Abstract
Methylation of histones is central to chromatin regulation, and thus novel mechanisms regulating genome function can be revealed through the discovery of new histone methyl marks. Here, we identify Set5 as the first histone H4 methyltransferase in budding yeast, which monomethylates the important H4 lysine residues 5, 8 and 12. Set5’s enzymatic activity functions together with the global chromatin-modifying complexes COMPASS and NuA4 to regulate cell growth and stress responses.
A major regulatory mechanism of chromatin function is the dynamic covalent post-translational modification (PTM) of histone N-terminal tails, which controls the structure and accessibility of chromatin to influence diverse processes such as gene expression and DNA repair1. One critical histone modification is the methylation of lysine (K) residues, catalyzed by lysine methyltransferases (KMTs), which add a mono-, di- or trimethyl mark. The most extensively characterized Saccharomyces cerevisiae KMTs are the evolutionarily conserved enzymes Set1, Set2 and Dot1, which methylate lysines 4, 36 and 79 of histone H3, respectively2. Proteomic studies highlight the existence of numerous methylation marks on the other core histones (H4, H2A, and H2B) for which the KMTs remain to be discovered3,4, suggesting that new mechanisms of genome regulation will be revealed through the identification and analysis of the enzymes that generate these histone marks.
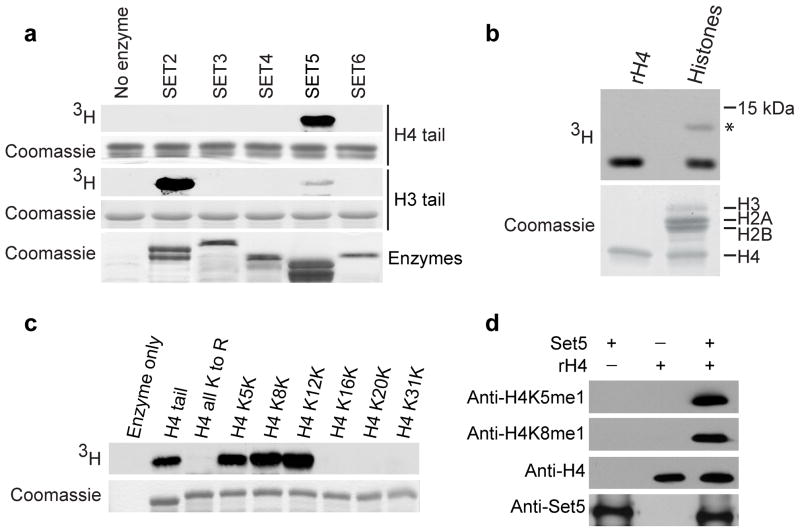
To identify new methylation events at chromatin, we performed a screen for activity of yeast candidate KMTs on histone tails. In vitro methylation assays of the uncharacterized KMTs (Supplementary Methods, Supplementary Table 1) revealed robust methylation on the histone H4 tail by the enzyme Set5 (Fig. 1a). Set5 also methylated full-length recombinant H4 and native H4 present in bulk purified histones (Fig. 1b). Set5 showed weak activity on purified nucleosomes in vitro, although it was able to methylate H4 in chromatin fractions from yeast (data not shown). The N-terminal tail of H4 contains 6 lysine (K) residues (Supplementary Fig. 1a), none of which were known targets of methyltransferases in budding yeast. To map the sites of methylation, a library of H4 tail constructs in which all lysines have been mutated to arginine except at one position was generated. Set5 only methylated H4 tails that maintained a lysine at position 5, 8 or 12 (Fig. 1c, Supplementary Fig. 1b). Additionally, Set5 was able to methylate H2A at lysines 4 and 7 (Supplementary Fig. 1c), although to a lesser extent than H4 (H2A represented by * in Fig. 1b).
Figure 1.
Identification of Set5 as an H4 methyltransferase. (a) Methylation assays with recombinant SET domain enzymes, H3 and H4 tails using 3H-SAM as the methyl donor. Autoradiograph and Coomassie (loading control) are shown. (b) Set5 methylates recombinant H4 (rH4) and H4 from native histones purified from calf thymus (* indicates H2A). (c) Set5 methylates lysines 5, 8 and 12. Methylation assay on H4 tails with lysines mutated to arginine (all K to R) or single lysines maintained and all other lysines mutated to arginine. (d) Monomethylation of H4K5 and H4K8 on rH4. Immunoblots with the indicated antibodies are shown.
Mass spectrometry analysis of an H4 peptide methylated by Set5 revealed that Set5 is able to monomethylate lysines 5, 8 and 12 (Supplementary Fig. 1d,e). We next raised antibodies against the H4K5me1 and H4K8me1 epitopes (anti-H4K5me1 and anti-H4K8me1, respectively), which showed high specificity in peptide dot blot assays (Supplementary Fig. 2). Using these antibodies, H4K5me1 and H4K8me1 were detected on recombinant H4 methylated by Set5 (Fig. 1d). Taken together, these data demonstrate that Set5 can monomethylate lysines 5, 8 and 12 of histone H4 in vitro.
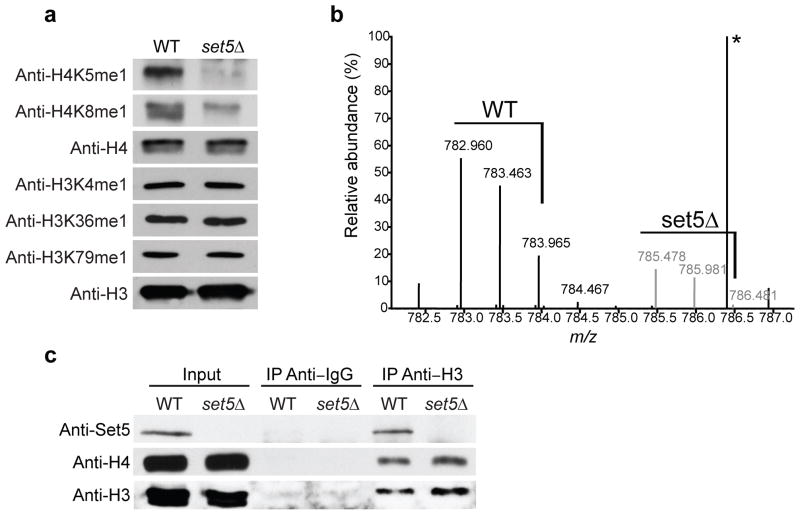
To determine if the H4 tail is monomethylated by Set5 in cells, we purified histones from wild type and set5Δ yeast and probed them with our monomethyl-specific antibodies. set5Δ yeast showed a specific decrease in the H4K5me1 and H4K8me1 signal compared to wild type (Fig. 2a). Quantitative mass spectrometry of the purified histones demonstrated a substantial decrease in the amount of monomethyl H4 species in set5Δ cells (Fig. 2b). The MS/MS spectrum revealed the existence of all three monomethyl marks, with H4K8me1 as the predominant species (Supplementary Fig. 3a). These data provide evidence that H4K5me1, H4K8me1 and H4K12me1 exist in vivo. Furthermore, we show that Set5 is required for H4K5me1 and H4K8me1 generation in cells.
Figure 2.
Set5 monomethylates H4 in cells. (a) Histones from WT and set5Δ yeast were subjected to immunoblotting with the indicated antibodies. (b) Quantitative mass spectrometry of purified histone H4 from WT (chemically-labeled with D0-propionyl; black), and from set5Δ (D5-propionyl; gray) shows a peptide corresponding to monomethylation of H4, amino acids 4–17. Relative abundance of this peptide decreases in the set5Δ cells (black versus gray signal; *non-specific peak). (c) Set5 co-precipitates with H3 and H4. Immunoprecipitation using anti-H3 or IgG was performed from WT and set5Δ cells; immunoblots are shown.
Our data suggest that Set5 might interact with chromatin to methylate H4. Consistent with this hypothesis, immunofluorescence revealed nuclear and cytoplasmic localization of tagged Set5 (Supplementary Fig. 3b). We generated an antibody against recombinant Set5 and confirmed that Set5 is present in both soluble and chromatin-enriched fractions of yeast lysates (Supplementary Fig. 3c), suggesting that Set5 associates with chromatin. Furthermore, co-immunoprecipitation experiments demonstrated an interaction between Set5 and histones H3 and H4 in vivo (Fig. 2c). Overall, these results argue that Set5 is a novel chromatin-associated methyltransferase that monomethylates histone H4 in cells.
Genome-scale studies performed with the commercial homozygous diploid Yeast Knockout (YKO) collection have reported defects in cellular stress responses and increased sporulation in the absence of Set55–8. However, we did not observe sensitivity of multiple independently generated set5Δ strains to the reported cellular stresses nor an increase in sporulation efficiency (Supplementary Fig. 4a,b; yeast strains listed in Supplementary Table 2). Further phenotypic analysis showed that Set5 does not appear to be regulated by or contribute to the control of the cell cycle (Supplementary Fig. 4c,d). Finally, deletion of SET5 resulted in only minor changes in global gene expression (data not shown), indicating that loss of Set5 alone does not drastically affect global transcription.
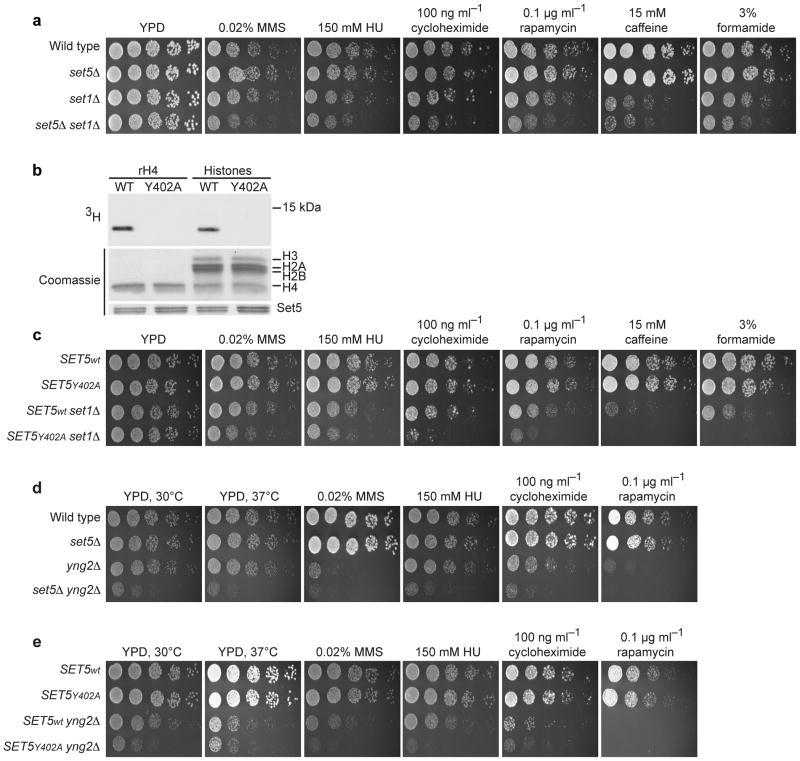
Chromatin function is frequently regulated by crosstalk between different histone-modifying complexes2. To determine if Set5 functionally interacts with other SET domain candidate KMTs, double mutants were generated and subjected to genotoxic and cellular stresses. Intriguingly, of the six SET domain-containing proteins analyzed, only a set5Δ set1Δ strain showed decreased fitness in the presence of stress compared to either single mutant (Fig. 3a, Supplementary Fig. 5a). Moreover, a set1Δ strain harboring an integrated point mutation of a conserved catalytic residue (Y402A) in the SET5 gene (Fig. 3b and Supplementary Fig. 5b,c) showed similar and even stronger phenotypes to that of set5Δ set1Δ cells, indicating that the functional genetic interaction between SET1 and SET5 is dependent on the catalytic activity of Set5 (Fig. 3c).
Figure 3.
Set5 functions with COMPASS and NuA4 complexes to regulate cell growth and stress responses. (a) Decreased fitness of set5Δ set1Δ upon stress. Four-fold serial dilutions of yeast were spotted and grown for 2–5 days at 30°C. (MMS: methylmethane sulfonate; HU: hydroxyurea) (b) Mutation of the conserved Y402 to alanine abolishes Set5’s catalytic activity. Methylation assay with wild type or Y402A Set5 on rH4 and native histones. (c) The catalytic activity of Set5 is required for normal growth of set1Δ cells under stress. Deletion (d) or catalytic inactivation (e) of SET5 in yng2Δ strains impairs cell growth.
Set1 is an integral member of the multi-subunit chromatin-modifying COMPASS complex that mediates methylation of histone H3 at lysine 4 (H3K4)9,10. We observed a subtle but reproducible growth defect upon stress in set5Δ cells lacking the COMPASS component Sdc1 (Supplementary Fig. 5d), providing additional evidence that SET5 may functionally interact with COMPASS. Interestingly, H3K4R-expressing yeast, which were unable to grow in the stress conditions tested except cycloheximide, showed slower growth upon deletion of SET5 in cycloheximide (Supplementary Fig. 5e). Together, these results suggest cooperation between methylation at H3K4 by Set1 and methylation at the H4 tail by Set5 to regulate cell growth under stress conditions.
Set5’s target residues on H4 are also subject to acetylation by the NuA4 complex, which plays important roles in cell growth and division11. Yeast lacking Yng2—a core component of NuA4—are compromised for global histone H4 acetylation but remain viable with slow growth12. Surprisingly, cells lacking both YNG2 and SET5, or YNG2 and Set5’s catalytic activity, were more severely impaired for growth than yng2Δ alone. A comparable growth defect was observed in double mutants under both normal and stress conditions (Fig. 3,d,e, Supplementary Fig. 5f). We postulate that methylation and acetylation of H4 either (1) exist in distinct histone subpopulations or (2) are present on different lysines of the same H4 tail, and thereby cooperate to promote cellular fitness. It is also possible that the functional interaction is dependent on other substrates of these enzymes. Regardless, these results demonstrate that Set5 and Yng2 function in parallel to promote optimal cell growth.
Consistent with our genetic interactions between SET5 and members of COMPASS and NuA4, impaired growth is observed when both COMPASS and NuA4 are mutated13. Combined mutation of their target histone residues results in a severe growth defect14,15 (Supplementary Fig. 5g). This phenotype is likely to be largely dependent on acetylation, but the existence of methylation at H4 lysines 5, 8 and 12 and SET5’s genetic interactions argue that methylation may contribute to H4 mutant phenotypes as well.
In summary, we demonstrate novel monomethylation events on histone H4 at lysines 5, 8 and 12, which are catalyzed by the first known H4 methyltransferase in budding yeast, Set5. We have implicated Set5’s function in cellular fitness and the response to stress together with two global chromatin-modifying complexes, COMPASS and NuA4. Thus, Set5-dependent methylation adds a new layer of regulation to chromatin-based signaling networks. This work expands our current knowledge of the epigenome, increasing the complexity of the array of known histone modifications, and highlights new mechanisms by which histone methylation impacts cellular fitness.
Supplementary Material
Acknowledgments
The authors are grateful to Ashby Morrison for critical discussions regarding the work and to members of the Gozani and Chua labs for insight. The authors also acknowledge Sharon Dent (The University of Texas MD Anderson Cancer Center), Scott Briggs (Purdue University) and Martha Cyert (Stanford University) for yeast strains and reagents. This work was supported in part by grants from the NIH to O.G. (R01 GM079641) and to B.A.G (DP2OD007447), and an NSF CAREER award (B.A.G.). G.M. was funded by the Postdoctoral Fellowship from the Ministerio de Ciencia e Innovación (Spain) and E.M.G acknowledges support from the Cancer Biology Program Postdoctoral Fellowship at Stanford University. N.L.Y was supported with an NIH F32 NRSA. O.G. is a recipient of an Ellison Senior Scholar in Aging Award.
Footnotes
Author Contributions
E.M.G., G.M. and O.G. conceived of and designed the experiments and wrote the manuscript. G.M. and E.M.G. contributed equally to the work and performed all biochemical and cellular experiments. Mass spectrometry analysis was performed by N.L.Y. and B.A.G.
References
- 1.Bannister AJ, Kouzarides T. Cell Res. 2011;21:381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Millar CB, Grunstein M. Nat Rev Mol Cell Biol. 2006;7:657–666. doi: 10.1038/nrm1986. [DOI] [PubMed] [Google Scholar]
- 3.Garcia BA, et al. J Biol Chem. 2007;282:7641–7655. doi: 10.1074/jbc.M607900200. [DOI] [PubMed] [Google Scholar]
- 4.Tan M, et al. Cell. 2011;146:1016–1028. doi: 10.1016/j.cell.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sinha H, et al. Genetics. 2008;180:1661–1670. doi: 10.1534/genetics.108.092932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hanway D, et al. PNAS. 2002;99:10605–10610. doi: 10.1073/pnas.152264899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Teixeira MC, Raposo LR, Mira NP, Lourenco AB, Sa-Correia I. Appl Environ Microbiol. 2009;75:5761–5772. doi: 10.1128/AEM.00845-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deutschbauer AM, Williams RM, Chu AM, Davis RW. PNAS. 2002;99:15530–15535. doi: 10.1073/pnas.202604399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miller T, et al. PNAS. 2001;98:12902–12907. doi: 10.1073/pnas.231473398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krogan NJ, et al. J Biol Chem. 2002;277:10753–10755. doi: 10.1074/jbc.C200023200. [DOI] [PubMed] [Google Scholar]
- 11.Choy JS, Tobe BT, Huh JH, Kron SJ. J Biol Chem. 2001;276:43653–43662. doi: 10.1074/jbc.M102531200. [DOI] [PubMed] [Google Scholar]
- 12.Nourani A, et al. Mol Cell Biol. 2001;21:7629–7640. doi: 10.1128/MCB.21.22.7629-7640.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krogan NJ, et al. PNAS. 2004;101:13513–13518. doi: 10.1073/pnas.0405753101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma XJ, Wu J, Altheim BA, Schultz MC, Grunstein M. PNAS. 1998;95:6693–6698. doi: 10.1073/pnas.95.12.6693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Durrin LK, Mann RK, Kayne PS, Grunstein M. Cell. 1991;65:1023–1031. doi: 10.1016/0092-8674(91)90554-c. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.