Abstract
A remarkable feature of many small non-coding RNAs (sRNAs) of Escherichia coli and Salmonella is their accumulation in the stationary phase of bacterial growth. Several stress response regulators and sigma factors have been reported to direct the transcription of stationary phase-specific sRNAs, but a widely conserved sRNA gene that is controlled by the major stationary phase and stress sigma factor, σS (RpoS), has remained elusive. We have studied in Salmonella the conserved SdsR sRNA, previously known as RyeB, one of the most abundant stationary phase-specific sRNAs in E. coli. Alignments of the sdsR promoter region and genetic analysis strongly suggest that this sRNA gene is selectively transcribed by σS. We show that SdsR down-regulates the synthesis of the major Salmonella porin OmpD by Hfq-dependent base pairing; SdsR thus represents the fourth sRNA to regulate this major outer membrane porin. Similar to the InvR, MicC and RybB sRNAs, SdsR recognizes the ompD mRNA in the coding sequence, suggesting that this mRNA may be primarily targeted downstream of the start codon. The SdsR-binding site in ompD was localized by 3′-RACE, an experimental approach that promises to be of use in predicting other sRNA–target interactions in bacteria.
INTRODUCTION
10 years ago, when several pioneering screens discovered a plethora of small non-coding RNAs (sRNAs) in Escherichia coli, it was noted with surprise that many of these sRNAs accumulated in the stationary rather than exponential phase of growth (1–5). This observation was in remarkable contrast to the common growth rate-independent expression of house-keeping RNAs and cis-encoded antisense RNAs of plasmids and phages (6–8) and fueled speculation that the new sRNAs belong to defined stress regulons. That is, stationary phase was increasingly being understood as a growth phase wherein bacteria prepare themselves for hard times, up-regulating numerous stress response regulators including the alternative sigma factor, σS. Encoded by the rpoS gene in E. coli (9), σS is a major stress sigma factor in many enterobacteria (10–12). Its activity sharply increases not only as cells cease to grow, but also under a variety of other stress conditions such as heat or osmotic shock (13,14). The combined results of gene fusion and global transcriptome studies have suggested that σS determines, directly or indirectly, the expression of ∼10% of all protein-encoding E. coli genes, many of which have key functions in stress survival (15,16).
A decade has passed and several of these differentially expressed sRNAs have indeed been assigned to various important regulons of E. coli and Salmonella. Examples include the RyhB and IsrE sRNAs as members of the iron-responsive Fur regulon (17–19); MicA and RybB, which are activated by the envelope stress sigma factor, σE (20–25); CyaR, whose transcription is governed by the cAMP–CRP complex (26–28); ArcZ and FnrS, which respond to oxygen availability via the ArcA/B or Fnr systems (29–31); and MgrR, which is a member of the Mg2+-responsive PhoP/Q regulon (32). The strict regulation of these sRNAs by their cognate transcription factors is often accompanied by the presence of an optimal binding site in the sRNA promoter region, and in fact some of these sRNAs are the most highly regulated genes in their respective regulons (20).
It has been speculated that every major transcriptional regulon contains at least one conserved sRNA gene (33). Regarding the σS regulon, several sRNAs have been reported to accumulate in stationary phase and to show greatly diminished expression in an rpoS deletion strain, as one would expect for an σS-transcribed sRNA. However, none of them is widely conserved: the 105 nt GadY sRNA, which acts to stabilize the oppositely encoded gadX mRNA (34), is found in a few E. coli strains only and IsrE (∼100 nt), a homologue of the widely conserved RyhB sRNA (iron starvation), is specific to Salmonella (18). Thus, a σS-dependent sRNA which is conserved throughout the enterobacterial clade had yet to be identified from among the nearly 100 sRNA species currently known in E. coli and Salmonella.
Here, we report the characterization of Salmonella SdsR (sigma S-dependent sRNA), a conserved sRNA that is a member of the σS regulon. SdsR was originally reported under the name RyeB as an abundant, stationary phase-specific, ∼100 nt sRNA transcribed from the pphA-yebY intergenic region of E. coli (1,2). Its expression was shown to be inversely correlated with that of SraC (a.k.a. RyeA, Tpke79, IS091), a ∼250 nt sRNA that is transcribed from the opposite strand (1–5). The extensive antisense complementarity of SdsR and SraC suggests mutual processing by the double strand-specific endoribonuclease, RNase III (2). Whether such co-processing would be of any functional relevance has remained unresolved, as have the cellular functions of SdsR and SraC.
In contrast with the poor conservation of the sraC sequence, sdsR genes can be predicted in many enterobacteria (35), as though sdsR and not sraC has been maintained by selection. In addition, SdsR has repeatedly been pulled down with the sRNA chaperone Hfq in both E. coli and Salmonella (1,36–38). This Hfq relationship suggests that SdsR functions as a post-transcriptional regulator of gene expression because Hfq-associated sRNAs commonly regulate trans-encoded target mRNAs by short base pairing interactions, resulting in the repression or activation of targets at the levels of translation, RNA stability or both (39).
The mRNA targets of Hfq-associated sRNAs, as inferred from the global Hfq coIP data, encode proteins that have functions in many cellular pathways (36,38,40). One such prominent target is the rpoS mRNA itself, whose expression is directly activated by several Hfq-binding sRNAs (DsrA, RprA, ArcZ) and repressed by OxyS (31,41–45). Another prominent group of targets are mRNAs of proteins with envelope localization; almost a third of all characterized E. coli/Salmonella sRNAs repress porins and outer membrane proteins (OMPs) (46–48). Although many of these sRNAs accumulate in stationary phase, none of them is known to depend on σS.
This article presents evidence for a novel link between σS, Hfq and sRNAs, revealing SdsR as an σS-dependent repressor of porin synthesis in Salmonella. Protein analysis of a Salmonella strain over-expressing SdsR detected down-regulation of OmpD, a very abundant OMP in several enterobacteria (49). The ompD mRNA has been a hotspot for regulation by Hfq-associated sRNAs, and was recently shown to be repressed by the conserved RybB and MicC sRNAs, as well as the virulence region-specific InvR sRNA (50–54). Using an experimental 3′-RACE approach for target site prediction, followed by mutational analysis, we have determined that SdsR represses the ompD mRNA by means of a short stretch of complementarity in the coding sequence, yet at a different site than each of the other three repressors. In light of the many stress conditions that activate σS, we hypothesize that SdsR contributes to the post-transcriptional control of ompD under such conditions.
MATERIALS AND METHODS
DNA/RNA oligonucleotides and plasmids
Sequences of all oligonucleotides employed in this study are listed in Supplementary Table S1. All plasmids used in this study are summarized in Supplementary Table S2. Plasmid pBAD-SdsR (pKP19-8) was constructed as pBAD-RybB in ref. (21), but using primers JVO-0902/JVO-0903 to amplify the complete sdsR gene from its transcriptional start site to 30 bp downstream of the terminator T-stretch from genomic DNA. The same insert was used for the pZE12-luc-derived pPL-SdsR (pKF68-3) following the cloning strategy described in ref. (55). For plasmids expressing different SdsR variants from the pLLacO promoter, pKF68-3 served as a template for PCR amplification with the primer pairs JVO-7159/pLLacO C (pPL-SdsR +7; pKF97-1), JVO-7161/pLLacO C (pPL-SdsR +19; pKF99-1) and JVO-7163/JVO-4731 (pPL-SdsR*; pKF101-26) and self-ligation was carried out as in ref. (56). Similarly, pPL-SdsR-TMA (pKF105-1) was constructed by self-ligation of a PCR-product of JVO-7224/pLLacO C on pFS135. A single nucleotide exchange (C46G) was inserted in the ompD complementation plasmid (insert: 493 bp upstream of the translational start site to 89 bp after the stop codon) with primers JVO-7225/JVO-7328 to obtain pKF109-1. For the sdsR/sraC, complementation plasmid psdsR (pVP203-1), a fragment carrying both sdsR and sraC (comprising 198 bp upstream of the transcriptional start site of SdsR to 353 bp downstream of the terminator T-stretch), was amplified using primers JVO-0051/JVO-0052 and genomic DNA; the product was treated with XbaI/XhoI and ligated into an equivalently digested low-copy version of pZE12 in which the origin had been swapped with pSC101* [pVP003; (57)]. To exchange the cytosine at position −13 in the sdsR promoter, pVP203-1 served as the template for PCR amplification with the primer pairs JVO-4262/JVO-4265 (psdsR C-13G; pKF106-2), JVO-4263/JVO-4265 (psdsR C-13A; pKF107-1) and JVO-4264/JVO-4265 (psdsR C-13T; pKF108-3), and the resulting DNA fragments were self-ligated. Competent E. coli TOP10 were used for all cloning purposes.
Bacterial strains
A complete list of bacterial strains employed in this study is provided in Supplementary Table S3. The Salmonella enterica serovar Typhimurium strain SL1344 (JVS-0007), the E. coli MC4100 derivative relA+ (JVS-5105) and the attenuated Shigella flexneri BS176 (JVS-0012) are referred to as wild-type strains and were used for mutant construction. Phage P22 or P1 transduction (using standard protocols) was employed to transfer each single chromosomal modification to a fresh Salmonella or E. coli wild-type background, respectively, as well as to obtain strains carrying multiple mutations. Single mutant derivatives were constructed by the λRed recombinase one-step inactivation method using pKD4 as the template. To eliminate the KanR cassette of λRed-derived mutants, cells were transformed with the FLP recombinase expression plasmid pCP20 (58). Mutant susceptibility to kanamycin and loss of the temperature-sensitive FLP expression plasmid were tested. Briefly, for JVS-9251 (SL1344 ΔPsraC) and JVS-9312 (MC4100 ΔPsraC), Salmonella or E. coli wild-type cells carrying the pKD46 helper plasmid were transformed with the DNA fragment to be integrated, amplified from pKD4, using JVO-7742/JVO-7743 (Salmonella) or JVO-7859/JVO-7860 (E. coli), and insertion of the KanR marker gene was verified by PCR using JVO-0051/JVO-0902 or JVO-1043/JVO-2390, respectively. The single copy transcriptional sdsR-lacZ fusion was constructed as in ref. (59). The sdsR mutant strain (deletion of the sdsR gene downstream of nucleotide +6 with respect to the transcriptional start site) was obtained by the λRed recombinase protocol using a DNA fragment generated by PCR amplification (JVO-6533/JVO-6534) on pKD4. Obtained clones were tested by PCR (JVO-0052/JVO-0903) and transformed with pCP20. Upon verifying loss of the KanR cassette, mutants were transformed in the presence of pCP20 with pKG136 and colonies were screened for the integration of lacZ-Y downstream of the sdsR promoter (JVO-0052/pMC874-lac). To construct the chromosomal single-nucleotide exchange in ompD and an isogenic WT strain, SL1344 ΔompD (JVS-0735) carrying pKD46 was transformed with a PCR product from JVO-5738/JVO-5739 using pKF109-1 or pVP42-3, respectively, as templates. Integration of the CmR cassette-containing fragment upstream of the complete ompD gene, including its own promoter, was verified by PCR using JVO-0802/JVO-2192. P22 lysates of these strains were used to transduce JVS-8827 (SL1344 ΔsdsR) to obtain JVS-9155 (SL1344 CmR::ompD*ΔsdsR) and JVS-9154 (SL1344 CmR::ompDΔsdsR), respectively. Similarly, chromosomal integration of the constitutive PLtet-O1 promoter upstream of the Salmonella ompD gene was constructed by λRed-mediated recombination using a DNA fragment generated by PCR amplification (JVO-2191/JVO-2192) on pVP192-1 as template. Transformants were screened by PCR (JVO-0802 and JVO-2192), and a P22 lysate of a positive clone was used to transduced JVS-1574 (to obtain JVS-9488), JVS-8726 (to obtain JVS-9491) and JVS-8798 (to obtain JVS-9655).
Bacterial growth conditions
Cells were grown aerobically in Luria Broth (LB) medium at 37°C unless stated otherwise. A final concentration of 0.2% l-arabinose was added to cultures to induce expression from pBAD-derived plasmids. Where appropriate, liquid and solid media were supplemented with antibiotics at the following concentrations: 100 µg/ml ampicillin, 50 µg/ml kanamycin, 20 µg/ml chloramphenicol and 15 µg/ml tetracycline. To apply osmotic shock, cells were cultured at 37°C in M9 minimal medium [1× M9 salts, 2 mM MgSO4, 0.1 mM CaCl2, 0.4% (v/v) glycerol, supplemented with thiamine (0.5 µg/ml), l-histidine (40 µg/ml) and cas-amino acids (0.2%)] to an OD600 of 0.3. Cultures were split and NaCl was added to one batch at a final concentration of 0.3 M. To apply heat shock, cells were grown in LB at 30°C to an OD600 of 0.3. Cultures were split and growth was continued at either 30°C or 44°C. Stringent response was induced by addition of serine hydroxamate (SHX; final concentration: 100 µM) to cells during exponential growth (OD600 of 0.15) in Nutrient Broth (Difco, #234000) supplemented with 0.75 mM l-serine. Envelope stress was induced by polymyxin B (final concentration: 5 µg/ml) in cells grown in LB to early stationary phase (OD600 of 1.5).
In vitro RNA synthesis and determination of SdsR in vivo copy number
For synthesis of the SdsR in vitro transcript, ∼200 ng of a DNA fragment amplified from Salmonella gDNA (employing primer pair JVO-7023/JVO-7025) served as template in a T7 transcription reaction using the Megascript kit (Ambion). The correct size and integrity of the RNA were confirmed on a denaturing polyacrylamide gel. To estimate the in vivo abundance of SdsR (copy number per cell), total RNA corresponding to 0.5 OD of wild-type Salmonella were compared to serial dilutions of the SdsR in vitro transcript (0.5/1/2.5/5/10 and 20 ng) by northern blot and hybridization with an SdsR riboprobe.
Protein sample analysis
To prepare total protein samples, cells were collected by centrifugation (2 min; 16 000 g; 4°C) and resuspended in 1× SLB (Fermentas) to a final concentration of 0.01 OD/µl. For 11% SDS–PAGE, 0.1 OD were loaded per lane and gels were stained overnight with Coomassie Blue. To analyse protein levels by western blot, 0.02 OD (detection of OmpD-GFP fusion proteins and OMPs) or 0.1 OD (detection of RpoS and ribosomal protein S1) were loaded per lane and resolved by SDS–PAGE, after which proteins were transferred to PVDF membranes as described in ref. (57). GFP fusion proteins and RpoS were detected using commercially available antibodies directed against GFP (1:5000; mouse; Roche) and RpoS (1:1000; mouse; #W0009, Neoclone), repectively. The major OMPs, and ribosomal protein S1, were detected using an antiserum recognizing OmpC/F/D/A (1:20 000; rabbit; provided by R. Misra) and an anti-S1 antibody (1:5000; rabbit; provided by M. Springer), respectively. Anti-mouse or anti-rabbit secondary antibodies conjugated with horseradish peroxidase (1:10 000; GE Healthcare) were used in all cases. Signals were visualized using the Western Lightning reagent (PerkinElmer) and an ImageQuant LAS 4000 CCD camera (GE Healthcare).
RNA isolation and northern blot analysis
Total bacterial RNA was isolated from culture aliquots using TRIzol reagent (Invitrogen) and analysed by northern blot as previously described (55). Briefly, for sRNA and mRNA detection, 5 or 10 µg of total RNA was resolved on 4–6%/7 M urea polyacrylamide gels and electroblotted. Membranes were hybridized with gene-specific 5′-end-labelled DNA-oligonucleotides or riboprobes at 42°C or 65°C, respectively, in Roti-Hybri-Quick hybridization solution (Roth) and washed in three subsequent steps with SSC wash buffers supplemented with 0.1% SDS (5×/1×/0.5× SSC or 2×/1×/0.5×, respectively). Signals were determined on a Typhoon FLA 7000 phosphorimager (GE Healthcare) and band intensities quantified with AIDA software (Raytest, Germany). SraC, SdsR and its variants, the TMA chimera, ompD mRNA and 5S rRNA, were detected by the 5′-end-labelled oligonucleotides JVO-2390, JVO-1032, JVO-0396, JVO-4314 and JVO-0322, respectively. Riboprobes were synthesized by T7-mediated in vitro transcription of ∼200 ng of template DNA (amplified on gDNA with JVO-7692/JVO-7693 or JVO-0902/JVO-0997 for osmY mRNA or SdsR sRNA, respectively) in the presence of 32P-α-UTP with the MAXIscript kit (Ambion).
3′-RACE
3′-RACE experiments were carried out following the protocols in refs. (5) and (52) with a few modifications. Briefly, 7.5 μg of total DNA-free RNA was dephosphorylated with 10 U of calf intestine alkaline phosphatase (CIP; #M0290, New England Biolabs) in the presence of 1× NEB buffer 3 in a total volume of 25 µl at 37°C for 1 h. Following P:C:I extraction, the RNA was precipitated from the aqueous phase together with 250 pmol of RNA adapter E1 and 15 µg GlycoBlue (#AM9515, Ambion) using three volumes of 30:1 ethanol:sodium acetate (pH 6.5) mix. For ligation of the RNA linker, the pellet was resuspended in H2O and allowed to dissolve for 10 min at 65°C, after which a 20 µl reaction containing 20 U T4 RNA ligase (#M0204, New England Biolabs), 1× T4 RNA ligase buffer, 10% (v/v) DMSO and 10 U SUPERaseIn RNase Inhibitor (#AM2694, Ambion) was incubated at 16°C overnight. The ligated RNA was P:C:I-extracted and precipitated with three volumes of 30:1 ethanol:sodium acetate (pH 6.5) mix. The RNA was reverse transcribed for 5 min at 50°C and 60 min at 55°C in the presence of adapter E1-specific oligo E3RACE (39 pmol) using 200 U SuperscriptIII reverse transcriptase (#18080-093, Invitrogen) in a 20 µl reaction mix containing 1× FS buffer, 2 mM dNTPs, 5 mM DTT and 10 U SUPERaseIn RNase Inhibitor. Template RNA was digested by RNase H. To identify ompD-specific fragments, 1 µl aliquots of the RT reaction were used as templates in a PCR reaction with 1 mM each linker-specific primer, E3RACE and gene-specific primer, JVO-2678 (binds at the transcriptional start of ompD mRNA), 1.25 U Taq-DNA polymerase (#M0267, NEB), 1× ThermoPol buffer and 1.5 mM dNTPs. Cycling conditions were as follows: 95°C for 5 min; 35 cycles of 95°C for 40 s, 58°C for 40 s, and 72°C for 50 s and 72°C for 5 min). The PCR products were resolved on 3.5% agarose gels. Selected bands were purified and subcloned using the TOPO TA Cloning Kit (pCR 2.1-TOPO, Invitrogen) as recommended by the manufacturer. Inserts of obtained clones were amplified by PCR (M13fwd/M13rev) and analysed by sequencing.
β-galactosidase assay
Levels of β-galactosidase expressed from the single-copy transcriptional sdsR-lacZ (JVS-8717) or osmY-lacZ (JVS-9145) fusions were assayed from three biological replicates as follows: at selected timepoints, cells were collected by centrifugation (2 min, 16 000 g, 4°C) and resuspended in Z-Buffer to a final concentration of 1 OD/ml. After the addition of 0.15 vol. equiv. chloroform and 0.1 vol. equiv. 0.1% SDS, samples were vigorously vortexed for 15 s and stored on ice. In a microtiter plate, 200 µl of each cell lysate were mixed with 40 µl ONPG (40 mg/ml) and the absorbances at OD405 and OD600 determined at 28°C over time (0–45 min) with a Victor3 plate reader (1420 Multilabel Counter, Perkin Elmer). Relative β-galactosidase levels were determined at timepoints at which the absorbance of o-nitrophenol (OD405) increased linearly with time and was within the linear response range of the detector. Absorbance at OD600 was measured to control for the amounts of cell debris.
Sequence retrieval and alignments
Information for sequence alignments was collected using BlastN searches (http://www.ncbi.nlm.nih.gov/sutils/genom_table.cgi) of the following genome sequences (accession numbers are given in parentheses): Salmonella typhimurium LT2 (NC_003197), Salmonella typhi Ty2 (NC_004631), Citrobacter koseri ATCC BAA-895 (NC_009792), E. coli K12 (NC_000913), S. flexneri 2a str 301 (NC_004337), Enterobacter Sp.638 (NC_009436), Cronobacter turicensis z30232 (NC_013282), Klebsiella pneumoniae 342 (NC_011283), Serratia proteamaculans 568 (NC_009832), Yersinia pestis KIM (NC_005088), Yersinia enterolitica subsp. enterolitica 8081 (NC_008800), Dickeya dadantii Ech 703 (NC_012880), Pantoea ananatis LMG 20103 (NC_013956), Sodalis glossinidius str. ‘morsitans' (NC_007712), Erwinia pyrifoliae Ep1/96 (NC_003197), Photorhabdus luminescens subsp. laumondii TT01 (NC_005126) and Xenorhabdus nematophila ATCC 19061 (NC_014228). Alignments were made using MultAlin (http://multalin.toulouse.inra.fr/multalin/multalin.html).
RESULTS
Expression of SdsR but not SraC is conserved in diverse enterobacteria
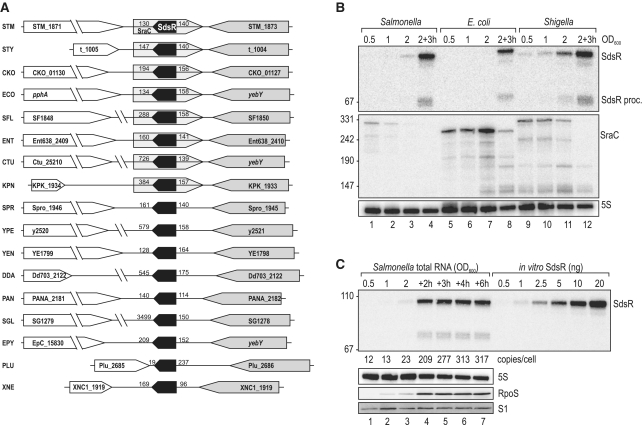
The sdsR gene was originally discovered in E. coli downstream of the conserved yebY gene on the minus strand of the chromosome, and had predicted homologues in several other species. Here, inspection of available genome sequences identified sdsR genes in many γ-proteobacteria, representative examples of which are shown in Figure 1A. In contrast, the sraC gene encoded on the plus strand was only found in a subset of these organisms (Figure 1A, Supplementary Figure S1).
Figure 1.
(A) Genomic context of the sdsR gene in various Enterobacteria. Synteny analysis of the sdsR/sraC genes in various enterobacterial species revealed partial conservation of the locus. In all cases sdsR is positioned downstream of yebY or homologues; the flanking gene at the 3′-end is variable. Distances to flanking genes are indicated in bp. STM: Salmonella typhimurium; STY: Salmonella typhi; CKO: Citrobacter koseri; ECO: Escherichia coli; SFL: Shigella flexneri; ENT: Enterobacter; CTU: Cronobacter turicensis; KPN: Klebsiella pneumoniae; SPR: Serratia proteamaculans; YPE: Yersinia pestis; YEN: Yersinia enterolitica; DDA: Dickeya dadantii; PAN: Pantoea ananatis; SGL: Sodalis glossinidius; EPY: Erwinia pyrifoliae; PLU: Photorhabdus luminescens; XNE: Xenorhabdus nematophila. (B) Expression levels of SdsR and SraC sRNAs in Salmonella, E. coli and Shigella. Northern blot analysis of total RNA isolated from wild-type Salmonella (JVS-1574), E. coli (JVS-5105) and Shigella (JVS-0012) cells grown to OD600 of 0.5, 1.0, 2.0 and 3 h after cells had reached an OD600 of 2.0. SdsR and SraC sRNAs were detected by radio-labelled oligo probes directed against the conserved sRNA sequences. 5S rRNA levels were determined to confirm equal loading. (C) SdsR copy number over growth. SdsR levels at various timepoints (OD600 of 0.5, 1.0, 2.0 and 2, 3, 4 or 6 h after cells had reached an OD600 of 2.0) in wild-type Salmonella were compared by northern blotting to signals of in vitro transcribed SdsR in indicated amounts. SdsR was detected using a riboprobe; probing for 5S RNA confirmed equal loading. Expression of RpoS at different growth stages was determined by western blotting. Detection of ribosomal protein S1 was used as loading control.
To determine how expression patterns of the sdsR-sraC locus compared among related bacteria, we probed RNA samples of E. coli, Salmonella and Shigella grown in rich medium from exponential to stationary phase (Figure 1B). The three species uniformly showed a highly stationary phase-specific accumulation of full-length SdsR and a faint, ∼70 nt processing product (3′-end of the sRNA), consistent with previous observations in E. coli (1,2). In contrast, probing of SraC yielded a mosaic picture: the SraC species of Shigella and Salmonella were ∼35 nt longer than their E. coli counterpart, and their expression peaked much earlier during growth. The difference in RNA size reflects the fact that the mapped −10 box of the sraC promoter element in E. coli (5) is located ∼35 bp further upstream in Shigella and Salmonella, while the 3′-end is conserved (Supplementary Figure S1). All in all, there is considerable variability in both the sequence and the expression of SraC, in contrast with the uniformity of SdsR, which argues that SdsR is the primary sRNA expressed by this locus.
SdsR is abundant and transcribed by σS
A previous RNomics (cDNA shot-gun cloning) approach revealed SdsR to be among the most abundant sRNAs in stationary phase E. coli cells (2). Here, we have quantitatively corroborated this finding by determining the in vivo copy number of SdsR at various stages of growth in Salmonella. To this end, cellular levels of SdsR RNA from different growth phases were compared on a northern blot along with an in vitro-synthesized SdsR transcript (Figure 1C). This analysis shows that SdsR RNA levels increase almost 30-fold from exponential to stationary phase and that when accumulation plateaus about 3 h after the cells reach an OD600 of 2.0 (early stationary phase) the sRNA is present in approximately 300 copies/cell.
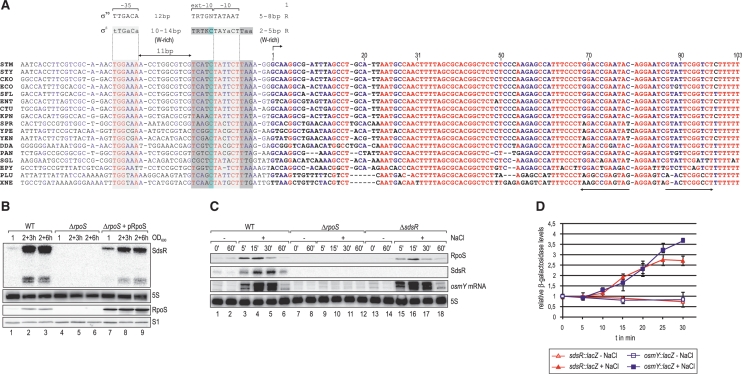
Given the high stationary phase specificity of sdsR expression, we hypothesized that the sdsR promoter is controlled by a transcription factor that is active in the late phase of growth. Our alignment of enterobacterial sdsR promoter sequences failed to reveal an extended conserved motif indicative of a transcription factor binding site; only the −10 and −35 boxes recognized by either the vegetative σ70 or the alternative σS factor are conserved (Figure 2A). Importantly, however, albeit these two sigma factors can recognize identical −10 and −35 consensus motifs in vitro (60), there are several promoter features that favour selective transcription by σS in vivo (61). A hallmark of σS selectivity is a conserved C at position −13 relative to the transcriptional start site, which is present in ∼70% of all experimentally confirmed σS-dependent promoters (61). Indeed, a C at position −13 is conserved in all sdsR promoter sequences (Figure 2A). The sdsR promoter meets additional criteria of σS selectivity: a 3 nt-long A/T-rich discriminator immediately downstream of the −10 box; a preference for nucleotides −8C and −14G/T; and a shorter spacer (16 instead of 17 bp) between the −10 and −35 boxes compared to σ70 (Figure 2A).
Figure 2.
(A) Non-redundant alignment of the sdsR gene including the upstream promoter region. All nucleotides are coloured regarding their degree of conservation (red: high conservation; blue: partial conservation; black: little or no conservation). σ70 or σS-specific promoter consensus motifs (61) are indicated above the alignment (W: A/T; R: A/G; Y: C/T; K: T/G); the putative −10 and the −35 sites of the sdsR promoter are boxed in light grey, σS-specific extensions of the −10 element are marked in dark grey and the conserved cytosine residue at position −13 is boxed in light blue. ‘+1’ marks the transcriptional start site, and the transcribed SdsR sequence is given in bold letters. The ρ-independent terminator is indicated by arrows. (B) SdsR sRNA is not detectable in the absence of RpoS. SdsR levels were determined by northern blotting of RNA isolated at indicated times over growth from Salmonella wild-type and rpoS mutant cells (JVS-5487) carrying either a control vector (pRH800) or a plasmid constitutively expressing E. coli RpoS (pRL40.1). RpoS expression was monitored by western blotting, and loading was controlled by probing for ribosomal protein S1. (C) SdsR sRNA and osmY mRNA are rapidly induced under osmotic stress in wild-type but not in rpoS mutant bacteria. Salmonella wild-type, ΔrpoS and ΔsdsR (JVS-8827) were grown in supplemented M9 medium to an OD600 of 0.3 when cells were split and osmotic shock was induced in one aliquot by addition of NaCl to a final concentration of 0.3 M while the other half was left untreated. Total RNA samples withdrawn prior to and at selected timepoints after NaCl addition were analysed by northern blotting; SdsR and osmY mRNA were detected using riboprobes, 5S levels were determined as loading control. Expression of RpoS was controlled by western blotting, detection of ribosomal protein S1 was used to control equal loading. (D) Transcriptional activity at sdsR and osmY promoters upon osmotic shock. Promoter activities of Salmonella sdsR-lacZ (JVS-8717; red triangles) and osmY-lacZ (JVS-9145; blue squares) fusions were determined after osmotic stress was induced as in (C) by measuring relative β-galactosidase activities over 30 min in culture samples with (filled symbols) or without NaCl added (open symbols). Error bars represent the standard deviation calculated from three biological replicates.
To test experimentally whether or not transcription of sdsR requires σS, we compared SdsR levels among wild-type (WT) Salmonella, an isogenic rpoS deletion mutant (ΔrpoS), and the mutant strain complemented with a plasmid carrying the E. coli rpoS gene driven by a constitutive Ptac promoter (62). Figure 2B shows that the SdsR RNA was entirely absent in ΔrpoS bacteria (lanes 4–6), but expression was restored upon ectopic expression of σS in the mutant (lanes 7–9). Western blot detection of σS protein in the same samples revealed a positive correlation of SdsR and σS levels in the WT strain (Figures 1C, 2B, lanes 1–3). Thus, σS and not σ70 is required to express the sdsR gene.
The fact that the plasmid-expressed σS was equally abundant at all tested growth stages, while SdsR expression remained largely stationary phase specific (Figure 2B, lanes 7–9), argues that the activity rather than solely the intracellular concentration of σS governs SdsR expression. It is known that selective σS transcription at many promoters requires the alarmone guanosine tetraphosphate (ppGpp), abundance of which is low in exponential phase (63). To determine whether ppGpp plays a role in SdsR expression as well, we probed the sRNA in a Salmonella ΔrelAΔspoT strain (64) that lacks both of the known ppGpp synthetases RelA and SpoT (65,66). Compared to WT, the absence of ppGpp reduced the up-regulation of SdsR through stationary phase but had no impact on σS protein levels (Supplementary Figure S2), suggesting that ppGpp and σS are essential for the efficient transcription of sdsR.
We further investigated the σS specificity of the sdsR promoter by testing the mutability of the characteristic cytosine residue at position −13. This nucleotide is predicted to impart a preference for σS over σ70 by engaging in favourable interactions with residues K173 of σS and unfavourable interactions with E458 in σ70 (67,68). C-13 was changed to A, G or T in an sdsR gene on a low-copy plasmid. Northern blot probing of ΔsdsR cells complemented with these plasmids and grown to stationary phase revealed lower expression levels for the sRNA mutants, which was most pronounced for the T-13 mutant that resulted in a ∼4-fold reduction compared to WT (Supplementary Figure S3). The sensitivity of this position to mutation supports our conclusion that the sdsR promoter is recognized by σS.
SdsR shows σS-dependent regulation under stress
To further investigate the σS-dependence of SdsR expression, we determined its induction under several stress conditions known to activate σS (13). First, we grew WT, ΔrpoS and ΔsdsR Salmonella to exponential phase and added sodium chloride to induce osmotic shock. Western blots confirmed the expected rapid increase in cellular σS levels in the two rpoS-positive strains, i.e. WT and ΔsdsR (Figure 2C, upper panel, lanes 3–6 and 15–18). Concomitantly, the salt treatment promoted a rapid accumulation of SdsR in wild-type Salmonella, but not in the ΔrpoS strain (Figure 2C, second panel from top, lanes 3–6 and 9–12). The osmotic shock regulation of sdsR qualitatively matches that of osmY, a known highly σS-dependent gene (9,69); our northern blot analysis shows that RNA levels of both SdsR and osmY peak ∼15–30 min post salt challenge (Figure 2C, second and third panel from top, lanes 4 and 5). Furthermore, the same trend was observed when the above strains were subjected to heat-shock (Supplementary Figure S4), which is another stress known to trigger σS-dependent gene expression (62).
To investigate the effect of osmotic and heat shock on the transcription of sdsR, we integrated a lacZ reporter gene downstream of the chromosomal sdsR promoter and measured transcriptional activity upon salt and heat stress (Figure 2D and Supplementary Figure S5). Analogous to the above experiments, we used a lacZ fusion to the osmY promoter as a positive control for σS-dependent gene expression. Under both conditions, the sdsR and osmY promoters exhibited almost identical activation within 30 min after stress induction. In agreement with our northern blot analyses (Figure 2C and Supplementary Figure S4), activation of the sdsR and osmY promoters was more rapid upon osmotic than heat shock (Figure 2D and Supplementary Figure S5). In summary, the results strongly suggest that in Salmonella, SdsR belongs to the group of cellular transcripts whose synthesis is very tightly controlled by σS, and this may hold true for all the enterobacteria in which this sRNA gene is present (Figure 2A).
SdsR is a repressor of OmpD synthesis
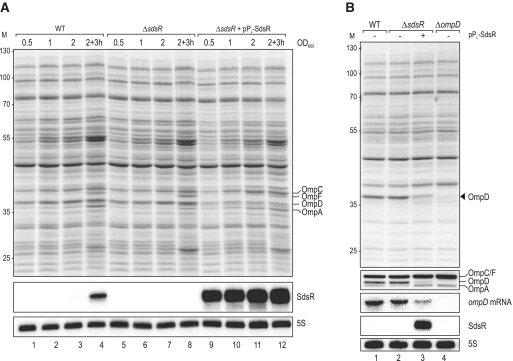
To identify a biological role of SdsR in Salmonella, we uncoupled its expression from σS by cloning the sdsR gene on a plasmid downstream of a constitutive PL promoter [synthetic PLlacO variant; (70)]. Protein profiles of Salmonella WT and ΔsdsR strains, carrying this SdsR expression plasmid or a control vector, were analysed by 1D SDS page in different growth phases, an approach that was previously successful at identifying targets of Hfq-dependent sRNAs (34,45,53,71). Whereas a comparison of the WT and ΔsdsR strains did not reveal obvious differences in total protein patterns (Figure 3A, lanes 1–8), the overexpression of SdsR depleted an abundant protein whose size of ∼40 kDa was indicative of it being the major Salmonella porin, OmpD (72) (Figure 3A, lanes 9–12). To test this prediction, we probed the expression of ompD at both the mRNA (northern blot) and the protein (western blot) levels with the same set of strains as above, but additionally included a Salmonella ΔompD strain as control. Figure 3B shows that SdsR indeed strongly reduces the level of OmpD protein without impacting other major OMPs, and that this reduction is accompanied by a down-regulation of the ompD mRNA, suggesting that SdsR targets OmpD production at the levels of transcription or mRNA rather than protein stability.
Figure 3.
(A) Proteome changes upon SdsR over-expression in Salmonella. Whole-cell protein patterns of wild-type and ΔsdsR Salmonella carrying either a control vector (pJV300) or the constitutive SdsR-expression plasmid pPL-SdsR (pKF68-3) grown in LB were compared by separation of total cell lysates from several conditions (OD600 of 0.5 (lanes 1, 5, 9); 1.0 (lanes 2, 6, 10); 2.0 (lanes 3, 6, 11); 3 h after cells had reached an OD600 of 2.0 (lanes 4, 9, 14)) by 11% SDS–PAGE; the gel was stained for abundant proteins with Coomassie Blue. Sizes of co-migrating marker proteins are marked at the left in kDa. Positions of the major porins OmpC, OmpF, OmpD and OmpA are indicated. SdsR expression was determined by northern blotting of RNA isolated from the same cultures and loading was controlled by probing for 5S RNA. (B) Verification of specific OmpD repression by SdsR. Wild-type, ΔsdsR and ΔompD (JVS-0735) Salmonella transformed with either the control vector or pPL-SdsR were grown to an OD600 of 2.0, and OmpD protein levels were analysed by SDS-PAGE (top panel) and western blotting using an antiserum detecting the major Salmonella porins as indicated (second panel). Northern blot analysis (three lower panels) of the same strains revealed reduced ompD mRNA steady-state levels in cells over-expressing SdsR. 5S RNA served as loading control.
Post-transcriptional repression of ompD mRNA by SdsR requires RNase E
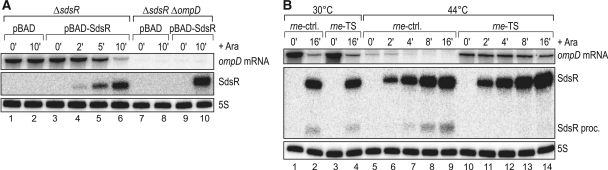
Given that SdsR is an Hfq-associated RNA (1,36–38), we considered that SdsR might repress ompD at the post-transcriptional level. To test this, we expressed SdsR from a plasmid-borne l-arabinose-controlled PBAD promoter (plasmid pBAD-SdsR) and monitored the down-regulation of chromosomal ompD mRNA within 10 min of induction in early stationary phase (OD600 of 1.5). These experiments were carried out in Salmonella ΔsdsR and ΔsdsRΔompD backgrounds, comparing the effects of induction of pBAD-SdsR to that of a pBAD control vector. The induction of SdsR causes a ∼10-fold decrease in ompD mRNA levels within 10 min of adding arabinose (Figure 4A, lanes 3–6), while the inducer itself has no effect on the ompD mRNA, as evident from the pBAD control samples (Figure 4A, lanes 1–2). Importantly, the intrinsic half-life of the ompD mRNA in this growth phase exceeds 10 min (57), from which follows that SdsR directly or indirectly reduces the stability of this target, and thus acts at the post-transcriptional level.
Figure 4.
(A) Pulse expression of SdsR sRNA from an inducible PBAD promoter results in rapid decrease of ompD mRNA levels. Salmonella ΔsdsR and ΔsdsR ΔompD (JVS-8434) cells carrying either the control vector pBAD (pKP8-35) or the expression plasmid pBAD-SdsR (pKP19-8) were grown to an OD600 of 1.5 and total RNA samples were collected prior to and at indicated timepoints after l-arabinose addition (0.2% final concentration). Both ompD mRNA and SdsR sRNA levels were detected by northern blotting. (B) SdsR requires RNase E for ompD mRNA decay. Salmonella rne-ctrl. deleted for rybB, micC, invR and sdsR sRNA genes (JVS-9549) and its isogenic rne-ts strain (JVS-9550) were transformed with pBAD-SdsR and grown at 30°C to an OD600 of 1.5 when cultures were split. Growth was continued for 30 min at 30°C or, to inactivate RNase E in rne-ts, at 44°C prior to arabinose-induced expression of SdsR for 10 min. Levels of ompD mRNA, SdsR RNA and 5S RNA (loading control) were determined by northern blot analysis of total RNA.
Our previous work on this target identified the major endoribonuclease RNase E as an essential co-factor for ompD repression by MicC sRNA (52). To address whether SdsR also requires this nuclease to repress ompD, we employed a temperature-sensitive mutant (rne-TS) of this otherwise essential enzyme (73,74), evaluating pBAD-SdsR mediated down-regulation of the ompD mRNA in the presence and absence of functional RNase E. To exclude any complementation of regulation by the other known ompD regulators, the Salmonella rne-TS and its isogenic WT strain were rendered null mutants of all four relevant sRNA genes (ΔsdsR, ΔmicC, ΔrybB and ΔinvR). These strains were then transformed with pBAD-SdsR and grown at the permissive temperature (30°C) to early stationary phase (OD600 of 1.5), at which point the cultures were split into two parts, and incubation was continued for additional 30 min at either the permissive or the non-permissive (44°C) temperature. Subsequently, SdsR expression was induced and changes in SdsR and ompD mRNA levels were determined over the course of 16 min. The permissive temperature, i.e. the presence of functional RNase E, allows repression of ompD mRNA by SdsR to equal degrees in the wild-type and rne-TS strains (Figure 4B, lanes 1–4). In contrast, growth at the non-permissive temperature abolishes the SdsR-mediated ompD repression in the rne-TS strain (Figure 4B, lanes 10–14) even though the sRNA does accumulate to the same levels as in the wild-type strain, in which case it fully retains its ability to deplete ompD mRNA (lanes 5–9). Interestingly, inactivation of RNase E also abolishes the accumulation of the processed SdsR variant which lacks the first 30 nt of the 5′-end (2). Altogether, the above results strongly suggest that SdsR acts upon the ompD mRNA, most likely by base pairing.
SdsR regulates ompD mRNA through binding in the CDS
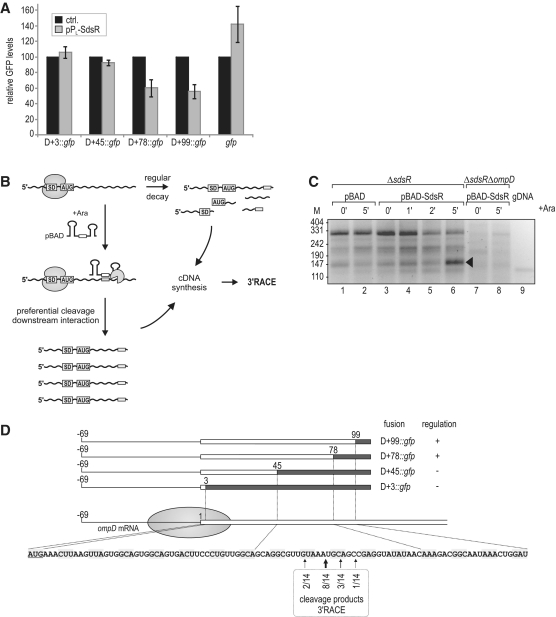
To further confirm the post-transcriptional regulation of ompD and locate a potential binding site for SdsR, we used a well-established GFP reporter system in which the sRNA and its target are co-expressed from constitutive promoters on compatible plasmids (55). In a first attempt, we used a so-called D+3 reporter construct in which the 5′ UTR (untranslated region) and start codon of ompD (positions −69 to +3 relative to AUG; A is +1) are fused to the N-terminus of gfp (52). Coexpression of SdsR had no effect on OmpD::GFP levels in this fusion (Figure 5A), suggesting that the 5′-UTR of ompD was insufficient to recapitulate the repression observed with the full-length mRNA. Our previous work has determined a ‘5 codon window’ in the early CDS of bacterial mRNAs wherein base-pairing sRNAs can repress the translational initiation of their targets by antisense competition with the 30S ribosomal subunit (50). To determine if SdsR operates within this window, a D+45 reporter, which includes the first 15 codons of ompD (52), was tested; yet again, SdsR failed to bring about a reduction in OmpD::GFP levels (Figure 5A).
Figure 5.
(A) Regulation of OmpD-GFP reporter fusions by SdsR. Salmonella ΔsdsR ΔompD cells carrying the control vector or pPL-SdsR were co-transformed with low-copy plasmids expressing gfp alone or a series of translational ompD::gfp fusions spanning the complete 5′-UTR plus an increasing number of nucleotides of the ompD coding sequence (D+3::gfp; D+45::gfp; D+78::gfp; D+99::gfp; see Supplementary Table S2 for details on plasmids) as depicted in (D). Whole-protein samples were collected from cells grown to an OD600 of 2.0, and regulation of reporter fusions was determined by signal quantification on western blots. Relative GFP levels in the presence of the control plasmid (black bars; set to 100) or the constitutive pPL-SdsR (grey bars); errors indicate standard deviation from three biological replicates. (B) Schematic illustration of the 3′-RACE approach employed for target site determination. (C) 3′-RACE analysis of ompD mRNA fragments enriched upon SdsR pulse expression. cDNA was prepared from total RNA of ΔsdsR cells as well as the ΔsdsR ΔompD control strain prior to and at indicated timepoints after SdsR induction from an inducible PBAD promoter. Salmonella genomic DNA (gDNA) served as a control template. DNA fragments were recovered from the indicated band of ∼150 bp (lane 6), and ompD 3′-ends were determined by sequencing of subcloned fragments. (D) Location of ompD 3′-ends obtained by 3′-RACE analysis. The ompD::gfp reporter plasmids and their regulation by SdsR (see Figure 5A) are represented schematically. The filled circle indicates the approximate coverage of ompD mRNA by the 30S ribosomal subunit binding to the RBS. Position as well as frequency of enriched break-down products determined by 3′-RACE (Figure 5C) are shown below the ompD CDS.
Notably, all three other sRNAs known to repress ompD at the post-transcriptional level bind the ompD mRNA in the CDS (51–54), and in fact MicC binds to a region as far downstream as codons 23–26 (52). Consequently, we also tested available D+78 (codon 26) and D+99 (codon 33) reporter fusions (52). Both reporters displayed a significant reduction in protein levels (Figure 5A), which we estimate to be 3-fold in both cases, correcting for the fact that SdsR, for reasons unknown, up-regulated 1.4-fold a GFP reporter with a control insert (Figure 5A). Altogether, these results indicate that SdsR targets ompD between the 15th and 26th codon.
A 3′-RACE approach maps the SdsR target site in ompD
To further investigate the binding of SdsR to the CDS of ompD, we took advantage of an observation we had previously made with MicC sRNA (52). Specifically, we discovered that MicC promotes target cleavage 4–5 nucleotides downstream of its binding site in ompD, generating a stable mRNA intermediate whose 3′-end revealed the approximate location of the base pairing region. Since both MicC and SdsR repress the ompD mRNA by means of RNase E, we postulated that such a proximal cleavage event may also help localize the SdsR site in this target. To this end, we used a 3′-RACE (rapid amplification of cDNA ends) approach to determine the termini of ompD mRNA fragments that accumulate transiently upon inducing expression of the sRNA from pBAD-SdsR (Figure 5B). DNA-free RNA was prepared from Salmonella ΔsdsR or ΔsdsRΔompD strains carrying the sRNA expression or control plasmids at various timepoints prior to and after adding arabinose (Figure 5C). Following 5′ dephosphorylation of the RNA samples, ligation of a 3′-RNA linker, and reverse transcription, cDNA ends were amplified using an ompD-specific forward and a 3′ adaptor-specific reverse primer. Separation of the PCR products on a high percentage agarose gel identified a ∼150 bp DNA fragment enriched in the sample taken 5 min after SdsR expression (lane 6), which appeared to be specific since it was not observed in the ompD deletion strain (lanes 7–8). This band was purified and analysed by sequencing after sub-cloning. All 14 sequenced inserts mapped to ompD with 3′-ends centring closely around the first guanosine of the 23rd codon (Figure 5D). The detected 3′-ends fully support the results obtained by using the GFP reporter system, which suggested that SdsR targets ompD between codons 15 and 26 (Figure 5A).
Identification of the SdsR-binding site on ompD mRNA
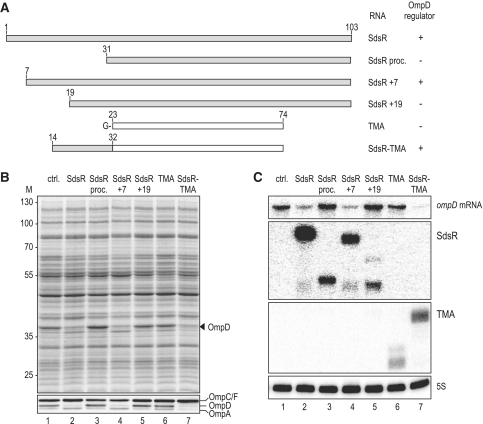
Although the above binding site window of ompD mRNA may seem narrow, standard algorithms for target site searches, such as RNAhybrid (75), failed to predict a robust SdsR–ompD interaction, especially when querying the conserved region of the sRNA, which typically serves as the target recognition domain of Hfq-associated antisense regulators (39). Thus, we took the opposite approach and narrowed down the binding site in the sRNA by overexpressing various fragments of it. First, we evaluated whether the naturally occurring, 5′ processed sRNA species, SdsR+31, lacking nucleotides 1–30 [Figures 4B and 6A; (2)] was able to regulate ompD. Under conditions where the overexpressed full-length SdsR totally depleted the OmpD protein in Salmonella, compared to the sRNA control vector (Figure 6B, lane 2 versus 1), this processed SdsR+31 RNA had no effect on OmpD abundance (lane 3). Similarly, an SdsR+19 variant was unable to regulate ompD (lane 5), although given the weaker accumulation, this SdsR variant may not be fully informative as to whether or not nucleotides more upstream in the 5′ region of SdsR are required for its activity on ompD (Figure 6C). However, an SdsR+7 variant lacking only the seven 5′ terminal nucleotides represses OmpD synthesis to the same degree as does full-length SdsR (Figure 6B, lane 4).
Figure 6.
(A) Schematic representation indicating size and composition of SdsR variants or chimeras tested for their impact on OmpD expression. TMA designates a 5′ shortened variant of the unrelated MicA sRNA (Truncated MicA). For sequences of the respective constructs, see Supplementary Figure S6. (B) The SdsR 5′-end is required for ompD regulation. Salmonella ΔsdsR transformed with a control vector or plasmids constitutively expressing versions of SdsR (as depicted in A; pPL-SdsR; pPL-SdsR proc.; pPL-SdsR +7; pPL-SdsR +19; pPL-SdsR-TMA; pPL-TMA; see Supplementary Table S2 for details on plasmids) were grown to an OD600 of 2.0. Total protein samples were analysed by SDS-PAGE (upper panel) and specific deregulation of OmpD was confirmed by western blotting (lower panel). (C) Expression of ompD mRNA in the presence of control RNAs or the various SdsR constructs used in (B) was determined by northern blotting; probing of 5S rRNA served as loading control.
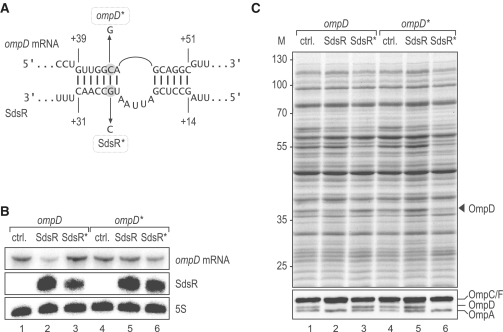
To narrow the binding region further, we fused nucleotides 14–32 of SdsR to the 5′-end of TMA RNA (Figure 6A and Supplementary S6), a scaffold for RNA fusions that has previously enabled us to pinpoint target binding domains of other Hfq-associated sRNAs (50–52). Remarkably, the resulting chimeric SdsR-TMA sRNA is as potent as full-length SdsR in its ability to down-regulate OmpD (Figure 6B, compare lanes 2 and 7), whereas expression of the parental TMA RNA has no such effect (lane 6). Altogether, the data obtained with SdsR truncation mutants indicate that SdsR nucleotides 14–32 (Figure 6) and nucleotides +40–60 of the ompD CDS (Figure 5) are crucial for regulation. When these shorter sequences were queried, RNAhybrid was able to predict an imperfect SdsR–ompD interaction composed of two short (6 and 7 bp) helices that flank a 5-nt bulge in SdsR (Figure 7A). To validate the predicted RNA duplex in vivo, we introduced compensatory point mutations in both partners (Figure 7A): a G to C change at nucleotide 26 yields the SdsR* variant, and a C to G change at position +46 of the chromosomally encoded ompD mRNA yields the compensatory ompD* mutant. The northern blot analysis presented in Figure 7B shows that the point mutations do not compromise the expression of SdsR or ompD mRNA. Singly, each point mutation abrogated the repression of OmpD porin synthesis, at both the RNA and the protein levels (Figure 7B and C). In contrast, when the SdsR* and ompD* variants were both present, SdsR-mediated OmpD repression was restored (Figures 7B and C, lane 6). These results provide further evidence supporting the conclusion that SdsR regulates this major porin directly, by forming a short duplex with its mRNA.
Figure 7.
(A) Predicted duplex forming between SdsR sRNA and ompD mRNA. Point mutations to generate the compensatory ompD* and SdsR* alleles are indicated.(B) Compensatory base pair exchanges validate the SdsR–ompD interaction. Salmonella ΔsdsR ompD* mutant (JVS-9155) or isogenic ΔsdsR ompD (JVS-9154) cells carrying plasmids for the constitutive overexpression of either SdsR (pKF68-3) or SdsR* (pKF101-26), respectively, were grown to an OD600 of 2.0. Expression levels of ompD/ompD* mRNAs and SdsR/SdsR* sRNAs were determined by northern blot analysis of total RNA; probing of 5S rRNA served as loading control.(C) OmpD levels with respect to SdsR or SdsR* expression were analysed by SDS–PAGE (upper panel) and western blot (lower panel) using total protein samples prepared in parallel to the RNA samples in (B). OmpD protein is indicated by an arrow.
Differential sRNA control of ompD mRNA under major stress conditions
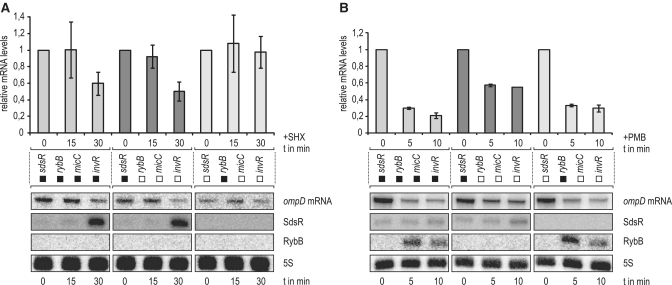
Of the four sRNAs which are now known to repress ompD, RybB and SdsR appeared redundant as both of them accumulate in stationary phase; in addition, their transcriptional activators (σE and σS, respectively) are induced by certain stresses such as heat, osmotic or acid stress in parallel (76–79). We reasoned, however, that other conditions such as amino acid starvation or membrane perturbation by bacteriocidal polymyxins activate either σE or σS, and therefore require repression of ompD by one or the other sRNA.
To address whether RybB and SdsR sRNAs controlled ompD post-transcriptionally under major stress conditions in a non-redundant fashion, we treated a Salmonella strain expressing ompD from a constitutive promoter with either the amino acid analogue serine hydroxamate (SHX) to induce the stringent response (σS), or polymyxin B to induce envelope stress (σE), and assessed down-regulation of the ompD mRNA decay in wild-type Salmonella and two mutants which lacked three of the four sRNAs, and expressed only either RybB or SdsR. As expected, SHX induced the expression of σS and SdsR but not RybB (Figure 8A and data not shown). The SHX treatment also caused a decrease of ompD mRNA levels, which required SdsR but none of the other three sRNAs in question. However, when we induced the σE pathway, most of the sRNA-dependent ompD mRNA decay could be accounted for by RybB, whereas SdsR had no role under this condition (Figure 8B). Thus, although these Hfq-dependent sRNAs accumulate in the same phase of growth in normal media, they can mediate ompD mRNA in response to specific conditions.
Figure 8.
(A) The stringent response triggers SdsR-specific ompD mRNA decay. Salmonella expressing ompD from a chromosomal constitutive PLtetO-1 promoter (JVS-9488) and derivative strains carrying either only sdsR or rybB of the four relevant sRNA genes (strains JVS-9491 or JVS-9655, respectively) were treated with serine hydroxamate (SHX) to induce the stringent response. Total RNA samples withdrawn from cultures prior to and 15 or 30 min after SHX addition were analysed by northern blotting. SdsR but not RybB was strongly induced upon 30 min of growth in the presence of SHX; ompD mRNA levels decreased concomitantly in strains carrying a functional sdsR allele (see left and middle panels) but remained stable in the absence of sdsR (right panel). Bars represent relative ompD mRNA levels as determined from the quantification of northern blots normalized by probing for 5S RNA; error bars indicate the standard deviation from three independent biological replicates. Black or white boxes mark the presence or absence, respectively, of the indicated sRNA genes in the strains used. (B) RybB promotes selective ompD mRNA repression during membrane stress. The envelope stress response was induced by addition of the antimicrobial peptide polymyxin B (PMB) with the same set of Salmonella strains as above. Northern blot analysis of total RNA samples prepared from cells collected prior to and 5 or 10 min after PMB addition revealed strong induction of RybB but not SdsR. Rapid decay of ompD mRNA was observed to the same extent in WT and the strain only expressing RybB (see left and right panel), but down-regulation was markedly reduced in its absence (see middle panel).
DISCUSSION
The study of the σS system over the years has revealed much as to how cells coordinate a transcriptional response to conditions of nutrient deprivations and stress; it has also contributed to our current appreciation of the important roles that Hfq and sRNAs have in connecting transcriptional and post-transcriptional levels of gene regulation. Following the seminal discovery that the rpoS mRNA itself is regulated by Hfq in Salmonella and E. coli (80,81), the activities of four Hfq-associated sRNAs that act upon this transcription factor target were investigated (see Figure 9A) (12,82). These sRNAs function under different conditions, i.e. low temperature or osmotic shock [DsrA, RprA; (42,83)], or extreme oxygen levels [ArcZ, OxyS; (31,45)], but each of them is conserved across enterobacteria, arguing that sRNA-mediated signal transduction to the rpoS gene is an elementary feature of the σS system.
Figure 9.
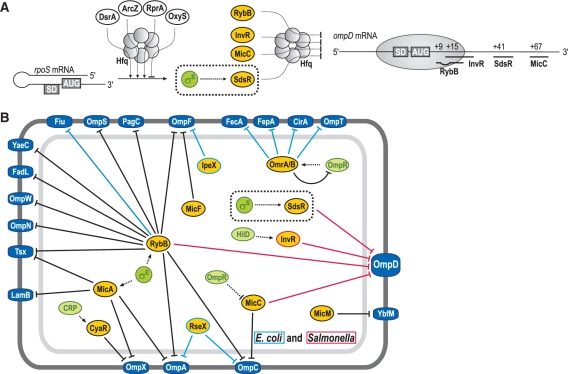
(A) Schematic display of regulatory RNAs affecting rpoS and ompD expression post-transcriptionally. sRNA regulators controlling rpoS mRNA are depicted in white ellipses, those that regulate ompD mRNA expression are shown in yellow. Hfq is indicated as a circled hexamer, σS as a green circle. Positioning of the 30S ribosomal subunit and pairing-sites of RybB, InvR, SdsR and MicC sRNAs on the ompD CDS are shown on the right. (B) Network of Hfq-dependent sRNA regulating outer membrane protein synthesis in E. coli and Salmonella. Transcriptional regulators are represented as green, sRNAs as yellow and OMPs as dark blue circles, respectively. Black lines mark sRNAs and regulatory functions common to both species while light blue or red lines denote sRNAs or regulation specific to E. coli or Salmonella, respectively. Note that a gene similar to ompD is generally present in E. coli and referred to as nmpC. However, in many E. coli strains including strain K12—our reference here—the NmpC/OmpD porin is not expressed due to an insertion element (84,85); consequently we indicate sRNA-mediated regulation of OmpD as specific to Salmonella, although the nmpC mRNA was also shown to be a RybB target in E. coli (24).
This wealth of conserved upstream riboregulators contrasted with a paucity of conserved sRNAs that acted immediately downstream in the σS cascade (see above). The sdsR gene investigated here is conserved in a broad range of enterobacterial species, and so are the structural and sequence elements of its promoter region that are known to impart σS selectivity [Figure 1A and ref. (61)]. This close-to-consensus structure of the sdsR promoter is reminiscent of the MicA, RybB and VrrA sRNAs that were recently reported to be members of the σE regulon (21–23,25,86). Under conditions of envelope stress, including stationary phase, the MicA and RybB sRNAs are highly and specifically transcribed by σE to repress many envelope-related mRNAs; they thereby indirectly endow σE, which is intrinsically restricted to gene activation, with the opposite repressor function (87). Whether such a division of labour applies to σS and SdsR as well remains to be seen. Different from MicA and RybB, which facilitate feedback regulation of σE (21,22,51), SdsR does not seem to modulate σS activity, at least under the conditions tested here (i.e. the levels of σS are the same in WT and ΔsdsR; Figure 2C). Perhaps SdsR impacts the dynamics or threshold of the σS response, similar to the recently identified role of Spot42 sRNA in cAMP-CRP mediated catabolite repression (88); addressing this will require a higher resolution picture of stress-induced gene expression changes in the presence or absence of sdsR. In this report, we have identified and validated the ompD mRNA as a direct target of SdsR, and will focus our considerations below on the mechanistic aspects of its regulation.
OmpD is regulated by four Hfq-dependent small RNAs
OmpD is Salmonella's most abundant porin, present at >105 molecules/cell (72). The ompD gene is tightly regulated at both the transcriptional and the post-transcriptional level (49), perhaps because even a slight increase in OmpD protein levels can trigger cell lysis (V. Pfeiffer and J. Vogel, unpublished results). Hfq is a key factor in the controlled repression of OmpD: Salmonella Δhfq strains typically overexpress porins and other envelope proteins (57,89) and this overexpression is very pronounced with OmpD (57). The importance of controlling OmpD synthesis was further demonstrated in elegant work by Bossi et al. (90) who showed that a secondary deletion of ompD fully reversed the induction of the envelope stress response that is caused by a Δhfq mutation.
Although Hfq alone binds the ompD mRNA with high affinity (21,36,57), repression results from the base pairing ability of cognate Hfq-associated sRNAs. Our identification of the fourth sRNA to repress ompD supports the notion that controlling this porin is of paramount importance to Salmonella. These sRNAs seem needed most as cells enter and progress through stationary phase. That is, MicC (regulated by OmpR) and InvR (regulated by HilD) transiently accumulate in early stationary phase (52,53,91). RybB and SdsR, which are transcribed by σE and σS, respectively, become highly abundant in deep stationary phase [(21) and this work]. However, we have shown that although the latter two sRNAs accumulate in the the same phase of growth in normal media, they can mediate ompD mRNA in response to specific conditions, i.e. the envelope stress response or stringent response, respectively.
Interestingly, OmpD now has four confirmed sRNA repressors, the same number as the related and abundant OmpC protein: in E. coli, the ompC mRNA is the target of IpeX (92), MicC (91), RseX (93) and RybB (24,25). In the ever growing network of sRNA-based porin regulation in Salmonella and E. coli (see Figure 8B for a compilation) many sRNAs are controlled by transcription factors that respond to envelope perturbations. SdsR is the first sRNA that has been shown to be transcribed by a general stress sigma factor, and it may therefore represent a fail-safe mechanism to repress the major porin of Salmonella.
Target regulation in the CDS
The CDS in bacterial messengers used to be considered as refractory to efficient sRNA targeting owing to the strong helicase activity of the elongating ribosome that can melt the relatively short sRNA–mRNA duplexes with ease (94). However, this view was challenged recently by our discovery of the 10 nucleotide-binding site of MicC sRNA well within the CDS of ompD [codons 23–26; (52)], well downstream of the first five codons where mRNAs are generally sensitive to antisense inhibition of translation initiation. Targeting the deeper CDS has since been shown to be a more common mechanism than previously thought; for example, the recently mapped sRNA–target interactions, ArcZ-tpx (95), RybB-fadL (51) and SgrS-manX (96) all fall in the CDS.
The exact manner in which SdsR represses the ompD mRNA has yet to be understood. By analogy with our model for MicC-ompD regulation (52), SdsR may not impair translation but instead act to induce RNase E-dependent cleavage of this messenger. The strict requirement for RNase E (Figure 4C), and the detection of a sRNA site-proximal mRNA cleavage by 3′-RACE, supports the notion that SdsR and MicC may act by a similar mechanism. It is puzzling, nevertheless, that all four sRNAs bind ompD exclusively within a rather narrow window in the CDS (see Figure 8A). One speculation is that an Hfq-binding site in the ompD CDS recruits sRNAs exclusively to this region. Another is that sRNAs might be unable to target the 5′-UTR of ompD because of its tight secondary structure. Altogether, the ompD mRNA lends itself as a model for understanding why certain mRNAs are targeted in the CDS and not in the 5′-UTR, where most sRNAs seem to operate. It will also be interesting to determine if RNase E is actively recruited to the CDS, as has been reported for sRNAs that block translation in the 5′-UTR (97,98).
Localization of sRNA sites by 3′-RACE
Although computational predictions for sRNA-binding sites have improved with additional information regarding what is required for sRNA–mRNA interactions (99), they are still plagued by high false positive rates. Experimental data on mRNA cleavage upon sRNA binding may complement or supersede theory, as shown here with 3′-RACE. We previously showed that MicC facilitates ompD mRNA degradation by promoting RNase E-mediated cleavage within the CDS, only a few nucleotides downstream of the interaction site (52). Similarly, SdsR requires RNase E to repress ompD mRNA in the CDS (Figures 4B and 5A). Using a 3′-RACE approach, we identified ompD mRNA fragments that transiently accumulated following SdsR induction (Figure 5C), and with heterogeneous 3′-ends ranging from ompD positions +55 to +65 (Figure 5D). These results, together with genetic analysis of the SdsR 5′-end (Figure 6), allowed us to narrow down the SdsR–ompD interaction site (Figure 7A). Positions of enriched ompD 3′-ends mapped mostly to the 8th-nt downstream of the proximal end of the SdsR interaction site (Figures 5D and 7A), which is somewhat longer than the intervening four nucleotide patch observed for the MicC–ompD interaction (52).
We have successfully applied this method for the experimental prediction of other sRNA sites, e.g. the RybB site in fadL mRNA (V. Pfeiffer and J. Vogel, unpublished results), suggesting that 3′-RACE is a promising method for binding site discovery with respect to the target. Furthermore, genetic modification of the cellular RNA degradation machinery may stabilize the desired 3′-ends and improve this analytical approach. Specifically, the educated inactivation of 3′-5′ exonucleases, such as PNPase, or a disruption of degradosome assembly may stabilize those mRNA degradation products that indicate nearby sRNA binding sites.
The role of SraC
An intriguing feature of SdsR is its chromosomal origin within the oppositely encoded sraC sRNA gene. In contrast to SdsR (see Figures 1A and 2B), sraC sequences are far less conserved and only found in about half of the organisms evaluated here (Supplementary Figure S1). Apparently, sraC sequence conservation is restricted to the region overlapping sdsR; especially the sraC promoter elements display significant variation between species, e.g. a potential −10 box in both Salmonella and Shigella is located about 35 nt upstream the respective site in E. coli (see Supplementary Figure S1). In accordance with this finding, we observed a ∼35 nt longer version of SraC for Salmonella and Shigella when compared to E. coli on northern blots (see Figure 1C). The expression patterns of SraC and SdsR vary inversely in all three species, i.e. SraC disappears as SdsR accumulates at the onset of stationary phase. The timing of SdsR expression is strictly dependent on the activity of σS, and the low levels of SdsR expression present during the exponential growth phase of Shigella might be assigned to the almost constitutive expression of σS in this species (not shown).
Earlier work in E. coli demonstrated that as a consequence of base pairing between SraC and SdsR, a shorter ∼150 nt version of SraC becomes detectable and that accumulation of this species is dependent on the activity of the double strand-specific ribonuclease RNase III (2). Since deletion of the sdsR gene from the chromosome would automatically also interrupt sraC, we compared SraC levels between WT cells and an rpoS mutant, i.e. an indirect sdsR mutant in both Salmonella and E. coli. In addition, we monitored SdsR expression in Salmonella and E. coli strains in which the sraC promoter had been deleted but the sdsR gene left intact. As expected, we found SdsR levels to be only slightly increased in the absence of SraC (∼1.5-fold; Supplementary Figure S7, lanes 4 and 8 in either panel) in both Salmonella and E. coli. In contrast, in the absence of RpoS, and thus of SdsR, SraC no longer disappears with the onset of stationary phase but rather appears to be constitutively expressed, albeit at low levels (Supplementary Figure S7, lanes 1–4 versus 9–12 in both panels). Thus, SdsR seems to play a significant role in the control of SraC expression; however, given the latter's low conservation among other enterobacterial species, and the variation in the location of its promoter elements, we assume that SraC is not essential for the control of SdsR expression, especially when cells enter stationary phase.
CONCLUSION
A picture has emerged that Salmonella and E. coli possess a couple of dozen ‘core sRNAs’ that are conserved at least throughout the enterobacterial clade. Many of them associate with the Hfq protein, and some individually govern large post-transcriptional regulons that represent as many as 1% of all Salmonella genes (56). SdsR clearly belongs to this set of core sRNAs, and at the same time is a member of the vast σS network which is important for stress control in many enterobacteria. There is indeed evidence that SdsR plays a broader role in the σS network than regulating just the OmpD porin. First, SdsR is abundant in all three organisms investigated, accumulating to ∼300 copies per cell (Figure 2); other Hfq-associated regulators with similar or higher copy numbers, such as ArcZ and OxyS, have been shown to act on multiple targets (45,95,100,101). Second, SdsR recognizes ompD through nucleotides that precede the most highly conserved region of the sRNA (compare Figures 1A and 7A), the latter of which would typically be the region of Hfq-associated sRNAs that interacts with targets (39), arguing that key targets of SdsR have yet to be identified. Third, preliminary microarray analyses have predicted that SdsR may directly repress a dozen more Salmonella mRNAs, including ones encoding proteins that function in global stress responses, by which means SdsR may substantially expand the regulatory scope of σS at the post-transcriptional level (Fröhlich, K.S. and Vogel, J., manuscript in preparation). Studies of SdsR promise to be particularly informative as to how Hfq and sRNAs intertwine the DNA and RNA levels of a general bacterial stress response. Future studies should also address a potential link with horizontal gene transfer (102): during Salmonella evolution, this sdsR locus has repeatedly seen the integration of exogenous DNA and it continues to act as an attachment site for contemporary bacteriophages.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online: Supplementary Figures S1–S7; Supplementary Tables 1–3; Supplementary References (103–105).
FUNDING
Work in the Vogel lab is supported by the DFG (German Research Council) as part of the priority program SPP1258 Sensory and Regulatory RNAs in Prokaryotes (Grant Vo875/2-2). AAH received a post-doctoral fellowship of the Alexander v. Humboldt Foundation. Funding for open access charge: DFG and University of Wuerzburg.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank S. Altuvia for helpful comments on the manuscript. Furthermore, we thank R. Hengge and J. Slauch for providing plasmids, R. Misra and M. Springer for antibodies and J. Casadesus, S. Altuvia, L. Bossi, T. Nyström and K. Tedin for strains.
REFERENCES
- 1.Wassarman KM, Repoila F, Rosenow C, Storz G, Gottesman S. Identification of novel small RNAs using comparative genomics and microarrays. Genes Dev. 2001;15:1637–1651. doi: 10.1101/gad.901001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vogel J, Bartels V, Tang TH, Churakov G, Slagter-Jager JG, Huttenhofer A, Wagner EG. RNomics in Escherichia coli detects new sRNA species and indicates parallel transcriptional output in bacteria. Nucleic Acids Res. 2003;31:6435–6443. doi: 10.1093/nar/gkg867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rivas E, Klein RJ, Jones TA, Eddy SR. Computational identification of noncoding RNAs in E. coli by comparative genomics. Curr. Biol. 2001;11:1369–1373. doi: 10.1016/s0960-9822(01)00401-8. [DOI] [PubMed] [Google Scholar]
- 4.Chen S, Lesnik EA, Hall TA, Sampath R, Griffey RH, Ecker DJ, Blyn LB. A bioinformatics based approach to discover small RNA genes in the Escherichia coli genome. Biosystems. 2002;65:157–177. doi: 10.1016/s0303-2647(02)00013-8. [DOI] [PubMed] [Google Scholar]
- 5.Argaman L, Hershberg R, Vogel J, Bejerano G, Wagner EG, Margalit H, Altuvia S. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr. Biol. 2001;11:941–950. doi: 10.1016/s0960-9822(01)00270-6. [DOI] [PubMed] [Google Scholar]
- 6.Wassarman KM, Zhang AX, Storz G. Small RNAs in Escherichia coli. Trends in Microbiology. 1999;7:37–45. doi: 10.1016/s0966-842x(98)01379-1. [DOI] [PubMed] [Google Scholar]
- 7.Wagner EG, Simons RW. Antisense RNA control in bacteria, phages, and plasmids. Annu. Rev. Microbiol. 1994;48:713–742. doi: 10.1146/annurev.mi.48.100194.003433. [DOI] [PubMed] [Google Scholar]
- 8.Brantl S. Antisense RNAs in plasmids: control of replication and maintenance. Plasmid. 2002;48:165–173. doi: 10.1016/s0147-619x(02)00108-7. [DOI] [PubMed] [Google Scholar]
- 9.Lange R, Hengge-Aronis R. Identification of a central regulator of stationary-phase gene expression in Escherichia coli. Mol. Microbiol. 1991;5:49–59. doi: 10.1111/j.1365-2958.1991.tb01825.x. [DOI] [PubMed] [Google Scholar]
- 10.Navarro Llorens JM, Tormo A, Martinez-Garcia E. Stationary phase in gram-negative bacteria. FEMS Microbiol. Rev. 2010;34:476–495. doi: 10.1111/j.1574-6976.2010.00213.x. [DOI] [PubMed] [Google Scholar]
- 11.Nystrom T. Stationary-phase physiology. Annu. Rev. Microbiol. 2004;58:161–181. doi: 10.1146/annurev.micro.58.030603.123818. [DOI] [PubMed] [Google Scholar]
- 12.Battesti A, Majdalani N, Gottesman S. The RpoS-Mediated General Stress Response in Escherichia coli. Annu. Rev. Microbiol. 2011;65:189–213. doi: 10.1146/annurev-micro-090110-102946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hengge-Aronis R. Interplay of global regulators and cell physiology in the general stress response of Escherichia coli. Curr. Opin. Microbiol. 1999;2:148–152. doi: 10.1016/S1369-5274(99)80026-5. [DOI] [PubMed] [Google Scholar]
- 14.Hengge-Aronis R. Signal transduction and regulatory mechanisms involved in control of the sigma(S) (RpoS) subunit of RNA polymerase. Microbiol. Mol. Biol. Rev. 2002;66:373–395. doi: 10.1128/MMBR.66.3.373-395.2002. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maciag A, Peano C, Pietrelli A, Egli T, De Bellis G, Landini P. In vitro transcription profiling of the {sigma}S subunit of bacterial RNA polymerase: re-definition of the {sigma}S regulon and identification of {sigma}S-specific promoter sequence elements. Nucleic Acids Res. 2011;39:5338–5355. doi: 10.1093/nar/gkr129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weber H, Polen T, Heuveling J, Wendisch VF, Hengge R. Genome-wide analysis of the general stress response network in Escherichia coli: sigmaS-dependent genes, promoters, and sigma factor selectivity. J. Bacteriol. 2005;187:1591–1603. doi: 10.1128/JB.187.5.1591-1603.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Masse E, Vanderpool CK, Gottesman S. Effect of RyhB small RNA on global iron use in Escherichia coli. J. Bacteriol. 2005;187:6962–6971. doi: 10.1128/JB.187.20.6962-6971.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Padalon-Brauch G, Hershberg R, Elgrably-Weiss M, Baruch K, Rosenshine I, Margalit H, Altuvia S. Small RNAs encoded within genetic islands of Salmonella typhimurium show host-induced expression and role in virulence. Nucleic Acids Res. 2008;36:1913–1927. doi: 10.1093/nar/gkn050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Masse E, Gottesman S. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl Acad. Sci. USA. 2002;99:4620–4625. doi: 10.1073/pnas.032066599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mutalik VK, Nonaka G, Ades SE, Rhodius VA, Gross CA. Promoter strength properties of the complete sigma E regulon of Escherichia coli and Salmonella enterica. J. Bacteriol. 2009;191:7279–7287. doi: 10.1128/JB.01047-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Papenfort K, Pfeiffer V, Mika F, Lucchini S, Hinton JC, Vogel J. SigmaE-dependent small RNAs of Salmonella respond to membrane stress by accelerating global omp mRNA decay. Mol. Microbiol. 2006;62:1674–1688. doi: 10.1111/j.1365-2958.2006.05524.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thompson KM, Rhodius VA, Gottesman S. SigmaE regulates and is regulated by a small RNA in Escherichia coli. J. Bacteriol. 2007;189:4243–4256. doi: 10.1128/JB.00020-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Udekwu KI, Wagner EG. Sigma E controls biogenesis of the antisense RNA MicA. Nucleic Acids Res. 2007;35:1279–1288. doi: 10.1093/nar/gkl1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gogol EB, Rhodius VA, Papenfort K, Vogel J, Gross CA. Small RNAs endow a transcriptional activator with essential repressor functions for single-tier control of a global stress regulon. Proc. Natl Acad. Sci. USA. 2011;108:12875–12880. doi: 10.1073/pnas.1109379108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johansen J, Rasmussen AA, Overgaard M, Valentin-Hansen P. Conserved small non-coding RNAs that belong to the sigmaE regulon: role in down-regulation of outer membrane proteins. J. Mol Biol. 2006;364:1–8. doi: 10.1016/j.jmb.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 26.Johansen J, Eriksen M, Kallipolitis B, Valentin-Hansen P. Down-regulation of outer membrane proteins by noncoding RNAs: unraveling the cAMP-CRP- and sigmaE-dependent CyaR-ompX regulatory case. J. Mol. Biol. 2008;383:1–9. doi: 10.1016/j.jmb.2008.06.058. [DOI] [PubMed] [Google Scholar]
- 27.Papenfort K, Pfeiffer V, Lucchini S, Sonawane A, Hinton JC, Vogel J. Systematic deletion of Salmonella small RNA genes identifies CyaR, a conserved CRP-dependent riboregulator of OmpX synthesis. Mol. Microbiol. 2008;68:890–906. doi: 10.1111/j.1365-2958.2008.06189.x. [DOI] [PubMed] [Google Scholar]
- 28.De Lay N, Gottesman S. The Crp-activated small noncoding regulatory RNA CyaR (RyeE) links nutritional status to group behavior. J. Bacteriol. 2009;191:461–476. doi: 10.1128/JB.01157-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boysen A, Moller-Jensen J, Kallipolitis B, Valentin-Hansen P, Overgaard M. Translational regulation of gene expression by an anaerobically induced small non-coding RNA in Escherichia coli. J. Biol. Chem. 2010;285:10690–10702. doi: 10.1074/jbc.M109.089755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Durand S, Storz G. Reprogramming of anaerobic metabolism by the FnrS small RNA. Mol. Microbiol. 2010;75:1215–1231. doi: 10.1111/j.1365-2958.2010.07044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mandin P, Gottesman S. Integrating anaerobic/aerobic sensing and the general stress response through the ArcZ small RNA. EMBO J. 2010;29:3094–3107. doi: 10.1038/emboj.2010.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moon K, Gottesman S. A PhoQ/P-regulated small RNA regulates sensitivity of Escherichia coli to antimicrobial peptides. Mol. Microbiol. 2009;74:1314–1330. doi: 10.1111/j.1365-2958.2009.06944.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gottesman S. Micros for microbes: non-coding regulatory RNAs in bacteria. Trends Genet. 2005;21:399–404. doi: 10.1016/j.tig.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 34.Opdyke JA, Kang JG, Storz G. GadY, a small-RNA regulator of acid response genes in Escherichia coli. J. Bacteriol. 2004;186:6698–6705. doi: 10.1128/JB.186.20.6698-6705.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hershberg R, Altuvia S, Margalit H. A survey of small RNA-encoding genes in Escherichia coli. Nucleic Acids Res. 2003;31:1813–1820. doi: 10.1093/nar/gkg297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sittka A, Lucchini S, Papenfort K, Sharma CM, Rolle K, Binnewies TT, Hinton JC, Vogel J. Deep sequencing analysis of small noncoding RNA and mRNA targets of the global post-transcriptional regulator, Hfq. PLoS Genet. 2008;4:e1000163. doi: 10.1371/journal.pgen.1000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sittka A, Sharma CM, Rolle K, Vogel J. Deep sequencing of Salmonella RNA associated with heterologous Hfq proteins in vivo reveals small RNAs as a major target class and identifies RNA processing phenotypes. RNA Biol. 2009;6:266–275. doi: 10.4161/rna.6.3.8332. [DOI] [PubMed] [Google Scholar]
- 38.Zhang A, Wassarman KM, Rosenow C, Tjaden BC, Storz G, Gottesman S. Global analysis of small RNA and mRNA targets of Hfq. Mol. Microbiol. 2003;50:1111–1124. doi: 10.1046/j.1365-2958.2003.03734.x. [DOI] [PubMed] [Google Scholar]
- 39.Vogel J, Luisi BF. Hfq and its constellation of RNA. Nat. Rev. Microbiol. 2011;9:578–589. doi: 10.1038/nrmicro2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chao Y, Vogel J. The role of Hfq in bacterial pathogens. Curr. Opin. Microbiol. 2010;13:24–33. doi: 10.1016/j.mib.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 41.Lease RA, Cusick ME, Belfort M. Riboregulation in Escherichia coli: DsrA RNA acts by RNA:RNA interactions at multiple loci. Proc. Natl Acad. Sci. USA. 1998;95:12456–12461. doi: 10.1073/pnas.95.21.12456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Majdalani N, Chen S, Murrow J, St John K, Gottesman S. Regulation of RpoS by a novel small RNA: the characterization of RprA. Mol. Microbiol. 2001;39:1382–1394. doi: 10.1111/j.1365-2958.2001.02329.x. [DOI] [PubMed] [Google Scholar]
- 43.Majdalani N, Cunning C, Sledjeski D, Elliott T, Gottesman S. DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proc. Natl Acad. Sci. USA. 1998;95:12462–12467. doi: 10.1073/pnas.95.21.12462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Soper T, Mandin P, Majdalani N, Gottesman S, Woodson SA. Positive regulation by small RNAs and the role of Hfq. Proc. Natl Acad. Sci. USA. 2010;107:9602–9607. doi: 10.1073/pnas.1004435107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Altuvia S, Weinstein-Fischer D, Zhang A, Postow L, Storz G. A small, stable RNA induced by oxidative stress: role as a pleiotropic regulator and antimutator. Cell. 1997;90:43–53. doi: 10.1016/s0092-8674(00)80312-8. [DOI] [PubMed] [Google Scholar]
- 46.Vogel J, Papenfort K. Small non-coding RNAs and the bacterial outer membrane. Curr. Opin. Microbiol. 2006;9:605–611. doi: 10.1016/j.mib.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 47.Guillier M, Gottesman S, Storz G. Modulating the outer membrane with small RNAs. Genes Dev. 2006;20:2338–2348. doi: 10.1101/gad.1457506. [DOI] [PubMed] [Google Scholar]
- 48.Valentin-Hansen P, Johansen J, Rasmussen AA. Small RNAs controlling outer membrane porins. Curr. Opin. Microbiol. 2007;10:152–155. doi: 10.1016/j.mib.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 49.Santiviago CA, Toro CS, Hidalgo AA, Youderian P, Mora GC. Global regulation of the Salmonella enterica serovar typhimurium major porin, OmpD. J. Bacteriol. 2003;185:5901–5905. doi: 10.1128/JB.185.19.5901-5905.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bouvier M, Sharma CM, Mika F, Nierhaus KH, Vogel J. Small RNA binding to 5′ mRNA coding region inhibits translational initiation. Mol. Cell. 2008;32:827–837. doi: 10.1016/j.molcel.2008.10.027. [DOI] [PubMed] [Google Scholar]
- 51.Papenfort K, Bouvier M, Mika F, Sharma CM, Vogel J. Evidence for an autonomous 5′ target recognition domain in an Hfq-associated small RNA. Proc. Natl Acad. Sci. USA. 2010;107:20435–20440. doi: 10.1073/pnas.1009784107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pfeiffer V, Papenfort K, Lucchini S, Hinton JC, Vogel J. Coding sequence targeting by MicC RNA reveals bacterial mRNA silencing downstream of translational initiation. Nat. Struct. Mol. Biol. 2009;16:840–846. doi: 10.1038/nsmb.1631. [DOI] [PubMed] [Google Scholar]
- 53.Pfeiffer V, Sittka A, Tomer R, Tedin K, Brinkmann V, Vogel J. A small non-coding RNA of the invasion gene island (SPI-1) represses outer membrane protein synthesis from the Salmonella core genome. Mol. Microbiol. 2007;66:1174–1191. doi: 10.1111/j.1365-2958.2007.05991.x. [DOI] [PubMed] [Google Scholar]
- 54.Balbontin R, Fiorini F, Figueroa-Bossi N, Casadesus J, Bossi L. Recognition of heptameric seed sequence underlies multi-target regulation by RybB small RNA in Salmonella enterica. Mol. Microbiol. 2010;78:380–394. doi: 10.1111/j.1365-2958.2010.07342.x. [DOI] [PubMed] [Google Scholar]
- 55.Urban JH, Vogel J. Translational control and target recognition by Escherichia coli small RNAs in vivo. Nucleic Acids Res. 2007;35:1018–1037. doi: 10.1093/nar/gkl1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sharma CM, Papenfort K, Pernitzsch SR, Mollenkopf HJ, Hinton JC, Vogel J. Pervasive post-transcriptional control of genes involved in amino acid metabolism by the Hfq-dependent GcvB small RNA. Mol. Microbiol. 2011;81:1144–1165. doi: 10.1111/j.1365-2958.2011.07751.x. [DOI] [PubMed] [Google Scholar]
- 57.Sittka A, Pfeiffer V, Tedin K, Vogel J. The RNA chaperone Hfq is essential for the virulence of Salmonella typhimurium. Mol. Microbiol. 2007;63:193–217. doi: 10.1111/j.1365-2958.2006.05489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ellermeier CD, Janakiraman A, Slauch JM. Construction of targeted single copy lac fusions using lambda Red and FLP-mediated site-specific recombination in bacteria. Gene. 2002;290:153–161. doi: 10.1016/s0378-1119(02)00551-6. [DOI] [PubMed] [Google Scholar]
- 60.Gaal T, Ross W, Estrem ST, Nguyen LH, Burgess RR, Gourse RL. Promoter recognition and discrimination by EsigmaS RNA polymerase. Mol. Microbiol. 2001;42:939–954. doi: 10.1046/j.1365-2958.2001.02703.x. [DOI] [PubMed] [Google Scholar]
- 61.Typas A, Becker G, Hengge R. The molecular basis of selective promoter activation by the sigmaS subunit of RNA polymerase. Mol. Microbiol. 2007;63:1296–1306. doi: 10.1111/j.1365-2958.2007.05601.x. [DOI] [PubMed] [Google Scholar]
- 62.Lange R, Hengge-Aronis R. The cellular concentration of the sigma S subunit of RNA polymerase in Escherichia coli is controlled at the levels of transcription, translation, and protein stability. Genes Dev. 1994;8:1600–1612. doi: 10.1101/gad.8.13.1600. [DOI] [PubMed] [Google Scholar]
- 63.Kvint K, Farewell A, Nystrom T. RpoS-dependent promoters require guanosine tetraphosphate for induction even in the presence of high levels of sigma(s) J. Biol. Chem. 2000;275:14795–14798. doi: 10.1074/jbc.C000128200. [DOI] [PubMed] [Google Scholar]
- 64.Tedin K, Norel F. Comparison of DeltarelA strains of Escherichia coli and Salmonella enterica serovar Typhimurium suggests a role for ppGpp in attenuation regulation of branched-chain amino acid biosynthesis. J. Bacteriol. 2001;183:6184–6196. doi: 10.1128/JB.183.21.6184-6196.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hernandez VJ, Bremer H. Escherichia coli ppGpp synthetase II activity requires spoT. J. Biol. Chem. 1991;266:5991–5999. [PubMed] [Google Scholar]
- 66.Xiao H, Kalman M, Ikehara K, Zemel S, Glaser G, Cashel M. Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J. Biol. Chem. 1991;266:5980–5990. [PubMed] [Google Scholar]
- 67.Lee SJ, Gralla JD. Open complex formation in vitro by sigma38 (rpoS) RNA polymerase: roles for region 2 amino acids. J. Mol. Biol. 2003;329:941–948. doi: 10.1016/s0022-2836(03)00369-3. [DOI] [PubMed] [Google Scholar]
- 68.Becker G, Hengge-Aronis R. What makes an Escherichia coli promoter sigma(S) dependent? Role of the -13/-14 nucleotide promoter positions and region 2.5 of sigma(S) Mol. Microbiol. 2001;39:1153–1165. doi: 10.1111/j.1365-2958.2001.02313.x. [DOI] [PubMed] [Google Scholar]
- 69.Muffler A, Barth M, Marschall C, Hengge-Aronis R. Heat shock regulation of sigmaS turnover: a role for DnaK and relationship between stress responses mediated by sigmaS and sigma32 in Escherichia coli. J. Bacteriol. 1997;179:445–452. doi: 10.1128/jb.179.2.445-452.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lutz R, Bujard H. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 1997;25:1203–1210. doi: 10.1093/nar/25.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Urban JH, Papenfort K, Thomsen J, Schmitz RA, Vogel J. A conserved small RNA promotes discoordinate expression of the glmUS operon mRNA to activate GlmS synthesis. J. Mol. Biol. 2007;373:521–528. doi: 10.1016/j.jmb.2007.07.035. [DOI] [PubMed] [Google Scholar]
- 72.Lee DR, Schnaitman CA. Comparison of outer membrane porin proteins produced by Escherichia coli and Salmonella typhimurium. J. Bacteriol. 1980;142:1019–1022. doi: 10.1128/jb.142.3.1019-1022.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Apirion D. Isolation, genetic mapping and some characterization of a mutation in Escherichia coli that affects the processing of ribonuleic acid. Genetics. 1978;90:659–671. doi: 10.1093/genetics/90.4.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Figueroa-Bossi N, Valentini M, Malleret L, Fiorini F, Bossi L. Caught at its own game: regulatory small RNA inactivated by an inducible transcript mimicking its target. Genes Dev. 2009;23:2004–2015. doi: 10.1101/gad.541609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rouviere PE, De Las Penas A, Mecsas J, Lu CZ, Rudd KE, Gross CA. rpoE, the gene encoding the second heat-shock sigma factor, sigma E, in Escherichia coli. EMBO J. 1995;14:1032–1042. doi: 10.1002/j.1460-2075.1995.tb07084.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Raina S, Missiakas D, Georgopoulos C. The rpoE gene encoding the sigma E (sigma 24) heat shock sigma factor of Escherichia coli. EMBO J. 1995;14:1043–1055. doi: 10.1002/j.1460-2075.1995.tb07085.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Muller C, Bang IS, Velayudhan J, Karlinsey J, Papenfort K, Vogel J, Fang FC. Acid stress activation of the sigma(E) stress response in Salmonella enterica serovar Typhimurium. Mol. Microbiol. 2009;71:1228–1238. doi: 10.1111/j.1365-2958.2009.06597.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bianchi AA, Baneyx F. Hyperosmotic shock induces the sigma32 and sigmaE stress regulons of Escherichia coli. Mol. Microbiol. 1999;34:1029–1038. doi: 10.1046/j.1365-2958.1999.01664.x. [DOI] [PubMed] [Google Scholar]
- 80.Muffler A, Fischer D, Hengge-Aronis R. The RNA-binding protein HF-I, known as a host factor for phage Qbeta RNA replication, is essential for rpoS translation in Escherichia coli. Genes Dev. 1996;10:1143–1151. doi: 10.1101/gad.10.9.1143. [DOI] [PubMed] [Google Scholar]
- 81.Brown L, Elliott T. Efficient translation of the RpoS sigma factor in Salmonella typhimurium requires host factor I, an RNA-binding protein encoded by the hfq gene. J. Bacteriol. 1996;178:3763–3770. doi: 10.1128/jb.178.13.3763-3770.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Beisel CL, Storz G. Base pairing small RNAs and their roles in global regulatory networks. FEMS Microbiol. Rev. 2011;34:866–882. doi: 10.1111/j.1574-6976.2010.00241.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sledjeski DD, Gupta A, Gottesman S. The small RNA, DsrA, is essential for the low temperature expression of RpoS during exponential growth in Escherichia coli. EMBO J. 1996;15:3993–4000. [PMC free article] [PubMed] [Google Scholar]
- 84.Prilipov A, Phale PS, Koebnik R, Widmer C, Rosenbusch JP. Identification and characterization of two quiescent porin genes, nmpC and ompN, in Escherichia coli BE. J. Bacteriol. 1998;180:3388–3392. doi: 10.1128/jb.180.13.3388-3392.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Highton PJ, Chang Y, Marcotte WR, Jr, Schnaitman CA. Evidence that the outer membrane protein gene nmpC of Escherichia coli K-12 lies within the defective qsr’ prophage. J. Bacteriol. 1985;162:256–262. doi: 10.1128/jb.162.1.256-262.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Song T, Mika F, Lindmark B, Liu Z, Schild S, Bishop A, Zhu J, Camilli A, Johansson J, Vogel J, et al. A new Vibrio cholerae sRNA modulates colonization and affects release of outer membrane vesicles. Mol. Microbiol. 2008;70:100–111. doi: 10.1111/j.1365-2958.2008.06392.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gogol EB, Rhodius VA, Papenfort K, Vogel J, Gross CA. Small RNAs endow a transcriptional activator with essential repressor functions for single-tier control of a global stress regulon. Proc. Natl Acad. Sci. USA. 2011;108:12875–12880. doi: 10.1073/pnas.1109379108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Beisel CL, Storz G. The base-pairing RNA spot 42 participates in a multioutput feedforward loop to help enact catabolite repression in Escherichia coli. Mol. Cell. 2011;41:286–297. doi: 10.1016/j.molcel.2010.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Figueroa-Bossi N, Lemire S, Maloriol D, Balbontin R, Casadesus J, Bossi L. Loss of Hfq activates the sigma-dependent envelope stress response in Salmonella enterica. Mol. Microbiol. 2006;62:838–852. doi: 10.1111/j.1365-2958.2006.05413.x. [DOI] [PubMed] [Google Scholar]
- 90.Bossi L, Maloriol D, Figueroa-Bossi N. Porin biogenesis activates the sigma(E) response in Salmonella hfq mutants. Biochimie. 2008;90:1539–1544. doi: 10.1016/j.biochi.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 91.Chen S, Zhang A, Blyn LB, Storz G. MicC, a second small-RNA regulator of Omp protein expression in Escherichia coli. J. Bacteriol. 2004;186:6689–6697. doi: 10.1128/JB.186.20.6689-6697.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Castillo-Keller M, Vuong P, Misra R. Novel mechanism of Escherichia coli porin regulation. J. Bacteriol. 2006;188:576–586. doi: 10.1128/JB.188.2.576-586.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Douchin V, Bohn C, Bouloc P. Down-regulation of porins by a small RNA bypasses the essentiality of the regulated intramembrane proteolysis protease RseP in Escherichia coli. J. Biol. Chem. 2006;281:12253–12259. doi: 10.1074/jbc.M600819200. [DOI] [PubMed] [Google Scholar]
- 94.Takyar S, Hickerson RP, Noller HF. mRNA helicase activity of the ribosome. Cell. 2005;120:49–58. doi: 10.1016/j.cell.2004.11.042. [DOI] [PubMed] [Google Scholar]
- 95.Papenfort K, Said N, Welsink T, Lucchini S, Hinton JC, Vogel J. Specific and pleiotropic patterns of mRNA regulation by ArcZ, a conserved, Hfq-dependent small RNA. Mol. Microbiol. 2009;74:139–158. doi: 10.1111/j.1365-2958.2009.06857.x. [DOI] [PubMed] [Google Scholar]
- 96.Rice JB, Vanderpool CK. The small RNA SgrS controls sugar-phosphate accumulation by regulating multiple PTS genes. Nucleic Acids Res. 2011;39:3806–3819. doi: 10.1093/nar/gkq1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Prevost K, Desnoyers G, Jacques JF, Lavoie F, Masse E. Small RNA-induced mRNA degradation achieved through both translation block and activated cleavage. Genes Dev. 2011;25:385–396. doi: 10.1101/gad.2001711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Morita T, Maki K, Aiba H. RNase E-based ribonucleoprotein complexes: mechanical basis of mRNA destabilization mediated by bacterial noncoding RNAs. Genes Dev. 2005;19:2176–2186. doi: 10.1101/gad.1330405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Backofen R, Hess WR. Computational prediction of sRNAs and their targets in bacteria. RNA Biol. 2010;7:33–42. doi: 10.4161/rna.7.1.10655. [DOI] [PubMed] [Google Scholar]
- 100.Argaman L, Altuvia S. fhlA repression by OxyS RNA: kissing complex formation at two sites results in a stable antisense-target RNA complex. J. Mol. Biol. 2000;300:1101–1112. doi: 10.1006/jmbi.2000.3942. [DOI] [PubMed] [Google Scholar]
- 101.Tjaden B, Goodwin SS, Opdyke JA, Guillier M, Fu DX, Gottesman S, Storz G. Target prediction for small, noncoding RNAs in bacteria. Nucleic Acids Res. 2006;34:2791–2802. doi: 10.1093/nar/gkl356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Balbontin R, Figueroa-Bossi N, Casadesus J, Bossi L. Insertion hot spot for horizontally acquired DNA within a bidirectional small-RNA locus in Salmonella enterica. J. Bacteriol. 2008;190:4075–4078. doi: 10.1128/JB.00220-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cherepanov PP, Wackernagel W. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene. 1995;158:9–14. doi: 10.1016/0378-1119(95)00193-a. [DOI] [PubMed] [Google Scholar]
- 104.Sanden AM, Prytz I, Tubulekas I, Forberg C, Le H, Hektor A, Neubauer P, Pragai Z, Harwood C, Ward A, et al. Limiting factors in Escherichia coli fed-batch production of recombinant proteins. Biotechnol. Bioeng. 2003;81:158–166. doi: 10.1002/bit.10457. [DOI] [PubMed] [Google Scholar]
- 105.Zychlinsky A, Prevost MC, Sansonetti PJ. Shigella flexneri induces apoptosis in infected macrophages. Nature. 1992;358:167–169. doi: 10.1038/358167a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.