Abstract
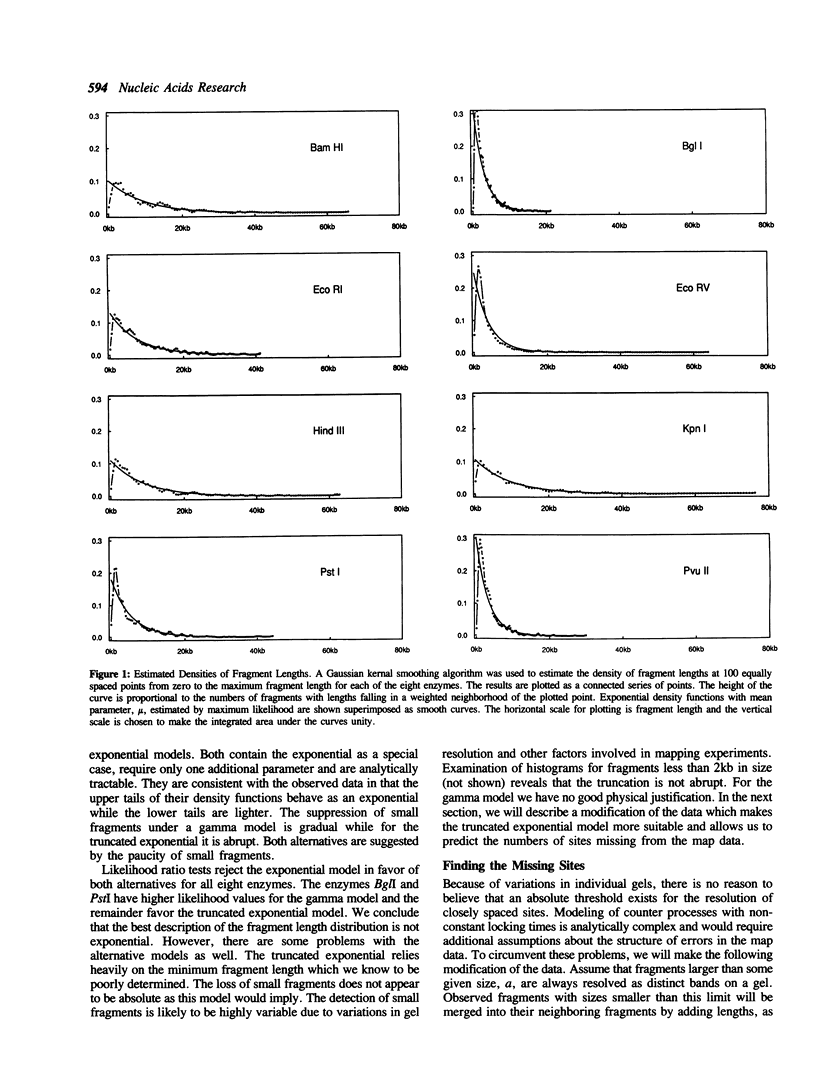
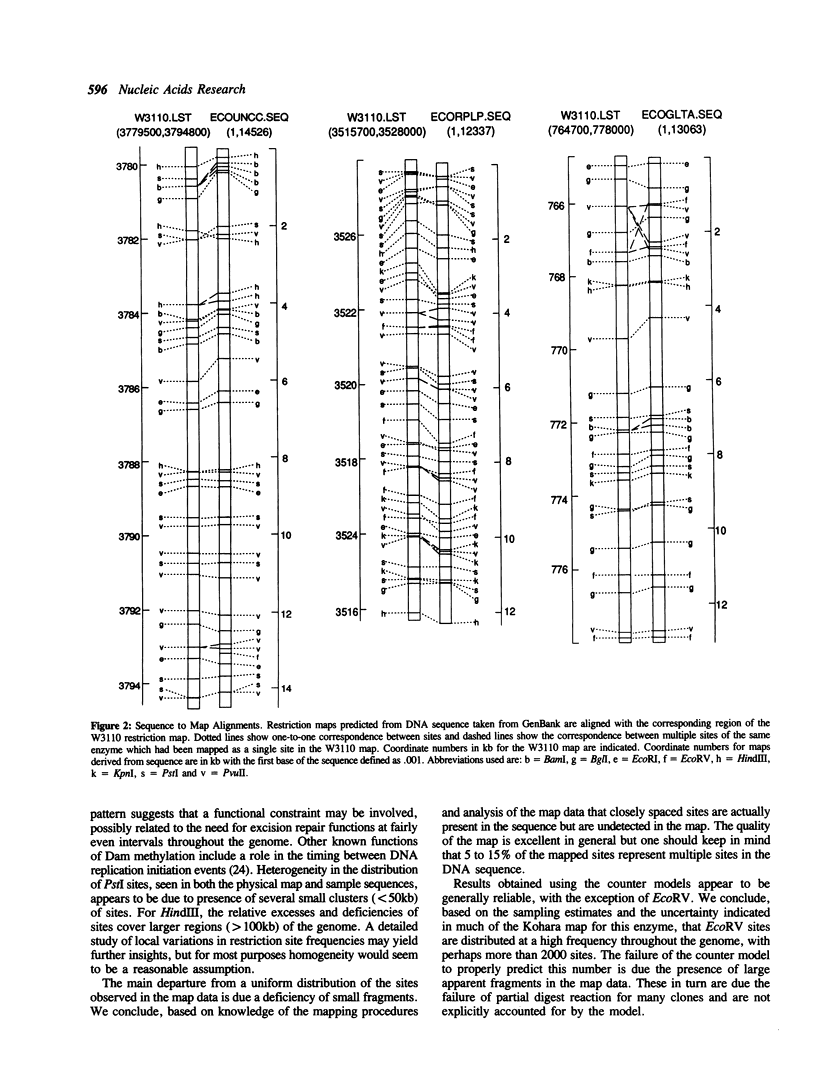
A statistical analysis of physical map data for eight restriction enzymes covering nearly the entire genome of E. coli is presented. The methods of analysis are based on a top-down modeling approach which requires no knowledge of the statistical properties of the base sequence. For most enzymes, the distribution of mapped sites is found to be fairly homogeneous. Some heterogeneity in the distribution of sites is observed for the enzymes Pstl and HindIII. In addition, BamHI sites are found to be more evenly dispersed than we would expect for random placement and we speculate on a possible mechanism. A consistent departure from a uniform distribution, observed for each of the eight enzymes, is found to be due to a lack of closely spaced sites. We conclude from our analysis that this departure can be accounted for by deficiencies in the physical map data rather than non-random placement of actual restriction sites. Estimates of the numbers of sites missing from the map are given, based both on the map data itself and on the site frequencies in a sample of sequenced E. coli DNA. We conclude that 5 to 15% of the mapped sites represent multiple sites in the DNA sequence.
Full text
PDF






Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bakker A., Smith D. W. Methylation of GATC sites is required for precise timing between rounds of DNA replication in Escherichia coli. J Bacteriol. 1989 Oct;171(10):5738–5742. doi: 10.1128/jb.171.10.5738-5742.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardi G., Olofsson B., Filipski J., Zerial M., Salinas J., Cuny G., Meunier-Rotival M., Rodier F. The mosaic genome of warm-blooded vertebrates. Science. 1985 May 24;228(4702):953–958. doi: 10.1126/science.4001930. [DOI] [PubMed] [Google Scholar]
- Bishop D. T., Williamson J. A., Skolnick M. H. A model for restriction fragment length distributions. Am J Hum Genet. 1983 Sep;35(5):795–815. [PMC free article] [PubMed] [Google Scholar]
- Kohara Y., Akiyama K., Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987 Jul 31;50(3):495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- McClelland M., Jones R., Patel Y., Nelson M. Restriction endonucleases for pulsed field mapping of bacterial genomes. Nucleic Acids Res. 1987 Aug 11;15(15):5985–6005. doi: 10.1093/nar/15.15.5985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messer W., Noyer-Weidner M. Timing and targeting: the biological functions of Dam methylation in E. coli. Cell. 1988 Sep 9;54(6):735–737. doi: 10.1016/s0092-8674(88)90911-7. [DOI] [PubMed] [Google Scholar]
- Waterman M. S. Frequencies of restriction sites. Nucleic Acids Res. 1983 Dec 20;11(24):8951–8956. doi: 10.1093/nar/11.24.8951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterman M. S., Smith T. F., Katcher H. L. Algorithms for restriction map comparisons. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):237–242. doi: 10.1093/nar/12.1part1.237. [DOI] [PMC free article] [PubMed] [Google Scholar]


