There are two alternate hypotheses for the mechanism of BMV subgenomic RNA transcription. One posits that transcription commences by recognition of at least four key nucleotides in the subgenomic promoter by the replicase. The other posits that subgenomic transcription starts by binding of the replicase to a hairpin formed by the subgenomic promoter that resembles the minus strand promoter hairpin SLC. This study has determined the three-dimensional structure of the subgenomic promoter hairpin using NMR and found that it is a stable hairpin that forms a pseudo-triloop structure similar to the promoter for genomic RNA synthesis. These similarities show that the replicase recognizes structure rather than sequence-specific motifs in both promoters.
Keywords: NMR spectroscopy, pseudo-triloop, brome mosaic virus, BMV, RNA-dependent RNA-polymerase, RNA replication
Abstract
In brome mosaic virus, both the replication of the genomic (+)-RNA strands and the transcription of the subgenomic RNA are carried out by the viral replicase. The production of (−)-RNA strands is dependent on the formation of an AUA triloop in the stem–loop C (SLC) hairpin in the 3′-untranslated region of the (+)-RNA strands. Two alternate hypotheses have been put forward for the mechanism of subgenomic RNA transcription. One posits that transcription commences by recognition of at least four key nucleotides in the subgenomic promoter by the replicase. The other posits that subgenomic transcription starts by binding of the replicase to a hairpin formed by the subgenomic promoter that resembles the minus strand promoter hairpin SLC. In this study, we have determined the three-dimensional structure of the subgenomic promoter hairpin using NMR spectroscopy. The data show that the hairpin is stable at 30°C and that it forms a pseudo-triloop structure with a transloop base pair and a nucleotide completely excluded from the helix. The transloop base pair is capped by an AUA triloop that possesses an extremely well packed structure very similar to that of the AUA triloop of SLC, including the formation of a so-called clamped-adenine motif. The similarities of the NMR structures of the hairpins required for genomic RNA and subgenomic RNA synthesis show that the replicase recognizes structure rather than sequence-specific motifs in both promoters.
INTRODUCTION
Brome mosaic virus (BMV) is a (+)-strand RNA plant virus that belongs to the Bromoviridae family (Rao 2001). The genome of BMV consists of three RNA strands—RNA1, RNA2, and RNA3—that encode four proteins. Three of these proteins, including the replicase that is responsible for replication of the viral genome, are translated directly from the genomic (+)-strand RNAs after the virus enters the host cell. A fourth protein, the coat protein (Kao and Sivakumaran 2000), is translated from RNA4, which is coterminal with the 3′ half of RNA3. Using (+)-strands as a template, the replicase synthesizes the corresponding (−)-strands. The replicase subsequently uses the (−)-strand RNAs as templates not only for the synthesis of additional full-length (+)-strand genomic (g) RNA but also for the synthesis of the shorter subgenomic (sg) RNA4.
The promoter region for (−)-strand synthesis is located in the 3′-untranslated regions of the three (+)-strand RNAs and includes a 5′-AUA-3′ triloop closed by a C:G base pair in the stem–loop C (SLC) hairpin. The high-resolution NMR structure of the SLC hairpin determined by Tinoco and coworkers shows a rigid triloop with A3 stacking inside the helix and U2 placed on top of A3 (Kim et al. 2000). The nucleobase of A1 adopts a so-called clamped adenine motif where it is pushed out of the helical stack and into the major groove. Here it is projected toward the backbone in the 5′-direction, its position stabilized by hydrogen bonds from its amino protons to a phosphate group (Kim et al. 2000). Combined structural and mutational analysis on the SLC hairpin indicates that the formation of a compact triloop structure with a C:G closing base pair and, in particular, the clamped adenine motif are crucial for the ability to initiate synthesis of (−)-strand RNAs (Kim and Tinoco 2001).
sgRNA synthesis, whether occurring through internal initiation on a full-length (−)-strand RNA or by end-to-end synthesis on a prematurely terminated (−)-strand, requires recognition of the subgenomic promoter by the viral replicase (Miller and Koev 2000; Sztuba-Solinska et al. 2011). The subgenomic promoter for BMV RNA4 synthesis consists of an A/U-rich sequence, a poly-U tract, and a 20-nucleotide (nt) core promoter (Miller et al. 1985; Marsh et al. 1988).
Based on in vitro data, Kao and coworkers posited that the replicase recognizes at least four key nucleotides in the subgenomic promoter, followed by an induced fit in which some of the nucleotides base pair prior to the initiation of (+)-strand sgRNA synthesis (Sivakumaran et al. 2004). On the other hand, Olsthoorn and coworkers suggested that the subgenomic promoter forms a short hairpin in (−)-strand RNA3. This preformed stable hairpin then serves as a landing pad for the replicase, and the recognition of the replicase is dictated by the promoter's three-dimensional structure (Haasnoot et al. 2000, 2002). Subsequent in vivo studies by Kao and coworkers show a more complex picture (Sivakumaran et al. 2004). The four key nucleotides are required for replicase binding but the stem of the hairpin is not, yet the stem is required for RNA synthesis as wild-type RNA4 levels could only be maintained by using constructs with an intact hairpin stem.
The hairpin of the core promoter, as suggested by Olsthoorn and coworkers, forms a stem–loop structure with a 5′-C1-AUA-G5-A-3′ hexaloop, which might fold into a pseudo-triloop structure through the formation of a transloop C1:G5 base pair (Haasnoot et al. 2002). This folding topology could potentially leave the central AUA part free to form an AUA triloop, which could lead to structurally similar promoters for the synthesis of both (−)-strand RNAs and sgRNA4. In vitro transcription efficiencies of shorter RNA constructs mimicking the core subgenomic promoter region of the (−)-strand RNA3, and mutants thereof, support the proposed formation of a pseudo-triloop. A single mutation turning the transloop C1:G5 base pair into either a C:C or a G:G mismatch, which might hinder the formation of the pseudo-triloop structure, results in a complete loss of transcription by the replicase. A substitution of the transloop C:G base pair with a G:C base pair, which could preserve the pseudo-triloop structure, does not abolish transcription, although the efficiency is only 17% of the wild type. Additionally the deletion of A6 in the hexaloop, which is equivalent to the formation of a regular AUA triloop, as seen in the SLC hairpin of the (+)-strand RNAs, slightly enhances the transcription level to 127% of the wild type (Haasnoot et al. 2000). In addition, comparable transcription efficiencies of the (−)-strand RNA3 are observed in vivo for the wild-type subgenomic promoter and a revertant, which can fold into a regular AUA triloop (Smirnyagina et al. 1994).
Here we present the structure of the BMV subgenomic promoter hairpin determined in solution by NMR spectroscopy. The hairpin forms a pseudo-triloop with a trans-loop C:G base pair and a bulged-out adenine. We compare the structure with the SLC hairpin and correlate it with biochemical data. To further elucidate the impact of the extra adenosine in the pseudo-triloop in the subgenomic hairpin compared with a normal triloop, we also investigated the del-A mutant that forms the usual AUA triloop. In addition to NMR spectroscopy, ultraviolet (UV) melting analysis was used to compare the thermodynamic parameters of the pseudo-triloop and triloop hairpins with the transloop G:C, C:C, and G:G base pair mutants. Finally, we discuss our results in the context of the two alternate hypotheses for replicase recognition and sgRNA synthesis.
RESULTS
Thermodynamic comparison of the BMV subgenomic promoter and mutants
In Figure 1, the secondary fold of the subgenomic pseudo-triloop hairpin (WT) is shown together with the mutant hairpins that we have investigated; the triloop with the bulged-out adenine, A11, deleted (del-A), and M2, M3, and M4 with the transloop C:G base pair replaced with G:C, C:C, and G:G, respectively.
FIGURE 1.
The sequence and nucleotide numbering scheme of the wild-type (WT) subgenomic hairpin and the four mutants investigated. The terminal G:C base pair was added to the native subgenomic hairpin to support the formation of a stable RNA hairpin.
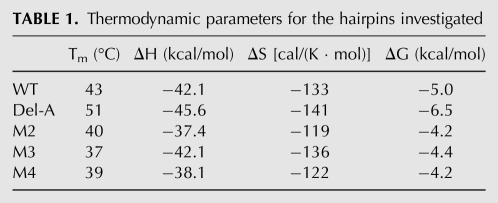
The UV absorbance of the five hairpins recorded as a function of temperature enabled us to determine their melting temperatures (Tm) and extract thermodynamical parameters using a two-state model (Table 1; Supplemental Material). The deletion of the bulged-out base, A11, induces a notable stabilization of the hairpin as evidenced by the Tms of 43°C and 51°C and Gibbs’ free energies of −5.0 kcal/mol and −6.5 kcal/mol for the WT and del-A hairpins, respectively. The difference in stability arises from a more negative enthalpy in del-A (−45.6 kcal/mol compared with −42.1 kcal/mol). Changes to the transloop C:G base pair result in a decrease in stability. In two cases, M2 and M4, a large enthalpic penalty is paid, which only partially is offset by an entropic gain. For M3, the destabilization is caused by a less-favorable entropic term. It is striking that the switch to a G:C closing base pair, M2, results in destabilization comparable to that observed for a complete abolishment of the transloop Watson–Crick base-pairing, M3 and M4.
TABLE 1.
Thermodynamic parameters for the hairpins investigated
NMR analysis of the WT and del-A hairpins
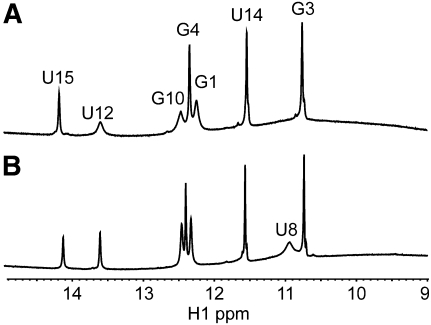
Chemical shift assignments of the 1H, 13C, and 31P resonances were performed by standard methods on a natural abundance sample (see Supplemental Material) (Varani et al. 1996). Chemical shifts are sensitive markers of changes in local structure, and consequently, chemical shift differences observed between the WT and del-A hairpins will be the first sign of differences in their local structure. The similarity between the imino regions of the WT and del-A hairpins in the one-dimensional 1H NMR spectra recorded at 5°C shows that the overall fold, with the formation of the four Watson–Crick base pairs and one G:U base pair in the stem, is identical (Fig. 2). Both the triloop closing base pair (C6:G10) in the del-A hairpin and the corresponding transloop C6:G10 base pair in the pseudo-triloop of the WT hairpin are formed. However, in the pseudo-triloop, the imino signals of G10 and U12, belonging to the two base pairs adjacent to the unpaired A11, are broadened relative to the triloop. When the temperature is increased to 25°C, the imino resonance signal of G10 in the WT pseudo-triloop disappears completely. The fast exchange implies a more frequent opening of the C6:G10 transloop base pair. Thus the pseudo-triloop of the WT hairpin is destabilized somewhat by the inclusion of A11 compared with the AUA-triloop in the del-A hairpin. Some notable proton chemical shift differences are observed for the 3 nt in the AUA triloops (up to 0.29 ppm for U8H1′) (see Supplemental Material). Minor chemical shift differences observed for A5 and C6 suggest that the stacking in the A5:U12 and C6:G10 base pair steps is only slightly different in the two hairpins, and virtually unchanged chemical shifts in the lower stem parts of the hairpins show that the structure of the stem is unaffected by the presence of A11.
FIGURE 2.
The imino region of 1H NMR spectra recorded at 5°C in H2O. (A) The WT hairpin. (B) The del-A hairpin.
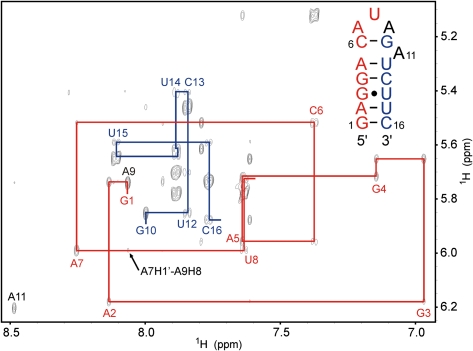
Both the WT and the del-A hairpins show similar NOEs, characteristic of an A-type helical stem, with strong NOEs between G3H1 and U14H3 confirming the formation of the G:U wobble base pair. In the WT hairpin, the helical characteristics of the NOE connectivities are extended to include the A5-C6 to G10-U12 steps of the helix (Fig. 3), implying that A11 is completely excluded from the interior of the helix and confirming the formation of a pseudo-triloop. The chemical shift of A11H8 is 8.48 ppm, which is very close to its so-called reference value, the chemical shift value when the adenine base is not affected by neighboring ring currents (Cromsigt et al. 2001). This is further evidence that A11 is completely destacked. The NOE patterns observed in the AUA loop parts of the two hairpins are very similar. If internucleotide NOEs that are not measurable because of peak overlap are excluded, only three NOEs from the WT hairpin were not observed for the del-A mutant and two NOEs from the del-A mutant were not observed for the WT hairpin. These five NOEs all belong to the group of very weak NOEs.
FIGURE 3.
The aromatic to H1′ region of the 300-msec NOESY of the WT hairpin. The sequential H1′↔H8/H6 NOE connectivity pathways observed through both sides of the helix are indicated. The A7H1′↔A9H8 cross peak discussed in the text is marked with an arrow. The aromatic and H1′ chemical shifts of G1 and A9 are both completely degenerate.
To assess the sugar conformations, DQF-COSY and TOCSY spectra were analyzed to determine the 3JH1′-H2′ coupling constants. In the WT hairpin with a pseudo-triloop, U8 adopts a C2′-endo conformation, while C6, A7, A9, G10, and A11 all show features of an equilibrium between C3′-endo and C2′-endo sugar puckers (see Supplemental Material). In the del-A hairpin, the sugar pucker pattern is similar except that G10 has a pure C2′-endo pucker and the equilibrium for A7 is shifted somewhat toward C2′-endo as indicated by a change in the 3JH1′-H2′ coupling constant from 5.0 Hz to 6.8 Hz. As expected, C3′-endo sugar puckers were observed for all helical nucleotides in the lower stem of the two helices, except for the terminal nucleotides G1 and C16, which are in an equilibrium between the C3′-endo and C2′-endo puckers.
The glycosidic torsion angle χ can be judged from the intensity of the intranucleotide H1′↔H8/H6 NOE in a short-mixing-time NOESY as the H1′-to-H8/H6 distance depends only on this angle and the chemical shifts of H2′ and H3′ protons (Cromsigt et al. 2001). A low-intensity NOE implies the helical anti conformation and a high-intensity NOE implies a syn conformation while a syn conformation shifts the H2′ and H3′ chemical shifts to higher values. For both hairpins, anti conformations were observed for all nucleotides except for A9 and G10 in the WT hairpin and A9 in the del-A hairpin (Fig. 3). These nucleotides displayed more intense H1′↔H8 NOEs than other nucleotides; however, a plot of the distance as a function of the NOE−6 showed that the intensity was too low to account for a pure syn conformation where one would anticipate an intensity on par with the cytosine H5↔H6 NOEs. In addition, the H2′ and H3′ chemical shifts of these nucleotides, between 4.60 and 4.95 ppm, reside in the high end of the normal range of chemical shifts covered by H2′ and H3′ in RNA. Thus, fast syn/anti exchanges appear likely rather than fixed syn or anti glycosidic angles for these nucleotides. Such an equilibrium is feasible as the energy barrier to interconversion is low for nucleotides, especially purines, with a C2′-endo sugar pucker (Foloppe et al. 2002).
The WT and del-A hairpin structures
The large and comparable number of restraints between the two hairpins enabled us to determine structures of high and similar quality (Table 2). Although the number of restraints per residue is of comparable size in the two hairpins, the heavy atom RMSD value of the WT hairpin is about twice that of the del-A hairpin. As seen in Table 2, this difference comes from the larger variation in the pseudo-triloop part of the WT molecule.
TABLE 2.
Structural statistics for the 20 lowest energy structures of the WT and del-A hairpins
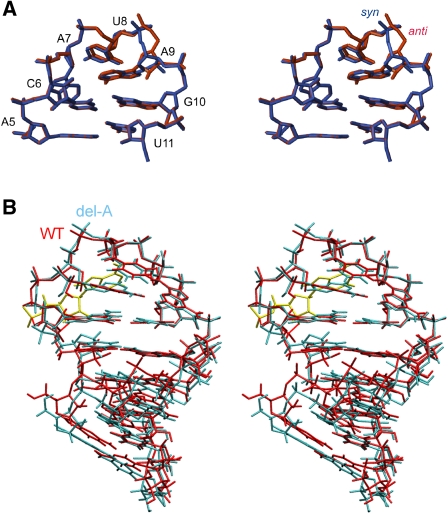
The sgRNA promoter consists of a 5-bp stem closed by a CAUAGA hexaloop that folds into pseudo-triloop with a transloop Watson–Crick base pair between C6 and G10 (Fig. 4). This base pair stacks on top of the A5:U12 base pair, leaving A11 completely bulged out of the helix. The position of A11 is not well-defined in the structure, but NOEs from A11H8 to A9H1′, G10H1′, and G10H4′ locate its nucleobase, transiently at least, in the minor groove pointing toward the AUA triloop part. The overall fold of the pseudo-triloop is similar to the structure of an AUA triloop. The nucleobase of A9 is inside the loop, where it stacks on top of the G10 nucleobase. U8 spans across A9 with its nucleobase pointing toward the major groove. The nucleobase of A7 forms the clamped adenine motif, where it is excluded from the interior of the loop and pushed into the major groove face pointing toward the phosphate group of A5. This position is stabilized by hydrogen bonds from the two amino protons of A7 to phosphate oxygens of A5 and C6, respectively. An additional hydrogen bond between the 2′-hydroxy group of A7 and the phosphate group of U8 is also observed. For the WT structure, these hydrogen bonds are observed in 12, 16, and 15 structures, respectively, of the 20 lowest-energy structures. In contrast, in the del-A hairpin, with the proper triloop structure, they occur in all of the 20 lowest-energy structures. A further noticeable, albeit minor, difference is a small change in the stacking geometry of A9's nucleobase upon G10. This is evidenced in the relatively large chemical shift differences between the hairpins observed for H1′, H4′, and H5′ (0.27 − 0.29 ppm) in the U8 ribose sugar, which is positioned right above the nucleobase of A9 and thus is strongly influenced by its ring current and, consequently, its relative position. The slightly different position of the nucleobase of A9 in the pseudo-triloop disrupts the hydrogen bond between its amino protons and A7N3 in contrast to the triloop structure, where this hydrogen bond is formed. The less-fixed position of A7's nucleobase is consistent with a shift in the C2′-endo/C3′-endo sugar equilibrium for this nucleotide.
FIGURE 4.
(A) Superposition of the ten structures with lowest energy calculated for the WT hairpin. (B) Stereoview of the structure with lowest energy. Hydrogen bonds from A7's amino group to phosphate oxygens are shown with green lines.
Both the C2′-endo/C3′-endo equilibrium of the ribose sugar and the syn/anti equilibrium of the glycosidic angle of A9 indicate conformational heterogeneity of the backbone at the 3′-side of the AUA triloop. The glycosidic angle of A9 is in neither a pure anti nor a pure syn conformation as indicated by the intermediate intensity of the H8↔H1′ intranucleotide NOEs and the H2′ and H3′ chemical shifts observed for both hairpins. Interestingly, structures calculated with the glycosidic angle of A9 restrained to either the syn or the anti conformation converge to identical loop structures. Rather than a change in the position of the A9 nucleobase, which stacks on top of G10, the backbone adjusts itself to accommodate the change in the glycosidic angle (Fig. 5A). Beyond the part flanking A9 and the bulged-out nucleotide A11, the backbone of the pseudo-triloop is rather well defined (Fig. 4).
FIGURE 5.
(A) Stereoview of an overlay of two calculated variants of the AUA triloop of the del-A hairpin. In blue is the χ-angle of A9 constrained to a syn conformation; in red, χ constrained to an anti conformation. (B) Stereoview of an overlay of the lowest energy structure of the WT (red) and del-A (blue) hairpins. The bulged-out adenine, A11, of the WT hairpin is shown in yellow. The structures are superimposed on the triloop and closing base pair.
The overall AUA loop structure determined for the del-A hairpin is identical to the WT (Fig. 5B). A superposition of the stem parts of the WT and del-A hairpins shows perfect convergence between the two stem parts, but the tip of the WT's pseudo-triloop is tipped slightly toward the major groove compared with the triloop in the del-A hairpin. The slight displacement of the pseudo-triloop originates from G10's dislocation toward the major groove, caused by the bulged A11 nucleotide, which increases the twisting of the A5:U12–C6:G10 step from ∼37° in the del-A hairpin to ∼42° in the WT hairpin. In addition, the backbone dynamics of G10 are dissimilar between the two hairpins as evidenced by the sugar pucker changing from a C2′-endo/C3′-endo equilibrium in the WT hairpin to a pure C2′-endo conformation in the del-A hairpin.
DISCUSSION
We here present the NMR structure of the subgenomic promoter of the BMV, which is the core promoter in the transcription of the sgRNA4 strand by the replicase (Smirnyagina et al. 1994). The hexaloop at the tip of the hairpin forms a pseudo-triloop with a transloop C6:G10 Watson–Crick base pair that stacks on top of the stem and is capped by an AUA triloop while A11 is excluded completely from the helix. The structural similarity between the subgenomic hairpin and the SLC hairpin, necessary in the promotion of the (+)-RNA strand transcription by the replicase, is striking. The NMR structure of the SLC hairpin (Protein Data Bank code: 1esh) (Kim et al. 2000) has already been determined, but we chose to determine the AUA triloop structure de novo with the stem of the subgenomic hairpin to achieve two completely comparable structures in terms of stem sequence, structure determination protocol, and quality of NMR data. A comparison of the SLC hairpin with the del-A hairpin shows an overall equivalent fold of the AUA triloop, although U8 seems to be better stacked upon A9 and the backbone is generally less well-defined in the del-A hairpin. The assignment of a higher number of internucleotide NOEs (40 in del-A compared with 30 in SLC) and the reassignment of two H5′ sugar protons led to the determination of an AUA triloop structure with a lower overall energy and less NOE and torsion angle violations.
A comparison of the replicase transcription efficiency (Kim et al. 2000) and the structural studies done of SLC hairpin mutants (Kim and Tinoco 2001) indicates that, in particular, the formation of the clamped adenine motif is important for efficient recruitment of the replicase. A disruption of this structural feature leads to a dramatic decrease in the transcription efficiency, whereas structural heterogeneity on the 3′ side of the triloop is tolerated. The introduction of the extrahelical adenosine in the loop of the subgenomic promoter hairpin, compared with the SLC hairpin, has only minor effects on the structure of the C-AUA-G part of the loop as proven by the small chemical shift, NOE, and sugar pucker differences between the WT and del-A hairpins. However, the bulged-out adenine base has a conspicuous influence on the stability of the pseudo-triloop, lowering it by 1.5 kcal/mol relative to the triloop (Table 1). The lower stability of the pseudo-triloop is clearly visible in the NMR spectra and the final structural ensemble. In the subgenomic promoter, the 2 bp flanking the bulged-out adenosine are weakened as shown by the broadening of the imino signals. In addition, a hydrogen bond between A7N3 and A9H61 is not formed at all, and three hydrogen bonds fixing the position of A7 are observed with lower frequency in the ensemble of structures calculated for the WT hairpin compared with the del-A hairpin. The difference in detailed loop structure is evidenced directly in the spectra as we observe a weak A7H1′↔A9H8 NOE in the WT hairpin but not in the del-A hairpin despite similar spectral quality (Fig. 3).
From a biological point of view, it is highly pertinent if the pseudo-triloop structure forms at the temperature used for in vitro transcription experiments, which is 28°C–30°C. The imino proton of G10 shows an exchange broadened peak in the spectrum recorded at 5°C, and in the spectrum recorded at 25°C, it is broadened beyond observation. This, however, is not proof that the C6:G10 transloop base pair is not formed at 25°C as Watson–Crick base pairs can be formed even though the imino proton is not observed (Luy and Marino 2000). In the D2O spectra recorded at 25°C, numerous cross-strand and inter-loop NOEs from nonexchangeable protons are observed. We performed two test calculations to probe the structure at 25°C. First, we excluded the C6:G10 base-pairing restraints and, second, we excluded all restraints from exchangeable protons, retaining only restraints directly observed at 25°C. Both these calculations converged to families of structures similar to the structures calculated with all restraints included, albeit convergence was slightly poorer and, most crucially, the pseudo-triloop and clamped adenine motif were formed in both sets of structures. Thus, the subgenomic promoter, via the numerous hydrogen bonds and efficient nucleobase stacking, forms an unusually well-packed and rigid pseudo-triloop structure at 25°C.
Two alternate hypotheses have been proposed on the mechanism by which sgRNA synthesis is initiated. As outlined in the introduction, one hypothesis is that the replicase recognizes four key residues, G4, C6, A7, and G10 (our numbering), followed by an induced fit between the replicase and the promoter RNA (Adkins and Kao 1998) but, in vivo, the formation of a stem is also necessary for replication (Sivakumaran et al. 2004). The other hypothesis promulgates the formation of a hairpin in the subgenomic promoter that is structurally equivalent to the genomic promoter (Haasnoot et al. 2000, 2002). Subgenomic (+)-strand RNA4 synthesis would then be initiated by a mechanism similar to genomic (−)-strand RNA synthesis from either a full-length or a prematurely terminated (−)-strand RNA3.
Our structure shows that a hairpin is indeed formed in the core promoter sequence at the temperature of in vitro experiments and that the tip of it structurally resembles the core promoter for genomic (–)-strand RNA synthesis with only minor structural differences. Thus it seems very likely that the replicase uses the same mechanism for both subgenomic and genomic promoter recognition, i.e., recognition of the clamped adenine structural motif rather than recognition of a specific RNA sequence. In this respect, BMV does not differ from the related alfalfa mosaic virus (AMV), for which it has been shown that the genomic promoter could substitute the subgenomic promoter in vitro and in vivo (Olsthoorn et al. 2004). Interestingly, the AMV promoters for sgRNA and gRNA synthesis form a hairpin with a regular triloop but can be functionally replaced by hairpins with a pseudo-triloop, like HBV ɛ (Flodell et al. 2002, 2006; Haasnoot et al. 2003), HIV-1 TAR (Haasnoot et al. 2003) and the BMV subgenomic promoter (RCL Olsthoorn, unpubl.).
Our structure can also be reconciled with the four key nucleotides as determined by Kao and coworkers (Adkins and Kao 1998) as C6 and G10 are the two nucleotides forming the transloop base pair and A7 is the adenine that creates the clamped adenine motif. Mutation of these residues would corrupt the pseudo-triloop and clamped adenine motifs. The fourth key residue, G4, is not involved in formation of the pseudo-triloop at the tip of the hairpin but might serve to stabilize the stem of the hairpin through formation of the G4:C13 base pair. That this G is not recognized in a specific manner is evident as the base pair can be switched to a C:G pair and still retain template activity in vitro (Haasnoot et al. 2002) and in vivo (Sivakumaran et al. 2004), and in the genomic promoter there is a U:A pair at this position. The tip of the SLC hairpin of the genomic promoter is more stable than that of the subgenomic promoter hairpin, explaining why a U:A base pair might be tolerated at this position. In the delineation of the specific chemical moieties needed to elicit replicase activity in the four key nucleotides, only the chemical groups that hydrogen bond in a Watson–Crick base pair were indispensable for C6 and G10 (Siegel et al. 1998). Interestingly, removal of G10's 2′-hydroxy group increased the replicase activity to 145%. This chemical alteration would preorganize G10 for a C2′-endo sugar pucker, which is indeed the pucker we and Tinoco and coworkers observe experimentally in both the subgenomic and genomic hairpins (Kim et al. 2000).
Thermodynamic analysis supports the claim that changes to the four key residues compromise the entire tip motif. When the transloop C:G Watson–Crick base pair in the pseudo-triloop is replaced by C:C or G:G mismatches, we observed a decrease of the stability of the hairpin (Table 1). Biological data show that a loss of the transloop C:G base-pairing by introduction of a C:C or a G:G mismatch leads to a complete loss of the subgenomic hairpin's promoter activity (Haasnoot et al. 2000), and as the transloop base-pairing is required to form the AUA triloop at the tip of the hairpin, it is obvious to conclude that the formation of the important clamped adenine motif is abolished in these mutants. Replacement of the transloop C:G base pair with a G:C base pairs leads to a substantial decrease in stability, owing to unfavorable enthalpy. The strong relationship to triloop structures can explain this destabilization, as the general trend is higher stability of loops closed by a C:G base pair as compared with a G:C base pair (Shu and Bevilacqua 1999). Nonetheless, as some biological activity is retained, 17% of the WT, it appears the compactness of the loop is, at least partly, conserved.
As the pseudo-triloop at the tip of the subgenomic promoter hairpin can be replaced with a regular AUA triloop without compromising template activity, it is not obvious what the biological significance of the bulged A is. The subgenomic promoter hairpin is less stable than the genomic promoter hairpin (SLC), and the bulged A destabilizes it by a further 1.5 kcal/mol. As the promoters for genomic (−)-strand RNAs and sgRNAs are likely to be initiated by a similar mechanism, the highly regulated levels and timing of BMV genomic (−)-strand RNA and sgRNA must be due to other factors than the promoter itself. The viral coat protein is involved in the regulation of RNA translation through interactions with the B-box RNA element that directs replicase assembly and the SLC (Zhu et al. 2007; Yi et al. 2009a,b; Kao et al. 2011). There is also some indication that the coat protein is required for efficient template activity (Gopinath et al. 2005). Given the structural similarity between the SLC and the subgenomic promoter hairpin, it is conceivable that the coat protein also binds to this hairpin, although it is still not known what the effect of the bulged A is. It is, however, tempting to link the difference in hairpin stability between the genomic and subgenomic promoters, exacerbated by the bulged A, to the mechanism by which the replicase, with or without the assistance of the coat protein, regulates between gRNA and sgRNA transcription.
To summarize, we have shown that the core subgenomic promoter from BMV forms a hairpin with a pseudo-triloop and a clamped adenine motif at the tip. Thus it seems likely that sgRNA synthesis is initiated by a mechanism similar to gRNA synthesis, as proposed by Olsthoorn and coworkers (Haasnoot et al. 2000, 2002). This is also consistent with the later results from Kao and coworkers who showed that in addition to the four key nucleotides proposed, the stem of the hairpin is also required for in vivo RNA4 synthesis (Sivakumaran et al. 2004). Furthermore, our structure and thermodynamic analysis supports that three of the key residues identified by Kao and coworkers are essential to form the pseudo-triloop and clamped adenine motifs. It still remains an open question whether the sgRNA synthesis is initiated internally from a full-length (−)-strand RNA3 or from a prematurely terminated one as both models are compatible with the formation of the subgenomic promoter hairpin. This question will have to await further biological studies.
MATERIALS AND METHODS
UV spectroscopy
The five RNA strands were purchased gel-purified from Dharmacon RNA technologies. The five Tm samples were all prepared to a final concentration of 10 mM NaCl, 10 mM phosphate buffer, 1 mM EDTA, and ∼5 μM of the given RNA strand. To ensure a folding into hairpins, the samples were heated for 5 min to 90°C followed by snap cooling on ice. The absorbance of the samples at 260 nm were measured from 10°C to 90°C with a 1°C/min increase, in a PerkinElmer Lambda 35 spectrophotometer equipped with a thermo regulated Peltier Element. The Tms were deduced from the first derivatives of the absorbance curves, and the thermodynamic parameters were extracted from a two-state analysis of the absorbance curves (Mergny and Lacroix 2003).
NMR spectroscopy
After a desalting step on 1-kDa filters, each RNA oligomer was dissolved in 500 μL of water containing 10 mM sodium phosphate buffer (pH 7), 50 mM sodium chloride, and 2 mM sodium EDTA, to a final RNA concentration of 1–2 mM. For both samples, 100-msec NOESY, 300-msec NOESY, 90-msec TOCSY, DQF-COSY, 1H-13C HSQC, and 1H-31P COSY spectra were acquired at 25°C in 99,96% D2O. We acquired 250 msec WATERGATE-NOESY spectra at 5°C in 10% D2O/90% H2O. All spectra were recorded on a Varian Inova 500 MHz spectrometer. The NMR data were processed with NmrPipe (Delaglio et al. 1995), and the spectra were visualized and assigned in SPARKY 3 (University of California, San Francisco, CA).
Assignment and NMR restraints
The assignment of 1H, 13C, and 31P chemical shifts was performed by standard methods (Varani et al. 1996). Distance restraints between nonexchangeable protons were qualitatively grouped as either strong (1.8–3.0 Å), medium (1.8–4.5 Å), weak (3.0–6.0 Å), or very weak (3.0–7.0 Å) depending on the NOE intensity compared with the H5-H6 (2.4 Å) NOE in the 100-msec and 300-msec NOESY spectra. The distance restraints originating from exchangeable protons in the 300-msec WATERGATE-NOESY spectra were grouped into distance intervals of either 2.0–6.0 Å or 3.0–7.0 Å. Where slow imino proton exchange and NOEs indicated base-pairing, three or two hydrogen bond restraints (1.74–2.10 Å) were placed on each G:C, A:U, or G:U base pair, respectively.
If no H1′↔H2′ cross peak was observed in the TOCSY spectrum (3JH1′-H2′ < 2 Hz), the nucleotide was restrained to a C3′-endo sugar pucker conformation. Residues with an observable H1′↔H2′ cross peak were not restrained, unless if 3JH1′-H2′ > 8 Hz when the nucleotide was restrained to a C2′-endo sugar conformation (Wijmenga and van Buuren 1998).
For phosphate groups in the stem part of the molecules, apart from those facing the closing base pair of the loop, the α and ζ torsion angles were restrained to standard A-type helical values if their 31P chemical shift was within the narrow range expected for a standard A-type helix. If four bond H4′-P couplings were observed in the 1H-31P COSY, torsion angle restrains were applied to ensure a near planar W-conformation between the two atoms (see Supplemental Material) (Sarma et al. 1973). No additional torsion angle restraints were placed on the sugar–phosphate backbone.
If a weak H1′↔H8/H6 NOE was observed, the χ angle was restrained to an anti conformation according to the sugar conformation. Where a stronger H1′↔H8/H6 was observed, though not strong enough to be a pure syn conformation, no restrains were applied on the χ angle. These cases coincided with H2′ and H3′ chemical shifts at the high end of the normal range for nucleotides with anti conformations, as discussed in the Results section.
Structure calculation
An extended unfolded RNA strand was built, and starting from this structure, 100 calculations were performed with different initial atom velocities in XPLOR version 3.851 (Brunger 1996) with the forcefield described by Cornell et al. (1995). The protocol followed a torsion angle dynamics, with heating from 300K to 20,000K in 60 psec and then a cooling from 20,000K to 1000K in 270 psec, with a time step of 2.5 fsec. Subsequently, a molecular dynamics cooling for 1000K to 200K in 50 psec with a time step of 0.5 fsec was carried out. Finally 1000 steps of Powell minimization was performed. The force constant of the dihedral restraints was set to 50 kcal mol−1 rad−2, and the force constant on the NOE restraints were varied from 5 to 150 kcal mol−1 Å−2. The electrostatic contribution was applied under the Verlet dynamics and the final minimization step with an 1/r dependency.
The 20 structures with the lowest overall energy where selected, viewed, compared, and analyzed in MOLMOL (Koradi et al. 1996). Helix parameters were analyzed using CURVES 5.1 (Lavery and Sklenar 1988).
DATA DEPOSITION
Atomic coordinates have been deposited in the Protein Data Bank with accession codes 2LP9 (WT) and 2LPA (del-A).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank the Danish National Research Foundation and the Danish Centre for Scientific Computing for financial support. P.P. was supported by a Marie Curie Early Stage Research Training Fellowship of the European Community's Sixth Framework Programme under contract no. MEST-CT-2004-504018. We thank Prof. C. Cheng Kao for valuable suggestions in the writing process.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.029918.111.
REFERENCES
- Adkins S, Kao CC 1998. Subgenomic RNA promoters dictate the mode of recognition by bromoviral RNA-dependent RNA polymerases. Virology 252: 1–8 [DOI] [PubMed] [Google Scholar]
- Brunger AT 1996. X-PLOR version 3.851. Yale University Press, New Haven, CT. [Google Scholar]
- Cornell WD, Cieplak P, Bayly I, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA 1995. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J Am Chem Soc 117: 5179–5197 [Google Scholar]
- Cromsigt JAMTC, Hilbers CW, Wijmenga SS 2001. Prediction of proton chemical shifts in RNA. J Biol NMR 21: 11–29 [DOI] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A 1995. NMRpipe: A multidimensional spectral processing system based on UNIX pipes. J Biol NMR 6: 277–293 [DOI] [PubMed] [Google Scholar]
- Flodell S, Schleucher J, Cromsigt J, Ippel H, Kidd-Ljunggren K, Wijmenga S 2002. The apical stem-loop of the hepatitis B virus encapsidation signal folds into a stable tri-loop with two underlying pyrimidine bulges. Nucleic Acids Res 30: 4803–4811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flodell S, Petersen M, Girard F, Zdunek J, Schleucher J, Kidd-Ljunggren K, Wijmenga SS 2006. Structure of the apical loop of the Hepatitis B virus encapsidation signal. Nucleic Acids Res 34: 4449–4457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foloppe N, Hartmann B, Nilsson L, MacKerell AD 2002. Intrinsic conformational energetics associated with the glycosyl torsion in DNA: A quantum mechanical study. Biophys J 82: 1554–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopinath K, Dragnea B, Kao C 2005. Interaction between Brome mosaic virus proteins and RNAs: effects on RNA replication, protein expression, and RNA stability. J Virol 79: 14222–14234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haasnoot PCJ, Brederode FT, Olsthoorn RCL, Bol JF 2000. A conserved hairpin structure in Alfamovirus and Bromovirus subgenomic promoters is required for efficient RNA synthesis in vitro. RNA 6: 708–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haasnoot PCJ, Olsthoorn RCL, Bol JF 2002. The Brome mosaic virus subgenomic promoter hairpin is structurally similar to the iron-responsive element and functionally equivalent to the minus-strand core promoter stem-loop C. RNA 8: 110–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haasnoot PC, Bol JF, Olsthoorn RC 2003. A plant virus replication system to assay the formation of RNA pseudotriloop motifs in RNA–protein interactions. Proc Natl Acad Sci 100: 12596–12600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao CC, Sivakumaran K 2000. Brome mosaic virus, good for an RNA virologist's basic needs. Mol Plant Pathol 1: 91–97 [DOI] [PubMed] [Google Scholar]
- Kao CC, Ni P, Hema M, Huang XL, Dragnea B 2011. The coat protein leads the way: An update on basic and applied studies with the Brome mosaic virus coat protein. Mol Plant Pathol 12: 403–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CH, Tinoco I 2001. Structural and thermodynamic studies on mutant RNA motifs that impair the specificity between a viral replicase and its promoter. J Mol Biol 307: 827–839 [DOI] [PubMed] [Google Scholar]
- Kim CH, Kao CC, Tinoco I 2000. RNA motifs that determine specificity between a viral replicase and its promoter. Nat Struct Biol 7: 415–423 [DOI] [PubMed] [Google Scholar]
- Koradi R, Billeter M, Wüthrich K 1996. MOLMOL: A program for display and analysis of macromolecular structures. J Mol Graph 14: 51–55 [DOI] [PubMed] [Google Scholar]
- Lavery R, Sklenar H 1988. The definition of generalized helicoidal parameters and for axis curvature of irregular nucleic acids. J Biomol Struct Dyn 6: 63–91 [DOI] [PubMed] [Google Scholar]
- Luy B, Marino JP 2000. Direct evidence for Watson-Crick base pairs in a dynamic region of RNA structure. J Am Chem Soc 122: 8095–8096 [Google Scholar]
- Marsh LE, Dreher TW, Hall TC 1988. Mutational analysis of the core and modulator sequences of the BMV RNA3 subgenomic promoter. Nucleic Acids Res 16: 981–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mergny JL, Lacroix L 2003. Analysis of thermal melting curves. Oligonucleotides 13: 515–537 [DOI] [PubMed] [Google Scholar]
- Miller WA, Koev G 2000. Synthesis of subgenomic RNAs by positive-strand RNA viruses. Virology 273: 1–8 [DOI] [PubMed] [Google Scholar]
- Miller WA, Dreher TW, Hall TC 1985. Synthesis of Brome mosaic virus subgenomic RNA in vitro by internal initiation on (−)-sense genomic RNA. Nature 313: 68–70 [DOI] [PubMed] [Google Scholar]
- Olsthoorn RC, Haasnoot PC, Bol JF 2004. Similarities and differences between the subgenomic and minus-strand promoters of an RNA plant virus. J Virol 78: 4048–4053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao ALN. 2001. Bromoviruses. In Encyclopedia of plant pathology (ed. OC Maloy, TD Murray), pp. 155–158. John Wiley & Sons, Toronto. [Google Scholar]
- Sarma RH, Mynott RJ, Wood DJ, Hruska FE 1973. Determination of the preferred conformations constrained along the C4′-C5′ and C5′-O5′ bonds of β-5′-nucleotides in solution. Four-bond 31P-1H coupling. J Am Chem Soc 95: 6457–6459 [DOI] [PubMed] [Google Scholar]
- Shu Z, Bevilacqua PC 1999. Isolation and characterization of thermodynamically stable and unstable RNA hairpins from a triloop combinatorial library. Biochemistry 38: 15369–15379 [DOI] [PubMed] [Google Scholar]
- Siegel RW, Bellon L, Beigelman L, Kao CC 1998. Moieties in an RNA promoter specifically recognized by a viral RNA-dependent RNA polymerase. Proc Natl Acad Sci 95: 11613–11618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivakumaran K, Choi S-K, Hema M, Kao CC 2004. Requirements for Brome mosaic virus subgenomic RNA synthesis in vivo and replicase-core promoter interactions in vitro. J Virol 78: 6091–6101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnyagina E, Hsu YH, Chua N, Ahlquist P 1994. Second-site mutations in the Brome mosaic-virus RNA3 intercistronic region partially suppress a defect in coat protein messenger-RNA transcription. Virology 198: 427–436 [DOI] [PubMed] [Google Scholar]
- Sztuba-Solinska J, Stollar V, Bujarski JJ 2011. Subgenomic messenger RNAs: mastering regulation of (+)-strand RNA virus life cycle. Virology 412: 245–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varani G, Aboul-ela F, Allain FHT 1996. NMR investigation of RNA structures. Prog Nucl Magn Reson Spectrosc 29: 51–127 [Google Scholar]
- Wijmenga SS, van Buuren BNM 1998. The use of NMR methods for conformational studies of nucleic acids. Prog Nucl Magn Reson Spectrosc 32: 287–387 [Google Scholar]
- Yi G, Letteney E, Kim CH, Kao CC 2009a. Brome mosaic virus capsid protein regulates accumulation of viral replication proteins by binding to the replicase assembly RNA element. RNA 15: 615–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi GH, Vaughan RC, Yarbrough I, Dharmaiah S, Kao CC 2009b. RNA binding by the Brome mosaic virus capsid protein and the regulation of viral RNA accumulation. J Mol Biol 391: 314–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Gopinath K, Murali A, Yi GH, Hayward SD, Zhu H, Kao C 2007. RNA-binding proteins that inhibit RNA virus infection. Proc Natl Acad Sci 104: 3129–3134 [DOI] [PMC free article] [PubMed] [Google Scholar]