Abstract
The dualism of genetic predisposition and environmental influences, their interactions, and respective roles in shaping the phenotype have been a hot topic in biological sciences for more than two centuries. Heritable epigenetic variation mediates between relatively slowly accumulating mutations in the DNA sequence and ephemeral adaptive responses to stress, thereby providing mechanisms for achieving stable, but potentially rapidly evolving phenotypic diversity as a response to environmental stimuli. This suggests that heritable epigenetic signals can play an important role in evolutionary processes, but so far this hypothesis has not been rigorously tested. A promising new area of research focuses on the interaction between the different molecular levels that produce phenotypic variation in wild, closely-related taxa that lack genome-wide genetic differentiation. By pinpointing specific adaptive traits and investigating the mechanisms responsible for phenotypic differentiation, such study systems could allow profound insights into the role of epigenetics in the evolution and stabilization of phenotypic discontinuities, and could add to our understanding of adaptive strategies to diverse environmental conditions and their dynamics.
1. Introduction
Patterns and causes of biological variation have fascinated and challenged natural scientists for a long time. The Darwinian evolutionary theory highlights the importance of natural variation as raw material upon which selection processes can act, thereby increasing the fitness of locally adapted phenotypes [1]. Conceptual and technical developments since the late 19th century have greatly enhanced our understanding of some of the main mechanisms producing and maintaining biological variation, namely, genetic mutation and recombination [2]. However, natural selection acts upon phenotypic variation represented by the individual [3], which is delimited by its genetic constitution, but also shaped by its specific environment [4] and developmental processes [5]. The process of evolution is thus a result of complex interactions between various intrinsic and extrinsic factors [6].
Therefore, current evolutionary investigations should consider several levels of biological variation [7]. First, differences in the DNA sequence account for a great amount of biological variation: the genetic system defines the range of functional possibilities of each individual. However, these heritable differences translate into the phenotype only indirectly via the resulting RNA and protein products which mould the structure and function of an organism. Much progress has been made in recent years in identifying gene functions and candidate genes coding for important metabolic enzymes, but analyses of whole genomes remain a complex challenge. Even in organisms whose whole genome is sequenced, a large number of genes still remain uncharacterized [8]. The second important source of biological variation is fluctuation in rates of gene expression, resulting in phenotypic plasticity [9, 10]. Genes can be up- or downregulated in response to environmental conditions, such as temperature regimes or water supply, or intrinsic factors such as specific phenological or developmental stages [11]. This leads to temporary modifications of the phenotype, which are generally not passed on to the next generation [12, 13]. The third level, heritable epigenetic variation, via both specialized enzymology inducing structural modifications of the DNA (through DNA methylation, histone acetylation [14, 15]) and small interfering (si) RNA populations [16, 17], results in (meta) stable chromatin landscape differences. Epigenetic differences determine if and where particular genes or groups of genes are to be expressed, while the underlying DNA sequence remains identical [18]. Most of these differences are reversible developmental effects and they are part of the molecular processes underlying phenotypic plasticity in response to variation in the environment [19]. However, environmental change, severe stress or genomic shock events like hybridization or genome duplication can change the epigenetic configuration of an organism resulting in new phenotypes [20–26], and some of these alterations can be passed on to the next generations [27–30].
The molecular mechanisms underlying these components of phenotypic variation differ in their stability and in the time frames in which they confer phenotypic novelty. The genetic sequence is the most stable, evolving slowly through mutation and gradually accumulating changes over a large number of generations. In contrast, gene expression levels can be rapidly and continuously regulated within a very short time [11], much shorter than the generation length of an organism, and allow an almost instantaneous response of the individual to its environment within limits defined by its genetic constitution. Heritable epigenetic alterations act within an intermediate time horizon, since they can occur as an immediate and multilocus reaction to different kinds of external or intrinsic stimuli [23] but are not as ephemeral as plastic gene regulation and can affect the following generations [18].
It has long been established that mutations in DNA sequence are the primary raw material for evolutionary change [2]. The involvement of environmental influences in generating heritable biological variation is still debated [13, 22], as is the necessity of extending our modern evolutionary synthesis [31]. Accumulating evidence indicates that modifications of epigenetic signals are correlated with phenotypic variation within and among species [25, 32–34], placing epigenetic differentiation even in a macroevolutionary context. Latest developments regarding the potential role of phenotypic plasticity in driving diversification and speciation have been discussed elsewhere (e.g., [13, 35]). We are hereafter focusing on the impact of heritable epigenetic variation on the process of evolution and propose a research plan to address its evolutionary significance.
2. Potential Impact of Heritable Epigenetic Variation on Evolution
Empirical studies have demonstrated high levels of epigenetic variation within natural populations [25, 36–41]. While experiments have shown that environmental conditions can override epigenetic signals (e.g., [26, 42, 43]) and increase this variation, few recent studies indicate that natural selection can act directly or indirectly on epigenetic variation [25, 38, 39, 44], potentially leading to evolutionary divergence and adaptation. Altogether, epigenetic information provides an additional source of natural variation, which may be particularly important for survival of small populations lacking genetic variability [45] and/or occupying a fragmented landscape. Selectable epigenetic variation can enable genetically depauperate lineages to adapt [46] until genetic assimilation occurs (i.e., when environmentally induced phenotypic variation becomes fixed by secondary genetic control, e.g., after deamination of methylated cytosine to thymine [13, 47]). Thus, heritable epigenetic variation could pave the way for genetic adaptation.
The epigenetic sources of variation can be stochastic epimutations, but a major part of the epigenetic variation is triggered by stress or changes in the environment [3, 22, 48], that is, under circumstances when new phenotypes could be crucial for survival. Moreover, if conditions return to their original state, spontaneous back-mutation of epialleles can restore original phenotypes (e.g., in position-effect variegation [27]). In the light of epigenetic variation, the involvement of the environment in evolution becomes twofold: as a stimulant of variation and as the selector of adaptive variation.
At the interface between genotype and environment, the overall rate of epimutations is often much higher than that of genetic mutations [49], resulting in a more dynamic level of variation. Novel epigenetic modifications may originate simultaneously in several individuals in a population under stress, which will facilitate fixation. Despite the potentially high loss of epigenetic novelties by epigenetic reset [19], epimutations can reach equilibrium frequencies within populations rapidly, over less than a dozen generations if the environmental stress is maintained long enough [28]. In stark contrast to the expected incidence of genetic mutations, environmental fluctuations can trigger multiple epimutations in the same individual. This renders fast ecological adaptation affecting (complex) adaptive traits more plausible [50]. Hence, recombination is not necessarily a prerequisite for adaptive change, if the latter is driven from the epigenetic level. In addition, epigenetic mechanisms may partly defy well-understood population processes, such as allelic drift (due to potential maintenance of relatively constant epiallelic frequencies through environmental influence). Being more flexible and dynamic than DNA sequence information, variation in epigenetic signals could therefore act as major driving force in rapid adaptive processes.
Epigenetic variation can have extensive consequences, even in the absence of genetic variability [45, 50, 51]. Epigenetics may introduce, or reinforce in a back-coupling process with environmental stimuli, major changes that lead to strong phenotypic differentiation [52] until becoming a real reproductive barrier. Most phenotypic differences between species are genetically controlled, but epigenetic inheritance can be of particular importance for the initial development of phenotypic divergence [25]. If adaptive and maintained long enough, phenotypic discontinuities can become genetically locked and trigger species divergence [53]. Modelling studies suggest that epigenetic variation can promote population divergence by facilitating adaptive peak shifts, reducing genetic barriers represented by fitness valleys in the adaptive landscape [47]. Therefore, epigenetic novelties have been one of the mechanisms put forward for saltational speciation [29, 54], but empirical data is not yet available to support or reject such a hypothesis.
3. A Research Idea
Recently developed tools, in combination with traditional methods, can shed light on the complex interactions between genotype, epigenotype, and environment, and test for their individual contribution to phenotypic divergence and evolution. Evolutionary biologists could address the evolutionary relevance of heritable epigenetic polymorphisms by targeting closely related ecotypes or species (hereafter types) that show phenotypic differentiation without apparent genome-wide genetic divergence. Such types could be identified, for example, within asexual lineages or descendants of recent adaptive radiation events. We suggest a multifaceted research plan using an array of molecular techniques and field experiments to investigate whether epigenetics is involved in speciation by triggering phenotypic diversification.
3.1. Phenotypic Differentiation
As speciation is facilitated by the process of divergence, the first question to be addressed should be whether phenotypic variation in the study group is discrete or continuous. Phenotypic variation is a common feature of populations and species, and only a discontinuity in this variation may indicate incipient divergence and the onset of isolating mechanisms. Therefore, various morphological, anatomical, and physiological traits among populations of different types should be compared to test whether the types form well defined, distinct groups or whether the extreme phenotypes are linked by individuals with intermediate traits or combinations of characters. In addition, measurements and observations of environmental characteristics (e.g., microclimate, geology, soil, biological interactions) could identify limiting environmental factors, and relate them to anatomical, morphological, and physiological specializations.
If main discontinuities in phenotypic variation separate populations along type boundaries (e.g., by morphology or habitat preference), the uniformity within each group and constant difference between the groups might suggest a single origin of each type and subsequent dispersal (Figure 1). However, this seems rather unlikely in absence of genome-wide genetic divergence among the types. An alternative scenario could invoke repeated migration and iterative in situ formation of each type in alternative environments, with very strong and almost identical selection pressures acting upon different populations of each of the types.
Figure 1.
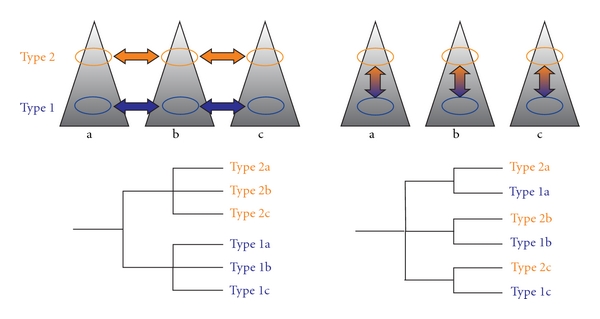
Putative relationships between populations of closely related alternative types (here exemplified with altitudinal differentiation), which lack apparent genome-wide divergence. Below the reflection of the relationships in hypothetical phylogenies is given. Left, single origin of each type, followed by dispersal to other geographical areas. Right, recurrent evolution of the types in several geographic regions under environmental influence.
3.2. Genetic and Epigenetic Differentiation
Singular versus multiple origin of each type should be tested by investigating the extent and structure of genome-wide genetic and epigenetic divergence within and among populations of both types. If populations cluster genetically in disagreement to the type (possibly determined by other factors, e.g., by geographic proximity), it may be hypothesized that their differentiation is underlaid by epigenetic mechanisms and that types have evolved several times in parallel. Alternatively, local high rates of gene flow combined with strong selection at a few adaptive genetic loci could hypothetically produce a similar pattern of highly porous genomes [55]. In such a case, a small number of adaptive (outlier) genetic loci of large effect should be responsible for the observed phenotypic differentiation. Outlier analyses [56–58] of genetic profiles provided, for example, by DNA fingerprinting techniques such as RAD (restriction site associated DNA) sequencing [59], microsatellites, or AFLP (amplified fragment length polymorphism [60]), could help identifying these loci or closely linked genomic regions. Positive selection will shape at target loci a significantly higher differentiation between populations of the alternative types than the genome-wide bulk of loci, while loci under purifying selection will show much lower differentiation [61]. On the other hand, if individuals of each group share type-specific epigenetic patterns and/or mRNA transcripts, differentiation could be mediated either by overall differences in the epigenome or by a few epialleles.
As epigenetic variation is not detectable in genomic surveys of sequence variation, dedicated investigations have to be employed to address it. In recent years, a variety of genome-wide approaches, including techniques involving next-generation sequencing, have been developed to comparatively profile epigenetic patterns in nonmodel organisms [62, 63]. Cost-effective comprehensive methods include, for example, fractioning the DNA using C0t filtration [64, 65] to enrich low-copy regions (mostly genes and their promoters) and sequence this genomic subsample by employing next generation methods and bisulfite sequencing. The latter is a process that converts unmethylated cytosines to uracils, which will then appear as thymines after sequencing [66]. Third-generation DNA sequencers, like the recently released single molecule real-time (SMRT) DNA sequencer could be employed for direct detection of DNA methylation [67] and thus enable much more profound study of both model and nonmodel epigenomes. Alternatively, genome-wide DNA methylation could be studied using isoschizomers [68, 69]. Similarly as for genetic dataset(s), the epigenetic information could be searched for general patterns of differentiation and for signatures of selection on individual (epi)loci [25, 44]. This should clarify if ecological and/or morphological divergence is dependent on just a few loci controlling traits for local adaptation, or if it is triggered by extensive differences. As the alternative types thrive in different environments, the selective pressures and their magnitude may vary across populations. Epigenetic signals will most often suffer from imperfect heritability; therefore, stronger selection will be needed to produce patterns that will be detected as outliers by statistical approaches.
To infer broad, genome-wide regulatory variation, in-depth quantitative gene expression analyses using next-generation sequencing (RNA-seq, [70–72]) could be performed searching for loci with significant expression differences between individuals of different types after growing them under uniform conditions to reduce the momentary-dependent noise in rates of expression. In addition, targeting posttranscriptional regulation, small RNA profiles could be compared using an smRNA-seq approach [63, 73, 74]. The different data types can finally be integrated in functional analyses (i.e., gene annotations) to identify correlated components that are part of the same regulatory network.
3.3. Heritability of Phenotypic Plasticity and Habitat Specificity
If the molecular basis of phenotypic differentiation and/or adaptation to divergent environments is identified within epigenetic rather than DNA sequence divergence, the next research step would be to investigate how stable the phenotypic divergence is. This will also help to assess the stage of speciation in which the group is at present. While facilitating population divergence and speciation [35], nonheritable phenotypic plasticity will trigger speciation only if the environmental conditions are stably different in the alternative localities [35] and gene flow is either infrequent or strongly opposed by natural selection. On the other hand, in the case of heritable phenotypic divergence that is fully stable even in the alternative environment, epigenetically triggered adaptation may have been already assimilated in the genetic code.
Reciprocal transplant experiments together with attempts to grow the different types under the same environment across several generations (i.e., between three and five as a minimum requirement) should be installed to determine the extent of phenotypic plasticity, and the ability of the different types to cope with altered environmental conditions. Growing individuals of the alternative types in a uniform environment across several generations may reveal the heritability of morphological and ecological characteristics within each of the types (“nature versus nurture”) [75]. Comparatively investigating relevant (epi)loci in transplanted individuals versus controls will pinpoint those patterns that are immediately disrupted by the environment, and those that persist or, alternatively, are not under the influence of the relevant limiting environmental differences. Integrating this information and comparing morphological, anatomical and physiological traits supplemented by a set of fitness components among transplants and controls will define the links between genotype, epigenotype and phenotype, together with providing additional information on the patterns of selection and their targets.
According to the mechanisms underlying the observed differentiation, at least two possible outcomes can be anticipated. If the morphological and/or ecophysiological differences are triggered by continuous but nonheritable responses to local environments (i.e., as a reaction norm [76]), there should be no phenotypic differences between the progeny of the two types when reared and grown under the same conditions. Such a scenario will not (yet) be relevant for speciation. On the other hand, if heritable epigenetic differences are involved, phenotypic divergence between individuals of the types should at least partly be retained in a common environment. In the latter case the morphology, anatomy, and physiological properties of the transplanted individuals should reflect their origin rather than their current environment. This may go to the extreme that individuals are maladapted and do not survive under alien environmental conditions.
The result of these experiments could simultaneously allow for inferring evolutionary and population dynamics within the study group. If individuals of alternative types can adapt phenotypically to the habitat of the other and develop the habitat-specific syndromes following transplant experiments, the possibility of frequent gene flow between populations of both types should be considered. This might as well explain the lack of overall differentiation, as it prevents lineage sorting and hampers or slows down speciation. On the contrary, low fitness (i.e., poor performance and high mortality) of individuals in the native habitat of the alternative type may point to a differentiation that is strong enough to prevent gene flow between populations. In this case, we may be observing a process of ongoing speciation, where differentiation starts at the epigenetic level, triggering profound changes leading to segregation in terms of habitat, phenology, and/or biological interactions. Divergent selection may reinforce this environmentally induced specialization/niche segregation and bring about reproductive isolation. This will eventually result in virtual isolation of gene pools, and ultimately give way to stronger overall differentiation by accumulation of genetic differences due to the stochastic effects of drift.
4. Heliosperma pusillum Group: An Example of an Appropriate Study System
Heliosperma pusillum and allied taxa from the carnation family (Caryophyllaceae) contain a variety of morphologically different taxa (Figure 2) with distinct ecology, which are altitudinally or geographically isolated, but genetically intermixed (Figure 3) and do not represent independent evolutionary lineages [78]. Molecular phylogenetic studies based on AFLPs [77] and sequences of several nuclear and chloroplast regions [77–79], show that genetic divergence within the group is generally shallow, many taxa seem to be polyphyletic, and geographically allied taxa often share the same genetic constitution. We hypothesize that they either (i) represent fixed ecotypes, that is, differ subtly in their DNA coding regions with major phenotypic effects, or (ii) result from middle- to short-term adaptive (epigenetic) processes, perhaps under the influence of the environment and independent of actual changes in DNA sequence. All of them are perennial caespitose herbs that inhabit rocky habitats and shallow caves in mountain ranges of southern Europe [78, 80], mostly on calcareous substrates.
Figure 2.

Low-elevation Heliosperma veselskyi and high-elevation H. pusillum are differentiated morphologically and ecologically. Particularly conspicuous is the dense indumentum of sticky glandular hairs on H. veselskyi in comparison to the glabrous leaves and stems of H. pusillum (Photographs: M. Sonnleitner).
Figure 3.
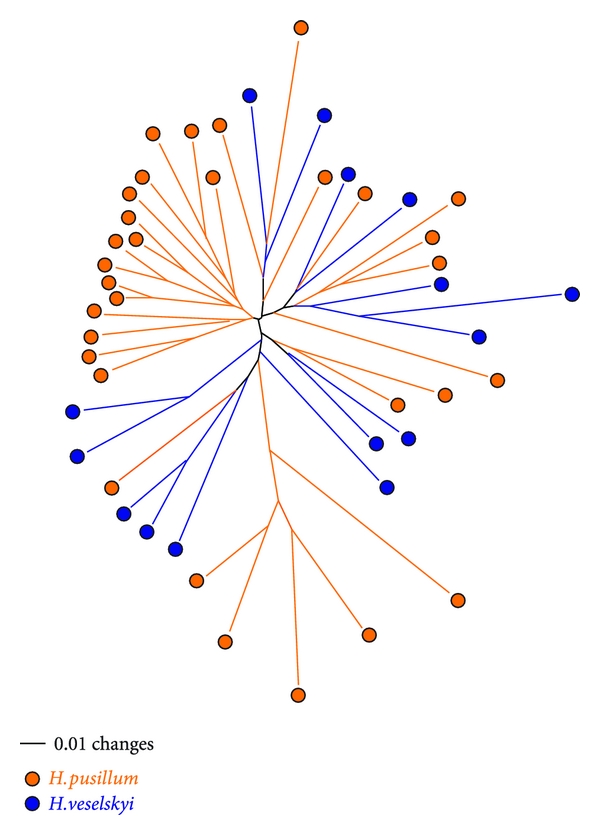
Genetic analyses do not support separation of higher-altitude Heliosperma pusillum (orange) and lower-altitude H. veselskyi (dark blue), but rather indicate an inextricable relationship between the two taxa. Unrooted neighbor joining tree based on Nei-Li distances calculated with PAUP from AFLP profiles [77].
Different authors [78, 81] have subdivided this complex into two ecologically and morphologically distinct groups of taxa: a higher elevation group occurring in damp, open habitats and among rocks above the timberline and a lower elevation group inhabiting canyons and gorges as well as shallow caves and cliff overhangs with rather dry soils, high atmospheric moisture and poor light conditions below the timberline. The higher elevation group, including H. albanicum, H. pudibundum, and H. pusillum s.str., differs from the lower elevation group by narrower, glabrous or sparsely hairy leaves and often unicellular glands as well as longer seed papillae [78, 81]. By contrast, plants of lower elevations share a denser indumentum with long multicellular glandular hairs and are often sticky (Figure 2). Generally, morphological variation is much higher in the lower elevation group, which contains several narrowly distributed taxa [78, 82]. Most of them are endemics of the Balkan Peninsula; only H. veselskyi is restricted to the southeastern Alps. The origin and evolution of the lower and higher elevation groups and the relationships between them are still poorly understood. Recent molecular phylogenetic studies [78] (see also Figure 3) indicate that neither higher nor lower elevation groups are actually monophyletic, but rather inextricably intermingled with each other, indicating that one of the groups evolved multiple times from the other. Mechanisms involved in the phenotypic diversification of the two groups, the morphological convergence within each group, and the stability of this phenotypic divergence remain unknown, but preliminary evidence suggests that morphological features remain constant in a common garden, at least in the first generation. The H. pusillum complex is suitable for (epi) genomic and transcriptomic analyses, because all taxa have a relatively small genome (1C = 1.32 pg [83]) and so far no polyploid cytotypes have been found (2n = 2x = 24). In addition, they can be easily grown from seeds and have short generation times, which make them optimally suitable for common garden and transplantation studies.
5. Synthesis and Outlook
Although the possibility of epigenetic inheritance has now been established [7, 18, 27, 30, 84] and we are increasingly understanding the full extent of its role in producing phenotypic variation [19, 25, 39, 40], little research has been done to systematically study the role of heritable epigenetic variation for speciation. Incorporation of epigenetics into evolutionary models and empirical studies is only now starting to be attempted (e.g., [28, 49]); however, more empirical information from natural populations is needed for accurate modelling of epigenetic dynamics. Indeed, the prevalence of alternative stable epialleles in natural populations, and their significance to phenotypic divergence, ecological interactions and selection in real-world contexts remain too little explored [3, 41, 53]. The limited relevant data available indicate a stochastic nature of epigenetic variation, which is continuously being shaped by the influence of the environment, and further tuned through natural selection [25, 38, 39]. Therefore, the epigenetic aspect of natural variation may contribute to evolution in a fashion similar to genetics, but much more rapidly. Implying heritability of adaptive (i.e., selected) traits, epigenetic inheritance is not a contradiction of the Darwinian evolutionary synthesis [31], but rather a complex augmentation of the classic view on genetic inheritance, particularly as genotype and epigenotype interact to produce a broad array of short- and long-term heritable combinations.
The recently available possibility to profile the epigenome and transcriptome of nonmodel organisms in a high-throughput manner [62, 63, 85] enables thorough investigation of some of the most challenging hypotheses in a modern evolutionary framework, such as achieving and maintaining stable divergence through epigenetic differences. The acquired knowledge also impacts several related domains, from conservation to theoretical evolutionary biology. Investigating recent adaptive radiations with epigenetic markers may be particularly informative. Most traits of ecological significance tend to be continuous or quantitative and appear to be governed by many genes, each of little effect, but with cumulative power [86], resulting in a complex picture of factors and mechanisms acting upon the phenotype. Using appropriate study systems it is now possible to interrogate the links between ecological divergence and many regulatory alterations of small effect or singular major epigenetic switches. In addition, such investigations are expected to pinpoint new loci that are sensitive to epigenetic modification and unravel information on the rates of spontaneous epimutations in natural populations and their stability over time.
Currently accumulating data will offer valuable clues on the establishment of broad regulatory determinants of functional diversity in natural populations. The early evidence we currently hold urges complementing our gene- and genome-centred evolutionary view with a substantial consideration of epigenetic factors when seeking to understand population processes that drive adaptation and divergence [3, 53, 87]. Using modern technologies, future research will identify the exact molecular mechanisms triggering relevant phenotypic divergence and reproductive isolation. We will soon be able to infer the corresponding selection pressures that are responsible for the presence of a particular individual/a particular species in its specific habitat. Understanding how new plant species form and adapt to novel ecological niches is crucial to advance our knowledge of evolutionary processes active at the population level driving adaptation and speciation. An increased knowledge of organismic adaptation strategies is also of outstanding importance in the current context of widespread environmental challenges. It may be a key for predicting effects of climate change and managing biodiversity in a sustainable manner.
Acknowledgments
The authors are grateful to Brigitta Erschbamer, Karl Hülber, Gilbert Neuner, Dieter Reich, Christina Richards and two anonymous reviewers for insightful comments. They thank Michaela Sonnleitner for providing skillful photographs. Financial support from the “Verein zur Förderung der wissenschaftlichen Ausbildung und Tätigkeit von Südtirolern an der Landesuniversität Innsbruck” for R. Flatscher and from the Austrian Science Foundation (FWF; project no. P22260-B16), and from the European Commission (PERG07-GA-2010-268462) for O. Paun is acknowledged.
References
- 1.Darwin C. On the Origin of Species. London, UK: John Murray; 1859. [Google Scholar]
- 2.Mayr E. What Evolution Is. London, UK: Weidenfeld & Nicolson; 2002. [Google Scholar]
- 3.Rapp RA, Wendel JF. Epigenetics and plant evolution. New Phytologist. 2005;168(1):81–91. doi: 10.1111/j.1469-8137.2005.01491.x. [DOI] [PubMed] [Google Scholar]
- 4.Jablonka E, Lamb MJ. Evolution in Four Dimensions: Genetic, Epigenetic, Behavioral, and Symbolic Variation in the History of Life. Cambridge, UK: The MIT Press; 2005. [Google Scholar]
- 5.Müller GB. Evo-devo: extending the evolutionary synthesis. Nature Reviews Genetics. 2007;8(12):943–949. doi: 10.1038/nrg2219. [DOI] [PubMed] [Google Scholar]
- 6.Pigliucci M. An extended synthesis for evolutionary biology. Annals of the New York Academy of Sciences. 2009;1168:218–228. doi: 10.1111/j.1749-6632.2009.04578.x. [DOI] [PubMed] [Google Scholar]
- 7.Bonduriansky R, Day T. Nongenetic inheritance and its evolutionary implications. Annual Review of Ecology, Evolution and Systematics. 2009;40:103–125. [Google Scholar]
- 8.Benfey PN, Mitchell-Olds T. From genotype to phenotype: systems biology meets natural variation. Science. 2008;320(5875):495–497. doi: 10.1126/science.1153716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pigliucci M. Phenotypic Plasticity: Beyond Nature and Nurture. John Hopkins University Press; 2001. [Google Scholar]
- 10.Schlichting CD, Smith H. Phenotypic plasticity: linking molecular mechanisms with evolutionary outcomes. Evolutionary Ecology. 2002;16(3):189–211. [Google Scholar]
- 11.Nicotra AB, Atkin OK, Bonser SP, et al. Plant phenotypic plasticity in a changing climate. Trends in Plant Science. 2010;15(12):684–692. doi: 10.1016/j.tplants.2010.09.008. [DOI] [PubMed] [Google Scholar]
- 12.Pigliucci M. Phenotypic plasticity. In: Pigliucci M, Müller GB, editors. Evolution—The Extended Synthesis. Cambridge, UK: The MIT Press; 2010. pp. 355–378. [Google Scholar]
- 13.Schwander T, Leimar O. Genes as leaders and followers in evolution. Trends in Ecology and Evolution. 2011;26(3):143–151. doi: 10.1016/j.tree.2010.12.010. [DOI] [PubMed] [Google Scholar]
- 14.Turner BM. Histone acetylation and an epigenetic code. BioEssays. 2000;22(9):836–845. doi: 10.1002/1521-1878(200009)22:9<836::AID-BIES9>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 15.Kalisz S, Purugganan MD. Epialleles via DNA methylation: consequences for plant evolution. Trends in Ecology and Evolution. 2004;19(6):309–314. doi: 10.1016/j.tree.2004.03.034. [DOI] [PubMed] [Google Scholar]
- 16.Großhans H, Filipowicz W. Molecular biology: the expanding world of small RNAs. Nature. 2008;451(7177):414–416. doi: 10.1038/451414a. [DOI] [PubMed] [Google Scholar]
- 17.Kawaji H, Hayashizaki Y. Exploration of small RNAs. PLoS Genetics. 2008;4(1):3–8. doi: 10.1371/journal.pgen.0040022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jablonka EVA, Raz GAL. Transgenerational epigenetic inheritance: prevalence, mechanisms, and implications for the study of heredity and evolution. Quarterly Review of Biology. 2009;84(2):131–176. doi: 10.1086/598822. [DOI] [PubMed] [Google Scholar]
- 19.Richards CL, Bossdorf O, Pigliucci M. What role does heritable epigenetic variation play in phenotypic evolution? BioScience. 2010;60(3):232–237. [Google Scholar]
- 20.Finnegan EJ. Epialleles—a source of random variation in times of stress. Current Opinion in Plant Biology. 2002;5(2):101–106. doi: 10.1016/s1369-5266(02)00233-9. [DOI] [PubMed] [Google Scholar]
- 21.Paun O, Fay MF, Soltis DE, Chase MW. Genetic and epigenetic alterations after hybridization and genome doubling. Taxon. 2007;56(3):649–656. [PMC free article] [PubMed] [Google Scholar]
- 22.Turner BM. Epigenetic responses to environmental change and their evolutionary implications. Philosophical Transactions of the Royal Society B. 2009;364(1534):3403–3418. doi: 10.1098/rstb.2009.0125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Angers B, Castonguay E, Massicotte R. Environmentally induced phenotypes and DNA methylation: how to deal with unpredictable conditions until the next generation and after. Molecular Ecology. 2010;19(7):1283–1295. doi: 10.1111/j.1365-294X.2010.04580.x. [DOI] [PubMed] [Google Scholar]
- 24.Jackson S, Chen ZJ. Genomic and expression plasticity of polyploidy. Current Opinion in Plant Biology. 2010;13(2):153–159. doi: 10.1016/j.pbi.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paun O, Bateman RM, Fay MF, Hedrén M, Civeyrel L, Chase MW. Stable epigenetic effects impact adaptation in allopolyploid orchids (Dactylorhiza: Orchidaceae) Molecular Biology and Evolution. 2010;27(11):2465–2473. doi: 10.1093/molbev/msq150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pecinka A, Dinh HQ, Baubec T, Rosa M, Lettner N, Scheid OM. Epigenetic regulation of repetitive elements is attenuated by prolonged heat stress in Arabidopsis . Plant Cell. 2010;22(9):3118–3129. doi: 10.1105/tpc.110.078493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Richards EJ. Inherited epigenetic variation—revisiting soft inheritance. Nature Reviews Genetics. 2006;7(5):395–401. doi: 10.1038/nrg1834. [DOI] [PubMed] [Google Scholar]
- 28.Slatkin M. Epigenetic inheritance and the missing heritability problem. Genetics. 2009;182(3):845–850. doi: 10.1534/genetics.109.102798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jablonka E, Lamb MJ. Transgenerational epigenetic inheritance. In: Pigliucci M, Müller GB, editors. Evolution—The Extended Synthesis. Cambridge, UK: The MIT Press; 2010. pp. 137–174. [Google Scholar]
- 30.Paszkowski J, Grossniklaus U. Selected aspects of transgenerational epigenetic inheritance and resetting in plants. Current Opinion in Plant Biology. 2011;14:195–203. doi: 10.1016/j.pbi.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 31.Pigliucci M. Do we need an extended evolutionary synthesis? Evolution. 2007;61(12):2743–2749. doi: 10.1111/j.1558-5646.2007.00246.x. [DOI] [PubMed] [Google Scholar]
- 32.Ha M, Pang M, Agarwal V, Chen ZJ. Interspecies regulation of microRNAs and their targets. Biochimica et Biophysica Acta. 2008;1779(11):735–742. doi: 10.1016/j.bbagrm.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ha M, Ng DW, Li WH, Chen ZJ. Coordinated histone modifications are associated with gene expression variation within and between species. Genome Research. 2011;21(4):590–598. doi: 10.1101/gr.116467.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hollister JD, Smith LM, Guo YL, Ott F, Weigel D, Gaut BS. Transposable elements and small RNAs contribute to gene expression divergence between Arabidopsis thaliana and Arabidopsis lyrata . Proceedings of the National Academy of Sciences of the United States of America. 2011;108(6):2322–2327. doi: 10.1073/pnas.1018222108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pfennig DW, Wund MA, Snell-Rood EC, Cruickshank T, Schlichting CD, Moczek AP. Phenotypic plasticity’s impacts on diversification and speciation. Trends in Ecology and Evolution. 2010;25(8):459–467. doi: 10.1016/j.tree.2010.05.006. [DOI] [PubMed] [Google Scholar]
- 36.Vaughn MW, Tanurdžić M, Lippman Z, et al. Epigenetic natural variation in Arabidopsis thaliana . PLoS Biology. 2007;5(7):1617–1629. doi: 10.1371/journal.pbio.0050174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Woo HR, Richards EJ. Natural variation in DNA methylation in ribosomal RNA genes of Arabidopsis thaliana . BMC Plant Biology. 2008;8, article 92 doi: 10.1186/1471-2229-8-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Herrera CM, Bazaga P. Epigenetic differentiation and relationship to adaptive genetic divergence in discrete populations of the violet Viola cazorlensis . New Phytologist. 2010;187(3):867–876. doi: 10.1111/j.1469-8137.2010.03298.x. [DOI] [PubMed] [Google Scholar]
- 39.Herrera CM, Bazaga P. Untangling individual variation in natural populations: ecological, genetic and epigenetic correlates of long-term inequality in herbivory. Molecular Ecology. 2011;20(8):1675–1688. doi: 10.1111/j.1365-294X.2011.05026.x. [DOI] [PubMed] [Google Scholar]
- 40.Massicotte R, Whitelaw E, Angers B. DNA methylation: a source of random variation in natural populations. Epigenetics. 2011;6(4):422–428. doi: 10.4161/epi.6.4.14532. [DOI] [PubMed] [Google Scholar]
- 41.Richards EJ. Natural epigenetic variation in plant species: a view from the field. Current Opinion in Plant Biology. 2011;14(2):204–209. doi: 10.1016/j.pbi.2011.03.009. [DOI] [PubMed] [Google Scholar]
- 42.Verhoeven KJF, Jansen JJ, van Dijk PJ, Biere A. Stress-induced DNA methylation changes and their heritability in asexual dandelions. New Phytologist. 2010;185(4):1108–1118. doi: 10.1111/j.1469-8137.2009.03121.x. [DOI] [PubMed] [Google Scholar]
- 43.Lira-Medeiros CF, Parisod C, Fernandes RA, Mata CS, Cardoso MA, Ferreira PCG. Epigenetic variation in mangrove plants occurring in contrasting natural environment. PLoS ONE. 2010;5(4) doi: 10.1371/journal.pone.0010326. Article ID e10326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Paun O, Bateman RM, Fay MF, et al. Altered gene expression and ecological divergence in sibling allopolyploids of Dactylorhiza (Orchidaceae) BMC Evolutionary Biology. 2011;11(1, article 113) doi: 10.1186/1471-2148-11-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Castonguay E, Angers B. The key role of epigenetics in the persistence of asexual lineages. doi: 10.1155/2012/534289. Genetics Research International. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scoville AG, Barnett LL, Bodbyl-Roels S, Kelly JK, Hileman LC. Differential regulation of a MYB transcription factor is correlated with transgenerational epigenetic inheritance of trichome density in Mimulus guttatus . New Phytologist. 2011;191(1):251–263. doi: 10.1111/j.1469-8137.2011.03656.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pál C, Miklós I. Epigenetic inheritance, genetic assimilation and speciation. Journal of Theoretical Biology. 1999;200(1):19–37. doi: 10.1006/jtbi.1999.0974. [DOI] [PubMed] [Google Scholar]
- 48.Halfmann R, Lindquist S. Epigenetics in the extreme: prions and the inheritance of environmentally acquired traits. Science. 2010;330(6004):629–632. doi: 10.1126/science.1191081. [DOI] [PubMed] [Google Scholar]
- 49.Tal O, Kisdi E, Jablonka E. Epigenetic contribution to covariance between relatives. Genetics. 2010;184(4):1037–1050. doi: 10.1534/genetics.109.112466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Johannes F, Porcher E, Teixeira FK, et al. Assessing the impact of transgenerational epigenetic variation on complex traits. PLoS Genetics. 2009;5(6) doi: 10.1371/journal.pgen.1000530. Article ID e1000530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reinders J, Wulff BBH, Mirouze M, et al. Compromised stability of DNA methylation and transposon immobilization in mosaic Arabidopsis epigenomes. Genes and Development. 2009;23(8):939–950. doi: 10.1101/gad.524609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Svanbäck R, Pineda-Krch M, Doebeli M. Fluctuating population dynamics promotes the evolution of phenotypic plasticity. American Naturalist. 2009;174(2):176–189. doi: 10.1086/600112. [DOI] [PubMed] [Google Scholar]
- 53.Bossdorf O, Richards CL, Pigliucci M. Epigenetics for ecologists. Ecology Letters. 2008;11(2):106–115. doi: 10.1111/j.1461-0248.2007.01130.x. [DOI] [PubMed] [Google Scholar]
- 54.Bateman RM, DiMichele WA. Generating and filtering major phenotypic novelties: neoGoldschmidtian saltation revisited. In: Cronk QCB, Bateman RM, Hawkins JA, editors. Developmental Genetics and Plant Evolution. London, UK: Taylor & Francis; 2002. pp. 109–159. [Google Scholar]
- 55.Kane NC, King MG, Barker MS, et al. Comparative genomic and population genetic analyses indicate highly porous genomes and high levels of gene flow between divergent Helianthus species. Evolution. 2009;63(8):2061–2075. doi: 10.1111/j.1558-5646.2009.00703.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Beaumont MA, Balding DJ. Identifying adaptive genetic divergence among populations from genome scans. Molecular Ecology. 2004;13(4):969–980. doi: 10.1111/j.1365-294x.2004.02125.x. [DOI] [PubMed] [Google Scholar]
- 57.Nielsen R. Molecular signatures of natural selection. Annual Review of Genetics. 2005;39:197–218. doi: 10.1146/annurev.genet.39.073003.112420. [DOI] [PubMed] [Google Scholar]
- 58.Nosil P, Funk DJ, Ortiz-Barrientos D. Divergent selection and heterogeneous genomic divergence. Molecular Ecology. 2009;18(3):375–402. doi: 10.1111/j.1365-294X.2008.03946.x. [DOI] [PubMed] [Google Scholar]
- 59.Baird NA, Etter PD, Atwood TS, et al. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS ONE. 2008;3(10) doi: 10.1371/journal.pone.0003376. Article ID e3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vos P, Hogers R, Bleeker M, et al. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Research. 1995;23(21):4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Foll M, Gaggiotti O. A genome-scan method to identify selected loci appropriate for both dominant and codominant markers: a Bayesian perspective. Genetics. 2008;180(2):977–993. doi: 10.1534/genetics.108.092221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hirst M, Marra MA. Next generation sequencing based approaches to epigenomics. Briefings in Functional Genomics. 2010;9(5-6):455–465. doi: 10.1093/bfgp/elq035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schmitz RJ, Zhang X. High-throughput approaches for plant epigenomic studies. Current Opinion in Plant Biology. 2011;14(2):130–136. doi: 10.1016/j.pbi.2011.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Peterson DG, Schulze SR, Sciara EB, et al. Integration of cot analysis, DNA cloning, and high-throughput sequencing facilitates genome characterization and gene discovery. Genome Research. 2002;12(5):795–807. doi: 10.1101/gr.226102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Peterson DG, Wessler SR, Paterson AH. Efficient capture of unique sequences from eukaryotic genomes. Trends in Genetics. 2002;18(11):547–550. doi: 10.1016/s0168-9525(02)02764-6. [DOI] [PubMed] [Google Scholar]
- 66.Henderson IR, Chan SR, Cao X, Johnson L, Jacobsen SE. Accurate sodium bisulfite sequencing in plants. Epigenetics. 2010;5(1):47–49. doi: 10.4161/epi.5.1.10560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Flusberg BA, Webster DR, Lee JH, et al. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nature Methods. 2010;7(6):461–465. doi: 10.1038/nmeth.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Madlung A, Masuelli RW, Watson B, Reynolds SH, Davison J, Comai L. Remodeling of DNA methylation and phenotypic and transcriptional changes in synthetic Arabidopsis allotetraploids. Plant Physiology. 2002;129(2):733–746. doi: 10.1104/pp.003095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Baurens FC, Bonnot F, Bienvenu D, Causse S, Legavre T. Using SD-AFLP and MSAP to assess CCGG methylation in the banana genome. Plant Molecular Biology Reporter. 2003;21(4):339–348. [Google Scholar]
- 70.Lister R, O’Malley RC, Tonti-Filippini J, et al. Highly integrated single-base resolution maps of the epigenome in Arabidopsis . Cell. 2008;133(3):523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Research. 2008;18(9):1509–1517. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tang F, Barbacioru C, Wang Y, et al. mRNA-seq whole-transcriptome analysis of a single cell. Nature Methods. 2009;6(5):377–382. doi: 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- 73.Kasschau KD, Fahlgren N, Chapman EJ, et al. Genome-wide profiling and analysis of Arabidopsis siRNAs. PLoS Biology. 2007;5(3, article e57) doi: 10.1371/journal.pbio.0050057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang X. The epigenetic landscape of plants. Science. 2008;320(5875):489–492. doi: 10.1126/science.1153996. [DOI] [PubMed] [Google Scholar]
- 75.Raj A, van Oudenaarden A. Nature, nurture, or chance: stochastic gene expression and its consequences. Cell. 2008;135(2):216–226. doi: 10.1016/j.cell.2008.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Aubin-Horth N, Renn SCP. Genomic reaction norms: using integrative biology to understand molecular mechanisms of phenotypic plasticity. Molecular Ecology. 2009;18(18):3763–3780. doi: 10.1111/j.1365-294X.2009.04313.x. [DOI] [PubMed] [Google Scholar]
- 77.Frajman B, Flatscher R, Surina B, Schönswetter P. Evolutionary history and phylogeographic patterns in the Heliosperma pusillum group (Caryophyllaceae): conflicting signals of plastid, low-copy nuclear sequences and AFLP fingerprints. unpublished. [Google Scholar]
- 78.Frajman B, Oxelman B. Reticulate phylogenetics and phytogeographical structure of Heliosperma (Sileneae, Caryophyllaceae) inferred from chloroplast and nuclear DNA sequences. Molecular Phylogenetics and Evolution. 2007;43(1):140–155. doi: 10.1016/j.ympev.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 79.Frajman B, Eggens F, Oxelman B. Hybrid origins and homoploid reticulate evolution within Heliosperma (Sileneae, Caryophyllaceae) - A multigene phylogenetic approach with relative dating. Systematic Biology. 2009;58(3):328–345. doi: 10.1093/sysbio/syp030. [DOI] [PubMed] [Google Scholar]
- 80.Chater AO, Walters SM, Akeroyd JR. Silene L. In: Tutin TG, et al., editors. Flora Europaea. Cambridge, UK: Cambridge University Press; 1993. pp. 191–211. [Google Scholar]
- 81.Neumayer H. Die Frage der Gattungsabgrenzung innerhalb der Silenoideen. Verhandlungen der Zoologisch-Botanischen Gesellschaft in Wien. 1923;72:53–59. [Google Scholar]
- 82.Niketić M, Stevanović V. A new species of Heliosperma (Caryophyllaceae) from Serbia and Montenegro. Botanical Journal of the Linnean Society. 2007;154(1):55–63. [Google Scholar]
- 83.Temsch EM, Temsch W, Ehrendorfer-Schratt L, Greilhuber J. Heavy metal pollution, selection, and genome size: the species of the Žerjav study revisited with flow cytometry. Journal of Botany. 2010;2010:11 pages. Article ID 596542. [Google Scholar]
- 84.Feng S, Jacobsen SE, Reik W. Epigenetic reprogramming in plant and animal development. Science. 2010;330(6004):622–627. doi: 10.1126/science.1190614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fazzari MJ, Greally JM. Introduction to epigenomics and epigenome-wide analysis. Methods in Molecular Biology. 2010;620:243–265. doi: 10.1007/978-1-60761-580-4_7. [DOI] [PubMed] [Google Scholar]
- 86.Stapley J, Reger J, Feulner PGD, et al. Adaptation genomics: the next generation. Trends in Ecology and Evolution. 2010;25(12):705–712. doi: 10.1016/j.tree.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 87.Richards EJ. Population epigenetics. Current Opinion in Genetics and Development. 2008;18(2):221–226. doi: 10.1016/j.gde.2008.01.014. [DOI] [PubMed] [Google Scholar]


