Abstract
Objective
Broadly neutralizing antibodies (bNt Abs) against HIV-1 are rarely produced during natural infection, and efforts to induce such Abs by vaccination have been unsuccessful. Thus, elucidating the nature and cellular origins of bNt Abs is a high priority for vaccine research. As the bNt monoclonal Abs (MAbs) 2F5, 4E10 and 2G12 have been reported to bind select autoantigens, we investigated whether these MAbs display a broader range of autoreactivity and how their autoreactivity compares with that of pathogenic autoAbs.
Methods
An autoantigen microarray comprising 106 connective tissue disease-related autoantigens and control antigens was developed and used, in combination with ELISAs, to compare the reactivity profiles of MAbs 4E10, 2F5 and 2G12 to those of four pathogenic autoAbs derived from patients with antiphospholipid-syndrome (APS), and to serum from a patient with systemic lupus erythematosus (SLE).
Results
The APS MAbs and SLE serum reacted strongly with multiple autoantigens on the microarray, whereas anti-HIV-1 MAb reactivity was limited mainly to HIV-1-related antigens. The APS autoAbs reacted strongly with CL, yet only 4E10 bound CL at high concentrations; both 2F5 and 4E10 bound their HIV-1 epitopes with a 2–3-log higher apparent affinity than CL. Moreover, the polyreactivity of 4E10, but not CL15, could be blocked with dried milk.
Conclusion
The reactivity profiles of bNt anti-HIV-1 MAbs are fundamentally distinct from those of pathogenic autoAbs that arise from dysregulated tolerance mechanisms. This suggests that the limited polyreactivity observed for the bNt MAbs, and for HIV-1-Nt Abs in general, may arise through alternative mechanisms, such as extensive somatic mutation due to persistent antigen selection during chronic infection.
Keywords: AIDS vaccine, autoantibodies, autoantigen microarray, broadly neutralizing antibodies, HIV
Introduction
Induction of rare broadly neutralizing (bNt) antibodies (Abs) able to block cellular infection by a variety of HIV-1 isolates remains the elusive goal of vaccine design. Currently, passive transfer with bNt Abs is the only means of conferring protection against viral challenge in animal models [1–4]. Six of the eight bNt monoclonal Abs (MAbs) identified to date have unusually long third complementarity-determining regions of the heavy chain (CDR-H3s) (b12, 2F5, 4E10, 447–52D, PG9/16 and HJ16 [5]; D. Corti, Bellinzona, Switzerland, personal communication), and all are highly mutated, prompting the question of whether these features are necessary for broad neutralization. MAbs 2F5 and 4E10 recognize adjacent linear epitopes in the membrane-proximal external region of gp41 (MPER; reviewed in [6]). Structural studies of these Abs in complex with peptides have revealed that their CDR-H3s make marginal contact, if any, with their epitopes [7,8]; yet, mutational studies have shown the hydrophobic tips of their CDR-H3s are required for viral neutralization [9–13].
In addition to their MPER epitopes, MAbs 2F5 and 4E10 have been reported to bind self antigens, including cardiolipin and other membrane lipids [9,14–18]. It has been proposed that 2F5 and 4E10 are polyreactive autoAbs, produced by plasma cells whose autoreactive precursors emerged due to a loss of tolerance checkpoints during development of the naive B-cell repertoire [19,20]. Evidence supporting this hypothesis includes the reactivity of 2F5 and 4E10 with self-antigens [14–19]; 4E10’s weak lupus anticoagulant activity [14,21]; the long CDR-H3s of these Abs, given that tolerance checkpoints may play a role in restricting CDR-H3 length [22]; and the disruption of B-cell development in IgH knock-in mice bearing the 2F5 variable region [23]. More recently, it has been shown that the hydrophobic tip of 2F5’s and 4E10’s CDR-H3 is required for weak interaction with lipid [9–13], and that residues of synthetic MPER peptides that are critical for interaction with MAbs 2F5 and 4E10 face into detergent micelles and lipid bilayers [24,25], leading to a proposed binding mechanism in which membrane surfaces are ‘scanned’ for MPER residues, which are then extracted from the viral membrane [15,25–27]. Based on these studies, it has been suggested that polyreactive, lipid-binding Abs are initially selected from the naive repertoire and later recruited into the anti-HIV-1 response by viral antigens [28]. Furthermore, one study has shown that serum anti-cardiolipin Ab titers are strongly correlated in chronically infected patients with anti-MPER Ab titers and neutralization breadth [29]. The proposal that HIV-1-Nt Abs arise from polyreactive and/or autoreactive precursors that are normally absent in the naive repertoire has fueled speculation that conventional vaccination strategies are unlikely to elicit bNt Abs, as selection for polyreactive Abs [28] or disruption of immunological tolerance [20] would be required for their development.
Serum reactivity in clinical anti-cardiolipin assays is associated with antiphospholipid syndrome (APS), an autoimmune, thrombogenic condition and systemic lupus erythematosus (SLE) [30]. In contrast, the clinical significance of reactivity with cardiolipin and other phospholipids that is commonly observed for serum Abs produced during HIV-1 infection is unclear [30,31]. The antiphospholipid Abs that develop during infection by HIV-1 and other pathogens [32] differ in several respects from those arising in APS and SLE: they are often transient [31,32], do not cross-react with β2-glycoprotein I [30,31] and are not associated with lupus anticoagulant activity or thrombosis [31]. Despite their putative cardiolipin reactivity, sustained passive infusion of MAbs 2F5 and 4E10 is well tolerated, with no incidents of adverse effects or thrombotic complications [21,33], and an alternate hypothesis to explain the mechanism of lipid reactivity of 2F5 has been described [10,34]. Thus, to further clarify the polyreactive and autoimmune nature of bNt anti-HIV-1 MAbs, we compared their reactivity profiles on an extensive panel of autoantigens with those of autoreactive, thrombogenic MAbs and serum produced by a patient with active SLE.
Methods
Materials
The NIH AIDS Research and Reference Reagent Program provided 15-mer synthetic peptides derived from the lab-adapted HIV-1 strain, MN (>80% purity; sequences available online). Synthetic 2F5 peptide [NH3+-EQELLELDKWASLWSGK(Biotin)GC-CONH2] and E4.6 peptide [NH3+-LHEESMDKWSNLMQCCT AEGK(Biotin)-CONH2] were from NeoMPS (San Diego, California, USA), synthetic 4E10 peptide [NH3+- SLWNWFDITNWLWYISGK(Biotin)GC-CONH2] was from the University of British Columbia’s NAPS Unit Peptide Synthesis Laboratory (Vancouver, Canada) and synthetic H3N6 peptide (NH3+-AEPAENNWFML TYFLAAEGC-CONH2) was from EZBiolab Inc. (Westfield, Indiana, USA); all were more than 95% pure. MAbs CL1, CL15, IS2 and IS4 were provided by P. Chen (UCLA) and Abs 2F5, 4E10 and 2G12 by R. Kunert and H. Katinger (University of Agricultural Sciences, Vienna). Recombinant gp41 and gp120 envelope proteins (MN strain) were from ImmunoDiagnostics (Woburn, Massachusetts, USA). Cardiolipin, BSA, oval-bumin (OVA), p-nitrophenyl phosphate (pNPP) and 2,2′-azino-bis-3-ethylbenzthiazoline-6-sulfonic acid (ABTS) were from Sigma-Aldrich (St Louis, Missouri, USA), nonfat dried milk from Bio-Rad (Hercules, California, USA) and Tween-20 from GE Healthcare (Piscataway, New Jersey, USA). Sources of microarray antigens are listed in Supplementary Table S1, http://links.lww.com/QAD/A142.
Titration ELISA
Protocols were adapted from an optimized procedure [14] (B.F. Haynes, Duke University, personal communication). Wells of 96-well microtiter plates (Corning Inc., Corning, New York, USA) were coated with 400 ng MN peptide in 50 μl PBS, pH 7.4 and dried at 55°C for 4 h; 2 μg cardiolipin in 50 μl methanol and dried at 55°C for 45 min; and protein [50 ng gp41 in 50 μl PBS, or 50 μl PBS containing 2% (w/v) BSA, dried milk, or OVA] with overnight incubation at 4°C. Wells were blocked at 37°C for 2 h with 50 μmol/l carbonate–bicarbonate buffer, pH 9.6, containing 3% BSA, then washed three times with PBS containing 0.05% (v/v) Tween-20 (PBST). Serial dilutions of 4E10, 2F5 and CL15 IgGs were prepared in Ab diluent [PBST containing 3% BSA and 2% (v/v) fetal calf serum (FCS)]; 50-μl aliquots were incubated in each well for 1 h at room temperature (RT). Wells were washed four times with PBST and then 50 μl alkaline phosphatase-conjugated goat antihuman IgG (Fab-specific; Pierce, Rockford, Illinois, USA), diluted 1 : 1000 in Ab diluent, was added for 1 h at RT. Wells were washed four times and developed using a pNPP tablet dissolved at 1 mg/ml in 100 μl 50 μmol/l carbonate–bicarbonate buffer containing 2 mmol/l MgCl2. After 45 min, absorbance at 405 nm was measured using a VersaMax Microplate Reader (Molecular Devices, Sunnyvale, California, USA).
Direct ELISA
Direct ELISAs were performed as described [35] with the following modifications. Wells were coated with 4E10, H3N6 or MN peptides (400 ng), autoantigens (1 μg), double-stranded DNA (2 μg) or cardiolipin (2 μg in 35 μl methanol) and dried at 55°C for 1 h and then other wells were coated overnight at 4°C with 50 ng gp41 or 2% BSA, dried milk or OVA. MAbs were diluted in TBS, pH 7.2, containing 0.1% Tween-20 and either 5% BSA, 5% dried milk, 50% FCS or 5% FCS as indicated.
Autoantigen microarrays
Purified autoantigens (0.2 mg/ml in PBS) were deposited in triplicate on nitrocellulose-coated FAST slides (Whatman, Florham Park, New Jersey, USA) using a VersArray ChipWriter Pro Robotic Arrayer (Bio-Rad). Arrays were blocked in dilution buffer (PBS, 5% FCS, 0.1% Tween-20) overnight at 4°C. Serum samples were diluted 1 : 300 in dilution buffer; MAbs were diluted to 35, 7 and 1.5 μg/ml (representing 50, 10 and 2% of total serum IgG, respectively, given a typical serum IgG concentration of 21 mg/ml) and then diluted 1 : 300; diluted Ab samples were incubated on slides for 1 h at 4°C. Arrays were washed twice in dilution buffer and then probed with a 1 : 2000 dilution of Cy-3-conjugated goat antihuman IgG/IgM (Jackson ImmunoResearch, West Grove, Pennsylvania, USA) for 1 h at 4°C. After two washes in dilution buffer, followed by one in PBS and then in water, arrays were spun dry and imaged using a GenePix 4000B microarray scanner (Molecular Devices). Protocols are available at http://utzlab.stanford.edu/protocols/.
Data analysis and heat map generation
Scanned array images were analyzed using Axon GenePix Pro 6.0 software (Molecular Devices). Median fluorescence intensity (MFI) values of less than 10 digital fluorescence units (DFUs) were set to 10 DFUs exactly. MFI values minus background from three replicate spots were averaged for each antigen. Heat maps were generated in TMEV Multiple Experiment Viewer v4.5.1 software by setting the highest intensity value for a given Ab–antigen interaction as 1.0 and normalizing the intensity of other Abs to this value, generating a percentage scale.
Significance analysis of microarrays
Significance analysis of microarrays (SAM) was performed in TMEV Multiple Experiment Viewer v4.5.1 software to identify significant differences in array reactivities. Raw MFI values were log2 transformed and two-class unpaired analyses were performed to compare each HIV-1 MAb to the APS MAb CL15 across all three dilutions tested. For each comparison, 20 permutations were calculated and threshold parameters were selected such that the false discovery rate was less than 0.05. Antigens revealed by SAM to be bound significantly differently were selected for cluster analysis. A hierarchical clustering algorithm using a Euclidian distance metric and complete linkage method was applied to order array features into clusters based on similarities in Ab–antigen reactivities.
Results
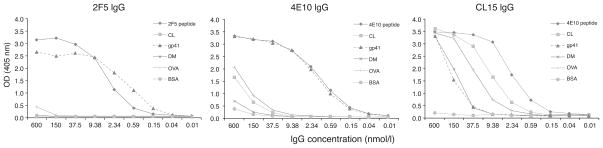
We first compared the cardiolipin reactivity of MAbs 2F5 and 4E10 with that of the pathogenic APS MAb, CL15 [36], using published ELISA conditions [14] (Fig. 1). In agreement with other works [21,37], cardiolipin binding by MAb 2F5 was undetectable, whereas binding by 4E10 occurred only at high concentrations; in this assay, 4E10 IgG bound cardiolipin and OVA approximately equally. There was more than a 2-log difference in the concentration of 4E10 MAb required to produce a given signal on 4E10 peptide (or gp41) and that required to produce the same signal on cardiolipin. In contrast, CL15 saturated all antigens tested, with the exception of BSA. Although MAbs 4E10 and CL15 bound the 4E10 peptide at similar half-maximal concentrations, only 4E10 neutralized HIV-1 strain HxB2 (Supplementary Fig. S1, http://links.lww.com/QAD/A142; provided by M.B. Zwick, The Scripps Research Institute). These data demonstrate that MAbs 2F5 and 4E10 prefer their cognate gp41 epitopes to cardiolipin by orders of magnitude, compared with the pathogenic and highly polyreactive autoAb CL15.
Fig. 1. Comparison of autoreactivity and polyreactivity of MAbs 2F5, 4E10 and CL15 and dependence on Ab concentration.
MAbs 2F5, 4E10 and CL15 were titrated at the concentrations indicated in a direct ELISA against wells containing immobilized peptides bearing the 2F5 or 4E10 epitopes (400 ng each), recombinant gp41 (50 ng), cardiolipin (CL; 2 μg) or a nonspecific antigen, BSA, dried milk (DM) or ovalbumin (OVA) (2%). Data are representative of three independent experiments. MAb 2F5 does not bind CL or any nonspecific antigen significantly, whereas MAb 4E10 binds CL and OVA with apparent affinities that are at least two orders of magnitude smaller than that for gp41. MAb CL15 binds many antigens with high apparent affinity.
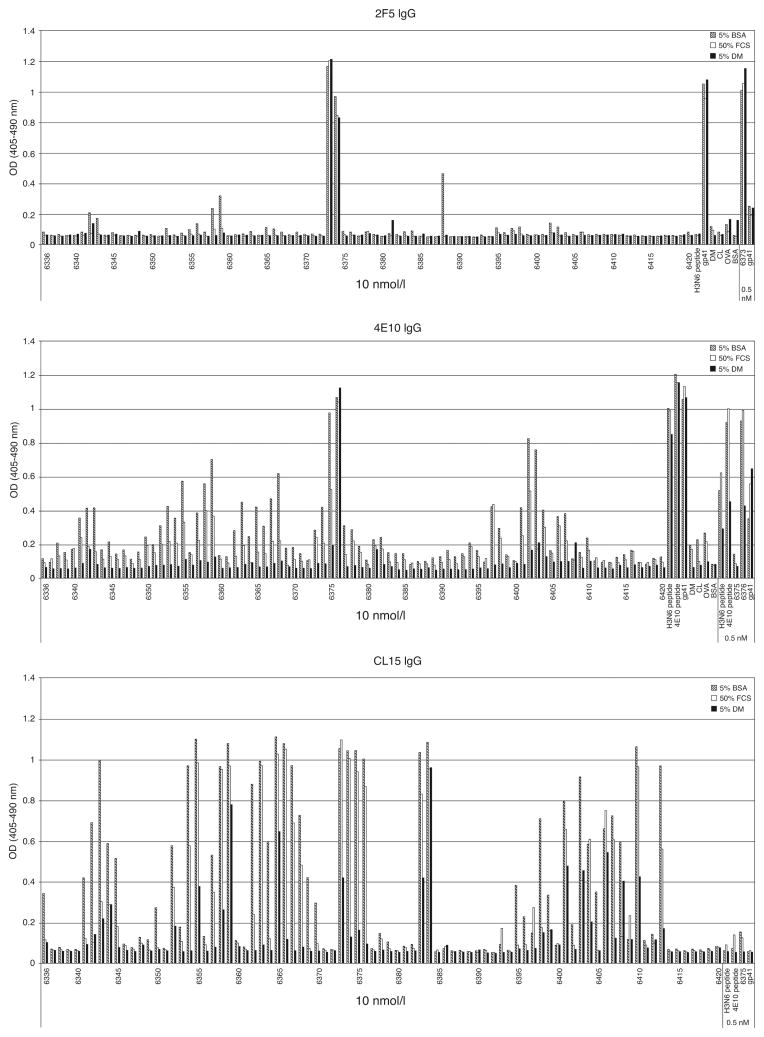
To further test the specificity of these MAbs, their binding was studied in ELISA to a panel of overlapping 15-mer peptides derived from HIV-1 envelope protein isolate MN (Fig. 2). When diluted in 5% BSA, MAb 2F5 bound specifically to peptides bearing its cognate HIV-1 epitope, and MAb 4E10 bound its cognate epitope better than the multiple peptides with which it weakly cross-reacted. In contrast, the APS autoAb CL15 bound promiscuously to more than half the assayed peptides and to OVA and dried milk. Dilution in 5% FCS or 5% dried milk abolished nonspecific binding of 4E10 but not of CL15. Linear regression analysis revealed that CL15 binding, but not 4E10 binding, was associated with a net negative charge on the peptides (Supplementary Fig. S2, http://links.lww.com/QAD/A142). These results are consistent with our previous studies in which both MAbs 2F5 and 4E10 selected peptides from phage-displayed libraries that share a core consensus sequence with their gp41 epitopes [38,39], whereas MAb CL15 selected peptides bearing completely unrelated sequences [40].
Fig. 2. Effects of complexity of Ab diluent on reactivity profiles of MAbs 2F5, 4E10 and CL15.
MAbs 2F5, 4E10 or CL15 (10 nmol/l or 66.7 μg/ml IgG) were diluted in 5% BSA, 50% fetal calf serum (FCS) or 5% dried milk (DM) and assayed on wells containing immobilized overlapping HIV-1 MN peptides (400 ng), 4E10 peptide (400 ng), H3N6 peptide (400 ng), recombinant gp41 (50 ng), cardiolipin (CL; 2 μg) or BSA, DM or ovalbumin (OVA) (2%). Each MAb was also tested at a 0.5 nmol/l concentration against positive-control antigens to verify that binding remained unaffected by choice of diluent. Low-level polyreactivity of MAb 2F5 was abrogated in FCS or DM, and that of MAb 4E10 was reduced in FCS and abrogated in DM; in contrast, MAb CL15 retained a promiscuous binding pattern even in DM.
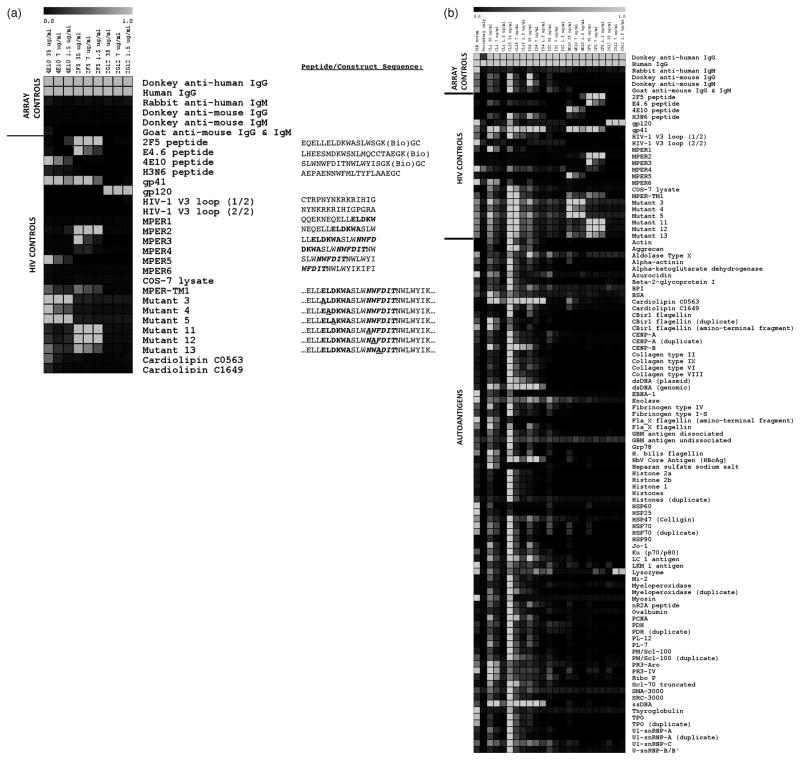
To compare the autoreactivity profiles of bNt anti-HIV-1 MAbs to those of pathogenic APS MAbs and SLE serum, we developed and validated an autoantigen microarray containing clinically relevant autoantigens targeted by Abs from patients with systemic autoimmune diseases [41], HIV-1 antigens and control antigens. For comparison, we analyzed the anti-MPER MAbs, 2F5 and 4E10, the anti-gp120 MAb 2G12 and four MAbs derived from APS patients: CL15, CL1 [36], IS2 [42] and IS4 [36]. In a previous study, cross-reactivity of MAbs 2F5, 4E10 and 2G12 against a panel of 400 mainly non-self, recombinant proteins was minimal [37]. However, we wanted to more comprehensively assess potential autoreactivity of these MAbs using an autoantigen microarray that had been previously validated with pathogenic autoantisera [43–49]. Figure 3a shows the reactivity profiles of MAbs 2F5, 4E10 and 2G12 against positive and negative control antigens, including synthetic peptides bearing the 2F5 and 4E10 epitopes, recombinant gp120 and gp41, overlapping MN peptides covering the gp41 MPER (MPER1-MPER6), cell lysates bearing gp41 fragments covering the MPER tethered to the cell surface by the gp41 transmembrane region [50], cell lysates bearing this gp41 fragment containing single Ala substitutions in the MPER sequence (mutants 3–5 and 11–13) and cell lysates alone. As expected, MAbs 2F5 and 4E10 each bound their cognate peptide epitopes [38,39], and 2G12 bound strongly to its carbohydrate epitope on gp120. As seen in Fig. 3a, MAb 2F5 reacted only with overlapping MPER peptides (MPER2 and MPER3) containing its core ELDKWA epitope (shown in bold). Similarly, MAb 4E10 bound only to peptide MPER5 containing its core NWF(D/N)ITepitope (shown in bold italics); no binding was observed to peptide MPER4, which contains the 4E10 core epitope but little additional C-terminal sequence, consistent with mapping studies [51]. In addition, both MAbs 2F5 and 4E10 bound to lysates of COS-7 cells transiently transfected with plasmids expressing MPER-TM1, a gp41 fragment covering part of the C-heptad repeat, the MPER, the transmembrane region and part of the cytoplasmic domain of HIV-1 JRCSF [50]. Binding was epitope-specific, as substitution of residues in either the 2F5 or 4E10 core epitope with Ala abrogated binding of the respective MAb (Fig. 3a). Thus, the specific binding patterns of anti-HIV-1 MAbs to their epitopes validated the sensitivity and specificity of the array platform.
Fig. 3. Autoantigen microarray analysis of reactivity profiles of bNt anti-HIV-1 MAbs, APS MAbs and SLE serum.
The HIV-1 bNt MAbs 2F5, 4E10 and 2G12, APS MAbs IS2, IS4, CL1 and CL15 (at indicated concentrations) and an SLE serum were used to probe slides. (a) Validation of the microarray platform using a panel of HIV-1 antigens and array control antigens. Positive control Abs include antihuman IgG (as all MAbs tested are isotype IgG) and human IgG (bound by the secondary Ab), whereas antihuman IgM and antimouse IgG/IgM are negative controls. The bNt anti-HIV-1 MAbs 2F5, 4E10 and 2G12 bind their cognate epitopes in the microarray but not to closely related HIV-1 antigens. Sequences of peptides and constructs are shown, with the core 2F5 and 4E10 epitopes in bold and bold italics, respectively; Ala substitutions in these epitopes are underlined. Cardiolipin is included as a control antigen because of conflicting reports of binding by MAbs 2F5 and 4E10; here only MAb 4E10 bound cardiolipin, and only at high concentration (35 μg/ml). (b) Full heat-map representation of binding patterns of all MAbs and sera to all array antigens, including the control antigens in part (a) for comparison. All data are representative of three independent experiments. Color versions of both figures are included as Supplemental Fig. S7a and b and raw log2-transformed MFI values are presented in Supplemental Fig. S8, http://links.lww.com/QAD/A142.
As seen in Fig. 3b, the bNt MAbs 2F5, 4E10 and 2G12 bound their cognate HIV-1 epitopes but had limited reactivity with all autoantigens tested (shown in alphabetical order), whereas the APS autoAbs and SLE serum IgGs displayed extreme polyreactivity. Consistent with the ELISA results, binding of MAbs 2F5 and 4E10 to cardiolipin was minimal, and 4E10 reacted significantly with cardiolipin and other autoantigens only at the highest concentration tested (35 μg IgG/ml). In contrast, most APS MAbs tested (CL1, CL15 and IS4) displayed strong binding to cardiolipin even at lower concentrations. The cardiolipin reactivity of MAb 4E10 was not associated with binding to β2-glycoprotein I (a well characterized APS autoantigen), whereas two APS MAbs (CL1 and CL15) reacted with both antigens. Binding of MAb 2F5 to centromere protein B (CENP-B), whole histones or histone subunits was undetectable, conflicting with previous reports [14]; MAb CL15, however, was particularly histone-reactive, and MAbs CL15 and IS4 bound CENP-B. MAbs 2F5 and 2G12 bound lysozyme at lower concentrations but not at higher ones, casting doubt upon this result. Together, these data provide further evidence that the bNt anti-HIV-1 MAbs 2F5 and 2G12 are not especially autoreactive or polyreactive, whereas MAb 4E10 exhibits modest polyreactivity at concentrations reflecting high levels of serum Ab (the equivalent of 50% of the average serum concentration of total IgG in healthy individuals).
We used SAM, a well validated algorithm [52] that we and others have used in comparing serum autoAb reactivities [43–49], to identify differences in the reactivity profiles of MAbs 2F5, 4E10, 2G12 and CL15. Table 1 lists antigens that were bound significantly differently (q <0.05) by a given pair of MAbs. MAb CL15 bound significantly better than anti-HIV-1 MAbs to a variety of autoantigens such as cardiolipin, CENP-B and double-stranded DNA as well as control antigens such as OVA and BSA, and in some cases, even HIV-1 antigens. Importantly, the only antigens to which MAbs 2F5, 4E10 and 2G12 bound significantly better than CL15 were antigens bearing their cognate HIV-1 epitopes; the same was true for pairwise SAM comparisons between each HIV-1 MAb and the least polyreactive APS MAb, IS2 (data not shown). Furthermore, statistical comparison of MAbs 2F5 and 4E10 confirmed that the only antigens shown to be bound differently by these MAbs bore their respective cognate epitopes (Table 1). MAbs were also arranged by hierarchical clustering based on reactivity similarities [53]. In each case, MAbs 2F5, 4E10 and 2G12 belonged to a distinct clade from MAb CL15 and SLE serum (Supplementary Figs S3–S6, http://links.lww.com/QAD/A142); the same overall trend was observed when hierarchical clustering was conducted using all antigens (data not shown). Thus, the behavior of MAbs 2F5, 4E10 and 2G12 is unlike that of pathogenic APS autoAbs and polyclonal SLE sera, as demonstrated by their minimal reactivity with self antigens.
Table 1.
List of antigens shown by significance analysis of microarrays to be bound differently by indicated monoclonal antibodies.
| Comparison: 2F5 vs. CL15
|
Comparison: 4E10 vs. CL15
|
Comparison: 2G12 vs. CL15
|
Comparison: 2FS vs. 4E10
|
||||
|---|---|---|---|---|---|---|---|
| CL15 >2F5
|
CL15 >4E10
|
CL15 >2G12
|
4E10 >2F5
|
||||
| Antigen | q-value | Antigen | q-value | Antigen | q-value | Antigen | q-value |
| Goat anti-mouse IgG and IgM | <0.001 | Donkey anti-mouse IgM | <0.001 | Goat anti-mouse IgG and IgM | <0.001 | 4E10 peptide | <0.001 |
| H3N6 peptide | <0.001 | MPER1 | <0.001 | E4.6 peptide | <0.001 | H3N6 peptide | <0.001 |
| gp120 | <0.001 | MPER4 | <0.001 | H3N6 peptide | <0.001 | MPER5 | <0.001 |
| HIV-1 V3 loop (1/2) | <0.001 | gp120 | <0.001 | MPER4 | <0.001 | Mutant 3 | <0.001 |
| COS-7 lysate | <0.001 | HIV-1 V3 loop (1/2) | <0.001 | gp41 | <0.001 | Mutant 4 | <0.001 |
| Actin | <0.001 | COS-7 lysate | <0.001 | HIV-1 V3 loop (1/2) | <0.001 | Mutant 5 | <0.001 |
| Aggrecan | <0.001 | Mutant 11 | <0.001 | COS-7 lysate | <0.001 | ||
| Alpha-actinin | <0.001 | Actin | <0.001 | MPER-TM1 | <0.001 | ||
| Alpha-ketoglutarate dehydrogenase | <0.001 | Aggrecan | <0.001 | Mutant 12 | <0.001 | ||
| Beta-2-glycoprotein I | <0.001 | Alpha-ketoglutarate dehydrogenase | <0.001 | Mutant 13 | <0.001 | ||
| CBir1 flagellin (duplicate) | <0.001 | Beta-2-glycoprotein I | <0.001 | Actin | <0.001 | ||
| Cardiolipin C0563 | <0.001 | CBir1 flagellin (duplicate) | <0.001 | Aggrecan | <0.001 | ||
| CENP-A | <0.001 | CENP-A | <0.001 | Alpha-actinin | <0.001 | ||
| CENP-A (duplicate) | <0.001 | CENP-A (duplicate) | <0.001 | Alpha-ketoglutarate dehydrogenase | <0.001 | ||
| CENP-B | <0.001 | CENP-B | <0.001 | CBir1 flagellin (duplicate) | <0.001 | ||
| Collagen type II | <0.001 | Collagen type II | <0.001 | Cardiolipin C0563 | <0.001 | ||
| Collagen type VI | <0.001 | Collagen type VI | <0.001 | CENP-B | <0.001 | ||
| Collagen type VIII | <0.001 | Collagen type VIII | <0.001 | Collagen type II | <0.001 | ||
| Collagen type IX | <0.001 | Collagen type IX | <0.001 | Collagen type VI | <0.001 | ||
| dsDNA (plasmid) | <0.001 | dsDNA (plasmid) | <0.001 | Collagen type VIII | <0.001 | ||
| dsDNA (genomic) | <0.001 | dsDNA (genomic) | <0.001 | Collagen type IX | <0.001 | ||
| Fibrinogen type I-S | <0.001 | Fibrinogen type I-S | <0.001 | dsDNA (genomic) | <0.001 | ||
| Fibrinogen type IV | <0.001 | GBM antigen dissociated | <0.001 | dsDNA (plasmid) | <0.001 | ||
| GBM antigen dissociated | <0.001 | HbV Core Antigen (HBcAg) | <0.001 | Fibrinogen type I-S | <0.001 | ||
| Grp78 | <0.001 | Histone 2a | <0.001 | Fibrinogen type IV | <0.001 | ||
| H. bilis flagellin | <0.001 | Histone 2b | <0.001 | GBM antigen dissociated | <0.001 | ||
| HbV Core Antigen (HBcAg) | <0.001 | Histone 1 | <0.001 | HbV Core Antigen (HBcAg) | <0.001 | ||
| Histone 2a | <0.001 | Histones | <0.001 | Histone 2a | <0.001 | ||
| Histone 2 b | <0.001 | HSP47 (Colligin) | <0.001 | Histone 2b | <0.001 | ||
| Histone 1 | <0.001 | Ku (p70/p80) | <0.001 | Histone 1 | <0.001 | ||
| Histones | <0.001 | LC 1 antigen | <0.001 | Histones | <0.001 | ||
| HSP90 | <0.001 | Lysozyme | <0.001 | HSP47 (Colligin) | <0.001 | ||
| Jo-1 | <0.001 | Mi-2 | <0.001 | HSP90 | <0.001 | ||
| Ku (p70/p80) | <0.001 | Myeloperoxidase | <0.001 | Jo-1 | <0.001 | ||
| LC 1 antigen | <0.001 | Myeloperoxidase (duplicate) | <0.001 | Ku (p70/p80) | <0.001 | ||
| Mi-2 | <0.001 | Ovalbumin | <0.001 | LC 1 antigen | <0.001 | ||
| Myeloperoxidase (duplicate) | <0.001 | PCNA | <0.001 | Mi-2 | <0.001 | ||
| nR2A peptide | <0.001 | PDH (duplicate) | <0.001 | Myeloperoxidase (duplicate) | <0.001 | ||
| PCNA | <0.001 | PM/Scl-100 | <0.001 | nR2A peptide | <0.001 | ||
| PDH (duplicate) | <0.001 | PM/Scl-100 (duplicate) | <0.001 | PCNA | <0.001 | ||
| PL-12 | <0.001 | PR3-Aro | <0.001 | PDH (duplicate) | <0.001 | ||
| PL-7 | <0.001 | Scl-70 truncated | <0.001 | PL-7 | <0.001 | ||
| PM/Scl-100 | <0.001 | SRC-3000 | <0.001 | PL-12 | <0.001 | ||
| PM/Scl-100 (duplicate) | <0.001 | ssDNA | <0.001 | PM/Scl-100 | <0.001 | ||
| Ribo P | <0.001 | TPO | <0.001 | PM/Scl-100 (duplicate) | <0.001 | ||
| Scl-70 truncated | <0.001 | TPO (duplicate) | <0.001 | Scl-70 truncated | <0.001 | ||
| SRC-3000 | <0.001 | U1-snRNP-A | <0.001 | SRC-3000 | <0.001 | ||
| ssDNA | <0.001 | U1-snRNP-A (duplicate) | <0.001 | ssDNA | <0.001 | ||
| TPO | <0.001 | TPO | <0.001 | ||||
| TPO (duplicate) | <0.001 | TPO (duplicate) | <0.001 | ||||
| U1-snRNP-A | <0.001 | U1-snRNP-A | <0.001 | ||||
| U1-snRNP-A (duplicate) | <0.001 | U1-snRNP-A (duplicate) | <0.001 | ||||
| U1-snRNP-C | <0.001 | U1-snRNP-C | <0.001 | ||||
| 2F5 >CL15
|
4E10 >CL15
|
2G12 >CL15
|
2F5 >4E10
|
||||
|---|---|---|---|---|---|---|---|
| Antigen | q-value | Antigen | q-value | Antigen | q-value | Antigen | q-value |
| 2F5 peptide | <0.001 | 4E10 peptide | <0.001 | 2F5 peptide | <0.001 | ||
| E4.6 peptide | <0.001 | MPER5 | <0.001 | E4.6 peptide | <0.001 | ||
| MPER2 | <0.001 | MPER2 | <0.001 | ||||
| MPER3 | <0.001 | MPER3 | <0.001 | ||||
| MPER4 | <0.001 | ||||||
| Mutant 11 | <0.001 | ||||||
| Mutant 12 | <0.001 | ||||||
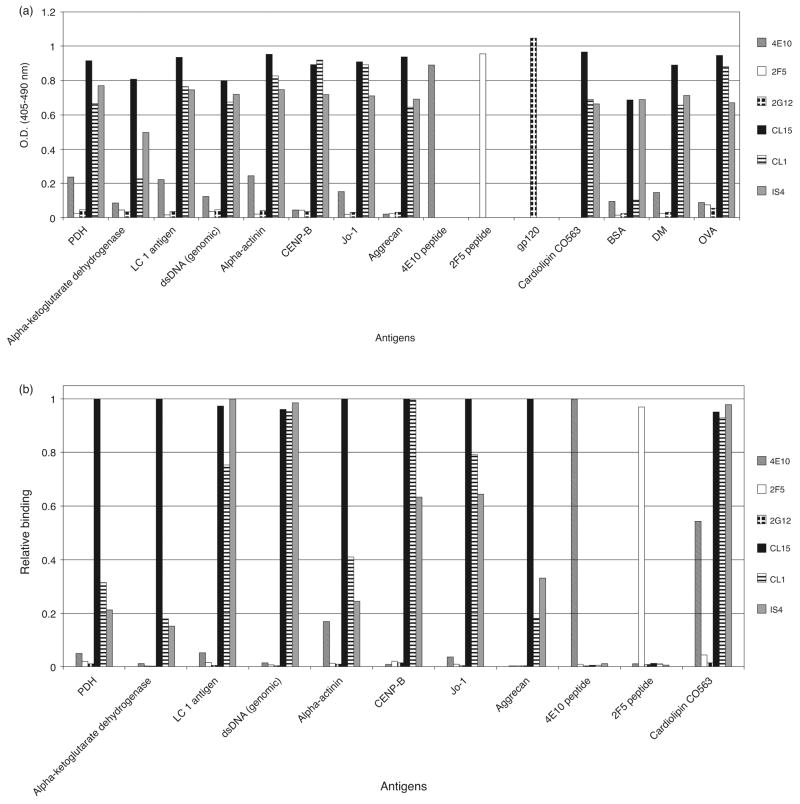
To validate reactivity patterns to selected autoantigens observed in the microarray, ELISAs were performed using antigens revealed by SAM to be bound differently by anti-HIV-1 MAbs and APS MAbs. Without exception, all differences between anti-HIV-1 and APS Abs observed by ELISA (Fig. 4a) were reflected in the microarray analysis (Fig. 4b), although the magnitudes of these differences were not always accurately replicated. In particular, two of the APS autoAbs, CL1 and IS4, appeared far more autoreactive in the ELISA compared to the microarray. However, overall binding trends indicated concordance of reactivity patterns, with the autoantigen microarray being more sensitive than ELISA in detecting differences in binding to a particular antigen.
Fig. 4. Validation of the autoantigen microarray for detecting differences between HIV-1 bNt Abs and APS autoAbs.
Side-by-side comparison of ELISA and microarray results for antigens that were bound significantly differently by anti-HIV-1 MAbs vs. APS MAbs. (a) MAbs 2F5, 4E10, 2G12, IS4, CL1 and CL15 (100 nmol/l) were diluted in 5% (v/v) fetal calf serum (FCS) and assayed against 1–2 μg of the indicated autoantigens immobilized in microplate wells. (b) Relative microarray binding of MAbs 2F5, 4E10, 2G12, IS4, CL1 and CL15 to selected autoantigens. For each antigen, the median fluorescence intensity (minus background) from the highest binding Ab in the microarray was set to 1.0, and all other Abs normalized to generate a scale based on the best-binding MAb.
Several ELISA experiments designed to gauge poly-reactivity and autoreactivity of bNt anti-HIV-1 MAbs have been performed using very high concentrations of Ab and no detergent [16–19,22,54–56]. Here, we tested several ELISA protocols to understand the effect of different assay conditions on the reactivity profiles of MAbs 2F5, 4E10 and CL15. We found that the modest polyreactivity of MAb 4E10 was markedly reduced or ablated by replacing BSA with FCS, and especially dried milk, in the Ab diluent; yet, this did not significantly affect binding to its cognate gp41 epitope (Fig. 2). This suggests that there may exist an Ab subspecies, perhaps in partially denatured form, whose activity is saturated in the presence of complex proteins and/or lipids existing in dried milk and serum. In contrast, dilution of MAb CL15 in FCS or dried milk only slightly reduced its polyreactivity. As their polyreactivity vanishes in assays that more closely approximate physiological conditions, we surmise that the polyreactivity of MAbs 2F5 and 4E10 is not biologically significant in vivo; this is supported by their lack of pathogenicity after passive infusion [21,33].
Discussion
Our data indicate that reactivity of MAbs 2F5 and 4E10 with cardiolipin is biologically insignificant compared with reactivity with their HIV-1 epitopes or with the cardiolipin reactivity of the pathogenic APS MAb, CL15. Moreover, the global reactivity patterns of MAbs 2F5, 4E10 and 2G12 with an extensive panel of clinically relevant autoantigens reveal that these Abs are not polyreactive autoAbs in the same sense as pathogenic APS or SLE autoAbs. These results support and extend those of Scherer et al. [37], who showed that MAbs 2F5 and 4E10 have very limited polyreactivity on a panel of antigens. The microarray data from these two experiments, assaying hundreds of autoantigens and unrelated proteins, have allowed us to draw strong conclusions regarding the polyreactivity and autoreactivity of bNt anti-HIV-1 MAbs. However, antigen microarray techniques have limitations in that a control antigen (gp120) was recognized by only one of its ‘cognate’ MAbs (2G12, but not b12) when printed on nitrocellulose slides; thus, b12, but not 2G12, was excluded from this analysis.
It is likely that the weak polyreactivity of bNt anti-HIV-1 Abs, in particular MAb 4E10, with self-antigens and nonself-antigens [14,15,21] is not evidence of a poly-reactive or autoreactive origin, but rather reflects normal, low level self-reactivity of Abs from the memory B-cell and plasma-cell repertoires [54]. Such reactivity could increase with chronic infection as a result of prolonged somatic mutation, and a lack of post-germinal center checkpoints for deleting weakly self-reactive or poly-reactive clones. Recently, Mouquet et al. [55] showed that somatic mutations and polyreactivity are high in anti-gp140 Abs cloned from the memory B cells of HIV-positive individuals, as compared with their non-gp140-binding Abs and with Abs from healthy controls. Although the in-vivo relevance of this polyreactivity –as measured by in-vitro assays in which Abs were diluted in PBS lacking blocking proteins or detergent [56] – is unclear, the increased polyreactivity of the anti-gp140 Abs is striking. Moreover, in only a few cases did reversion of somatic mutations to germline reduce polyreactivity, leading the authors to conclude that polyreactive precursors of these clones must have been present in the naive B-cell repertoire. However, as somatic mutations in CDR3, particularly in CDR-H3, are not easily identified, their contributions to polyreactivity may have been overlooked (N.B., this issue has been circumvented using TdT knockout mice, with the result that all Ab autoreactivity was abolished upon reversion to germline [57]). Similarly, the dysregulation of B-cell development observed when using highly mutated VH genes (such as MAb 2F5’s [22]) for knock-in experiments can be attributed to disruption of the pre-B-cell receptor, and thus is not necessarily good proof of tolerance induction [58].
We propose that the polyreactivity and self-reactivity observed among anti-HIV-1 MAbs and sera are generated during chronic viral infection, which drives both repeated rounds of somatic mutation and persistent antigen selection. In this scenario, extensive mutation, including insertions that lengthen CDR-H3, would broaden the reactivity profiles of clones that continue to retain affinity for antigen, and therefore to be recruited into the Ab response. Consistent with this hypothesis, breadth of serum neutralization is significantly correlated with viral load [59,60], reflecting antigen persistence, and where anti-MPER Nt Abs have been observed, with cardiolipin polyreactivity [29]. Although the possibility that low-level polyreactivity may be a general feature of highly mutated Abs against viral pathogens in persistent or repeated infection [61] awaits further investigation, it is certainly conceivable, given the shared genetic features of these Abs [5] and the role of somatic hypermutation in generating polyreactive and self-reactive specificities [54,57].
Regardless of their origins, our results support a fundamental distinction between bNt anti-HIV-1 MAbs and pathogenic autoAbs that arise in autoimmune states like APS or SLE. The majority of the bNt anti-HIV-1 MAbs isolated to date have atypically long CDR-H3s and extensive somatic mutations, but so do anti-HIV-1 Abs and antiviral Abs produced during chronic infection in general [5]. Elucidating the conditions under which bNt Abs arise will require a better understanding of the Abs comprising the naive and memory B-cell repertoires and mechanisms for selecting Ab-secreting plasma cells during chronic HIV-1 infection.
Supplementary Material
Acknowledgments
We thank the following people for their contribution to this work: P.P. Chen, R. Kunert and H. Katinger for providing antibodies; B.F. Haynes for the cardiolipin ELISA protocol; M.B. Zwick for testing the neutralizing activity of 4E10 and CL15 (The Scripps Research Institute); R. Gupta (Stanford University), M. Mai (Simon Fraser University; SFU), C. Lepik (SFU) and N. Sandhu (SFU) for technical assistance; M. Montero (SFU) for materials; F. Breden (SFU) for advice on statistical analyses; and G. J. Silverman (University of California, San Diego) for comments on the manuscript. This work was supported by NIH grants A1051614 (P.J.U.), A149111 (J.K.S.) and A10680201 (J.K.S.), the Canada Research Chair Program (J.K.S.), the NIH NRSA program (A.C.) and the CIHR Doctoral Research Award Program (K.A.H.). For the remaining authors none were declared.
H.S., S.S.T.W., A.C., P.J.U. and J.K.S. designed research; H.S., S.S.T.W. and A.C. performed research; H.S., K.A.H., P.J.U. and J.K.S. analyzed data and wrote the manuscript.
References
- 1.Hessell AJ, Poignard P, Hunter M, Hangartner L, Tehrani DM, Bleeker WK, et al. Effective, low-titer antibody protection against low-dose repeated mucosal SHIV challenge in macaques. Nat Med. 2009;15:951–954. doi: 10.1038/nm.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hessell AJ, Rakasz EG, Tehrani DM, Huber M, Weisgrau KL, Landucci G, et al. Broadly neutralizing monoclonal antibodies 2F5 and 4E10 directed against the human immunodeficiency virus type 1 gp41 membrane-proximal external region protect against mucosal challenge by simian-human immunodeficiency virus SHIVBa-L. J Virol. 2010;84:1302–1313. doi: 10.1128/JVI.01272-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mascola JR, Lewis MG, Stiegler G, Harris D, VanCott TC, Hayes D, et al. Protection of Macaques against pathogenic simian/human immunodeficiency virus 89. 6PD by passive transfer of neutralizing antibodies. J Virol. 1999;73:4009–4018. doi: 10.1128/jvi.73.5.4009-4018.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mascola JR, Stiegler G, VanCott TC, Katinger H, Carpenter CB, Hanson CE, et al. Protection of macaques against vaginal transmission of a pathogenic HIV-1/SIV chimeric virus by passive infusion of neutralizing antibodies. Nat Med. 2000;6:207–210. doi: 10.1038/72318. [DOI] [PubMed] [Google Scholar]
- 5.Breden F, Lepik C, Longo NS, Lipsky PE, Scott JK. Comparison of antibody repertoires produced by HIV-1 Infection, other chronic and acute infections, and systemic autoimmune disease. PLoS One. 2011;6:e16857. doi: 10.1371/journal.pone.0016857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Montero M, van Houten NE, Wang X, Scott JK. The membrane-proximal external region of the human immunodeficiency virus type 1 envelope: dominant site of antibody neutralization and target for vaccine design. Microbiol Mol Biol Rev. 2008;72:54– 84. doi: 10.1128/MMBR.00020-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cardoso RM, Zwick MB, Stanfield RL, Kunert R, Binley JM, Katinger H, et al. Broadly neutralizing anti-HIV antibody 4E10 recognizes a helical conformation of a highly conserved fusion-associated motif in gp41. Immunity. 2005;22:163–173. doi: 10.1016/j.immuni.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 8.Ofek G, Tang M, Sambor A, Katinger H, Mascola JR, Wyatt R, et al. Structure and mechanistic analysis of the antihuman immunodeficiency virus type 1 antibody 2F5 in complex with its gp41 epitope. J Virol. 2004;78:10724–10737. doi: 10.1128/JVI.78.19.10724-10737.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alam SM, Morelli M, Dennison SM, Liao HX, Zhang R, Xia SM, et al. Role of HIV membrane in neutralization by two broadly neutralizing antibodies. Proc Natl Acad Sci U S A. 2009;106:20234–20239. doi: 10.1073/pnas.0908713106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Julien JP, Huarte N, Maeso R, Taneva SG, Cunningham A, Nieva JL, et al. Ablation of the complementarity-determining region H3 apex of the anti-HIV-1 broadly neutralizing antibody 2F5 abrogates neutralizing capacity without affecting core epitope binding. J Virol. 2010;84:4136–4147. doi: 10.1128/JVI.02357-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ofek G, McKee K, Yang Y, Yang ZY, Skinner J, Guenaga FJ, et al. Relationship between antibody 2F5 neutralization of HIV-1 and hydrophobicity of its heavy chain third complementarity-determining region. J Virol. 2010;84:2955–2962. doi: 10.1128/JVI.02257-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu H, Song L, Kim M, Holmes MA, Kraft Z, Sellhorn G, et al. Interactions between lipids and human anti-HIV antibody 4E10 can be reduced without ablating neutralizing activity. J Virol. 2010;84:1076–1088. doi: 10.1128/JVI.02113-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zwick MB, Komori HK, Stanfield RL, Church S, Wang M, Parren PW, et al. The long third complementarity-determining region of the heavy chain is important in the activity of the broadly neutralizing antihuman immunodeficiency virus type 1 antibody 2F5. J Virol. 2004;78:3155–3161. doi: 10.1128/JVI.78.6.3155-3161.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haynes BF, Fleming J, St Clair EW, Katinger H, Stiegler G, Kunert R, et al. Cardiolipin polyspecific autoreactivity in two broadly neutralizing HIV-1 antibodies. Science. 2005;308:1906–1908. doi: 10.1126/science.1111781. [DOI] [PubMed] [Google Scholar]
- 15.Alam SM, McAdams M, Boren D, Rak M, Scearce RM, Gao F, et al. The role of antibody polyspecificity and lipid reactivity in binding of broadly neutralizing anti-HIV-1 envelope human monoclonal antibodies 2F5 and 4E10 to glycoprotein 41 membrane proximal envelope epitopes. J Immunol. 2007;178:4424–4435. doi: 10.4049/jimmunol.178.7.4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beck Z, Karasavvas N, Tong J, Matyas GR, Rao M, Alving CR. Calcium modulation of monoclonal antibody binding to phosphatidylinositol phosphate. Biochem Biophys Res Commun. 2007;354:747–751. doi: 10.1016/j.bbrc.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 17.Matyas GR, Beck Z, Karasavvas N, Alving CR. Lipid binding properties of 4E10, 2F5, and WR304 monoclonal antibodies that neutralize HIV-1. Biochim Biophys Acta. 2009;1788:660–665. doi: 10.1016/j.bbamem.2008.11.015. [DOI] [PubMed] [Google Scholar]
- 18.Matyas GR, Wieczorek L, Beck Z, Ochsenbauer-Jambor C, Kappes JC, Michael NL, et al. Neutralizing antibodies induced by liposomal HIV-1 glycoprotein 41 peptide simultaneously bind to both the 2F5 or 4E10 epitope and lipid epitopes. AIDS. 2009;23:2069–2077. doi: 10.1097/QAD.0b013e32832faea5. [DOI] [PubMed] [Google Scholar]
- 19.Haynes BF, Moody MA, Verkoczy L, Kelsoe G, Alam SM. Antibody polyspecificity and neutralization of HIV-1: a hypothesis. Hum Antibodies. 2005;14:59–67. [PMC free article] [PubMed] [Google Scholar]
- 20.Nabel GJ. Immunology. Close to the edge: neutralizing the HIV-1 envelope. Science. 2005;308:1878–1879. doi: 10.1126/science.1114854. [DOI] [PubMed] [Google Scholar]
- 21.Vcelar B, Stiegler G, Wolf HM, Muntean W, Leschnik B, Mehandru S, et al. Reassessment of autoreactivity of the broadly neutralizing HIV antibodies 4E10 and 2F5 and retrospective analysis of clinical safety data. AIDS. 2007;21:2161–2170. doi: 10.1097/QAD.0b013e328285da15. [DOI] [PubMed] [Google Scholar]
- 22.Wardemann H, Yurasov S, Schaefer A, Young JW, Meffre E, Nussenzweig MC. Predominant autoantibody production by early human B cell precursors. Science. 2003;301:1374–1377. doi: 10.1126/science.1086907. [DOI] [PubMed] [Google Scholar]
- 23.Verkoczy L, Diaz M, Holl TM, Ouyang YB, Bouton-Verville H, Alam SM, et al. Autoreactivity in an HIV-1 broadly reactive neutralizing antibody variable region heavy chain induces immunologic tolerance. Proc Natl Acad Sci U S A. 2010;107:181–186. doi: 10.1073/pnas.0912914107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schibli DJ, Montelaro RC, Vogel HJ. The membrane-proximal tryptophan-rich region of the HIV glycoprotein, gp41, forms a well defined helix in dodecylphosphocholine micelles. Biochemistry. 2001;40:9570–9578. doi: 10.1021/bi010640u. [DOI] [PubMed] [Google Scholar]
- 25.Sun ZY, Oh KJ, Kim M, Yu J, Brusic V, Song L, et al. HIV-1 broadly neutralizing antibody extracts its epitope from a kinked gp41 ectodomain region on the viral membrane. Immunity. 2008;28:52–63. doi: 10.1016/j.immuni.2007.11.018. [DOI] [PubMed] [Google Scholar]
- 26.Song L, Sun ZY, Coleman KE, Zwick MB, Gach JS, Wang JH, et al. Broadly neutralizing anti-HIV-1 antibodies disrupt a hinge-related function of gp41 at the membrane interface. Proc Natl Acad Sci U S A. 2009;106:9057–9062. doi: 10.1073/pnas.0901474106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dennison SM, Stewart SM, Stempel KC, Liao HX, Haynes BF, Alam SM. Stable docking of neutralizing human immunodeficiency virus type 1 gp41 membrane-proximal external region monoclonal antibodies 2F5 and 4E10 is dependent on the membrane immersion depth of their epitope regions. J Virol. 2009;83:10211–10223. doi: 10.1128/JVI.00571-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiao X, Chen W, Feng Y, Dimitrov D. Maturation pathways of cross-reactive HIV-1 neutralizing antibodies. Viruses. 2009;1 :802–817. doi: 10.3390/v1030802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gray ES, Taylor N, Wycuff D, Moore PL, Tomaras GD, Wibmer CK, et al. Antibody specificities associated with neutralization breadth in plasma from human immunodeficiency virus type 1 subtype C-infected blood donors. J Virol. 2009;83:8925–8937. doi: 10.1128/JVI.00758-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petrovas C, Vlachoyiannopoulos PG, Kordossis T, Moutsopoulos HM. Antiphospholipid antibodies in HIV infection and SLE with or without antiphospholipid syndrome: comparisons of phospholipid specificity, avidity and reactivity with beta2-GPI. J Autoimmun. 1999;13:347–355. doi: 10.1006/jaut.1999.0324. [DOI] [PubMed] [Google Scholar]
- 31.Abuaf N, Laperche S, Rajoely B, Carsique R, Deschamps A, Rouquette AM, et al. Autoantibodies to phospholipids and to the coagulation proteins in AIDS. Thromb Haemost. 1997;77 :856–861. [PubMed] [Google Scholar]
- 32.Loizou S, Singh S, Wypkema E, Asherson RA. Anticardiolipin, antibeta(2)-glycoprotein I and antiprothrombin antibodies in black South African patients with infectious disease. Ann Rheum Dis. 2003;62:1106–1111. doi: 10.1136/ard.62.11.1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trkola A, Kuster H, Rusert P, Joos B, Fischer M, Leemann C, et al. Delay of HIV-1 rebound after cessation of antiretroviral therapy through passive transfer of human neutralizing antibodies. Nat Med. 2005;11:615–622. doi: 10.1038/nm1244. [DOI] [PubMed] [Google Scholar]
- 34.Maeso R, Huarte N, Julien JP, Kunert R, Pai EF, Nieva JL. Interaction of anti-HIV type 1 antibody 2F5 with Phospholipid bilayers and its relevance for the mechanism of virus neutralization. AIDS Res Hum Retroviruses. 2011 doi: 10.1089/AID.2010.0265. [DOI] [PubMed] [Google Scholar]
- 35.van Houten NE, Henry KA, Smith GP, Scott JK. Engineering filamentous phage carriers to improve focusing of antibody responses against peptides. Vaccine. 2010;28:2174–2185. doi: 10.1016/j.vaccine.2009.12.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu M, Olee T, Le DT, Roubey RA, Hahn BH, Woods VL, Jr, et al. Characterization of IgG monoclonal anticardiolipin/anti-beta2GP1 antibodies from two patients with antiphospholipid syndrome reveals three species of antibodies. Br J Haematol. 1999;105:102–109. [PubMed] [Google Scholar]
- 37.Scherer EM, Zwick MB, Teyton L, Burton DR. Difficultiesin eliciting broadly neutralizing anti-HIV antibodies are not explained by cardiolipin autoreactivity. AIDS. 2007;21:2131–2139. doi: 10.1097/QAD.0b013e3282a4a632. [DOI] [PubMed] [Google Scholar]
- 38.Bahr SL. MSc thesis. Burnaby: Simon Fraser University; 2004. Peptide markers for the HIV-1 neutralizing antibody 4E10. [Google Scholar]
- 39.Menendez A, Chow KC, Pan OC, Scott JK. Human immunodeficiency virus type 1-neutralizing monoclonal antibody 2F5 is multispecific for sequences flanking the DKW core epitope. J Mol Biol. 2004;338:311–327. doi: 10.1016/j.jmb.2004.02.051. [DOI] [PubMed] [Google Scholar]
- 40.Visvanathan S, Scott JK, Hwang KK, Banares M, Grossman JM, Merrill JT, et al. Identification and characterization of a peptide mimetic that may detect a species of disease-associated anticardiolipin antibodies in patients with the antiphospholipid syndrome. Arthritis Rheum. 2003;48:737–745. doi: 10.1002/art.10836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robinson WH, DiGennaro C, Hueber W, Haab BB, Kamachi M, Dean EJ, et al. Autoantigen microarrays for multiplex characterization of autoantibody responses. Nat Med. 2002;8:295–301. doi: 10.1038/nm0302-295. [DOI] [PubMed] [Google Scholar]
- 42.Olee T, Pierangeli SS, Handley HH, Le DT, Wei X, Lai CJ, et al. A monoclonal IgG anticardiolipin antibody from a patient with the antiphospholipid syndrome is thrombogenic in mice. Proc Natl Acad Sci U S A. 1996;93:8606–8611. doi: 10.1073/pnas.93.16.8606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Garren H, Robinson WH, Krasulova E, Havrdova E, Nadj C, Selmaj K, et al. Phase 2 trial of a DNA vaccine encoding myelin basic protein for multiple sclerosis. Ann Neurol. 2008;63:611–620. doi: 10.1002/ana.21370. [DOI] [PubMed] [Google Scholar]
- 44.Hueber W, Kidd BA, Tomooka BH, Lee BJ, Bruce B, Fries JF, et al. Antigen microarray profiling of autoantibodies in rheumatoid arthritis. Arthritis Rheum. 2005;52:2645–2655. doi: 10.1002/art.21269. [DOI] [PubMed] [Google Scholar]
- 45.Richez C, Yasuda K, Bonegio RG, Watkins AA, Aprahamian T, Busto P, et al. IFN regulatory factor 5 is required for disease development in the FcgammaRIIB−/−Yaa and FcgammaR-IIB−/− mouse models of systemic lupus erythematosus. J Immunol. 2010;184:796–806. doi: 10.4049/jimmunol.0901748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Robinson WH, Fontoura P, Lee BJ, de Vegvar HE, Tom J, Pedotti R, et al. Protein microarrays guide tolerizing DNA vaccine treatment of autoimmune encephalomyelitis. Nat Biotechnol. 2003;21:1033–1039. doi: 10.1038/nbt859. [DOI] [PubMed] [Google Scholar]
- 47.Sekine H, Graham KL, Zhao S, Elliott MK, Ruiz P, Utz PJ, et al. Role of MHC-linked genes in autoantigen selection and renal disease in a murine model of systemic lupus erythematosus. J Immunol. 2006;177:7423–7434. doi: 10.4049/jimmunol.177.10.7423. [DOI] [PubMed] [Google Scholar]
- 48.Thibault DL, Chu AD, Graham KL, Balboni I, Lee LY, Kohlmoos C, et al. IRF9 and STAT1 are required for IgG autoantibody production and B cell expression of TLR7 in mice. J Clin Invest. 2008;118:1417–1426. doi: 10.1172/JCI30065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thibault DL, Graham KL, Lee LY, Balboni I, Hertzog PJ, Utz PJ. Type I interferon receptor controls B-cell expression of nucleic acid-sensing Toll-like receptors and autoantibody production in a murine model of lupus. Arthritis Res Ther. 2009;11:R112. doi: 10.1186/ar2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Montero M. PhD thesis. Burnaby: Simon Fraser University; 2007. Antigenicity and immunogenicity of the membrane proximal external region of the HIV-1 envelope protein gp41. [Google Scholar]
- 51.Brunel FM, Zwick MB, Cardoso RM, Nelson JD, Wilson IA, Burton DR, et al. Structure-function analysis of the epitope for 4E10, a broadly neutralizing human immunodeficiency virus type 1 antibody. J Virol. 2006;80:1680–1687. doi: 10.1128/JVI.80.4.1680-1687.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tusher VG, Tibshirani R, Chu G. Significance analysis of micro-arrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tiller T, Tsuiji M, Yurasov S, Velinzon K, Nussenzweig MC, Wardemann H. Autoreactivity in human IgG+memory B cells. Immunity. 2007;26:205–213. doi: 10.1016/j.immuni.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mouquet H, Scheid JF, Zoller MJ, Krogsgaard M, Ott RG, Shukair S, et al. Polyreactivity increases the apparent affinity of anti-HIV antibodies by heteroligation. Nature. 2010;467:591–595. doi: 10.1038/nature09385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tiller T, Meffre E, Yurasov S, Tsuiji M, Nussenzweig MC, Wardemann H. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J Immunol Methods. 2008;329:112–124. doi: 10.1016/j.jim.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guo W, Smith D, Aviszus K, Detanico T, Heiser RA, Wysocki LJ. Somatic hypermutation as a generator of antinuclear antibodies in a murine model of systemic autoimmunity. J Exp Med. 2010;207:2225–2237. doi: 10.1084/jem.20092712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Heltemes-Harris L, Liu X, Manser T. An antibody VH gene that promotes marginal zone B cell development and heavy chain allelic exclusion. Int Immunol. 2005;17:1447–1461. doi: 10.1093/intimm/dxh323. [DOI] [PubMed] [Google Scholar]
- 59.Doria-Rose NA, Klein RM, Daniels MG, O’Dell S, Nason M, Lapedes A, et al. Breadth of human immunodeficiency virus-specific neutralizing activity in sera: clustering analysis and association with clinical variables. J Virol. 2010;84:1631–1636. doi: 10.1128/JVI.01482-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Piantadosi A, Panteleeff D, Blish CA, Baeten JM, Jaoko W, McClelland RS, et al. Breadth of neutralizing antibody response to human immunodeficiency virus type 1 is affected by factors early in infection but does not influence disease progression. J Virol. 2009;83:10269–10274. doi: 10.1128/JVI.01149-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hangartner L, Zinkernagel RM, Hengartner H. Antiviral antibody responses: the two extremes of a wide spectrum. Nat Rev Immunol. 2006;6:231–243. doi: 10.1038/nri1783. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.