Abstract
Arthritis susceptibility genes were sought by analysis of differential gene expression between pristane-induced arthritis (PIA)-susceptible DA rats and PIA-resistant E3 rats. Inguinal lymph nodes of naïve animals and animals 8 days after pristane injection were analyzed for differential gene expression. mRNA expression was investigated by microarray and real-time PCR, and protein expression was analyzed by flow cytometry or ELISA. Twelve genes were significantly differentially expressed when analyzed by at least two independent methods, and an additional five genes showed a strong a tendency toward differential expression. In naïve DA rats IgE, the bone marrow stromal cell antigen 1 (Bst1) and the MHC class II β-chain (MhcII) were expressed at a higher level, and the immunoglobulin kappa chain (Igκ) was expressed at a lower level. In pristane-treated DA rats the MHC class II β-chain, gelatinase B (Mmp9) and the protein tyrosine phosphatase CL100 (Ptpn16) were expressed at a higher level, whereas immunoglobulins, the CD28 molecule (Cd28), the mast cell specific protease 1 (Mcpt1), the carboxylesterase precursor (Ces2), K-cadherin (Cdh6), cyclin G1 (Ccng1), DNA polymerase IV (Primase) and the tumour associated glycoprotein E4 (Tage) were expressed at a lower level. Finally, the differentially expressed mRNA was confirmed with protein expression for some of the genes. In conclusion, the results show that animal models are well suited for reproducible microarray analysis of candidate genes for arthritis. All genes have functions that are potentially important for arthritis, and nine of the genes are located within genomic regions previously associated with autoimmune disease.
Keywords: arthritis, differential gene expression, microarray, quantitative trait locus, rat
Introduction
In rheumatoid arthritis (RA) the peripheral joints are attacked by an autoimmune, chronic inflammatory process [1]. The cause of the disease is not known, but it is influenced by genetic as well as by environmental factors [2,3]. Despite recent success with new biological therapies, there is no cure for RA and there is no effective therapy for large groups of patients. It is therefore of great importance that we improve our understanding of the genetic basis of the disease as well as of the biological pathways that are responsible for its pathogenesis. Recently, new powerful techniques, based on microarrays, have been developed for analysis of gene expression [4]. These techniques can be used to analyze a large number of genes simultaneously and, when used for analysis in suitable experimental circumstances, provide valuable information on arthritis pathogenesis.
Direct comparison of RA patients with control groups is difficult because any differential gene expression in RA may be masked by genetic and/or environmental differences between individuals. However, animal models of RA are well suited for analysis of differential gene expression because it is possible to analyze many individuals in which genetic background, environmental exposure and disease stage can be controlled. One animal model with close similarity to the human disease is pristane-induced arthritis (PIA) in rats [5]. It is an erosive arthritis that specifically affects peripheral joints in a symmetrical manner; rheumatoid factors are elevated and it develops into a chronic, relapsing disease. In addition, it is associated with a strong acute inflammatory response [6] and fragments of cartilage oligomeric matrix protein are released into blood as a reflection of joint erosion [7]. Although autoantigens in PIA, as in RA, have not been identified, the disease is MHC class II associated, it is dependent on the activation of αβ T cells and it can be transferred. The DA rat is 100% susceptible to the disease whereas the E3 rat is resistant. The DA and E3 combination has been extensively analyzed for genetic susceptibility, and many loci that are linked to various inflammatory diseases have been identified [8,9]. In both PIA and experimental autoimmune encephalomyelitis it has been demonstrated that different loci control different disease subtrates, such as onset, severity and chronicity.
In the present study, inguinal lymph nodes from the PIA-susceptible DA rat and from the PIA-resistant E3 strain were analyzed for differential gene expression using Affymetrix technology (Affymetrix, Inc., Santa Clara, CA, USA). Both naïve rats and rats 8 days after pristane injection were studied. The postinjection time point was selected at 2–6 days before onset of PIA. The reason for this was that by then an immune reaction has started but no secondary effects of disease have yet occurred. The draining inguinal lymph nodes were chosen because we believe that they are important in the early phases of disease. Optimally, microarray analysis should be conducted in isolated populations of cells so that differential gene expression may be directly correlated with transcription of the genes; however, as discussed elsewhere [10], in complex diseases such as RA the important cell types are not known, and therefore analysis of a complex tissue increases the probability of analyzing differential gene expression in cells of importance for pathogenesis. By fluorescence activated cell sorting (FACS) analysis of the composition of the most important immunological cell types in the lymph nodes, we attempted to identify eventually apparent differential expressions caused by differences in cell numbers. Differential mRNA expressions in the lymph nodes were detected by Affymetrix and validated by custom-made oligomer glass arrays. Some of the detected genes were also analyzed by real-time PCR for confirmation of the microarray results. Finally, the mRNA expression levels of some of the genes were correlated with protein expression, which was determined either by FACS or ELISA. To investigate further an eventual role of the differentially expressed genes in arthritis pathogenesis, the genomic locations of the genes were compared with the locations of previously reported quantitative trait loci (QTLs) for arthritis and other autoimmune diseases.
Materials and method
Rats
DA and E3 rat strains (from Zentralinstitut für Versuchstierzucht, Hannover, Germany) were kept in the animal facility of Medical Inflammation Research with 12-hour light–dark cycles; they were housed in polystyrene cages containing wood shavings and were fed standard rodent chow and given free access to water. All experiments were conducted in male rats aged 8 weeks. The ethics committee for animal experiments at Lund University approved all experiments.
Arthritis induction
To induce the immune reaction that leads to arthritis in the DA rat, 150 μl pristane (2,6,10,14-tetramethylpentadecane) was injected intradermally at the base of the tail. The pristane was purchased from Aldrich (Milwaukee, WI, USA).
RNA isolation
The inguinal lymph nodes from both sides of untreated rats and rats 8 days after pristane injection were removed and stored in RNAlater™ (Ambion, Austin, TX, USA) at -80°C until homogenization. The homogenate was phenol : chloroform : isoamylalcohol (Ambion) extracted before total RNA isolation using the RNeasy kit (Qiagen, Valencia, CA, USA). The purity and concentration of RNA was determined by A260/280 analysis and the RNA quality was analyzed by agarose gel electrophoresis.
Affymetrix analysis
Total RNA (20 μg/chip) was synthesized to cRNA and labelled according to GeneChip® expression analysis (technical manual provided by Affymetrix). For pooled samples, total RNA from six rats was combined before labelling. Pooled samples were analyzed for naïve rats and rats 8 days after pristane injection. For the pristane-treated rats, chips with three individual rats per strain were also analyzed. The samples were hybridized to the rat genome chip RG_U34A, and washed and stained according to protocols indicated by Affymetrix.
Custom-made chip analysis
Custom-made, 50mer oligomer chips were produced by MWG Biotech AG (Ebersberg, Germany). For selection of the optimal oligomers, the bioinformatic service of MWG Biotech AG was used and the oligomers were chosen in nonpolymorphic regions. Total RNA was labelled by direct incorporation of Cy3 or Cy5 during cDNA synthesis. Briefly, 40 μg total RNA from naïve animals and 50 μg from pristane-treated animals were pre-annealed with a poly dT primer, mixed with reverse transcriptase, deoxynucleotides, RNase inhibitors and dUTP-Cy3 or dUTP-Cy5. After labelling, the RNA was hydrolyzed and the Cy3 and Cy5 labelled probes were washed, combined and hybridized to the chips according to the MWG Biotech AG manual http://www.mwg-biotech.com/. The hybridized chips were scanned in a laser confocal scanner (Virtek ChipReader™, Waterloo, Ontario, Canada). Five individual DA rats were compared with a pool of five E3 rats. For each comparison, a dye-swap experiment was done.
Taqman analysis
To examine the reliability of the microarray method, a selection of five genes, differentially expressed according to Affymetrix, were selected for Taqman analysis (Applied Biosystems, Foster City, CA, USA). The genes chosen were as follows: Primase, Ccng1, Ig rearranged γ-chain variable region, Mmp9 and Bst1. Primers and Taqman probes were chosen by the Primer Express™ 1.5 software (PE Applied Biosystems, CA, USA) and synthesized by PE Applied Biosystems. The probes were FAM6/TAMRA labelled. Five animals from each group were analyzed individually.
Flow cytometry analysis
The cell type composition and level of protein expression for some genes in the inguinal lymph nodes were analyzed by flow cytometry. Single cell suspensions were washed and stained for various phenotypes using FITC, PE, or biotinylated conjugated mouse antirat monoclonal antibodies purchased from Pharmingen (San Diego, CA, USA), American Type Culture Collection (Manassas, VA, USA) and kindly donated by Dr Torres-Nagel (Institute for Virology and Immunobiology, University of Wurzburg, Germany). The lymph nodes were analyzed for the total numbers of leucocytes (OX-1+), granulocytes (HIS48+), B cells (OX-33+), αβ T cells (R73+), CD4+ cells of αβ T cells (OX-35+gated on R73+), CD8+ cells of αβ T cells (OX8+ gated on R73+), γδ T cells (V65+, OX-19+), NK cells (3.2.3+, R73-) and NK T cells (3.2.3+, R73+). The differential geometric mean values were analyzed for RT1.B (OX-6+), RT1.D (OX-17+), CD28 (JJ319+), B7.2 (24F+), CD45RC (OX-22+), anti-IgM (MAR18.5+) and interferon-γ (DB-1). Acquisition was made using FACSort (Becton Dickinson, Franklin Lakes, NJ, USA), using the BD Cell-Quest™ Pro, Version 4.0.1 software (Becton Dickinson). Three individuals per time point and strain were analyzed.
Enzyme-linked immunosorbent assay analysis
The concentration of total IgG, total IgM and total IgE in plasma from DA and E3 rats on days 0 and 8 after pristane injection were analyzed by ELISA. Plasma from heparinized blood were separated by centrifugation and stored at -20°C until analysis. Total IgG and IgM were analyzed by coating with 5 μg/ml purified goat antirat IgG (Southern Biotechnology Associates Inc., Birmingham, AL, USA) or mouse antirat IgM (Pharmingen), followed by incubation with 10,000 × diluted plasma, secondary peroxidase conjugated goat antirat IgG or IgM (Jackson ImmunoResearch Inc., West Grove, PA, USA) and substrate ABTS® tablets (Roche Diagnostics GmbH, Mannheim, Germany). The total IgE ELISA followed the main protocol; plates were coated with 5 μg/ml mouse antirat IgE (Pharmingen), followed by incubation with 6000 × diluted plasma, 2 μg/ml biotin conjugated mouse antirat IgE secondary antibody (Pharmingen), extravidinperoxidase and substrate incubation. The ELISA findings were analyzed in a Titertek Multiskan® Plus ELISA plate reader (Molecular Devices Corporation, Sunnyvale, CA, USA) at 405 nm. Four rats per group were analyzed and all ELISA samples were run in duplicate.
Statistical analysis
The Affymetrix data were analyzed by the Microarray Suit Software Version 4.0. To normalize the chips the global scaling option was used. Statistical analysis of individual animal per strain chips was done using the dChip software [11]. The probe sets were sorted by the absolute t-statistic. The custom-made chips data were analyzed using the ImaGene™ software (BioDiscovery Inc., Los Angeles, CA, USA). The data were processed using the 'local background subtraction' and normalized using the 'all spot' option. The signal for each gene was calculated as the average signal of each spot of the three oligomers from both dye-swap chips. The mean signal from the five individual DA rats was divided by the corresponding E3 pool signal. These ratios were tested by one group t-test using the null hypothesis that the DA/E3 ratio was equal to one (i.e. no difference). Statistical analysis on the Taqman findings was done by comparison of the two groups of individual DA and E3 rats using Mann–Whitney U-test. Statistical analysis of the flow cytometry data was done by analysis of the cell numbers and expression of certain molecules between the groups of individual DA and E3 rats using Mann–Whitney U-test. Statistical analysis of the immunoglobulin ELISAs was done by calculating the average of the ELISA duplicates and analyzing these values between the individuals in the DA and E3 groups by Mann–Whitney U-test. For statistical analysis, the StatView software was used (SAS Institute Inc., Cary, NC, USA).
Differentially expressed genes: quantitative trait locus association
The mapping of the differentially expressed genes to QTLs for autoimmune disease was investigated. The genomic location of the genes were searched for using Map Viewer http://www.ncbi.nlm.nih.gov/mapview/map_search.cgi?chr=rat.inf. The genomic positions were compared with reported positions of QTLs for arthritis and multiple sclerosis using RatQTL http://ratmap.gen.gu.se/qtler/.
Results
Cell composition of the inguinal lymph nodes of the DA and E3 rat strains
To distinguish between true differential gene expression and apparent differential gene expression caused by difference in cell numbers, the cell compositions of the inguinal lymph nodes from the DA and E3 rats were investigated by flow cytometry. Several interesting differences between the DA and E3 rats were found (Table 1). In naïve rats (day 0), the number of granulocytes and NK T cells was significantly lower in the DA rats than in the E3 rats. Furthermore, the number of αβ T cells was significantly higher in the DA rats. These cells were probably CD4+ cells because this number was higher in the DA rat whereas no difference could be detected in the numbers of CD8+ T cells. Also, a marginally lower number of γδ T cells were detected in DA rats. Eight days after pristane injection, the numbers of NK T cells and γδ T cells were significantly lower in the DA rats than in the E3 rats, but the most striking difference in cell composition was the markedly lower number of B cells in DA rats 8 days after pristane injection. The low number of B cells contributed to the general lower level of immunoglobulin gene expression in the DA rats 8 days after pristane injection (see below), but no other clearcut correlation was observed between cell numbers and differential gene expression.
Table 1.
Total number of cells in lymph nodes in DA and E3 naïve rats and rats 8 days after pristane injection
| Total cell number (millions) | ||||||
| Cell type | DA day 0* | E3 day 0* | Difference between DA and E3 on day 0** | DA day 8 | E3 day 8 | Difference between DA and E3 on day 8** |
| Leucocytes | 10 ± 1.5 | 8 ± 0.79 | NC, P < 0.1 | 27 ± 3.6 | 45 ± 21 | NC, P < 0.3 |
| Granulocytes | 0.034 ± 0.01 | 0.052 ± 0.01 | D, P < 0.04 | 0.48 ± 0.14 | 0.92 ± 0.62 | NC, P < 0.5 |
| B cells | 1.8 ± 0.64 | 2.3 ± 0.29 | NC, P < 0.25 | 6.3 ± 1.2 | 24 ± 13 | D, P < 0.03 |
| αβ T cells | 7.9 ± 0.7 | 5.4 ± 0.3 | I, P < 0.02 | 20 ± 1.3 | 19 ± 2.8 | NC, P < 1 |
| CD4+/αβ T cells | 6.2 ± 0.5 | 4.3 ± 0.2 | MI, P < 0.08 | 14 ± 0.9 | 13 ± 1.7 | NC, P < 0.3 |
| CD8+/αβ T cells | 2.2 ± 0.2 | 1.7 ± 0.1 | NC, P < 0.2 | 6.1 ± 0.7 | 7.5 ± 1.4 | NC, P < 0.3 |
| γδ T cells | 0.024 ± 0.013 | 0.038 ± 0.0086 | MD, P < 0.1 | 0.048 ± 0.018 | 0.24 ± 0.16 | D, P < 0.03 |
| NK cells | 0.095 ± 0.034 | 0.12 ± 0.01 | NC, P < 0.4 | 0.46 ± 0.07 | 1.51 ± 1.0 | NC, P < 0.2 |
| NK T cells | 0.054 ± 0.019 | 0.12 ± 0.024 | D, P < 0.02 | 0.19 ± 0.035 | 2 ± 1.2 | D, P < 0.03 |
*Mean total cell number (in millions) ± standard deviation (SD) determined by fluorescence activated cell sorting (FACS) analysis. Data based on three DA and E3 naïve rats and three DA and E3 rats 8 days after pristane injection. **Difference in cell number: DA versus E3. P values calculated by Mann–Whitney nonparametric test. D, decrease; I, increase; MD, marginally decrease; MI, marginally increase; NC, no change.
Experimental design
First, the mRNA expression was analyzed by Affymetrix in pooled samples of naïve animals, and in both pooled and individual samples for pristane-treated rats. However, the results from the pooled and individual samples of pristane-treated animals were not regarded as independent observations because the biological samples were partly the same. An independent method, employing custom-made oligomer glass chips, was used to confirm the differential mRNA expression of both naïve and pristane-treated animals. In these custom-made chips there were spotted oligomers for 170 genes, which had been found to be differentially expressed in various Affymetrix analyses or were regarded as potentially interesting for arthritis. (For the complete list of genes on these chips, see Additional file 5.) Sequences of the selected oligomers are available on request from the corresponding author.
All annotated genes differentially expressed with a change greater than threefold in the pristane-treated rats and approximately half of the genes differentially expressed in the naïve rats were represented on the chips. The chips were used to analyze individual DA rats and compare them with a pool of E3 rats. For the pristane-treated rats a selection of five genes was also analyzed by real-time PCR. The differential mRNA expression of some genes of interest was compared with the expression of the encoded protein using either FACS or ELISA. The statistic significance threshold values for differential expression with the individual methods were set as follows: Affymetrix pooled samples, fold change > 3.0; Affymetrix individual samples (among the 30 most probably differentially expressed genes), t-test P < 0.002; and custom-made chips, real time PCR, FACS and ELISA, P < 0.05. To be regarded as differentially expressed between the DA and E3 rat, a gene should differ in the same direction between two different biological samples (taken at two time points) when analyzed by at least two different methods. These genes are denoted 'S' in the Tables 3 and 4. An additional set of genes showed a strong tendency toward differential expression. These genes were differentially expressed in two biological samples and fulfilled the threshold values for significance for one method and fulfilled the following threshold value for at least one additional method: pooled sample Affymetrix fold change values, >1.9; Affymetrix individual samples (among the 30 most probably differentially expressed genes), t-test P < 0.002; and custom-made chip, real-time PCR, FACS and ELISA, P < 0.1. These genes are denoted 'T' in Tables 3 and 4.
Table 3.
Genes differentially expressed in naïve (day 0) DA versus E3 rats
| Accession no. | Gene ID | Significance | Affymetrix | Custom-made | FACS | ELISA |
| Upregulated | ||||||
| X56596 | MHC class II antigen RT1.B-1 β-chain (MhcII) | T | I (56.8) | NC | I (1.2); P < 0.08 | ND |
| X00923 | IgE heavy chain (Ig ε) | S | I (3.6) | I (1.9); P < 0.1 | ND | I (1.4); P < 0.05 |
| D49955 | Bone marrow stromal cell antigen 1 (Bst1) | T | I (3.7) | I (2.0); P < 0.1 | ND | ND |
| Downregulated | ||||||
| S81289 | IgM κ chain variable region (CDR1 to CDR3 region; Ig κ) | S | D (-3.6) | D (-1.6);P < 0.0005 | NC | ND |
To be regarded as differentially expressed a gene must be differentially expressed in two biological samples analyzed by a minimum of two independent methods. Differential expression values in bold text are statistically significant by that particular method. Genes significantly differentially expressed by a minimum of two independent methods are denoted 'S'. Another subset of genes showed a strong tendency toward differential expression; these are denoted 'T'. ELISA, enzyme linked immunosorbent assay; FACS, fluorescence activated cell sorting; NC, no change (for these genes, no statistically significant differential expression could be shown); ND, not determined.
Table 4.
Genes differentially expressed in DA versus E3 rats 8 days after pristane injection
| Affymetrix | |||||||
| Accession no. | Gene ID | Significance | Pooled | Individual | Taqman | Custom-made | FACS |
| Upregulated | |||||||
| X56596 | MHC class I antigen RT1.B-1 β-chain (MhcII) | S | I (47.4) |
I (43.4); P < 0.0001 |
ND | I (2.8); P < 0.1 |
I (1.9); P < 0.03 |
| U24441 | Metalloproteinase 9 (Mmp9) | T | I (5) |
I (1.5); P < 0.001 |
I (1.9); P < 0.08 |
NC‡ | ND |
| S81478 | CL100 protein tyrosine phosphatase (Ptpn16) | T | I (2.4) | NC* | ND |
I (1.6); P < 0.02 |
ND |
| Downregulated | |||||||
| L07399 | (Hybridoma 56R-7) immunoglobulin rearranged γ-chain variable (Ig γ) |
S | D (-6.4) | NC* | ND |
D (-2.0); P < 0.02 |
ND |
| L07401 | (Hybridoma 57R-1) immunoglobulin rearranged γ-chain variable (Ig γ) |
S | D (-62.4) | NC* |
D (-2.1); P < 0.03 |
NC‡ | ND |
| M14434 | Immunoglobulin active κ-chain VJC region from immunocytoma IR2 (Ig κ) |
S | D (-34.7) | NC* | ND |
D (-1.9); P < 0.003 |
ND |
| X00923 | IgE heavy chain (Ig ε) | S | D (-28) | NC* | ND |
D (-2.0); P < 0.05 |
ND |
| S81289 | IgM κ-chain variable (Ig κ) | S | D (-9.3) | NC* | ND |
D (-2.5); P < 0.0003 |
ND |
| M15402 | Immunoglobulin active κ-chain VJ region from immunocytoma IR162 (Ig κ) |
S | D (-3.9) | NC* | ND |
D (-3.0); P < 0.006 |
ND |
| nm_013121 | CD28 (Cd28) | S | NC | NC* | ND |
D (-2.2); P < 0.01 |
D (-9.8); P < 0.03 |
| S69206 | Mast cell protease 1 (Mcpt1) | S | D (-4.7) | NC* | ND |
D (-2.1); P < 0.002 |
ND |
| AB010635 | Carboxylesterase precursor (Ces2) | S | D (-4.1) | NC* | ND |
D (-2.5) ; P < 0.003 |
ND |
| D25290 | K-cadherin (Cdh6) | S | D (-4.1) | NC* | ND |
D (-1.7) ; P < 0.01 |
ND |
| X70871 | Cyclin G1 (Ccng1) | S | D (-9.6) |
D (-4.8); P < 0.001 |
D† | NC‡ | ND |
| AJ011608 | DNA polymerase α-subunit IV (Primase) | S | D (-3.3) |
D (-2.6); P < 0.001 |
D (-3.5); P < 0.03 |
NC‡ | ND |
| L12025 | Tumour-associated glycoprotein E4 (Tage) | T | D (-3.1) | NC* | ND | D (-1.8); P < 0.1 |
ND |
To be regarded as differentially expressed a gene must be differentially expressed in two biological samples analyzed by a minimum of two independent methods. Differential expression values in bold text are statistically significant by that particular method. Genes significantly differentially expressed by a minimum of two independent methods are denoted 'S'. Another subset of genes showed a strong tendency toward differential expression; these are denoted 'T'. FACS, fluorescence activated cell sorting; NC, no change; ND, not determined. *These genes were not included among the 30 genes with highest probability of differential expression (t-test P < 0.002). †This gene was only detectable in the E3 rat. ‡For these genes, no statistically significant differential expression could be shown.
Differential gene expression between naïve DA and E3 rats
The differential mRNA expressions between naïve DA and E3 rats were analyzed and compared with protein cell surface expression or plasma protein levels. The numbers of statistically significant differentially expressed genes for the individual methods are indicated in Table 2. (The complete lists of genes are provided in Additional files 1, 6, 8 and 9.) Genes that were differentially expressed in two different biological samples of naïve rats, as observed using at least two independent methods, and fulfilling the threshold values for the 'S' group and 'T' group', as defined above, are shown in Table 3.
Table 2.
The number of statistically significant differentially expressed genes in naïve and pristane-treated rats according to the individual methods
| Method | Higher in naïve DA rats | Lower in naïve DA rats | Higher in pristane-treated DA rats | Lower in pristane-treated DA rats |
| Affymetrix (pooled samples) | 15 (8800) | 24 (8800) | 38 (8800) | 65 (8800) |
| Custom-made chips | 19 (170) | 2 (170) | 2 (170) | 23 (170) |
| Real-time PCR | 0 | 0 | 0 (5) | 3 (5) |
| FACS | 4 (7) | 3 (7) | 3 (7) | 2 (7) |
| ELISA | Total IgE and IgM (3) | 0 (3) | Total IgM (3) | Total IgG (3) |
To be regarded as differentially expressed with statistical significance, a gene had to fulfil the following threshold values: Affymetrix (pooled samples), differentially expressed with a fold change > 3.0; custom-made oligomer glass chips, differentially expressed with one group t-test P < 0.05; real time PCR, differential expression with Mann–Whitney P < 0.05; FACS, differential geometric mean values with a Mann–Whitney P < 0.05; ELISA, differential plasma concentrations with a Mann–Whitney P < 0.05. The values within parenthesis represent the total number of genes analyzed using the method. ELISA, enzyme linked immunosorbent assay; FACS, fluorescence activated cell sorting.
Only one gene, namely the IgM κ-chain variable region, exhibited significantly lower expression in DA rats. In addition, one gene – the IgE heavy chain – was significantly highly expressed in the DA rat, and the MHC class II antigen RT1.B β-chain and Bst1 showed a strong tendency toward higher expression. FACS analyses confirmed the higher expression of MHC class II antigen RT1.B β-chain in the DA rat, but failed to confirm the differential expression of the κ-chain (Table 3, Fig. 1). Furthermore, it was interesting to note that, although lymph nodes are not the main source for immunoglobulin secretion into plasma, the differential expression of IgE was reflected in the secretion of IgE into plasma (Table 3, Fig. 1).
Figure 1.
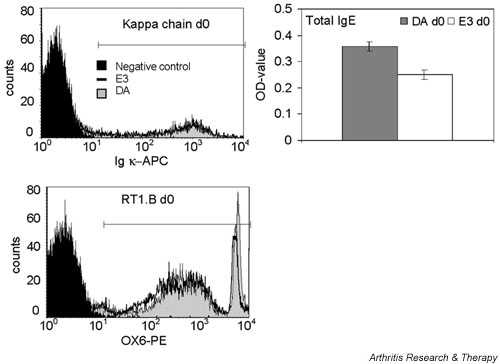
Differential expression of some proteins between naïve DA and E3 rats was investigated by fluorescence activated cell sorting (FACS) or ELISA. Differential κ-chain and MHC class II RT1.B expression was investigated using FACS. Inguinal lymph node cells from three DA and E3 rats were stained with anti-Igκ or OX-6 and analyzed on FACSort using the BD CellQuest™Pro Version 4.0.1 software (Becton Dickinson, Franklin Lakes, NJ, USA). In the histogram, the black peak represents the negative control, the filled grey peak the DA rats, and the unfilled peak the E3 rats. The horizontal line represent the area used for calculation of the geometric mean values. In the histogram the κ-chain and RT1.B expression of the DA and E3 rats with individual values closest to the group mean value are shown. The total plasma concentration of IgE was analyzed by ELISA. Plasma samples from four DA and E3 rats were analyzed. The data are presented as DA and E3 mean values, and the error bars represent the standard error of the mean.
Differential gene expression between DA and E3 rats 8 days after pristane injection
The differential mRNA expressions between DA and E3 rats 8 days after pristane injection were analyzed and compared with cell surface expression of some proteins and secretion of immunoglobulins into plasma. mRNA expression was analyzed by Affymetrix, custom-made chips and real-time PCR for a selection of genes. The protein expressions of cell surface molecules and the plasma concentrations of immunoglobulins were analyzed using FACS and ELISA. The numbers of significantly differentially expressed genes for the individual methods are indicated in Table 2. (The complete lists of genes are provided in Additional files 2, 3, 4, 7, 8 and 9.) Genes that were differentially expressed in two different biological samples of pristane-treated rats, as observed using at least two independent methods, and fulfilling the threshold values for the 'S' group and 'T' group, as defined above, are shown in Table 4.
One gene, namely the MHC class II RT1.B β-chain (MhcII), had significantly higher expression in the DA rat using the X56596 oligomer sets in Affymetrix; two additional genes – the gelatinase B (Mmp9) and the CL100 protein tyrosine phosphatase (Ptpn16) genes – exhibited a strong tendency toward higher expression. For the MHC class II RT1.B β-chain, Affymetrix analysis yielded a contradictory result when the 'U65217_i_at' probe set was used. Affymetrix indicated a highly significant lower expression in the DA rat than in the E3 rat, with unrealistic fold change values, whereas custom-made chips and FACS data indicated a higher expression (Table 4, Fig. 2). The reason for this was an unfortunate placement of the 'U65217_i_at' oligomers in a highly polymorphic part of the MHC gene (see below), and therefore only findings with the 'X56596_at' set were regarded as reliable. Furthermore, a number of genes were found to have significantly lower expression in the DA rat: six immunoglobulin genes, the costimulatory molecule Cd28, mast cell protease 1 (Mcpt1), K-cadherin (Cdh6), the carboxylesterase precursor (Ces2), and the cell cycle related genes cyclin G (Ccng1) and Primase. One more gene, the tumour-associated glycoprotein E4 gene (Tage), showed a tendency toward lower expression in the DA rat. The lower mRNA expression of immunoglobulins was reflected in plasma as a significantly lower total IgG concentration and a tendency toward lower IgE concentration (Table 4, Fig. 2), and flow cytometry confirmed the lower expression of Cd28. The lower expression level of immunoglobulins was most likely caused by the decreased number of B cells in the DA rat as compared with the E3 rat (Table 1).
Figure 2.
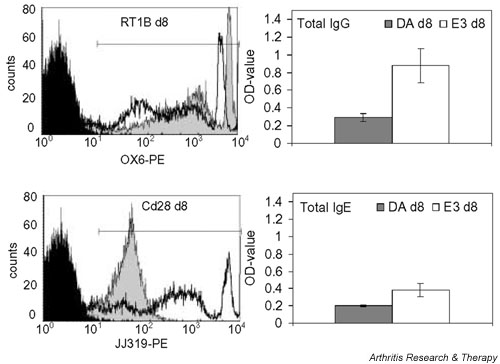
Differential expression of some proteins between pristane-treated DA and E3 rats was investigated by fluorescence activated cell sorting (FACS) or ELISA. Differential MHC class II RT1.B and Cd28 expression was investigated using FACS. Inguinal lymph node cells from three DA and E3 rats were stained with OX-6 or JJ319 and analyzed on FACSort using the BD CellQuest™ Pro Version 4.0.1 software (Becton Dickinson, Franklin Lakes, NJ, USA). In the histogram the black peak represents the negative control, the filled grey peak the DA rats, and the unfilled peak the E3 rats. The horizontal line represent the area used for calculation of the geometric mean values. In the histogram the RT1.B and Cd28 expression of the DA and E3 rats with individual values closest to the group mean value are shown. The total plasma concentration of IgG and IgE was analyzed by ELISA. Plasma samples from four DA and E3 rats were analyzed. The data are presented as DA and E3 mean values and the error bars represent the standard error of the mean.
Sequence alignment revealed problems with Affymetrix oligomer set
A discrepancy between Affymetrix and custom-made chip data for the MHC class II RT1.B β-chain and some other genes of immunological importance was observed. For the MHC class II RT1.B β-chain a simple but intriguing explanation was identified. The MHC, like many key genes of the immune system, is polymorphic; the DA rat is of the a-haplotype and the E3 rat is of the u-haplotype. To investigate the cause of the contradictory data observed for the MHC, careful sequence alignments were made. The Affymetrix and custom-made chip oligomer sets for U65217 (u-haplotype) and X56596 (l-haplotype) were aligned to the U65217, X56596 and the a-haplotype exon 2 (M76780) sequences. This revealed that the Affymetrix oligomers for the U65217 gene were all placed in the same region, covering a total of 44 bases. In this area there is a high degree of polymorphism between different haplotypes, making it unlikely that U65217 (u-haplotype) would also bind a-haplotype coded RNA. Accordingly, this would give the false result of a much lower expression of the MHC class II RT1.B β-chain in the DA rat than in the E3 rat. On the other hand, the Affymetrix oligomers for X56596 were spread out, covering 150 bases in a basically nonoverlapping manner, and the custom-made chip oligomer sets were placed so that at least two oligomers per gene were not overlapping. Accordingly, these oligomer sets are less sensitive to polymorphism and more likely to provide reliable results.
Differentially expressed genes: quantitative trait locus association
Thirteen unique genes fulfilled the threshold values for significance of differential expression or showed a strong tendency toward being differentially expressed. To further investigate the possibility that these genes play a vital role in arthritis pathogenesis, their genomic locations were compared with those of rat QTLs that have been reported to be associated with arthritis (PIA, collagen-induced arthritis, adjuvant-induced arthritis and oil-induced arthritis, anticollagen antibody titres), and experimental allergic encephalomyelitis. Interestingly, nine of the genes were found to be located within autoimmune disease QTLs, as identified previously in various rat crosses (Fig. 3). Four genes are within PIA QTLs: MhcII (the Pia1/Eae1/ Aia1/Cia1/Ciaa1 locus [8,9,12,13]), the immunoglobulin heavy chain (the Pia3/Eae9 locus [8,9]), the immunoglobulin κ-chain (the Pia5/Pia7/Cia13/Ciaa4/Cia3/Aia3 locus [8,12-16]) and Bst1 (the Piax locus [17]). In addition, five more genes (Tage, Ces2, Cd28, Ptpn16 and Ccng1) are mapped to QTLs of other autoimmune disease models (the Cia2 [13], Eae6 [9], Ciaa3 [15], Cia16/Eae3 [16,18] and Aia5 [19] loci, respectively).
Figure 3.
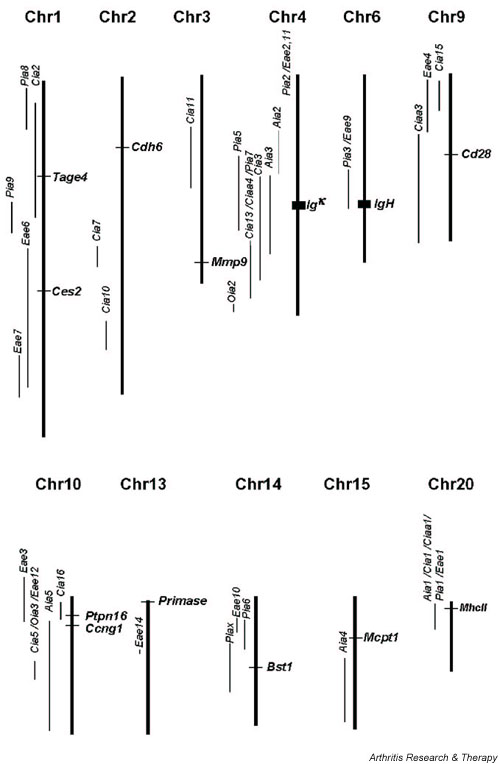
Thirteen genes were found to be differentially expressed between the DA and E3 rats before and/or 8 days after pristane injection. The genomic location of these genes (found using Map Viewer: http://www.ncbi.nlm.nih.gov/mapview/map_search.cgi?chr=rat.inf was mapped to various quantitative trait loci (QTLs) for autoimmune disease found in rat crosses. The figure shows QTLs on the same chromosome as the differentially expressed genes. The position of the flanks of the QTLs reported in ratQTL http://ratmap.gen.gu.se/qtler/ were used to set the borders.
Discussion
By using microarray and real-time PCR, we investigated differential gene expression in the draining inguinal lymph nodes between the arthritis-susceptible DA and arthritis-resistant E3 rats before and after pristane injection. This resulted in a list of 17 (13 different) genes that were differentially expressed or exhibited a strong tendency toward being so in at least two different biological samples analyzed using at least two independent methods. In order to produce reliable data, we conducted an initial scan with Affymetrix followed by statistical analysis and confirmation of the data with independent custom-made chips. Alternatively, statistical analysis could have been conducted by running a large number of Affymetrix chips, but this approach is rather expensive and would not have excluded artefacts such as those illustrated by the MHC class II gene example. In addition, Affymetrix often appears to yield unrealistic fold change values, which may be caused by an unfortunate placement of oligomers and/or the algorithm for calculating them. Custom-made chips, on the other hand, give more realistic fold change values and are not likely to produce the same artefacts because the selection of oligomers, labelling and scanning procedures are independent of Affymetrix. Accordingly, a strategy in which Affymetrix and custom-made chips are used in combination is likely to produce more reliable data than when either technique is used alone. In addition, real-time PCR was used to confirm the microarray data for a few genes. Clearly, this approach employing several methods diminishes the number of differentially expressed genes, but it is more likely to result in a list of genes that are truly differentially expressed. Efficient exclusion of false positives early in the process saves time and effort later.
The cell composition of the inguinal lymph nodes was analyzed to determine whether an apparent differential expression was caused by differential gene transcription or by an altered cell number. A strain dependent difference in a few cell types was observed in naïve rats. The DA rats had more αβ T cells than did the E3 rats, and had fewer granulocytes and NK T cells. By comparing the cell compositions of pristane-treated and naïve rats, several interesting conclusions could be made. The differences in numbers of some cell types (e.g. the NK T cells and γδ T cells) became more pronounced in pristane-treated rats. For other cell types (e.g. granulocytes and CD4+ αβ T cells) the difference in cell number had disappeared after pristane treatment, and for B cells a huge difference in number of B cells became apparent. The large differences in cell compositions of inguinal lymph nodes between naïve and pristane-treated rats argues for an important role for this tissue in the early events that take place after pristane challenge. Furthermore, the differential cell composition between the DA and E3 rats is interesting per se and highlights ongoing mechanisms. The disappearing difference in CD4+ αβ T cells is interesting because these cells are believed to play an important role in both adjuvant arthritis and RA [20-22]. The more pronounced difference in number of NK T cells and γδ T cells is in agreement with data from oil-induced arthritis resistant and susceptible rat strains [23]. Finally, the large difference in B cells between pristane-treated DA and E3 rats is highly interesting. Clearly, the E3 rat responds to pristane challenge with a substantial increase in the number of B cells; this reaction is much smaller in the DA rat. What consequences this will have for the rat's ability to counteract an arthritic inflammation is yet unknown and should be further investigated.
The differential gene expression of naïve rats is strain dependent and not necessarily related to arthritis. However, the MHC class II molecule has a huge record of correlation with arthritis [24,25], and a high density of this molecule on blood cells in early RA leads to greater disease activity later [26]. In addition, MHC haplotypes associated with RA are more densely expressed on the cell surface than haplotypes that are not associated [26]. Our data indicate that this is also true for the rat. The other genes that are expressed to a greater degree in naïve DA rat than in E3 rat, namely Bst1 and IgE, are not expressed to a higher degree in pristane-treated DA rats. The change in expression pattern indicates an involvement in arthritogenic mechanisms. The Bst1 molecule may be involved in the same mechanism as the recently cloned arthritis controlling gene Ncf1 [27] because it supports NADPH oxidase catalyzed superoxide generation [28], among several other interesting functions. In addition, a higher expression of Bst1 has been associated with severe RA in patients [29]. The lower expression of the immunoglobulin κ-chain in naïve animals could reflect a marginally lower B cell number between naïve rats, but this is not in agreement with a higher IgE mRNA and protein expression.
All molecules, except for the MHC class II RT1.B β-chain, that were found to be differentially expressed between pristane-treated DA and E3 rats were not differentially expressed between naïve rats. This strongly argues for that they are involved in the different responses of the two strains to pristane injection, which results in arthritis in the DA rat but not in the E3 rat. It is therefore tempting to speculate on their potential role in the early events that lead to arthritis.
Two genes, in addition to MHC, were expressed at a higher level in the pristane-treated DA rats, Mmp9 and Ptpn16. Mmp9 is a granulocyte-secreted type IV collagenase and Ptpn16 is induced by oxidative stress and inflammation [30,31] to act as a negative feedback regulator of mitogen-activated protein kinases [32]. Mmp9 has the potential to promote arthritis in many ways and its role in early autoimmune disease is supported by Mmp9 knockout mice, which show less susceptibility to experimental autoimmune encephalomyelitis [33]. The higher expression of the Ptpn16 gene, instead of directly promoting arthritis, could represent an unsuccessful attempt by the DA rat to switch off the pristane-induced reaction.
The largest group of differentially expressed genes in pristane-treated rats were expressed at a lower level in the DA rat. Several of the genes are intimately associated with an active immune defence, which indicates that the arthritis resistance of the E3 rat is not caused by lack of response but rather by a strong immune reaction against insults such as pristane that protects the body from autoimmune disease. The large difference in B cell numbers, and subsequent immunoglobulin expression, indicates that the response is B cell mediated, and several of the differentially expressed genes can provide clues as to the pathways involved. The Cd28 molecule, as a costimulator of T cells, may be essential for the (protective) B cell response of the E3 rat. Interestingly, this molecule has also been found to be downregulated in RA patients [34]. The lower expression of Mcpt1 [35] supports recent observations that mast cells are important for arthritis. Interestingly, at least two subgroups of mast cells with different roles in arthritis have been identified, and these subgroups are distinguished by their differential Mcpt1 expression [36]. The adhesion between immune cells also appears to be important for the process because the cell adhesion molecule Cdh6 [37] is expressed to a lower degree in the DA rat. In addition to a direct association with the immune system, a differential regulation of apoptosis between the DA and E3 rat is indicated because Ccng1 is differentially expressed. Ccng1 has been suggested as a negative feedback regulator of p53 [38], and therefore lower expression of Ccng1 in the DA rat would lead to more apoptosis in the lymph nodes just before arthritis onset. This stands in contradiction to observations in chronically inflamed synovium in which defective apoptosis was suggested to drive the inflammation [39]. The exact role played by Ccng1 in the cell cycle and various phases of arthritis must therefore be further investigated. Finally, the lower expression of a carboxylesterase in the DA rat may reflect another interesting issue – a defective drug and lipid metabolizing system. A defect in such a system would lead to an impaired ability to handle lipophilic, potentially harmful substances, which may lead to development of arthritis. If this turns out to be true, then it will nicely reflect how genetic and environmental factors interact in diseases such as arthritis.
Finally, 9 out of 13 identified candidate genes were mapped to genomic regions previously reported to be QTLs for autoimmune diseases. Although it is premature to suggest that the nine genes are candidate genes for these QTLs, the data indicate that gene expression profiling will be useful to assist in cloning of genes from QTLs. Most of the QTLs were isolated in congenic strains [40-45] and work is ongoing to clone the genes positionally. Gene expression profiling will clearly speed up the cloning of genes by assisting in selection of recombinants, and will give additional valuable information regarding the pathways in which the genes are operating.
Conclusion
We showed that a rather simple microarray analysis is useful in identifying interesting pathways and candidate genes that operate early in animal models of arthritis. Several of the identified genes could be mapped to QTLs, and therefore microarray analysis may be a valuable tool for cloning arthritis controlling genes and improving our understanding of their biological pathways. Therefore, microarray analysis of animal models has the potential to significantly improve our understanding of arthritis pathogenesis and contribute to better treatment of RA.
Competing interests
None declared.
Abbreviations
ELISA = enzyme-linked immunosorbent assay; FACS = fluorescence activated cell sorting; NK = natural killer; PCR = polymerase chain reaction; PIA = pristane-induced arthritis; QTL = quantitative trait locus; RA = rheumatoid arthritis.
Supplementary Material
Affymetrix data from pooled RNA samples, prepared from inguinal lymph nodes of six naïve DA rats and six naïve E3 rats. The differentially expressed genes are presented as DA versus E3 rats. All genes with a fold change >1.9 are presented. Genes with a fold change >3.0 are regarded as differentially expressed in this method, and genes with a fold change >1.9 but <3.0 are regarded as genes with a tendency toward differential expression. Genes marked with an 'x' are represented on the custom-made glass chip.
Affymetrix data from pooled RNA samples, prepared from inguinal lymph nodes of six DA rats and six E3 rats injected with 150 μl pristane 8 days before analysis. The differentially expressed genes are presented as DA versus E3 rats. All genes with a fold change >1.9 are presented. Genes with a fold change >3.0 are regarded as differentially expressed in this method, and genes with a fold change >1.9 but <3.0 are regarded as genes with a tendency toward differential expression. Genes marked with an 'x' are represented on the custom-made glass chip.
Affymetrix data from RNA samples, prepared from inguinal lymph nodes of three DA rats and three E3 rats injected with 150 μl pristane 8 days before analysis. The RNA samples were prepared and analyzed individually for these rats. The differentially expressed genes are presented as DA versus E3 rats. The Affymetrix data was analyzed statistically using the D-chip software and sorted on the absolute t-statistic. Fifteen genes with the highest probability of being downregulated and fifteen genes with the highest probability of being upregulated are presented. These thirty genes were regarded as differentially expressed by this method.
Taqman analysis was performed on five selected genes. Individual RNA samples from five pristane-injected DA and five E3 rats were analyzed. Differentially expressed genes with Mann–Whitney P < 0.05 are regarded as differentially expressed genes, and genes with P < 0.1 are regarded as genes with a tendency toward differential expression. *Cyclin G was expressed in all the E3 rats investigated, but it was not detectable in the DA rats.
The complete gene list of the custom-made glass oligomer chips. For each of these genes three different oligomers were constructed in the nonpolymorphic regions. The oligomer sequences are available on request.
Custom-made glass oligomer chip results from RNA samples, prepared from inguinal lymph nodes of five naïve DA rats and five naïve E3 rats. The DA rat RNA samples were prepared and analyzed individually in comparison with a pool of RNA from the five E3 rats. The differentially expressed genes are presented as DA versus E3 rats. The data were analyzed statistically using Statview software. All genes with t-test P < 0.05 are regarded as differentially expressed in this method, and genes with P > 0.05 but < 0.1 are regarded as genes with a tendency toward differential expression.
Custom-made glass oligomer chip results from RNA samples, prepared from inguinal lymph nodes of five DA rats and five E3 rats injected with 150 μl pristane 8 days before analysis. The DA rat RNA samples were prepared and analyzed individually in comparison with a pool of RNA from the five E3 rats. The differentially expressed genes are presented as DA versus E3 rats. The data were analyzed statistically using Statview software. All genes with t-test P < 0.05 are regarded as differentially expressed in this method, and genes with P > 0.05 but < 0.1 are regarded as genes with a tendency toward differential expression.
Fluorescence activated cell sorting (FACS) results. Three naïve and three pristane-treated individuals of each rat strain were analyzed by flow cytometry to determine whether differential detection on mRNA level was correlated with protein level. The differentially expressed genes are presented as DA versus E3 rats. The data were analyzed statistically using the Statview software. All genes with Mann–Whitney P < 0.05 are regarded as differentially expressed in this method, and genes with P > 0.05 but < 0.1 are regarded as genes with a tendency toward differential expression. D, decreased; I, increased; NC, no change (fold change value = 1).
ELISA result. Plasma from four naïve and pristane-treated individuals from each rat strain were analyzed by ELISA to determine whether the differential detection of immunoglobulins on mRNA level was correlated with the immunoglobulin concentrations in plasma. The data were analyzed statistically using the Statview software. All genes with Mann–Whitney P < 0.05 are regarded as differentially expressed in this method, and genes with P > 0.05 but < 0.1 are regarded as genes with a tendency toward differential expression. D, decreased; I, increased.
Acknowledgments
Acknowledgement
The authors wish to acknowledge Mrs Toth at the Department of Immunology in Rostock for technical assistance, the animal personnel at the animal facilities in Medical Inflammation Research in Lund, and Dr Ingemar Berglund at Arexis for sharing his expertise on Affymetrix analysis. This work was supported by grants from EU (EUROME QLG1-CT2001-01407) and Trygg Hansa, The Swedish Rheumatism Association, the Foundations of Crafoord, Professor Nanna Svartz, Nilsson-Ehle and King Gustaf V's 80-year Foundation. Dr Lena Wester is supported by a Marie Curie Fellowship (MCFH-1999-0130).
References
- Lee DM, Weinblatt ME. Rheumatoid arthritis. Lancet. 2001;358:903–911. doi: 10.1016/S0140-6736(01)06075-5. [DOI] [PubMed] [Google Scholar]
- Silman AJ, MacGregor AJ, Thomson W, Holligan S, Carthy D, Farhan A, Ollier WE. Twin concordance rates for rheumatoid arthrtis: results from a nationwide study. Br J Rheumatol. 1993;32:903–907. doi: 10.1093/rheumatology/32.10.903. [DOI] [PubMed] [Google Scholar]
- Aho K, Koskenvuo M, Tuominen J, Kaprio J. Occurrence of rheumatoid arthritis in a nationwide series of twins. J Rheumatol. 1986;13:899–902. [PubMed] [Google Scholar]
- Duggan DJ, Bittner M, Chen Y, Meltzer P, Trent JM. Expression profiling using cDNA microarrays. Nat Genet. 1999;21:10–14. doi: 10.1038/4434. [DOI] [PubMed] [Google Scholar]
- Vingsbo C, Sahlstrand P, Brun JG, Jonsson R, Saxne T, Holmdahl R. Pristane-induced arthritis in rats: a new model for rheumatoid arthritis with a chronic disease course influenced by both major histocompatibility complex and non-major histocompatibility complex genes. Am J Pathol. 1996;149:1675–1683. [PMC free article] [PubMed] [Google Scholar]
- Olofsson P, Nordquist N, Vingsbo-Lundberg C, Larsso A, Falkenberg C, Pettersson U, Åkerström B, Holmdahl R. Genetic links between the acute-phase response and arthritis development in rats. Arthritis Rheum. 2002;46:259–268. doi: 10.1002/1529-0131(200201)46:1<259::AID-ART10035>3.0.CO;2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vingsbo-Lundberg C, Saxne T, Olsson H, Holmdahl R. Increased serum levels of cartilage oligomeric matrix protein in chronic erosive arthritis in rats. Arthritis Rheum. 1998;41:544–550. doi: 10.1002/1529-0131(199803)41:3<544::AID-ART21>3.3.CO;2-R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vingsbo-Lundberg C, Nordquist N, Olofsson P, Sundvall M, Saxne T, Pettersson U, Holmdahl R. Genetic control of arthritis onset, severity and chronicity in a model for rheumatoid arthritis in rats. Nat Genet. 1998;20:401–404. doi: 10.1038/3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergsteinsdottir K, Yang HT, Pettersson U, Holmdahl R. Evidence for common autoimmune disease genes controlling onset, severity, and chronicity based on experimental models for multiple sclerosis and rheumatoid arthritis. J Immunol. 2000;164:1564–1568. doi: 10.4049/jimmunol.164.3.1564. [DOI] [PubMed] [Google Scholar]
- Eaves IA, Wicker LS, Ghandour G, Lyons P, Peterson LB, Todd JA, Glynne RJ. Combining mouse congenic strains and microarray gene expression analyses to study a complex trait: the NOD model of type 1 diabetes. Genome Res. 2002;12:232–243. 10.1101/gr.214102. Article published online before print in January 2002. [PubMed] [Google Scholar]
- Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci USA. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahito Y, Cannon GW, Gulko PS, Remmers EF, Longman RE, Reese VR, Wang J, Griffiths MM, Wilder RL. Localization of quantitative trait loci regulating adjuvant-induced arthritis in rats: evidence for genetic factors common to multiple autoimmune diseases. J Immunol. 1998;161:4411–4419. [PubMed] [Google Scholar]
- Remmers EF, Longman RE, Du Y, O'Hare A, Cannon GW, Griffiths MM, Wilder RL. A genome scan localizes five non-MHC loci controlling collagen-induced arthritis in rats. Nat Genet. 1996;14:82–85. doi: 10.1038/ng0996-82. [DOI] [PubMed] [Google Scholar]
- Nordquist N, Olofsson P, Vingsbo-Lundberg C, Petterson U, Holmdahl R. Complex genetic control in a rat model for rheumatoid arthritis. J Autoimmunity. 2000;15:1–8. doi: 10.1006/jaut.2000.0384. [DOI] [PubMed] [Google Scholar]
- Griffiths MM, Wang J, Joe B, Dracheva S, Kawahito Y, Shepard JS, Reese VR, McCall-Vining S, Hashiramoto A, Cannon GW, Remmers EF, Wilder RL. Identification of four new quantitative trait loci regulating arthritis severity and one new quantitative trait locus regulating autoanitbody production in rats with collagen-induced arthritis. Arthritis Rheum. 2000;43:1278–1289. doi: 10.1002/1529-0131(200006)43:6<1278::AID-ANR10>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- Furuya T, Salstrom JL, McCall-Vining S, Cannon GW, Joe B, Remmers EF, Griffiths MM, Wilder RL. Genetic dissection of a rat model for rheumatoid arthritis: significant gender influences on autosomal modifier loci. Hum Mol Genet. 2000;9:2241–2250. doi: 10.1093/oxfordjournals.hmg.a018915. [DOI] [PubMed] [Google Scholar]
- Lu S, Nordquist N, Holmberg J, Olofsson P, Pettersson U, Holmdahl R. Both common and unique susceptibility genes in different rat strains with pristane-induced arthritis. Eur J Hum Genet. 2002;10:475–483. doi: 10.1038/sj.ejhg.5200832. [DOI] [PubMed] [Google Scholar]
- Roth MP, Viratelle C, Dolbois L, Delverdier M, Borot N, Pelletier L, Druet P, Clanet M, Coppin H. A genome-wide search identifies two susceptible loci for experimental autoimmune encephalomyelitis on rat chromosomes 4 and 10. J Immunol. 1999;162:1917–1922. [PubMed] [Google Scholar]
- Joe B, Cannon GW, Griffiths MM, Dobbins DE, Gulko PS, Wilder RL, Remmers EF. Evaluation of quantitative trait loci regulating severity of mycobacterial adjuvant-induced arthritis in monocongenic and polycongenic rats: identification of a new regulatory locus on rat chromosome 10 and evidence of overlap with rheumatoid arthritis susceptibility loci. Arthritis Rheum. 2002;46:1075–1085. doi: 10.1002/art.10164. [DOI] [PubMed] [Google Scholar]
- Fox DA. The role of T cells in the immunopathogenesis of rheumatoid arthritis: new perspectives. Arthritis Rheum. 1997;40:598–609. doi: 10.1002/art.1780400403. [DOI] [PubMed] [Google Scholar]
- Holmdahl R, Goldschmidt TJ, Kleinau S, Kvick C, Jonsson R. Arthritis induced in rats with adjuvant oil is a genetically restricted, alpha beta T-cell dependent autoimmune disease. Immunol. 1992;76:197–202. [PMC free article] [PubMed] [Google Scholar]
- Pelegri C, Morante MP, Castellote C, Franch A, Castell M. Treatment with an anti-CD4 monoclonal antibody strongly ameliorates established rat adjuvant arthritis. Clin Exp Immunol. 1996;103:273–278. doi: 10.1046/j.1365-2249.1996.d01-624.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansson ÅM, Lorentzen JC, Bucht A. CD8+ cells suppress oil induced arthritis. Clin Exp Immunol. 2000;120:532–536. doi: 10.1046/j.1365-2249.2000.01241.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weyand CM, Goronzy JJ. Association of MHC and rheumatoid arthritis. HLA polymorphisms in phenotypic variants of rheumatoid arthritis. Arthritis Res. 2000;2:212–216. doi: 10.1186/ar90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKay K, Eyre S, Myerscough A, Milicic A, Barton A, Laval S, Barrett J, Lee D, White S, John S, Brown MA, Bell J, Silman A, Ollier W, Wordsworth P, Worthington J. Whole-genome linkage analysis of rheumatoid arthritis susceptibility loci in 252 affected sibling pairs in the United Kingdom. Arthritis Rheum. 2002;46:632–639. doi: 10.1002/art.10147. [DOI] [PubMed] [Google Scholar]
- Czerwony G, Alten R, Gromnica-Ihle E, Hagemann D, Reuter U, Sörensen H, Müller B. Differential surface expression of HLA-DRB1 and HLA-DRB4 among peripheral blood cells of DR4 positive individual. Hum Immunol. 1999;60:1–9. doi: 10.1016/S0198-8859(98)00096-2. [DOI] [PubMed] [Google Scholar]
- Olofsson P, Holmberg J, Tordsson J, Lu S, Åkerström B, Holmdahl R. Postitional identification of Ncf1 as a gene that regulates arthritis severity in rats. Nat Genet. 2003;33:25–32. doi: 10.1038/ng1058. [DOI] [PubMed] [Google Scholar]
- Shimaoka Y, Attrep JF, Hirano T, Ishihara K, Suzuki R, Toyosaki T, Ochi T, Lipsky PE. Nurse-like cells from bone marrow and synovium of patients with rheumatoid arthritis promote survival and enhance function of human B cells. J Clin Invest. 1998;102:606–618. doi: 10.1172/JCI3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BO, Ishihara K, Denno K, Kobune Y, Itho M, Muraoka O, Kaisho T, Sasaki T, Ochi T, Hirano T. Elevated levels of the soluble form of bone marrow stromal cell antigen 1 in the sera of patients with severe rheumatoid arthritis. Arthritis Rheum. 1996;39:629–637. doi: 10.1002/art.1780390414. [DOI] [PubMed] [Google Scholar]
- Tournier C, Thomas G, Pierre J, Jaquemin C, Pierre M, Saunier B. Mediation of arachidonic acid metabolites of the H2 O2-induced stimulation of mitogen-activated protein kinases (extracellular-signal-regulated kinase and c-jun NH2-terminal kinase) Eur J Biochem. 1997;244:587–595. doi: 10.1111/j.1432-1033.1997.00587.x. [DOI] [PubMed] [Google Scholar]
- Kasid U, Wang FH, Whiteside TL. Ionizing radiation and TNF-alpha stimulate gene expression of a Thr/Tyr-protein phosphatase HVH1 and inhibitory factor IkappaBalpha in human squamous carcinoma cells. Mol Cell Biochem. 1997;173:193–197. doi: 10.1023/A:1006892121504. [DOI] [PubMed] [Google Scholar]
- Sohaskey ML, Ferrell JE., Jr Activation of p42 mitogen-activated protein kinase (MAPK), but not c-jun NH2-Terminal kinase, induces phosphorylation and stabilization of MAPK phosphatase XCL100 in Xenopus oocytes. Mol Biol Cell. 2002;13:454–468. doi: 10.1091/mbc.01-11-0553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois B, Masure S, Hurtenbach U, Paemen L, Heremans H, van den Oord J, Sciot R, Meinhardt T, Hämmerling G, Opdenakker G, Arnold B. Resistance of young gelatinase B-deficient mice to experimental autoimmune encephalomyelitis and necrotizing tail lesions. J Clin Invest. 1999;104:1507–1515. doi: 10.1172/JCI6886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryl E, Vallejo AN, Weyand CM, Goronzy JJ. Down-regulation of CD28 expression by TNFα. J Immunol. 2001;167:3231–3228. doi: 10.4049/jimmunol.167.6.3231. [DOI] [PubMed] [Google Scholar]
- Lützelschwab C, Pejler G, Aveskogh M, Hellman L. Secretory granule proteases in rat mast cells. Cloning of 10 different serine proteases and a carboxypeptidase A from various rat mast cell populations. J Exp Med. 1997;185:13–29. doi: 10.1084/jem.185.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeil HP, Gotis-Graham I. Human mast cell subsets: distinct functions in inflammation? Inflamm Res. 2000;49:3–7. doi: 10.1007/PL00012386. [DOI] [PubMed] [Google Scholar]
- Munro SB, Duclos AJ, Jackson AR, Baines MG, Blaschuk OW. Charecterization of cadherins expressed by murine thymocytes. Cell Immunol. 1996;169:309–312. doi: 10.1006/cimm.1996.0123. [DOI] [PubMed] [Google Scholar]
- Ohtsuka T, Ryu H, Minamishima YA, Ryo A, Lee SW. Modulation of p53 and p73 levels by cyclin G: implication of a negative feedback regulation. Oncogene. 2003;22:1678–1687. doi: 10.1038/sj.onc.1206306. [DOI] [PubMed] [Google Scholar]
- Tak PP, Zvaifler NJ, Green DR, Firestein GS. Rheumatoid arthritis and p53: how oxidative stress might alter the course of inflammatory diseases. Immunol Today. 2000;21:78–82. doi: 10.1016/S0167-5699(99)01552-2. [DOI] [PubMed] [Google Scholar]
- Holmdahl R, Lorentzen JC, Lu S, Olofsson P, Wester L, Holmberg J, Pettersson U. Arthritis induced in rats with non-immunogenic adjuvants as models for rheumatoid arthritis. Immunol Rev. 2001;184:184–202. doi: 10.1034/j.1600-065x.2001.1840117.x. [DOI] [PubMed] [Google Scholar]
- Remmers EF, Joe B, Griffiths MM, Dobbins DE, Dracheva SV, Hashiramoto A, Furuya T, Salström JL, Wang J, Gulko PS, Cannon GW, Wilder RL. Modulation of multiple experimental arthritis models by collagen-induced arthritis quantitative trait loci isolated in congenic rat lines: different effects of non-major histocompatibility complex quantitative trait loci in males and females. Arthritis Rheum. 2002;46:2225–2234. doi: 10.1002/art.10439. [DOI] [PubMed] [Google Scholar]
- Joe B, Remmers EF, Dobbins DE, Salström JL, Furuya T, Dracheva S, Gulko PS. Genetic dissection of collagen-induced arthritis in chromosome 10 quantitative trait locus speed congenic rats: evidence for more than one regulatory locus and sex influences. Immunogenetics. 2000;51:930–944. doi: 10.1007/s002510000231. [DOI] [PubMed] [Google Scholar]
- Jagodic M, Kornek B, Weissert R, Lassman H, Olsson T, Dahlman I. Congenic mapping confirms a locus on rat chromosome 10 conferring strong protection against myelin oligodendrocyte glycoprotein-induced experimental autoimmune encephalomyelitis. Immunogenetics. 2001;53:410–415. doi: 10.1007/s002510100342. [DOI] [PubMed] [Google Scholar]
- Backdahl L, Ribbhammar U, Lorentzen JC. Mapping and functional characterization of rat chromosome 4 regions that regulate arthritis models and phenotypes in congenic strains. Arthritis Rheum. 2003;48:551–559. doi: 10.1002/art.10782. [DOI] [PubMed] [Google Scholar]
- Holm BC, Xu HW, Jacobsson L, Larsson A, Luthman H, Lorentzen JC. Rats made congenic for Oia3 on chromosome 10 become susceptible to squalene-induced arthritis. Hum Mol Genet. 2001;10:565–572. doi: 10.1093/hmg/10.6.565. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Affymetrix data from pooled RNA samples, prepared from inguinal lymph nodes of six naïve DA rats and six naïve E3 rats. The differentially expressed genes are presented as DA versus E3 rats. All genes with a fold change >1.9 are presented. Genes with a fold change >3.0 are regarded as differentially expressed in this method, and genes with a fold change >1.9 but <3.0 are regarded as genes with a tendency toward differential expression. Genes marked with an 'x' are represented on the custom-made glass chip.
Affymetrix data from pooled RNA samples, prepared from inguinal lymph nodes of six DA rats and six E3 rats injected with 150 μl pristane 8 days before analysis. The differentially expressed genes are presented as DA versus E3 rats. All genes with a fold change >1.9 are presented. Genes with a fold change >3.0 are regarded as differentially expressed in this method, and genes with a fold change >1.9 but <3.0 are regarded as genes with a tendency toward differential expression. Genes marked with an 'x' are represented on the custom-made glass chip.
Affymetrix data from RNA samples, prepared from inguinal lymph nodes of three DA rats and three E3 rats injected with 150 μl pristane 8 days before analysis. The RNA samples were prepared and analyzed individually for these rats. The differentially expressed genes are presented as DA versus E3 rats. The Affymetrix data was analyzed statistically using the D-chip software and sorted on the absolute t-statistic. Fifteen genes with the highest probability of being downregulated and fifteen genes with the highest probability of being upregulated are presented. These thirty genes were regarded as differentially expressed by this method.
Taqman analysis was performed on five selected genes. Individual RNA samples from five pristane-injected DA and five E3 rats were analyzed. Differentially expressed genes with Mann–Whitney P < 0.05 are regarded as differentially expressed genes, and genes with P < 0.1 are regarded as genes with a tendency toward differential expression. *Cyclin G was expressed in all the E3 rats investigated, but it was not detectable in the DA rats.
The complete gene list of the custom-made glass oligomer chips. For each of these genes three different oligomers were constructed in the nonpolymorphic regions. The oligomer sequences are available on request.
Custom-made glass oligomer chip results from RNA samples, prepared from inguinal lymph nodes of five naïve DA rats and five naïve E3 rats. The DA rat RNA samples were prepared and analyzed individually in comparison with a pool of RNA from the five E3 rats. The differentially expressed genes are presented as DA versus E3 rats. The data were analyzed statistically using Statview software. All genes with t-test P < 0.05 are regarded as differentially expressed in this method, and genes with P > 0.05 but < 0.1 are regarded as genes with a tendency toward differential expression.
Custom-made glass oligomer chip results from RNA samples, prepared from inguinal lymph nodes of five DA rats and five E3 rats injected with 150 μl pristane 8 days before analysis. The DA rat RNA samples were prepared and analyzed individually in comparison with a pool of RNA from the five E3 rats. The differentially expressed genes are presented as DA versus E3 rats. The data were analyzed statistically using Statview software. All genes with t-test P < 0.05 are regarded as differentially expressed in this method, and genes with P > 0.05 but < 0.1 are regarded as genes with a tendency toward differential expression.
Fluorescence activated cell sorting (FACS) results. Three naïve and three pristane-treated individuals of each rat strain were analyzed by flow cytometry to determine whether differential detection on mRNA level was correlated with protein level. The differentially expressed genes are presented as DA versus E3 rats. The data were analyzed statistically using the Statview software. All genes with Mann–Whitney P < 0.05 are regarded as differentially expressed in this method, and genes with P > 0.05 but < 0.1 are regarded as genes with a tendency toward differential expression. D, decreased; I, increased; NC, no change (fold change value = 1).
ELISA result. Plasma from four naïve and pristane-treated individuals from each rat strain were analyzed by ELISA to determine whether the differential detection of immunoglobulins on mRNA level was correlated with the immunoglobulin concentrations in plasma. The data were analyzed statistically using the Statview software. All genes with Mann–Whitney P < 0.05 are regarded as differentially expressed in this method, and genes with P > 0.05 but < 0.1 are regarded as genes with a tendency toward differential expression. D, decreased; I, increased.


