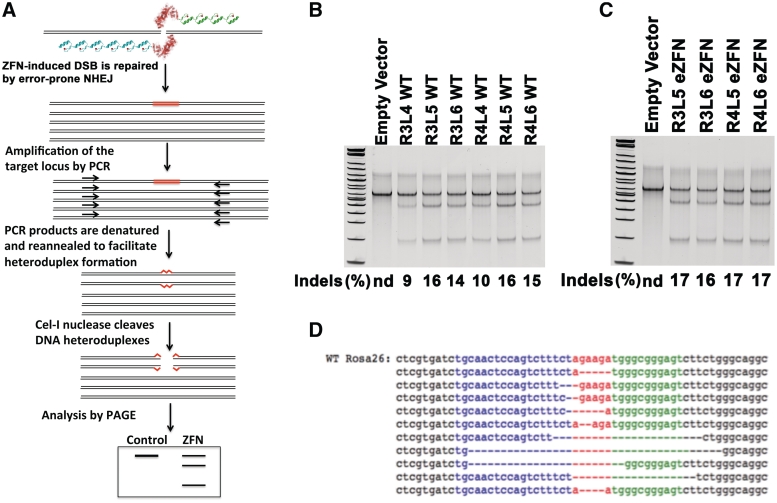
Figure 3.
Activity of the ROSA26 ZFNs at the endogenous target locus. (A) Schematic representation of the Surveyor assay. Genomic DNA from cells transfected with ZFNs is isolated and used in a PCR reaction that amplifies wild-type DNA and DNA cleaved by the ZFNs and repaired by error-prone non-homologous end joining (NHEJ). The PCR product is melted and reannealed to facilitate the formation of heteroduplexes, which can be detected as cleaved fragments of the PCR product upon digestion with Cel-I nuclease. (B) Level of endogenous allele modification induced by ROSA26 ZFNs with a wild-type FokI domain. Neuro2A cells were transfected with ZFNs containing 3, 4, 5 or 6 modules in the left and right array. After 3 days the genomic DNA was isolated and the Surveyor assay was performed. No gene modification was detected in cells electroporated with control plasmid, whereas significant levels of modification were detected in all samples treated with ZFNs. Although there was no significant difference in the activity of ZFN pairs containing R3 or R4 arrays by this assay, ZFN activity increases when the L5 module is added to the L4 array. (C) Level of endogenous allele modification induced by ROSA26 eZFNs. The level of endogenous allele modification by the eZFNs was significantly higher than modification by wild-type ZFNs regardless of the number of zinc finger modules in either array. (D) Sequences of indels at the ROSA26 target site following ZFN-mediated cleavage and repair by NHEJ.
Neuro2A cells were transfected with the ZFN pairs R3L4, R3L5, R3L6, R4L4, R4L5 and R4L6. After 3 days the genomic DNA was isolated and the Surveyor assay was performed (Figure 3B). No gene modification was detected in cells electroporated with control plasmid, whereas significant levels of modification were detected in all samples treated with ZFNs. There was no significant difference in the activity of ZFN pairs containing R3 or R4 arrays. However, the activity of ZFNs containing the L4 array (∼10% indels) increased to ∼16% indels when the L5 module was added, independent of the number of modules in the right ZFN. Addition of the L6 module to ZFN L5 slightly decreased the endonuclease activity of the ZFNs at the endogenous locus between 1 and 2%, which is within the variability of this assay. The rate of endogenous allele modification by the eZFNs with the R3L5, R3L6, R4L5 and R4L6 zinc finger arrays was between 16 and 17%, indicating that the obligate heterodimers with the Sharkey mutations may have slightly increased the endonuclease activity of the FokI domains at the genomic target site (Figure 3C). To confirm the modification of the endogenous ROSA26 locus by ZFNs, the target locus was amplified by PCR of the genomic DNA from cells treated with the R4L6 eZFN and ligated into a plasmid. Individual plasmids were screened by digestion with XbaI, which cuts within the ZFN target site, to identify plasmids containing indels. Plasmids that were not cut by XbaI were sequenced to verify the presence of indels resulting from error-prone repair of the double-strand break induced by the endonuclease activity of the ROSA26 ZFNs (Figure 3D).
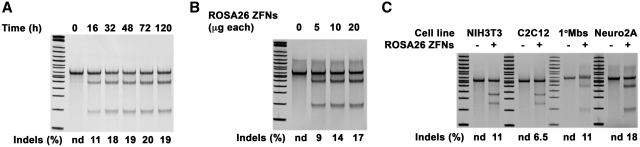
Neuro2A cells were transfected with ROSA26 R4L6 eZFNs and genomic DNA was isolated at 16, 32, 48, 72 and 120 h after transfection. The endonuclease activity of the ZFNs can be readily detected at 16 h (11% indels) and reaches near maximal levels of activity of ∼18% at 32 h. The percentage of gene modification remained effectively constant for 5 days. The results demonstrated a time-dependent increase in the endonuclease activity of the ROSA26 ZFNs at the target site, which persists over time indicating that this targeted genomic modification is well-tolerated by the cells (Figure 4A). The dependence of ZFN dose on gene modification was assessed by electroporating 5, 10 or 20 µg of each R4L6 eZFN into Neuro2A cells. Genomic DNA was collected 72 h after electroporation. The rates of endogenous gene modification were 9, 14, and 17%, demonstrating a dose-dependent effect of the amount of ZFN expression vector delivered to the cells on endonuclease activity at the target locus (Figure 4B).
Figure 4.
Levels of ROSA26 ZFN activity as a function of time, dose, and cell type. (A) Time-dependent activity of ROSA26 ZFNs. Neuro2A cells were transfected with ROSA26 R4L6 eZFNs and genomic DNA was isolated at 16, 32, 48, 72 and 120 h after transfection. The endonuclease activity of the ZFNs was readily detected at 16 h and reached near maximal levels of activity at 32 h. The percentage of gene modification remained effectively constant for 5 days. (B) Dose-dependent activity of ROSA26 ZFNs. DNA was isolated from Neuro2A cells transfected with 5, 10 or 20 μg of plasmid encoding ROSA26 ZFNs and the level of genomic modification was measured by the Surveyor assay. Targeted DNA cleavage increased with the dose of ROSA26 ZFN expression plasmid. (C) Cell type-dependent activity of ROSA26 ZFNs. The activity of the ROSA26 R4L6 eZFN was tested in a panel of murine cell lines, including NIH3T3 embryonic fibroblasts, C2C12 skeletal myoblasts, Neuro2A neuroblastoma cells, and primary skeletal myoblasts. The activity of the ZFNs was consistently highest in Neuro2A cells, however, significant levels of gene modification were detected in all cell lines tested.
Many cell type-dependent factors influence the activity of ZFNs (34). The activity of the ROSA26 R4L6 eZFN was tested in a panel of murine cell lines, including NIH3T3 embryonic fibroblasts, C2C12 skeletal myoblasts, Neuro2A neuroblastoma cells, and primary skeletal myoblasts (Figure 4C). Although the activity of the ZFNs was highest in Neuro2A cells with rates of endogenous gene modification of 18%, substantial rates of gene modification were detected in NIH3T3 cells and primary myoblasts (11%) and C2C12 cells (6.5%). These results suggest that the ROSA26 ZFNs can facilitate cell line engineering in a diverse variety of cell types.
Targeted gene addition at the ROSA26 locus
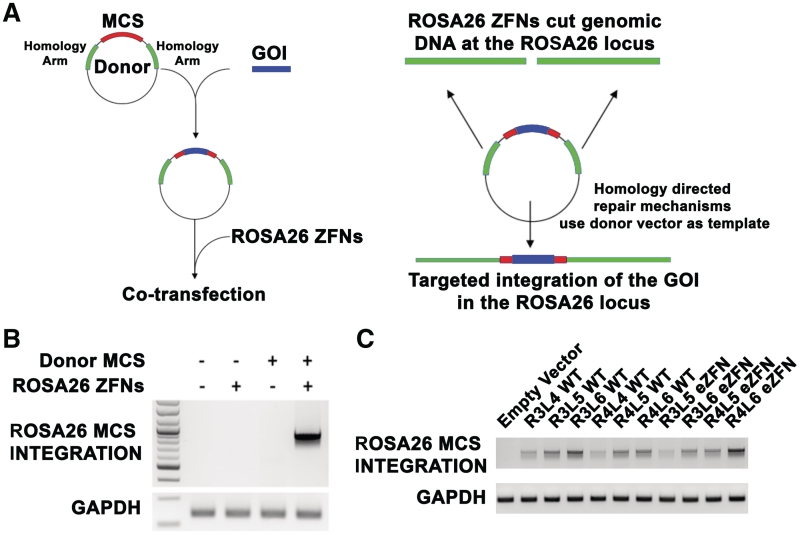
The endogenous cellular machinery can efficiently repair DNA by homologous recombination (HR) at sites of a double-strand break (8,9,23). For this reason, ZFNs are valuable tools to promote gene insertion at specific genomic target sites. To introduce DNA sequences at the target site, a donor vector was assembled that contains two 800 bp homology arms adjacent to the left and right of the ROSA26 target site separated by a 46 bp multiple cloning site (MCS) (Figure 5A). Specific integration of the MCS into the ROSA26 locus was identified by PCR with primers that bind in the MCS and in the ROSA26 genomic DNA beyond the homology arms. Only specific integration events generate a PCR product. No specific integration events were detected following delivery of an empty vector, the ROSA26 eZFNs, or the donor vector alone. However, when the L6R4 eZFNs were transfected together with the donor vector, the MCS derived from the donor vector was integrated into the ROSA26 locus at the target site (Figure 5B). This result was confirmed by sequencing the PCR product.
Figure 5.
Targeted homologous recombination directed by ROSA26 ZFNs. (A) Schematic representation of targeted integration via homologous recombination using ZFNs. The donor vector contains two 800 bp arms of homology with the ROSA26 locus adjacent to the ZFN target site. Genes of interest (GOI), such as the ZeocinR-EGFP cassette (ZeoEGFP), can be cloned into the multiple cloning site (MCS) between the two homology arms. When this vector is delivered together with the ZFNs, the endogenous repair mechanisms can use the donor vector as template to repair the double-strand break. This will result in integration of the gene of interest into the genome at the ZFN target site. (B) Integration of a 45 bp sequence into the ROSA26 locus using the ROSA26 R4L6 eZFN. Neuro2A cells were electroporated with control DNA, with ROSA26 R4L6 eZFN, with an empty donor vector alone or with ROSA26 R4L6 eZFN and the donor vector. PCR with primers that bind the MCS and the ROSA26 locus beyond the homology arms was performed. No specific integration events were detected following delivery of an empty vector, the ROSA26 eZFNs, or the donor vector alone. However when the ZFNs were transfected together with the donor vector, the MCS derived from the donor vector was integrated into the ROSA26 locus at the target site. (C) Integration of a 45 bp sequence into the ROSA26 locus using ZFNs of variably sized zinc finger arrays. Neuro2A cells were electroporated with expression plasmids for eZFN pairs containing 3, 4, 5 or 6 modules in the left and right arrays and the MCS donor vector. All the ZFN pairs were effective at mediating integration of the MCS into the genomic DNA at the correct target site, as determined by PCR with primers that bind the MCS and the ROSA26 locus beyond the homology arms.
To examine the ability of various ZFN pairs to integrate sequences of interest in the ROSA26 locus, Neuro2A cells were electroporated with R3L4, R3L5, R3L6, R4L4, R4L5 and R4L6 ZFNs and R3L5, R3L6, R4L5 and R4L6 eZFNs together with the donor vector that contains an MCS between the homology arms. Using PCR with primers that bind the MCS and the ROSA26 locus beyond the homology arms, we demonstrated that all the ZFN pairs built were effective at mediating integration of the MCS in the genomic DNA at the target site (Figure 5C).
The efficiency of targeted integration driven by HR decreases as the length of the inserted DNA sequence increases (31). Therefore, to characterize the efficiency with which the ROSA26 ZFNs facilitate addition of a complete gene at this locus, a ∼1.6 kb sequence consisting of an expression cassette for a ZeocinR-EGFP fusion protein driven by a CMV promoter was inserted between the homology arms of the donor vector described above. The empty vector, R4L6 ROSA26 eZFNs alone, donor vector alone, or donor vector together with the L6R4 ROSA26 eZFNs were electroporated into Neuro2A cells. Targeted gene addition was assayed by PCR of the genomic DNA with primers that bind in the ZeocinR-EGFP expression cassette and in the ROSA26 locus beyond the homology arms. The transgene was inserted correctly in the target site only when the donor vector was delivered with the ZFNs (Figure 6A). Sequencing of the PCR product confirmed the expected HR result.
ZeocinR-EGFP gene expression in our donor vector is driven by a CMV promoter and therefore EGFP can be readily detected in samples electroporated with donor vector regardless of the expression of the ROSA26 ZFNs. Because transient expression of EGFP is expected to decay over time and only expression from donor plasmid integrated in the genome will be sustained, EGFP expression was measured by flow cytometry over 25 days in cells transfected with donor vector only or with donor vector and ZFNs to estimate the efficiency of transgene integration (Figure 6B). The results show a rapid decrease in the levels of transient protein expression between days 8 and 11 in cells transfected with donor vector only. At day 14 and afterward, expression of EGFP was detectable in <1% of the cells. However, in cells transfected with donor vector and ROSA26 ZFNs, ∼10% of the cell population expressed EGFP after 25 days, confirming that site-specific double-strand breaks facilitate highly efficient targeted integration of donor sequences into the genome.
The ZeocinR-EGFP transgene confers resistance to the antibiotic Zeocin. Neuro2A cells transfected with the donor vector alone or cotransfected with the donor vector and R4L6 ROSA26 eZFNs were selected with Zeocin. After 2 weeks, clonal populations were isolated and analyzed for presence of the ZeocinR-EGFP transgene in the ROSA26 locus. All 12 clones contained the desired integration event when the donor vector and the ZFNs were cotransfected (Figure 6C), which is consistent with the >10-fold increase in cells stably expressing EGFP (Figure 6B). None of the 12 ZeocinR clones from cells transfected with donor vector alone contained targeted integration events, indicating these clones were the result of random plasmid integration. To determine whether the targeted clones were homozygous or heterozygous, PCR was performed with primers that amplify a 621 bp DNA fragment in the wild-type ROSA26 locus surrounding the ZFN target site. The presence of this PCR product together with a targeted integration PCR product indicates a heterozygous cell line. The results showed that all 12 cell lines studied were heterozygous (Figure 6C). Importantly, American Type Culture Collection (ATCC) reports the Neuro2A cell line to be hyperploid with an unstable karyotype. Consequently, it is likely that there are more than two ROSA26 alleles within each genome and the exact number of targeted alleles within each clone cannot be determined by this assay.
Each of the 24 clonal cell lines was analyzed by flow cytometry to determine the levels of EGFP expression. Clones derived from transfections with the donor vector alone displayed highly variable levels of EGFP expression between clones and within each clone, as demonstrated by wide histograms (Figure 6D). This is likely the result of chromosomal positional effects caused by random plasmid integration. In fact, one clone was resistant to Zeocin but did not express EGFP, indicating that only a fragment of the expression cassette was integrated. However, in the clones from cells treated with ZFNs and expression vector, the levels of EGFP expression were homogeneous between clones and less variable within a clone, as demonstrated by narrow and uniform histograms (Figure 6E). The dramatically reduced variability of gene expression achieved by ZFN-mediated gene targeting was quantified as a >2-fold decrease in the mean of the coefficient of variation of cellular fluorescence intensity of each clonal population and a >10-fold decrease in the standard deviation of the coefficient of variation for each clone (Figure 6F). Additionally the coefficient of variation of the mean fluorescence intensity of each clone decreased from 0.76 to 0.43 when ZFNs were added. These results underscore the benefits of transgenesis and isogenic cell line engineering at a defined locus.
DISCUSSION
The use of cell lines with identical genetic content is critical to ensuring uniform gene expression within a cell population. Methods for obtaining these isogenic cell lines are widely employed in basic science, biomedical research and biotechnological applications such as biopharmaceutical production. This is often achieved by random vector integration into the genome, either by viral transduction or plasmid transfection, and selection for clones that stably express the transgene cassette. However, because of highly variable levels of transgene expression following random integration (Figure 6D), the laborious process of screening clones for suitable expression levels is necessary. Alternatively, several commercial systems are available for targeted plasmid integration into mammalian genomes, such as the Flp-InTM and Jump-InTM systems marketed by Invitrogen which make use of the Flp recombinase (56) and phiC31 integrase (57), respectively. Zinc finger recombinases have also been used for targeted plasmid integration in mammalian cells (58–60). However, all of these approaches require that genomic landing pads containing the recombinase and integrase target sites be preintroduced into the genome. Therefore these systems have not yet been used to target endogenous genomic loci in otherwise unmodified cells. The ZFN technology utilized in this study provides a general method for cell line engineering that can be applied to any primary or established murine cell line. The highly specific gene targeting directed by the cellular HR machinery suggests that screening for targeted integration events will be unnecessary in many cell types. This work offers a murine analogue to previous protocols developed for similar purposes targeting the AAVS1 locus in human cell lines (32,34).
In addition to cell line engineering, gene targeting with the ROSA26 ZFNs may be useful for in vivo applications as well. A recent study demonstrated the efficacy of gene targeting in the liver of mice following in vivo administration of ZFNs and donor vectors (61). This study used transgenic mice carrying a human gene sequence and ZFNs that target this sequence. The ROSA26 ZFNs described here may provide a method for in vivo gene targeting of any mouse strain. This approach is valuable for understanding gene function in the context of a living organism and can be used as a preclinical model to evaluate gene therapy strategies based on therapeutic transgene integration at ‘safe harbor loci’ (32,34).
In this study, we developed several ZFNs containing different number of zinc finger modules targeting the ROSA26 locus in the mouse genome. Episomal and genomic reporter assays of ZFN activity showed that increasing the number of modules enhances the nuclease activity of the ZFN pairs at the target site. More importantly, assays for DNA repair at the endogenous locus demonstrated a correlation between additional modules and enhanced activity. In general, we have observed a common trend with ZFNs constructed with modular assembly reagents (39,40) that ZFNs with three zinc finger modules have little or no activity (unpublished results). However, ZFN activity is dramatically enhanced by the addition of a fourth, fifth, or sometimes sixth module (Figures 2B, C and 3C and unpublished results). The results are consistent with a recent report using ZFNs assembled by the same method (48). The addition of a fourth zinc finger module also increases the activity of zinc finger recombinases created by modular assembly (58,62). In a small number of cases we have observed that the addition of a fifth or sixth zinc finger module decreases overall ZFN activity (unpublished results), consistent with another recent study (54). Collectively, these results suggest that the number of modules on ZFN arrays may be an underappreciated parameter in determining the activity of modularly assembled ZFNs. A large scale study of the role of module number on ZFN activity and specificity is necessary to determine if these anecdotal observations are generally applicable to ZFN engineering by modular assembly.
Many methods have been described for the design, selection and assembly of zinc finger modules to generate ZFNs targeted to loci of interest (18,36–43). The modular assembly approach used here is particularly straightforward and can be used in combination with an online utility (39) to design ZFNs that can be synthesized de novo or constructed by recombinant DNA technology in <2 weeks (40). Other studies have reported a surprisingly low success rate with this method when evaluating only ZFNs consisting of three zinc finger modules (18,44), although many retain their intended sequence specificity (63). Therefore these studies are consistent with the results reported here and in other work (48) that four zinc finger modules are often necessary to generate functional ZFNs by modular assembly. The effect of increasing zinc finger module number on ZFN specificity and off-target activity remains to be determined, but it is noteworthy that many of the most active ZFNs in the published literature consist of four to six zinc finger modules (34,61,64). Collectively, all of these results are consistent with the hypothesis that the balance between specificity and affinity is more important than the precise number of zinc finger modules (54).
Our overall success rate in obtaining clones with targeted integrations was highly variable among different cell lines. Neuro2A cells appear to be particularly amenable to ZFN-mediated genome editing, similar to the highly efficient ZFN-mediated gene targeting of human DNA sequences routinely observed in K562 cells (34). It is important to note that both the rates of NHEJ and the efficiency of HR are cell type-specific. Although all of the targeted Neuro2A clones contained the desired integration event, in other cells lines the targeting frequency was lower. The reason for these different results is unclear; however the chromatin state, epigenetic effects, cell cycle rates, and the status of the cellular to HR machinery are cell type-dependent factors that likely affect ZFN-mediated gene targeting. Additionally, the frequency of random donor vector integration varies dramatically between cell types and affects the overall rate of gene targeting.
This study describes the development, testing and validation of several ZFN pairs and donor vectors to achieve specific integration of transgenes in the mouse ROSA26 locus, where they are homogeneously and uniformly expressed. We anticipate that this work will be relevant for many varied applications in the fields of cell line and mouse engineering and preclinical studies of gene and cell therapy.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures S1–S2.
FUNDING
The Hartwell Foundation and a Basil O'Connor Starter Scholar Award from the March of Dimes; the Duke Biomedical Engineering Howard G. Clark III Fellowship (to D.G.O.); the Howard Hughes Undergraduate Research Fellows Program (M.T.B.). Funding for open access charge: Duke University
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
Plasmids for ZFN assembly and SSA assays were provided by Carlos F. Barbas, III and David J. Segal. Primary skeletal myoblasts were provided by Terence A. Partridge.
REFERENCES
- 1.Koller BH, Smithies O. Altering genes in animals by gene targeting. Annu. Rev. Immunol. 1992;10:705–730. doi: 10.1146/annurev.iy.10.040192.003421. [DOI] [PubMed] [Google Scholar]
- 2.Capecchi MR. Gene targeting in mice: functional analysis of the mammalian genome for the twenty-first century. Nat. Rev. Genet. 2005;6:507–512. doi: 10.1038/nrg1619. [DOI] [PubMed] [Google Scholar]
- 3.Zambrowicz BP, Imamoto A, Fiering S, Herzenberg LA, Kerr WG, Soriano P. Disruption of overlapping transcripts in the ROSA beta geo 26 gene trap strain leads to widespread expression of beta-galactosidase in mouse embryos and hematopoietic cells. Proc. Natl Acad. Sci. USA. 1997;94:3789–3794. doi: 10.1073/pnas.94.8.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 5.Friedrich G, Soriano P. Promoter traps in embryonic stem cells: a genetic screen to identify and mutate developmental genes in mice. Genes Dev. 1991;5:1513–1523. doi: 10.1101/gad.5.9.1513. [DOI] [PubMed] [Google Scholar]
- 6.Rouet P, Smih F, Jasin M. Expression of a site-specific endonuclease stimulates homologous recombination in mammalian cells. Proc Natl Acad. Sci. USA. 1994;91:6064–6068. doi: 10.1073/pnas.91.13.6064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith J, Bibikova M, Whitby FG, Reddy AR, Chandrasegaran S, Carroll D. Requirements for double-strand cleavage by chimeric restriction enzymes with zinc finger DNA-recognition domains. Nucleic Acids Res. 2000;28:3361–3369. doi: 10.1093/nar/28.17.3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bibikova M, Beumer K, Trautman JK, Carroll D. Enhancing gene targeting with designed zinc finger nucleases. Science. 2003;300:764. doi: 10.1126/science.1079512. [DOI] [PubMed] [Google Scholar]
- 9.Porteus MH, Baltimore D. Chimeric nucleases stimulate gene targeting in human cells. Science. 2003;300:763. doi: 10.1126/science.1078395. [DOI] [PubMed] [Google Scholar]
- 10.Pavletich NP, Pabo CO. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A. Science. 1991;252:809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- 11.Desjarlais JR, Berg JM. Toward rules relating zinc finger protein sequences and DNA binding site preferences. Proc. Natl Acad. Sci. USA. 1992;89:7345–7349. doi: 10.1073/pnas.89.16.7345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nardelli J, Gibson T, Charnay P. Zinc finger-DNA recognition: analysis of base specificity by site-directed mutagenesis. Nucleic Acids Res. 1992;20:4137–4144. doi: 10.1093/nar/20.16.4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rebar EJ, Pabo CO. Zinc finger phage: affinity selection of fingers with new DNA-binding specificities. Science. 1994;263:671–673. doi: 10.1126/science.8303274. [DOI] [PubMed] [Google Scholar]
- 14.Choo Y, Klug A. Toward a code for the interactions of zinc fingers with DNA: selection of randomized fingers displayed on phage. Proc. Natl Acad. Sci. USA. 1994;91:11163–11167. doi: 10.1073/pnas.91.23.11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jamieson AC, Kim SH, Wells JA. In vitro selection of zinc fingers with altered DNA-binding specificity. Biochemistry. 1994;33:5689–5695. doi: 10.1021/bi00185a004. [DOI] [PubMed] [Google Scholar]
- 16.Wu H, Yang WP, Barbas CF., III Building zinc fingers by selection: toward a therapeutic application. Proc. Natl Acad. Sci. USA. 1995;92:344–348. doi: 10.1073/pnas.92.2.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greisman HA, Pabo CO. A general strategy for selecting high-affinity zinc finger proteins for diverse DNA target sites. Science. 1997;275:657–661. doi: 10.1126/science.275.5300.657. [DOI] [PubMed] [Google Scholar]
- 18.Maeder ML, Thibodeau-Beganny S, Osiak A, Wright DA, Anthony RM, Eichtinger M, Jiang T, Foley JE, Winfrey RJ, Townsend JA, et al. Rapid “open-source” engineering of customized zinc-finger nucleases for highly efficient gene modification. Mol. Cell. 2008;31:294–301. doi: 10.1016/j.molcel.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Segal DJ, Dreier B, Beerli RR, Barbas CF., III Toward controlling gene expression at will: selection and design of zinc finger domains recognizing each of the 5′-GNN-3′ DNA target sequences. Proc. Natl Acad. Sci. USA. 1999;96:2758–2763. doi: 10.1073/pnas.96.6.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dreier B, Beerli RR, Segal DJ, Flippin JD, Barbas CF., III Development of zinc finger domains for recognition of the 5′-ANN-3′ family of DNA sequences and their use in the construction of artificial transcription factors. J. Biol. Chem. 2001;276:29466–29478. doi: 10.1074/jbc.M102604200. [DOI] [PubMed] [Google Scholar]
- 21.Dreier B, Fuller RP, Segal DJ, Lund CV, Blancafort P, Huber A, Koksch B, Barbas CF., III Development of zinc finger domains for recognition of the 5′-CNN-3′ family DNA sequences and their use in the construction of artificial transcription factors. J. Biol. Chem. 2005;280:35588–35597. doi: 10.1074/jbc.M506654200. [DOI] [PubMed] [Google Scholar]
- 22.Segal DJ, Beerli RR, Blancafort P, Dreier B, Effertz K, Huber A, Koksch B, Lund CV, Magnenat L, Valente D, et al. Evaluation of a modular strategy for the construction of novel polydactyl zinc finger DNA-binding proteins. Biochemistry. 2003;42:2137–2148. doi: 10.1021/bi026806o. [DOI] [PubMed] [Google Scholar]
- 23.Urnov FD, Miller JC, Lee YL, Beausejour CM, Rock JM, Augustus S, Jamieson AC, Porteus MH, Gregory PD, Holmes MC. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- 24.Porteus MH, Carroll D. Gene targeting using zinc finger nucleases. Nat. Biotechnol. 2005;23:967–973. doi: 10.1038/nbt1125. [DOI] [PubMed] [Google Scholar]
- 25.Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat. Rev. Genet. 2010;11:636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- 26.Doyon Y, McCammon JM, Miller JC, Faraji F, Ngo C, Katibah GE, Amora R, Hocking TD, Zhang L, Rebar EJ, et al. Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nat. Biotechnol. 2008;26:702–708. doi: 10.1038/nbt1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meng X, Noyes MB, Zhu LJ, Lawson ND, Wolfe SA. Targeted gene inactivation in zebrafish using engineered zinc-finger nucleases. Nat. Biotechnol. 2008;26:695–701. doi: 10.1038/nbt1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Geurts AM, Cost GJ, Freyvert Y, Zeitler B, Miller JC, Choi VM, Jenkins SS, Wood A, Cui X, Meng X, et al. Knockout rats via embryo microinjection of zinc-finger nucleases. Science. 2009;325:433. doi: 10.1126/science.1172447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meyer M, de Angelis MH, Wurst W, Kuhn R. Gene targeting by homologous recombination in mouse zygotes mediated by zinc-finger nucleases. Proc. Natl Acad. Sci. USA. 2010;107:15022–15026. doi: 10.1073/pnas.1009424107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carbery ID, Ji D, Harrington A, Brown V, Weinstein EJ, Liaw L, Cui X. Targeted genome modification in mice using zinc-finger nucleases. Genetics. 2010;186:451–459. doi: 10.1534/genetics.110.117002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moehle EA, Rock JM, Lee YL, Jouvenot Y, DeKelver RC, Gregory PD, Urnov FD, Holmes MC. Targeted gene addition into a specified location in the human genome using designed zinc finger nucleases. Proc. Natl Acad. Sci. USA. 2007;104:3055–3060. doi: 10.1073/pnas.0611478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lombardo A, Genovese P, Beausejour CM, Colleoni S, Lee YL, Kim KA, Ando D, Urnov FD, Galli C, Gregory PD, et al. Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nat. Biotechnol. 2007;25:1298–1306. doi: 10.1038/nbt1353. [DOI] [PubMed] [Google Scholar]
- 33.Hockemeyer D, Soldner F, Beard C, Gao Q, Mitalipova M, DeKelver RC, Katibah GE, Amora R, Boydston EA, Zeitler B, et al. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat. Biotechnol. 2009;27:851–857. doi: 10.1038/nbt.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DeKelver RC, Choi VM, Moehle EA, Paschon DE, Hockemeyer D, Meijsing SH, Sancak Y, Cui X, Steine EJ, Miller JC, et al. Functional genomics, proteomics, and regulatory DNA analysis in isogenic settings using zinc finger nuclease-driven transgenesis into a safe harbor locus in the human genome. Genome Res. 2010;20:1133–1142. doi: 10.1101/gr.106773.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cui X, Ji D, Fisher DA, Wu Y, Briner DM, Weinstein EJ. Targeted integration in rat and mouse embryos with zinc-finger nucleases. Nat. Biotechnol. 2011;29:64–67. doi: 10.1038/nbt.1731. [DOI] [PubMed] [Google Scholar]
- 36.Isalan M, Klug A, Choo Y. A rapid, generally applicable method to engineer zinc fingers illustrated by targeting the HIV-1 promoter. Nat. Biotechnol. 2001;19:656–660. doi: 10.1038/90264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beerli RR, Barbas CF., III Engineering polydactyl zinc-finger transcription factors. Nat. Biotechnol. 2002;20:135–141. doi: 10.1038/nbt0202-135. [DOI] [PubMed] [Google Scholar]
- 38.Joung JK, Ramm EI, Pabo CO. A bacterial two-hybrid selection system for studying protein-DNA and protein-protein interactions. Proc. Natl Acad. Sci. USA. 2000;97:7382–7387. doi: 10.1073/pnas.110149297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mandell JG, Barbas CF., III Zinc finger tools: custom DNA-binding domains for transcription factors and nucleases. Nucleic Acids Res. 2006;34:W516–W523. doi: 10.1093/nar/gkl209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gonzalez B, Schwimmer LJ, Fuller RP, Ye Y, Asawapornmongkol L, Barbas CF., III Modular system for the construction of zinc-finger libraries and proteins. Nat. Protoc. 2010;5:791–810. doi: 10.1038/nprot.2010.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sander JD, Dahlborg EJ, Goodwin MJ, Cade L, Zhang F, Cifuentes D, Curtin SJ, Blackburn JS, Thibodeau-Beganny S, Qi Y, et al. Selection-free zinc-finger-nuclease engineering by context-dependent assembly (CoDA) Nat. Methods. 2011;8:67–69. doi: 10.1038/nmeth.1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sander JD, Zaback P, Joung JK, Voytas DF, Dobbs D. Zinc finger targeter (ZiFiT): an engineered zinc finger/target site design tool. Nucleic Acids Res. 2007;35:W599–W605. doi: 10.1093/nar/gkm349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim HJ, Lee HJ, Kim H, Cho SW, Kim JS. Targeted genome editing in human cells with zinc finger nucleases constructed via modular assembly. Genome Res. 2009;19:1279–1288. doi: 10.1101/gr.089417.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ramirez CL, Foley JE, Wright DA, Muller-Lerch F, Rahman SH, Cornu TI, Winfrey RJ, Sander JD, Fu F, Townsend JA, et al. Unexpected failure rates for modular assembly of engineered zinc fingers. Nat. Methods. 2008;5:374–375. doi: 10.1038/nmeth0508-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Orlando SJ, Santiago Y, DeKelver RC, Freyvert Y, Boydston EA, Moehle EA, Choi VM, Gopalan SM, Lou JF, Li J, et al. Zinc-finger nuclease-driven targeted integration into mammalian genomes using donors with limited chromosomal homology. Nucleic Acids Res. 2010;38:e152. doi: 10.1093/nar/gkq512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morgan JE, Beauchamp JR, Pagel CN, Peckham M, Ataliotis P, Jat PS, Noble MD, Farmer K, Partridge TA. Myogenic cell lines derived from transgenic mice carrying a thermolabile T antigen: a model system for the derivation of tissue-specific and mutation-specific cell lines. Dev. Biol. 1994;162:486–498. doi: 10.1006/dbio.1994.1103. [DOI] [PubMed] [Google Scholar]
- 47.Doyon Y, Vo TD, Mendel MC, Greenberg SG, Wang J, Xia DF, Miller JC, Urnov FD, Gregory PD, Holmes MC. Enhancing zinc-finger-nuclease activity with improved obligate heterodimeric architectures. Nat. Methods. 2011;8:74–79. doi: 10.1038/nmeth.1539. [DOI] [PubMed] [Google Scholar]
- 48.Guo J, Gaj T, Barbas CF., III Directed evolution of an enhanced and highly efficient FokI cleavage domain for zinc finger nucleases. J. Mol. Biol. 2010;400:96–107. doi: 10.1016/j.jmb.2010.04.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bhakta MS, Segal DJ. The generation of zinc finger proteins by modular assembly. Methods Mol. Biol. 2010;649:3–30. doi: 10.1007/978-1-60761-753-2_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guschin DY, Waite AJ, Katibah GE, Miller JC, Holmes MC, Rebar EJ. A rapid and general assay for monitoring endogenous gene modification. Methods Mol. Biol. 2010;649:247–256. doi: 10.1007/978-1-60761-753-2_15. [DOI] [PubMed] [Google Scholar]
- 51.Doyon Y, Choi VM, Xia DF, Vo TD, Gregory PD, Holmes MC. Transient cold shock enhances zinc-finger nuclease-mediated gene disruption. Nat. Methods. 2010;7:459–460. doi: 10.1038/nmeth.1456. [DOI] [PubMed] [Google Scholar]
- 52.Qiu P, Shandilya H, D'Alessio JM, O'Connor K, Durocher J, Gerard GF. Mutation detection using Surveyor nuclease. Biotechniques. 2004;36:702–707. doi: 10.2144/04364PF01. [DOI] [PubMed] [Google Scholar]
- 53.Miller JC, Holmes MC, Wang J, Guschin DY, Lee YL, Rupniewski I, Beausejour CM, Waite AJ, Wang NS, Kim KA, et al. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat. Biotechnol. 2007;25:778–785. doi: 10.1038/nbt1319. [DOI] [PubMed] [Google Scholar]
- 54.Shimizu Y, Sollu C, Meckler JF, Adriaenssens A, Zykovich A, Cathomen T, Segal DJ. Adding fingers to an engineered zinc finger nuclease can reduce activity. Biochemistry. 2011;50:5033–5041. doi: 10.1021/bi200393g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Szczepek M, Brondani V, Buchel J, Serrano L, Segal DJ, Cathomen T. Structure-based redesign of the dimerization interface reduces the toxicity of zinc-finger nucleases. Nat. Biotechnol. 2007;25:786–793. doi: 10.1038/nbt1317. [DOI] [PubMed] [Google Scholar]
- 56.O'Gorman S, Fox DT, Wahl GM. Recombinase-mediated gene activation and site-specific integration in mammalian cells. Science. 1991;251:1351–1355. doi: 10.1126/science.1900642. [DOI] [PubMed] [Google Scholar]
- 57.Calos MP. The phiC31 integrase system for gene therapy. Curr. Gene Ther. 2006;6:633–645. doi: 10.2174/156652306779010642. [DOI] [PubMed] [Google Scholar]
- 58.Gordley RM, Gersbach CA, Barbas CF., III Synthesis of programmable integrases. Proc. Natl Acad. Sci. USA. 2009;106:5053–5058. doi: 10.1073/pnas.0812502106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gaj T, Mercer AC, Gersbach CA, Gordley RM, Barbas CF., III Structure-guided reprogramming of serine recombinase DNA sequence specificity. Proc. Natl Acad. Sci. USA. 2011;108:498–503. doi: 10.1073/pnas.1014214108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gersbach CA, Gaj T, Gordley RM, Mercer AC, Barbas CF., III Targeted plasmid integration into the human genome by an engineered zinc-finger recombinase. Nucleic Acids Res. 2011;39:7868–7878. doi: 10.1093/nar/gkr421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li H, Haurigot V, Doyon Y, Li T, Wong SY, Bhagwat AS, Malani N, Anguela XM, Sharma R, Ivanciu L, et al. In vivo genome editing restores haemostasis in a mouse model of haemophilia. Nature. 2011;475:217–221. doi: 10.1038/nature10177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gersbach CA, Gaj T, Gordley RM, Barbas CF., III Directed evolution of recombinase specificity by split gene reassembly. Nucleic Acids Res. 2010;38:4198–4206. doi: 10.1093/nar/gkq125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lam KN, van Bakel H, Cote AG, van der Ven A, Hughes TR. Sequence specificity is obtained from the majority of modular C2H2 zinc-finger arrays. Nucleic Acids Res. 2011;39:4680–4690. doi: 10.1093/nar/gkq1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Perez EE, Wang J, Miller JC, Jouvenot Y, Kim KA, Liu O, Wang N, Lee G, Bartsevich VV, Lee YL, et al. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat. Biotechnol. 2008;26:808–816. doi: 10.1038/nbt1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.