Abstract
MicroRNAs (miRNAs) are involved in various biological processes and human diseases. The development of strong low-molecular weight inhibitors of specific miRNAs is thus expected to be useful in providing tools for basic research or in generating promising new therapeutic drugs. We have previously described the development of ‘Tough Decoy (TuD) RNA’ molecules, which achieve the long-term suppression of specific miRNA activity in mammalian cells when expressed from a lentivirus vector. In our current study, we describe new synthetic miRNA inhibitors, designated as S-TuD (Synthetic TuD), which are composed of two fully 2′-O-methylated RNA strands. Each of these strands includes a miRNA-binding site. Following the hybridization of paired strands, the resultant S-TuD forms a secondary structure with two stems, which resembles the corresponding TuD RNA molecule. By analyzing the effects of S-TuD against miR-21, miR-200c, miR-16 and miR-106b, we have elucidated the critical design features of S-TuD molecules that will provide optimum inhibitory effects following transfection into human cell lines. We further show that the inhibitory effects of a single transfection of S-TuD-miR200c are quite long-lasting (>7 days) and induce partial EMT, the full establishment of which requires 11 days when using a lentivirus vector that expresses TuD-miR200c continuously.
INTRODUCTION
MicroRNAs (miRNAs) are small (18–25 nt) non-coding RNAs that suppress the expression of target transcripts at the post-transcriptional level (1–3). There are more than 1500 miRNA species in the human genome (miRBase release 18), and these nucleic acids are now known to play important roles in various biological processes such as stemness, development, differentiation and the cellular defense responses to infection (4–7). As the aberrant high-level expression of certain miRNAs has now often been reported to be associated with cancers and human infectious diseases, analysis of the underlying molecular mechanisms is likely to become much more extensive (8,9). The development of reagents that can strongly suppress specific miRNAs has also generated much interest and will be important for both basic miRNA research and also as a possible therapeutic strategy.
Most synthetic low-molecular weight inhibitors of specific miRNAs are based on anti-miRNA antisense oligonucleotides (AMO), in which some or all of the ribonucleotides are modified to 2′-O-methylated RNA (10,11), locked nucleic acids (LNA)/DNA or 2′-methoxyethylated RNA to provide resistance to cellular nucleases and to increase affinity towards complementary miRNA sequences (12,13). In addition, the backbones of some AMOs are substituted with phosphorothioate (14). These modifications and substitutions can be used in combination and some AMOs have flanking sequences or are connected to lipids through the use of linkers (14,15). However, since these reagents will be diluted by successive cell divisions, and in some cases metabolized in the cytoplasm, their effects are transient in many cases. To achieve the long-term suppression of a specific miRNA, specialized plasmid- and virus-vectors carrying expression units for inhibitory RNA molecules have also been developed. These inhibitory RNA molecules include ‘antagomir’, ‘eraser’ and ‘sponge’ (16–18). We have also reported on such plasmid- or lentivirus-based vectors expressing inhibitory RNA targeting specific miRNAs, which we have termed Tough Decoy (TuD) RNA (19).
TuD RNAs have been already used for the basic research, for the identification of miRNA targets (20), and in the functional analysis of miRNA in cancer cells (21), in tumour formation in mice (22) and in myoblast differentiation (23). TuD RNAs are single RNA molecules with a complex secondary structure composed of four elements: a stem of 18 bp in length, two miRNA-binding sites (MBSs) that have a sequence complementary to that of a mature miRNA of interest, a stem–loop structure which connects these two MBSs, four linkers with 3 nt also connecting the two MBSs and flanking stems (Figure 1A). These elements provide efficient nuclear export, binding to the target miRNA, resistance to cellular nucleases and enhancement of the MBS accessibility to the target miRNA, respectively. By screening several alternative MBS sequences to optimize their decoy activity, we have further identified a highly potent TuD, the MBS of which has a 4 nt insertion between positions 10 and 11 from the 3′-end of the perfectly complementary sequence to the entire mature miRNA of interest, where the Ago2-containing RISC cleaves target mRNAs (24). When these TuD RNAs were expressed using lentivirus vectors, they were shown to be efficiently transported to the cytoplasm and exhibit strong inhibitory effects for more than 1 month.
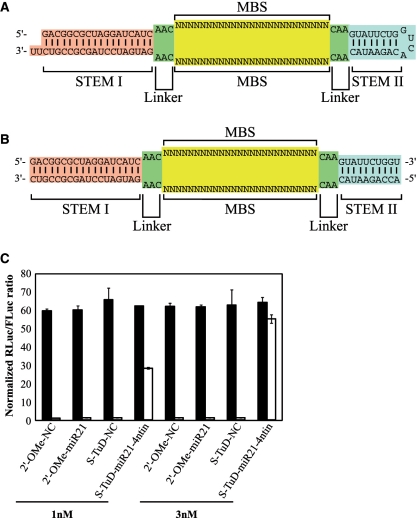
Figure 1.
Schematic representation of the structure of S-TuD RNA (A) and S-TuD (B). (C) Inhibitory effects of S-TuD-miR21-4ntin upon endogenous miR-21 activity. S-TuD or 2′-O-methylated RNA oligonucleotide-based antisense molecules were transfected into HCT-116 cells together with the Renilla luciferase miR-21 reporter (miR-21-RL) (open bars) or the untargeted control Renilla luciferase reporter (UT-RL) (black bars) as well as the Firefly luciferase reporter (FL) as a transfection control. After performing a dual luciferase assay, the expression levels were normalized to the ratio of the activity of miR-21-RL to that of FL in 1 nM S-TuD-NC transfected HCT-116 cells and are represented by the mean ± SD (n = 3).
Retro/lentivirus vectors carrying TuD, however, have a potential disadvantage in terms of their therapeutic application as this requires human gene therapy. In our current study, we aimed to develop low-molecular weight reagents that can retain miRNA inhibitory activity even after several rounds of cell division by mimicking the structural features of TuD RNA. We have developed a synthetic miRNA inhibitor composed of two strands of 2′-O-methylated RNA oligonucleotides, the structure of which is very similar to that of the corresponding TuD RNA and we have designated this as S-TuD (Synthetic TuD). We provide evidence that if appropriate MBSs are selected, S-TuDs targeting miR-21, -200 c, -16 and -106b retain potent inhibitory effects even when transfected at a low dose range of 1 nM–30 pM. We further show that a single transfection of S-TuD-miR200c with the appropriate MBSs can induce a partial epithelial–mesenchymal transition, indicating that the inhibitory effects of S-TuD can withstand dilution by several rounds of cell division.
MATERIALS AND METHODS
MiRNA inhibitors
For the preparation of S-TuD, a series of fully 2′-O-methylated RNA oligonucleotide pairs were synthesized as listed in Supplementary Table S1 and each pair was annealed prior to transfection. For the preparation of 5-FAM (Fluorescein-5-carboxamido)-labeled S-TuD, 5-FAM labeled 2′-O-methylated RNA oligonucleotide strands were synthesized and annealed with the unlabeled opposite 2′-O-methylated RNA oligonucleotide strands (Supplementary Table S1). MiRNA hairpin-inhibitor-miR-21 and miRNA hairpin-inhibitor-miR-106b were purchased from Thermo Scientific (miRIDIAN microRNA Hairpin inhibitors, IH-300492-05 and IH-300649-07, respectively). The anti-miR200c LNA and anti-miR106b LNA antisense oligonucleotide molecules were purchased from Exiqon (miRCURY LNA™ miRNA inhibitors 410126-00 and 426648-00, respectively). Antisense-miR106b was purchased from Life Technologies (AM10067). Technical information regarding the miRNA inhibitors described above is provided in Supplementary Table S2. Anti-miR21 PNA and negative control PNA antisense oligonucleotide molecules were supplied from Panagene (PI-1050 and PN-1001, respectively). Negative control LNA/DNA antisense oligonucleotide was synthesized by GeneDesign. Negative control 2′-O-methylated RNA and anti-miR21 2′-O-methylated RNA antisense oligonucleotides were synthesized by Hokkaido System Science. The 2′-O-methylated RNA AMOs and negative control LNA/DNA AMO sequences are listed in Supplementary Table S3.
Design of the S-TuD negative control sequence
We synthesized a massive number (∼1 billion) of random 25 nt RNA sequences in silico as candidate negative control molecules. Sequences with GC contents of between 20% and 80% were selected from this panel and were subsequently checked for their complementarity against 735 seed miRNA sequences (second to eighth region from the 5′-end) extracted from the entire human repertoire listed on miRBase Release 14. To exclude sequences which could potentially bind to human endogenous miRNAs, sequences that possessed six or more Watson–Crick type base pairs with any seed sequence were excluded from the list of candidates. A negative control sequence for S-TuD was arbitrarily chosen from a candidate group that had the smallest number of sequences that were complementary to the seed sequences of the entire human miRNA complement.
Plasmid construction
For the construction of luciferase reporter plasmids, the oligonucleotide pairs listed in Supplementary Table S4 were annealed and cloned into the XbaI–FseI sites of pGL4.74 (Promega, Madison, WI, USA) to generate pGL4.74-T21, pGL4.74-T200c, pGL4.74-T16 and pGL4.74-T106b, respectively. For the construction of the h7SK (human 7SK) promoter type TuD shuttle vector, we amplified a 0.3-kb human 7SK promoter fragment by PCR from human genomic DNA using the primers listed in Supplementary Table S5, followed by cloning into pCR2.1 (Invitrogen, Carlsbad, CA, USA). An oligo pair, listed in Supplementary Table S5, was annealed and cloned into this product via KpnI and HindIII sites to generate the ph7SK-TuD-shuttle. For the construction of TuD RNA expression cassettes, a series of oligonucleotide pairs were synthesized (Supplementary Table S6). Each oligo pair was annealed and cloned into the ph7SK-TuD-shuttle at the BsmBI site to generate h7SK-TuD-miR200c and h7SK-TuD-NC cassettes, from which 0.4-kb BamHI–EcoRI fragments were subcloned into the lentivirus vector pLSP to generate pLSP-h7SK-TuD-miR200c and pLSP-h7SK-TuD-NC (19), respectively.
Cell culture
The human colorectal adenocarcinoma cell line, HCT-116, was obtained from ATCC and cultured at 37°C in DMEM containing 10% fetal bovine serum (FBS).
RNA preparation and quantitative RT–PCR for mRNA
HCT-116 cells were seeded at 1 × 105 cells per well in six-well culture plates at 1 day prior to transfection. S-TuD-miR21-4ntin (0, 0.3, 1 or 10 nM) was transfected using Lipofectamine 2000 (Invitrogen) in accordance with the manufacturer's instructions. Poly(I)-poly(C) dsRNA (100 ng/ml, Sigma) was transfected as a positive control to induce interferon responses. Total RNA was prepared from HCT-116 cells just prior to transfection (0 h) and at 7 and 24 h after transfection using RNeasy (Qiagen). First strand cDNA was then synthesized using a SuperScript VILO cDNA synthesis kit (Invitrogen). Real-time RT–PCR was performed using the 7900 HT fast real-time PCR system (Applied Biosystems) with SYBR Green as a reporter. The data were normalized using GAPDH expression, and the levels expressed relative to the pre-transfected conditions (0 h). The sequences of the primers used for real-time PCR are listed in Supplementary Table S7.
Transfection and Luciferase assays
Cells were seeded at densities of 1 × 105 cells per well in 24-well plates in DMEM containing 10% FBS the day before transfection. The cells were then transfected in triplicate with Lipofectamine 2000 and 10 ng of Firefly luciferase plasmid pTK4.12 (Supplementary Figure S1A), 100 ng of RLuc target reporter plasmid and various concentrations of miRNA inhibitors (0.003 and 25 nM; Supplementary Figure S1B–S1F). We performed all assays at 48 h after the transfection using the dual luciferase assay on Glomax (Promega).
UV spectroscopy
Each S-TuD was dissolved in 10 mM sodium phosphate (pH 7.0) containing 10 mM NaCl. The UV-melting curves of 1.5 μM S-TuD at 260 nm were measured on a Shimazu UV-2450 UV–VIS spectrophotometer with a melting rate of 0.5°C/min.
MiR qRT–PCR
HCT-116 cells were seeded at 2 × 105 cells per well (six-well plates) in DMEM containing 10% FBS and transfected with 0.05 nM of S-TuD-miR106b-pf using the siPORT NeoFX transfection reagent (Ambion) according to the manufacturer's instructions. Total RNA was prepared from HCT-116 cells at 48 h after transfection using mirVana miRNA Isolation Kit (Applied Biosystems, CA, USA). Expression of mature miRNAs was determined by miR-qRT–PCR using miRNA-specific looped RT-primers and TaqMan probes as recommended by the manufacturer (Applied Biosystems). U6 snRNA was used as an internal control. PCR was performed in triplicate using the 7300 Real-Time PCR System (Applied Biosystems).
Oligonucleotides transfection, FACS analysis and sorting
HCT-116 cells were seeded at 2 × 105 cells per well (six-well plates) in DMEM containing 10% FBS and transfected with 10 nM of 5-FAM-labeled S-TuD-miR200c-pf or S-TuD-NC using the siPORT NeoFX transfection reagent (Ambion). To specifically isolate miRNA inhibitor-transfected cells, 5-FAM positive cells were sorted using FACS Aria (BD).
Virus transduction and Luciferase assays
HCT-116 cells were seeded at 1 × 105 cells per well in six-well plates in DMEM containing 10% FBS. After 24 h, the cells were transduced with each TuD RNA virus stock (3 × 105 TU) in the presence of 8 µg/ml of polybrene. After a further 24 h, the medium was then substituted with DMEM containing 10% FBS and puromycin (1 µg/ml). After 7 days of selection, the puromycin was removed from the medium.
Western blotting
Total proteins were extracted from cells using 1.5× SDS denaturing buffer and protein concentrations were measured using the Bio-Rad protein assay kit. The protein extracts were separated by 12% SDS–PAGE and transferred onto a PVDF membrane (Millipore). Immunoblotting was performed by incubating the membrane with antibodies against E-cadherin (ab76055, Abcam), vimentin (sc-6260, Santa Cruz), ZEB1 (ab64098, Abcam) and actin (612656, BD transduction) for 2 h at room temperature. Secondary antibodies conjugated with horseradish peroxidase were incubated with the membranes for 1 h at RT after three washes with phosphate-buffered saline with Tween-20. Signals were detected on an imaging analyzer LAS4000-EPUVm (FUJIFILM) using ECL reagent (Amersham) or Immunostar DL (WAKO).
RESULTS
Synthesis of two modified RNA strands that form a structure resembling TuD-miR21-4ntin after hybridization
We have previously reported the design of TuD RNAs targeting miR-21, miR-140 and miR-16. Among the several TuD-miR21s constructed, TuD-miR21 harboring a 4 nt insertion between positions 10 and 11 from the 3′-end of the perfectly complementary sequence to miR-21, where the RISC cleaves the target mRNAs (TuD-miR21-4ntin), exhibited the most potent inhibitory effects when expressed from a plasmid vector. A potential approach to the development of other potent TuDs was therefore to synthesize 2′-O-methylated RNA oligonucleotides that have the same sequence as TuD-miR21-4ntin. However, the synthesis of the full length molecule of TuD-miR21-4ntin (122 nt) in large amounts would not have been easy using current techniques. Considering also that the both sides of the two MBS sequences in a TuD molecule are flanked by two dsRNA stems, we decided to separate the entire TuD-miR21-4ntin molecule into two strands (60 nt each) by cutting the middle of the loop region so that the structure formed after the hybridization of both strands resembled that of TuD-miR21-4ntin (Figure 1B).
To test whether such hybridized 2′-O-methylated RNA strands would suppress endogenous miR-21 activity in HCT-116 cells (which express very high levels of endogenous miR-21), we transiently cotransfected them with dual luciferase reporters (DLR), which are composed of the Renilla luciferase reporter with or without the insertion of a 22-bp DNA sequence fully complementary to the mature miR-21 within the 3′-UTR (Supplementary Figure S1BC). The firefly luciferase reporter was used as a transfection control (Supplementary Figure S1A). As shown in Figure 1C, the hybridized 2′-O-methylated RNA inhibited endogenous miR-21 activity much more efficiently than the conventional 2′-O-methylated RNA oligonucleotide AMO, when 1–3 nM of these inhibitors were introduced. Appreciable inhibitory effects of the AMO, 2′-O-methylated RNA and PNA oligonucleotide, were only observed at a concentration of 50 nM (Supplementary Figure S2). We designated this 2′-O-methylated RNA-based inhibitor of miRNA with a similar structure to TuD as S-TuD (synthesized TuD) and decided to further optimize its inhibitory effects.
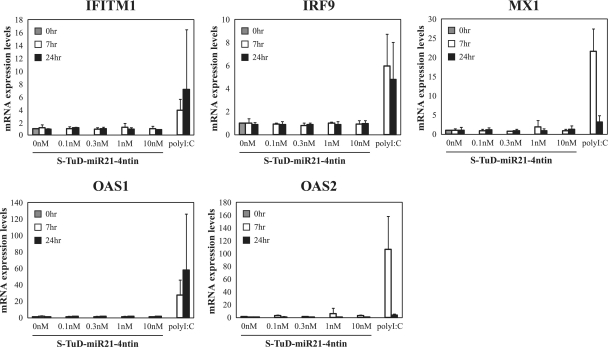
Since S-TuD harbors two regions with a complete double stranded structure (18 and 10 bp, respectively), we speculated that it might potentially trigger an interferon response in the introduced cells and thus disrupt any functional analysis of miRNAs. To test this possibility, we transfected HCT-116 cells with S-TuD-miR21-4ntin as well as Poly I:C as a positive control and the mRNA levels of the interferon responsive genes OAS1, OAS2, MX1, IRF9 and IFITM1 were determined at 0–24 h after transfection by quantitative RT–PCR (Figure 2). The results clearly showed that S-TuD does not induce an immune response in cell cultures even at the highest concentration used (10 nM) as judged by the interferon induction level. Essentially similar results were obtained when DLD-1 cells were used (data not shown).
Figure 2.
Measurement of the interferon response in cells transfected with S-TuD. S-TuD-miR21-4ntin was transfected into HCT-116 cells at several doses. Interferon response genes were then measured by qRT–PCR and are represented by the mean ± SD (n = 3). Poly(I)-poly(C) dsRNA (100 ng/ml) was used as a positive control.
Modulation of the MBS sequence of S-TuD to obtain optimum inhibitory effects
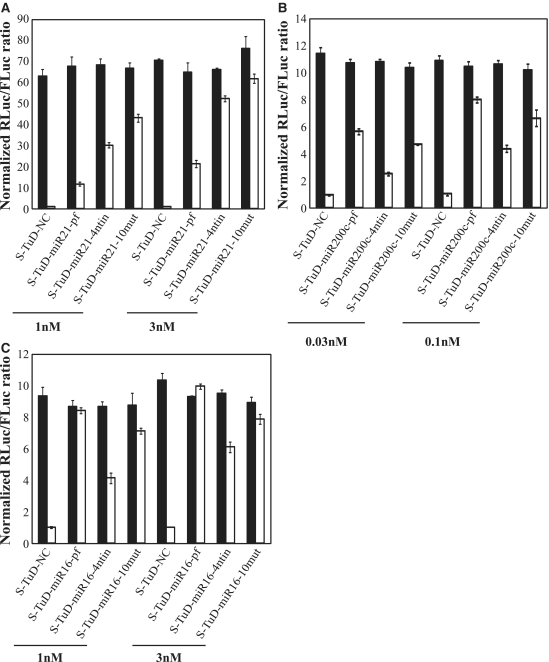
TuD-miR21-4ntin has two MBS regions which carry a 4 nt-insertion between positions 10 and 11 from the 3′-end of the perfectly complementary sequence to mature miR-21, where the RISCs cleave the target mRNAs. This insertion sequence is designed to form a 4-nt bulge when the target miRNA binds to the MBS of the corresponding TuD. Through the presence of this bulge, we previously demonstrated that TuD-miR21-4ntin efficiently escapes from this cleavage and has a much higher inhibitory effect than TuD-miR21-pf (TuD-miR21 harboring MBSs perfectly complementary to miR-21). Since S-TuD is composed of fully 2′-O-methylated RNA, we tested whether it would require this 4-nt insertion to avoid cleavage. We compared the inhibitory effects of S-TuD carrying MBSs with or without the 4-nt bulge, using S-TuDs targeting miR-21 and miR-200 c, respectively. Whereas S-TuD-miR21-4ntin inhibited miR-21 more efficiently than S-TuD-miR21-pf at the same dosage (Figure 3A), the inhibitory effects of 0.1 nM S-TuD-miR200c-pf were even greater than those exerted by 0.3 nM S-TuD-miR200c-4ntin (Figure 3B).
Figure 3.
Impact of MBS sequences on the inhibitory effects of S-TuD molecules. (A) Comparison of three types of S-TuD-miR21 (S-TuD-miR21-pf, S-TuD-miR21-4ntin, and S-TuD-miR21-10mut). HCT-116 cells were transfected using similar procedures to those shown in Figure 1C. After performing a dual luciferase assay, the expression levels were normalized to the ratio of the activity of miR-21-RL to that of FL in 1 nM S-TuD-NC transfected HCT-116 cells and are represented by the mean ± SD (n = 3). (B) Comparison of three types of S-TuD-miR200c (S-TuD-miR200c-pf, S-TuD-miR200c-4ntin, and S-TuD-miR200c-10mut) which were transiently transfected into HCT-116 cells together with the Renilla luciferase miR-200 c reporter (miR-200 c-RL; open bars) or the untargeted control Renilla luciferase reporter (UT-RL; black bars). In all cases, the Firefly luciferase reporter (FL) was cotransfected as a transfection control. After performing a dual luciferase assay, the expression levels were normalized to the ratio of the activity of miR-200c-RL to that of FL in 0.03 nM S-TuD-NC transfected HCT-116 cells and are represented by the mean ± SD (n = 3). (C) Comparison of three types of S-TuD-miR16 (S-TuD-miR16-pf, S-TuD-miR16-4ntin, and S-TuD-miR16-10mut) which were transiently transfected into HCT-116 cells together with the Renilla luciferase miR-16 reporter (miR-16-RL; open bars) or the untargeted control Renilla luciferase reporter (UT-RL; black bars). In each case, the Firefly luciferase reporter (FL) was cotransfected as a transfection control. Following a dual luciferase assay, the expression levels were normalized to the ratio of the activity of miR-16-RL to that of FL in 1 nM S-TuD-NC transfected HCT-116 cells and are represented by the mean ± SD (n = 3).
To resolve the apparent discrepancies found in the requirement of the bulge for higher S-TuD activity, we next examined the possible secondary structures of these S-TuDs using CentroidFold (http://www.ncrna.org/centroidfold) (25). CentroidFold can predict the secondary structure of a single native RNA molecule but not one composed of two strands of 2′-O-methylated RNAs. We therefore analyzed the TuD RNA that has the same MBS sequences as the corresponding S-TuD to obtain an approximate secondary structure (Supplementary Figure S3). When the base pair formation between the two MBSs in the S-TuD targeting miR-21 was compared, it was much stronger in S-TuD-miR21-pf than in S-TuD-miR21-4ntin. On the other hand, the interaction between the two MBSs of S-TuD-miR200c-4ntin was found to be higher than that of S-TuD-miR200c-pf. From these observations, we speculated that the accessibility to the target miRNA drastically decreases if the interaction between the two MBSs of one S-TuD molecule is very strong. To test this hypothesis, we attempted to introduce a point mutation in MBSs, which would reduce the heavy base pair formation between the two MBSs of S-TuD-miR21-pf without significantly affecting the binding activity. Regions close to the 5′- and 3′-ends of the MBS would then not easily form base pairs because the linkers that flank these regions were designed not to hybridize to each other. Point mutations at any positions complementary to the seed regions or the 3′-compensatory sites would also drastically reduce the miRNA inhibitory effects and point mutations at any positions complementary to the seed regions would affect target specificity (26). We therefore introduced a point mutation at position 10 from the 3′-end of both MBSs (S-TuD-miR21-10mut). Base pair formation between the two MBSs of S-TuD-miR21-10mut was greatly reduced compared with that of S-TuD-miR21-pf (Supplementary Figure S3). When the miRNA inhibitory effects of S-TuD-miR21-pf, S-TuD-miR21-4ntin and S-TuD-miR21-10mut were tested (Figure 3A), S-TuD-miR21-10mut was found to be much more efficient. In the case of miR-200c, however, S-TuD-miR200c-pf was found to be a more potent inhibitor than S-TuD-miR200c-10mut (Figure 3B). This is possibly because neither S-TuD-miR200c-pf nor S-TuD-miR200c-10mut forms significant base pairing between the two MBSs. In such cases, a point mutation would reduce the inhibitory effects of the S-TuD, because of a lower binding affinity between the miRNA and MBSs.
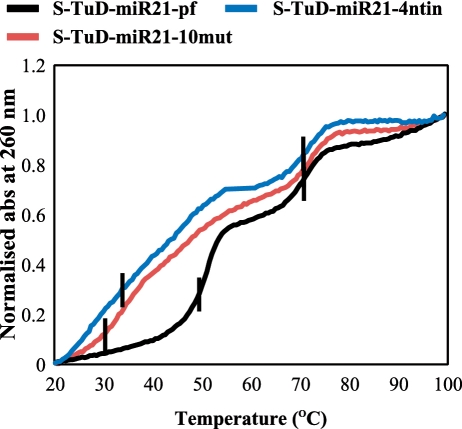
If the above hypothesis was correct, we expected to see a clear difference between the secondary structure of S-TuD-miR21-pf and S-TuD-miR21-10mut (or S-TuD-miR21-4ntin). To test this, we measured the UV-melting curves of these three S-TuDs at 260 nm. As shown in Figure 4, the curves of S-TuD-miR21-10mut and S-TuD-miR21-4ntin were very similar and had two Tm (melting temperature) values of around 30–34°C and 70°C. In contrast, Tm values of 50°C and 70°C were observed for S-TuD-miR21-pf. Since a Tm at 70°C was observed in all three of these S-TuDs, this would represent a dissociation of the 18 bp stem I. A Tm at 30–34°C for S-TuD-miR21-10mut and S-TuD-miR21-4ntin would mainly reflect the dissociation of the 10 bp stem II. The gradual increase in UV absorbance at around 30–45°C and the rather large and sharp sigmoid curve at 50°C observed for S-TuD-miR21-pf reflects the secondary structure formed between the two MBSs of S-TuD-miR21-pf in addition to the base pairing of stem II. These thermal denaturation analyses thus support the idea that the interaction between the two MBSs of S-TuD-miR21-pf is significantly higher than those of S-TuD-miR21-4ntin and S-TuD-miR21-10mut.
Figure 4.
Normalized UV melting curves for 1.5 μM concentrations of S-TuD-miR21-pf, S-TuD-miR21-4ntin and S-TuD-miR21-10mut in a 10 mM sodium phosphate (pH 7.0) buffer containing 10 mM NaCl. Melting was assessed by UV absorbance at 260 nm and a melting rate of 0.5°C/min. Vertical black bars indicate the Tm points.
Taken together, these results suggest that to design an efficient S-TuD molecule in general, the two MBS regions should not form base pairs to a threshold level. If an MBS that is perfectly complementary to miRNA cannot satisfy this requirement, the introduction of a point mutation in the middle region or a 4-nt insertion would be one possible way to improve the inhibitory effects of the S-TuD molecule in question, even with the possible loss of binding affinity to the miRNA. For example, in the case of S-TuD-miR16-pf, S-TuD-miR16-4ntin and S-TuD-miR16-10mut (Supplementary Figure S3), the MBSs of which do not form strong base pairing, S-TuD-miR16-pf was shown to be the most efficient inhibitor as expected, when tested by the miR-16 luciferase reporter system (Figure 3C). From these experimental results, we roughly expect that the threshold number of base pairs between the two MBSs (plus 3-nt linkers at their both ends) is about nine, as predicted by CentroidFold.
In our previous report, the length of stem I was designed to be more than 16 bp and fixed at 18 bp in most cases so that the TuD RNA molecules synthesized in the nuclei can be efficiently transported into the cytoplasm (19,27). Since S-TuD is directly introduced into the cytoplasm by transfection, a long stem I structure is likely not to be required. To test this, we synthesized two S-TuDs with shorter stem I lengths of 14 and 10 bp, (S-TuD-miR21-10mut-14 bp and S-TuD-miR21-10mut-10 bp, respectively) (Supplementary Figure S4A) and compared their potency against the parental S-TuD molecule, S-TuD-miR21-10mut. The inhibitory effects of S-TuD-miR21-10mut-14 bp were slightly reduced and those of S-TuD-miR21-10mut-10 bp were considerably lower when compared with S-TuD-miR21-10mut (Supplementary Figure S4B). This indicated that an 18 bp Stem I contributes to inhibitory potency. To further understand the molecular mechanisms underlying this phenomenon, we analyzed the UV-melting curves of these three S-TuDs at 260 nm (Supplementary Figure S4C). As described in the previous section, the two Tm values for S-TuD-miR21-10mut were calculated at 34°C and 70°C, which reflect the dissociation of the 10 bp length stem II and 18 bp length stem I, respectively.
The Tm at 32°C for S-TuD-miR21-10mut-stemI-14 bp and at 37°C for S-TuD-miR21-10mut-stemI-10 bp likely reflect the dissociation of a single 10 bp stem II and two 10 bp stems of Stems I and II, respectively. This result suggests that the two strands comprising S-TuD-miR21-10mut-stemI-10 bp would at least partly dissociate from each other in cells at 37°C, consistent with the observation that the inhibitory effects of this molecule are severely affected (See Supplementary Figure S4B). Furthermore, a Tm at 57°C for S-TuD-miR21-10mut-stemI-14 bp, reflecting the dissociation of a 14 bp stem I, reveals that its S-TuD structure is not fully thermodynamically stable. These results thus show that an 18 bp length stem I structure strongly contributes to the stabilization of the two stems that form the S-TuD structure and support the potent inhibitory activity of this molecule.
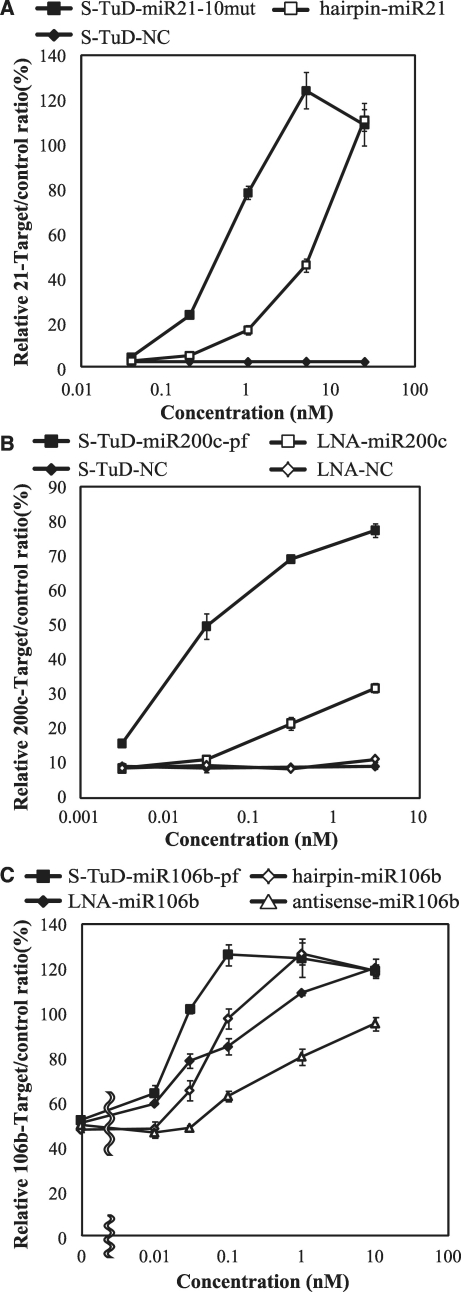
Dose dependency of S-TuD inhibitory activity
We next determined the dose–response curves of the S-TuDs targeting miR-21, miR-200 c and miR-106 b. The dose dependency of S-TuD-miR21-10mut as well as that of 2′-O-methylated RNA-based miRNA hairpin inhibitor-miR-21 was first determined (Figure 5A). S-TuD-miR21-10mut showed inhibitory effects at a concentration of 0.2 nM. At a concentration of 5 nM, the luciferase activity reached saturation, which almost corresponded to the levels of the reporter without the target sequence. On the other hand, miRNA hairpin inhibitor-miR-21 showed inhibitory effects at a concentration of 1 nM and saturation had not been reached as a concentration of 25 nM.
Figure 5.
Dose-dependency of S-TuD-miR-21-10mut (A), S-TuD-miR-200c-pf (B) or S-TuD-miR-106b-pf (C) miRNA inhibitory activities. HCT-116 cells were transfected using similar procedures to those shown in Figure 3. (A) A dual luciferase assay was performed 48 h after transfection into HCT-116 cells and the miRNA hairpin inhibitor-miR21 was used for comparison. The expression ratios of miR-21-RL/FL to UT-RL/FL are represented by the mean ± SD (n = 3). (B) A dual luciferase assay was performed 48 h after transfection into HCT-116 cells. The LNA-inhibitor-miR200c was used for comparison. The ratios of miR-200c-RL/FL to UT-RL/FL expression are represented by the mean ± SD (n = 3). (C) A dual luciferase assay was performed 48 h after transfection into HCT-116 cells. Antisense-miR-106b, miRNA hairpin inhibitor-miR106b and LNA-inhibitor-miR106b were used for comparison. The ratios of miR-106b-RL/FL to UT-RL/FL are represented by the mean ± SD (n = 3) as the expression levels.
We next determined the dose dependency of S-TuD-miR200c-pf and of the LNA oligonucleotide antisense inhibitor targeting miR-200 c (Figure 5B). S-TuD-miR200c-pf showed inhibitory effects at 0.003 nM which were saturated at 0.3 nM, whereas the effects of LNA-miR200c did not reach saturation even at 3 nM. Finally, we tested S-TuD-miR106b-pf as well as miRNA hairpin inhibitor and both LNA and 2′-O-methylated RNA antisense oligonucleotides, all of which are designed to target miR-106 b (Figure 5C). S-TuD-miR106b-pf showed inhibitory effects at 0.01 nM which reached saturation at 0.1 nM. LNA-miR106b also showed inhibitory effects at 0.01 nM, but its saturation point was not reached even at 1 nM. The miRNA hairpin inhibitor-miR-106 b showed inhibitory effects at 0.03 nM and reached saturation at 1 nM. 2′-O-methylated RNA antisense-miR106b oligonucleotides showed inhibitory effects at 0.1 nM but these remained low even at a concentration of 1 nM. We next determined levels of free miR-106b in cells introduced with these inhibitors using real-time PCR in a parallel cell culture treated with 0.05 nM of the inhibitors. At this dosage, relative target/control is expected to be significantly different among these four inhibitors. The free miR-106b levels determined are in the reverse order to the luciferase activity of these inhibitors (Supplementary Figure S5). We think the apparently high amount of free miR-106b even in S-TuD-miR106b-pf treated cells is partly reflecting that transfection efficiency of inhibitors at a low concentration (0.05 nM) into HCT116 cells would not be high. Taken together, these results show that S-TuDs inhibit their target miRNAs at very high efficiencies.
The effects of S-TuD RNA on other family members of the target miRNA
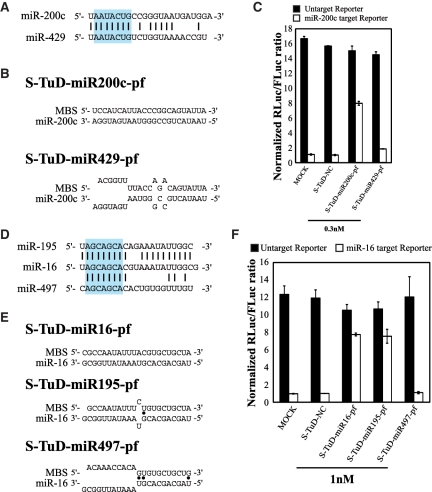
We next tested whether an S-TuD targeting a particular miRNA would exhibit inhibitory effects on other family members of this target molecule, i.e. miRNAs that share the same seed sequence (2–8 bases from the 5′-end). We first investigated the specificity of S-TuD using the miR-200 b/-200 c/-429 family as the target in HCT-116 cells, where the expression of miR-200 c is predominant (Supplementary Figure S6A). We expected that the expression of a reporter containing a sequence that is perfectly complementary to miR-200 c would accurately reflect the functional miR-200 c levels. We tested the inhibitory effects of S-TuD-miR200c-pf and S-TuD-miR429-pf (Figure 6A, B and Supplementary Figure S6B), respectively, and found that the inhibitory activity of S-TuD-miR200c-pf was high, whereas S-TuD-miR429-pf had only slight effects (Figure 6C). These results suggested that strong cross reactivity between miRNA targets is not detectable even when their seed sequences are identical.
Figure 6.
Inhibitory effects of S-TuD on related miRNAs. (A) miR-200c and -429 sequences and (D) miR-16,-195 and -497 sequences, and the degree of homology between them. Sequences shown in blue box are the heptameric seed sequence (2–8 nt from the 5′-end). Black bars indicated homologous nucleotides. Imperfect pairing between (B) miR-200 c and (E) miR-16 and MBSs of the corresponding S-TuD. Black dots indicate G-U pairs. (C, F) S-TuDs were transfected into HCT-116 cells together with the Renilla luciferase (C) miR-200c and (F) miR-16 reporter (open bars) or the untargeted control Renilla luciferase reporter (black bars) as well as the Firefly luciferase reporter as a transfection control. HCT-116 cells transfected with only luciferase vectors were also used as MOCK-transfected cells. After performing a dual luciferase assay, the expression levels were normalized to the ratio of the activity of (C) miR-200c-RL and (F) miR-16-RL to that of FL in S-TuD-NC transfected cells and are represented by the mean ± SD (n = 3).
Since these observations are somehow different from those obtained for TuD RNA, where significant cross reactivity was detected, we chose the miR-15 a/-15 b/-16/-195/-424/-497 family in HCT-116 cells for analysis as these molecules were previously tested for TuD RNA (19). Since the expression of miR-16 is predominant in HCT-116 cells (Supplementary Figure S6C) the miR-16 reporter mainly reflects the functional levels of this molecule in this experimental setting. We tested the inhibitory effects of S-TuD-miR16-pf, S-TuD-miR195-pf and S-TuD-miR497-pf, respectively (Figure 6D, E and Supplementary Figure S6D). The inhibitory activity of S-TuD-miR195-pf was equivalent to that of S-TuD-miR16-pf, whereas S-TuD-miR497-pf showed no effects (Figure 6F). These results confirmed that cross-reactivity among target miRNAs requires not only identical seed sequence but also extensive homology between the 3′-compensatory regions (26) of these molecules.
Duration of the inhibitory effects of S-TuD
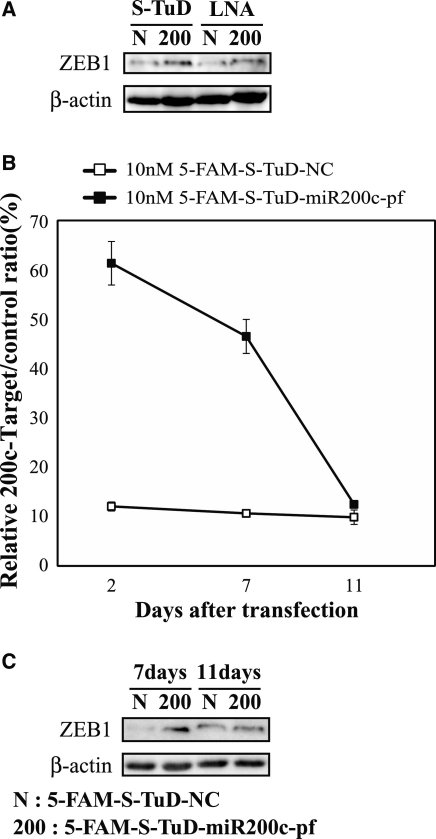
To confirm that S-TuD can efficiently increase the amount of endogenous proteins targeted by the miRNA, we transfected S-TuD-miR200c-pf or LNA-miR200c into HCT-116 cells and determined the expression levels of the ZEB1 protein which is an established target of miR-200c at 2 days after transfection (28). We observed that S-TuD-miR200c-pf elevated ZEB1 expression compared with S-TuD-NC and further that the elevated level was slightly higher than that of LNA-miR200c (Figure 7A).
Figure 7.
The inhibitory effects of S-TuD-miR200c-pf. (A) HCT-116 cells were transfected with S-TuD-miR200c-pf (200), S-TuD-NC (N), LNA-miR200c (200) and LNA-NC (N) at concentrations of 10 nM, respectively. From these transfected cells, total proteins were prepared 2 days after transfection. ZEB1 (top) and β-actin (bottom) were detected by western blotting. (B) Time course analysis of the inhibitory effects of S-TuD-miR200c-pf. HCT-116 cells were transfected with 5-FAM labeled S-TuD-miR200c-pf or S-TuD-NC at concentrations of 10 nM and 5-FAM positive cells were flow sorted 2 days after transfection. Sorted cells were maintained in culture and parallel cultures were prepared at each cellular passage for transfection with reporter genes. Dual luciferase assays were performed 2 days after transfection. The relative expression levels were normalized to those of 10 nM 5-FAM-S-TuD-NC transfected HCT-116 cells and are represented by the mean ± SD (n = 3). (C) The expression levels of ZEB1 in cells shown in (B). Total proteins from these sorted cells were prepared 7 and 11 days after transfection. ZEB1 (top) and β-actin (bottom) were detected by western blotting.
Given the high inhibitory effects of S-TuD against its target miRNA, we next analyzed the duration of these effects using S-TuD-miR200c-pf. We first synthesized 5-FAM-labeled S-TuD-miR200c-pf and S-TuD-NC, and confirmed that the 5-FAM label has no impact on the miRNA inhibitory effects of S-TuD (Supplementary Figure S7A). We transfected HCT-116 cells with 5-FAM-labeled S-TuD-miR200c-pf and 5-FAM-labeled S-TuD-NC, respectively, and then sorted 5-FAM-positive cells 2 days later and maintained them in culture (Supplementary Figure S7B–D). In the process of cellular passaging, parallel cultures were prepared for the transfection of luciferase reporter genes, the activity of which was assayed 2 days after transfection (Figure 7B). The inhibitory effects of 5-FAM-S-TuD-miR200c-pf were high at 2 days after the introduction of S-TuD and significant inhibitory effects were observed even at 7 days after transfection. The inhibitory effects of 5-FAM-S-TuD-miR200c-pf became marginal by 11 days. We also investigated the duration of the effects on the expression levels of ZEB1. As shown in Figure 7C, the expression levels of ZEB1 were increased at 7 days after S-TuD transfection, but these effects had disappeared at 11 days after transfection, consistent with the reporter assay results. These data indicate that the inhibitory effects of a single transfection of S-TuD are quite long-lasting (>7days) and persist even after several cell divisions (about seven cycles).
An epithelial–mesenchymal transition is induced by miR-200 c inhibition
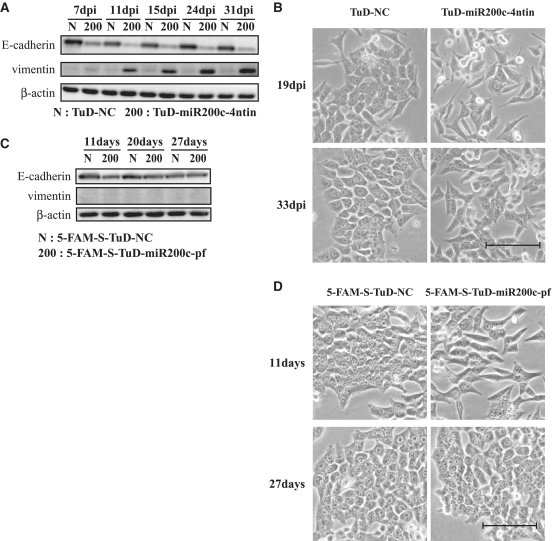
It has been reported previously that the epithelial–mesenchymal transition (EMT) is induced by inhibiting the activities of the miR-200 family (28). In this previous study, HCT-116 cells were transfected with 50 nM of miRNA inhibitor (LNA) five times every 3 days to establish the EMT. In our current study, we estimated the time required to establish the EMT by suppressing miR-200 family activity via the continuous expression of TuD in HCT-116 cells. We used a lentivirus vector system carrying TuD-miR200c expression cassettes driven by the human 7SK promoter which fully inhibited miR-200c in HCT-116 cells (Supplementary Figure S8AB). We next determined the expression levels of E-cadherin and vimentin, as epithelial and mesenchymal markers, respectively, in these cells (Figure 8A). At 7 days after the transduction of the vector carrying TuD-miR200c, the E-cadherin expression levels were significantly reduced but vimentin expression was only marginally detected. High levels of vimentin expression, concomitant with very low E-cadherin levels, were established by 11 days after TuD-miR200c transduction, indicating that the establishment of EMT requires continuous expression of TuD-miR200c for 8–11 days. In addition, morphological changes into fibroblastic cells were observed and were maintained for at least 1 month (Figure 8B).
Figure 8.
Induction of the EMT by the inhibition of miR-200 c. (A) A TuD RNA-expressing lentiviral vector was transduced into HCT-116 cells with an MOI of three followed by selection with puromycin. The expression levels of E-cadherin and vimentin, gene markers of the epithelium or mesenchyme, respectively, were then measured. Total proteins were prepared at 7, 11, 15, 24 and 31 days after transduction. E-cadherin (top) and vimentin (middle) and a β-actin loading control (bottom) were detected by western blotting. (B) Morphological status of HCT-116 cells transduced with TuD-miR200c-4ntin or TuD-NC expressing lentivirus at 19 and 33 days after transduction. Bar, 10 µm. (C) HCT-116 cells were transfected with 5-FAM-S-TuD-miR200c-pf or 5-FAM-S-TuD-NC, both at a concentration of 10 nM, and 5-FAM positive cells were sorted at 2 days after transfection. Total proteins were prepared 11, 20 and 27 days after transfection. E-cadherin (top), vimentin (middle) and β-actin (bottom) were detected by western blotting. (D) Morphological status of HCT-116 cells transfected with 5-FAM-S-TuD-miR200c-pf or 5-FAM-S-TuD-NC at 11 and 27 days after transfection. Bar, 10 µm.
Because the inhibitory effects of 5-FAM-S-TuD-miR200c-pf were found to be quite long-lasting and marginal inhibitory effects were detectable even at 11 days after the transfection of HCT-116 cells with this molecule, we evaluated whether EMT could be induced by a single transfection of 5-FAM-S-TuD-miR200c-pf at a 10 nM dose. 5-FAM positive cells were flow sorted and cultured. As shown in Figure 8C, E-cadherin expression was decreased in 5-FAM-S-TuD-miR200c-pf introduced cells at 11 days after transfection, when the larger cellular population assumed a fibroblastic morphology (Figure 8D). However, vimentin induction remained undetectable in these cells. These results indicate that EMT is only partially induced by 5-FAM-S-TuD-miR200c-pf when compared with the full EMT induction observed in HCT-116 cells harboring the TuD-miR200c expression lentivirus vector. Furthermore, the reduction of E-cadherin expression was recovered and this protein reached normal levels at 20–27 days after transfection. At the same time, the fibroblastic morphology of the cells transduced with 5-FAM-S-TuD-miR200c-pf returned fully to the parental epithelial cell appearance (Figure 8D). These results indicate that the E-cadherin reduction and fibroblastic morphology induced by a single transfection of 5-FAM-S-TuD-miR200c-pf is both transient and reversible.
DISCUSSION
In our current study, we have developed a novel synthetic miRNA inhibitor, S-TuD, that has a similar structure to that of TuD RNA, which we previously developed to be expressed from plasmid- or lentivirus vectors (19). Through a single transduction of an optimized S-TuD (S-TuD-miR200c-pf), sufficient miRNA inhibitory effects were obtained and shown to persist for 7 days (about seven cell divisions) and to partially introduce EMT. During the process of optimizing the MBS sequences in our S-TuD design, we found that the two MBS regions within a single S-TuD molecule should not form a threshold level of base pairing, ∼ 9 bp as predicted by CentroidFold (Figure 3 and Supplementary Figure S3). When an MBS that is completely complementary to the target miRNA sequence exceeds this threshold, we found that the introduction of a single mutation or 4 nt insertion in the middle region of the MBS is sufficient in many cases to abolish the base pairing without significantly affecting the affinity to the target miRNA. Although secondary structures composed of 2′-O-methylated RNA will differ from those comprising native RNA, we show from our analysis that the secondary structure predictions for three S-TuD-miR21 molecules by CentroidFold were very consistent with the melting curve analysis (Figure 4). Our prediction accuracy, however, will be increased much more if a secondary structure algorithm for 2′-O-methylated RNA becomes available.
Our present results show that the inhibitory effects of S-TuD-miR21 were drastically reduced with a stem I length of 10 bp, indicating that S-TuDs requires a long stem I to maintain a rigid structure, where two MBSs are bounded by two stable stems (stem I and II). In such S-TuDs, both ends of the two MBSs are so firmly fixed by the flanking stems that each MBS will not twist freely along with its strand axis. We therefore speculate that this conformational restriction contributes to an efficient access of the MBS to the target miRNA. These unique structural features of S-TuD as well as TuD may explain why S-TuD shows stronger inhibitory activity than other currently available miRNA inhibitors (their basic structural features are summarized in Supplementary Table S2)
In summary, S-TuD shows long-lasting inhibitory effects at very low dosages and has no detectable interferon inducing activity in culture. These characteristics of S-TuD are advantageous for its potential as a future therapeutic drug. Using recent evidence from pharmaceutical research focusing on siRNA, we would like to now develop an efficient drug delivery system for S-TuD and to reduce potential side effects of these molecules, such as unwanted cytokine responses, when they are introduced in vivo.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables S1–S7. Supplementary Figures S1–S8.
FUNDING
Grant-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Culture, Sports, Science and Technology, Japan (MEXT) [17016015]; by a Grant-in-Aid for Young Scientists (B) [22700873]; and by A-Step of the Japan Science and Technology Agency. Funding for open access charge: Grant in AID for Scientific Research on Priority Areas from MEXT.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to M. Hamada (University of Tokyo and AIST) for kind advice concerning CentroidFold. The authors thank K. Takagi for assistance with the quantitative RT–PCR. The authors also thank the IMSUT FACS Core Laboratory for assistance in using the FACS Aria. Super-computing resources were provided by the Human Genome Center, University of Tokyo, Japan (http://sc.hgc.jp/shirokane.html). The authors are additionally grateful to S. Kawaura and A. Kato for assistance in the preparation of this manuscript.
REFERENCES
- 1.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 3.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 4.Lin SL, Chang DC, Chang-Lin S, Lin CH, Wu DT, Chen DT, Ying SY. Mir-302 reprograms human skin cancer cells into a pluripotent ES-cell-like state. RNA. 2008;14:2115–2124. doi: 10.1261/rna.1162708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li QJ, Chau J, Ebert PJ, Sylvester G, Min H, Liu G, Braich R, Manoharan M, Soutschek J, Skare P, et al. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell. 2007;129:147–161. doi: 10.1016/j.cell.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 6.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 7.Lecellier CH, Dunoyer P, Arar K, Lehmann-Che J, Eyquem S, Himber C, Saib A, Voinnet O. A cellular microRNA mediates antiviral defense in human cells. Science. 2005;308:557–560. doi: 10.1126/science.1108784. [DOI] [PubMed] [Google Scholar]
- 8.Shimono Y, Zabala M, Cho RW, Lobo N, Dalerba P, Qian D, Diehn M, Liu H, Panula SP, Chiao E, et al. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell. 2009;138:592–603. doi: 10.1016/j.cell.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 10.Hutvagner G, Simard MJ, Mello CC, Zamore PD. Sequence-specific inhibition of small RNA function. PLoS Biol. 2004;2:E98. doi: 10.1371/journal.pbio.0020098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meister G, Landthaler M, Dorsett Y, Tuschl T. Sequence-specific inhibition of microRNA- and siRNA-induced RNA silencing. RNA. 2004;10:544–550. doi: 10.1261/rna.5235104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Orom UA, Kauppinen S, Lund AH. LNA-modified oligonucleotides mediate specific inhibition of microRNA function. Gene. 2006;372:137–141. doi: 10.1016/j.gene.2005.12.031. [DOI] [PubMed] [Google Scholar]
- 13.Davis S, Lollo B, Freier S, Esau C. Improved targeting of miRNA with antisense oligonucleotides. Nucleic Acids Res. 2006;34:2294–2304. doi: 10.1093/nar/gkl183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 15.Vermeulen A, Robertson B, Dalby AB, Marshall WS, Karpilow J, Leake D, Khvorova A, Baskerville S. Double-stranded regions are essential design components of potent inhibitors of RISC function. RNA. 2007;13:723–730. doi: 10.1261/rna.448107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scherr M, Venturini L, Battmer K, Schaller-Schoenitz M, Schaefer D, Dallmann I, Ganser A, Eder M. Lentivirus-mediated antagomir expression for specific inhibition of miRNA function. Nucleic Acids Res. 2007;35:e149. doi: 10.1093/nar/gkm971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sayed D, Rane S, Lypowy J, He M, Chen IY, Vashistha H, Yan L, Malhotra A, Vatner D, Abdellatif M. MicroRNA-21 targets Sprouty2 and promotes cellular outgrowths. Mol. Biol. Cell. 2008;19:3272–3282. doi: 10.1091/mbc.E08-02-0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat. Methods. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haraguchi T, Ozaki Y, Iba H. Vectors expressing efficient RNA decoys achieve the long-term suppression of specific microRNA activity in mammalian cells. Nucleic Acids Res. 2009;37:e43. doi: 10.1093/nar/gkp040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sakurai K, Furukawa C, Haraguchi T, Inada K, Shiogama K, Tagawa T, Fujita S, Ueno Y, Ogata A, Ito M, et al. MicroRNAs miR-199a-5p and -3p target the Brm subunit of SWI/SNF to generate a double-negative feedback loop in a variety of human cancers. Cancer Res. 2011;71:1680–1689. doi: 10.1158/0008-5472.CAN-10-2345. [DOI] [PubMed] [Google Scholar]
- 21.Hikichi M, Kidokoro M, Haraguchi T, Iba H, Shida H, Tahara H, Nakamura T. MicroRNA regulation of glycoprotein B5R in oncolytic vaccinia virus reduces viral pathogenicity without impairing its antitumor efficacy. Mol. Ther. 2011;19:1107–1115. doi: 10.1038/mt.2011.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu Z, Li Y, Takwi A, Li B, Zhang J, Conklin DJ, Young KH, Martin R. miR-301a as an NF-κB activator in pancreatic cancer cells. EMBO J. 2011;30:57–67. doi: 10.1038/emboj.2010.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gagan J, Dey BK, Layer R, Yan Z, Dutta A. MicroRNA-378 targets the myogenic repressor MyoR during myoblast differentiation. J. Biol. Chem. 2011;286:19431–19438. doi: 10.1074/jbc.M111.219006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sato K, Hamada M, Asai K, Mituyama T. CENTROIDFOLD: a web server for RNA secondary structure prediction. Nucleic Acids Res. 2009;37:W277–W280. doi: 10.1093/nar/gkp367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zeng Y, Cullen BR. Structural requirements for pre-microRNA binding and nuclear export by Exportin 5. Nucleic Acids Res. 2004;32:4776–4785. doi: 10.1093/nar/gkh824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.