Abstract
Biomacromolecules that are challenging for the usual structural techniques can be studied with atomic resolution by solid-state nuclear magnetic resonance. However, the paucity of >5 Å distance restraints, traditionally derived from measurements of magnetic dipole-dipole couplings between protein nuclei, is a major bottleneck that hampers such structure elucidation efforts. Here we describe a general approach that enables the rapid determination of global protein fold in the solid phase via measurements of nuclear paramagnetic relaxation enhancements (PREs) in several analogs of the protein of interest containing covalently-attached paramagnetic tags, without the use of conventional internuclear distance restraints. The method is demonstrated using six cysteine-EDTA-Cu2+ mutants of the 56-residue B1 immunoglobulin-binding domain of protein G, for which ~230 longitudinal backbone 15N PREs corresponding to ~10-20 Å distances were obtained. The mean protein fold determined in this manner agrees with the X-ray structure with a backbone atom root-mean-square deviation of 1.8 Å.
The determination of three-dimensional (3D) structures of biological macromolecules is critical to understanding their physiological function and is also the primary focus of structural genomics initiatives.1 Magic-angle spinning (MAS) solid-state nuclear magnetic resonance (NMR) provides a set of spectroscopic tools for structural studies of biological systems that cannot be induced to form sufficiently diffracting crystals and are also not amenable to solution NMR due to size limitations, such as certain membrane-bound polypeptides2,3 and amyloids.4,5 However, in spite of the remarkable recent progress in determining relatively high-resolution structural models for proteins with molecular weights up to ~20 kDa by MAS solid-state NMR,5-19 the routine generation of such structures continues to present considerable challenges.
Even for small to medium-size uniformly 13C,15N or uniformly 15N and extensively 13C labeled proteins—the latter generally prepared by using complementary [1,3-13C] and [2-13C]glycerol based expression media6 in order to achieve improved spectral resolution and magnetization transfer efficiencies6,15 by suppressing many one-bond 13C–13C dipolar and J-couplings—the 2D 13C-13C and 15N-13C chemical shift correlation spectra, which form the basis for the vast majority of the current solid-state NMR structure determination protocols, are typically highly congested and contain many tens to hundreds (or even thousands) of resonances. These spectra rely on magnetization transfers using radiofrequency pulse schemes such as transferred echo double resonance (TEDOR),20,21 proton driven spin diffusion (PDSD) or dipolar assisted rotational resonance (DARR),22 CHHC, NHHC or NHHN,23 or proton assisted recoupling (PAR),12 and report on the magnitudes of through-space 13C-13C, 15N-13C and 1H-1H magnetic dipole-dipole couplings, and thereby distances, among the protein nuclei. Though the use of ultrahigh-field (≥ ~20 Tesla) NMR spectrometers, additional spectral dimensions and tailored isotope labeling schemes (e.g., perdeuterated proteins containing sparsely distributed proton nuclei at a limited number of amide and methyl sites),18-20 as well as structure calculation algorithms that are able to deal with ambiguous distance restraints8,9 has certainly helped to alleviate some of the problems related to NMR signal overlap, the fundamental limitation in most solid-state NMR structure determination endeavors has been the relative scarceness of unambiguous long-range interatomic distance restraints. This is primarily because dipolar couplings between 1H, 13C and 15N spins, which scale with the inverse third power of the internuclear distance, become vanishingly small for distances in the ~5-10 Å regime and beyond (typical coupling magnitudes are on the order of a few Hertz to tens of Hertz). The detection and reliable quantification of these couplings is often further complicated by the fact that they are part of tightly coupled multiple spin networks. Consequently, correlations that encode information about the most structurally-relevant inter-residue dipolar couplings typically comprise only a small fraction of the observable resonances in multidimensional solid-state NMR datasets, and are also usually associated with some of the weakest cross-peak intensities.
In this article, we describe a general approach that has the potential to substantially accelerate the solid-state NMR protein structure determination process. In broad terms, this methodology involves the measurement of nuclear paramagnetic relaxation enhancements (PREs)24,25 in several point mutants of the protein of interest modified with covalently-attached paramagnetic tags, followed by the application of these PREs as structural restraints to establish the global protein backbone fold. The PRE phenomenon is well-known in magnetic resonance—the magnitude of the effect scales with the inverse sixth power of the electron-nucleus distance24,25 and it can be non-negligible for distances up to ~20-25 Å or longer depending on the observed nucleus and paramagnetic center (i.e., distances that are at least a factor of ~3-4 larger than those obtained by using traditional dipolar coupling based solid-state NMR techniques). Indeed, PRE-based methods of this kind, employing nitroxide spin labels and measurements of longitudinal or transverse amide 1H relaxation enhancements, have previously been successfully used to define the fold of soluble proteins with limited nuclear Overhauser effect (NOE) data.26-28
The latest advances in MAS NMR spectroscopy of paramagnetic proteins29-36 have laid the foundation for application of the PRE-based approach to the structural analysis of biological macromolecules in the solid phase. Using several 13C,15N-labeled analogs of a model 56-residue protein, B1 immunoglobulin binding domain of protein G (GB1), containing EDTA-Cu2+ tags attached to cysteine residues introduced into the protein especially for this purpose,37,38 we have recently demonstrated that residue-specific longitudinal PREs can be rapidly determined for most backbone amide 15N nuclei by multidimensional solid-state NMR methods.34,35 These 15N PREs, which correspond to ~10-20 Å 15N-Cu2+ distances, were found to be in good quantitative agreement with those predicted for protein structural models based on the wild-type (WT) GB1 fold. Here we show that a set of ~230 such PREs recorded for six Cys-EDTA-Cu2+ mutants of GB1 (i.e., ~4-5 restraints per residue), supplemented only by standard chemical shift based restraints on secondary structure and, importantly, in the absence of any dipolar coupling based internuclear distance restraints, is sufficient to derive the correct global fold for GB1 in a de novo fashion. Notably, since PRE restraints can be easily extracted with high-sensitivity from routine 2D or 3D solid-state NMR chemical shift correlation spectra, this methodology is expected to become widely applicable to larger proteins available in limited quantities.
Results
Determination of 15N paramagnetic relaxation enhancements in GB1 mutants by solid-state NMR spectroscopy
Residue-specific backbone amide 15N longitudinal PREs, Γ N1, were determined from measurements of longitudinal 15N relaxation rate constants using series of 2D 15N-13CO (NCO) spectra recorded with different values of the relaxation delay, τrelax (see Fig. 1a and Supplementary Fig. S1), as described in the Methods section. Relaxation data were collected for six pairs of GB1 mutants containing non-native Cys-EDTA-Cu2+ or Zn2+ sidechains incorporated at solvent exposed sites corresponding to residues N8, E19, K28, E42, D46 or T53 in the WT protein (Fig. 1b); all of these GB1 analogs exhibit the WT fold as judged by the similarity of solution and solid-state NMR backbone chemical shifts to those of WT GB1.33-35 For brevity, the GB1 mutants used in this study are labeled as 8EDTA-Cu2+/Zn2+, 19EDTA-Cu2+/Zn2+, 28EDTA-Cu2+/Zn2+, 42EDTA-Cu2+/Zn2+, 46EDTA-Cu2+/Zn2+ and 53EDTA-Cu2+/Zn2+, according to location of the Cys-EDTA-Cu2+/Zn2+ tag.
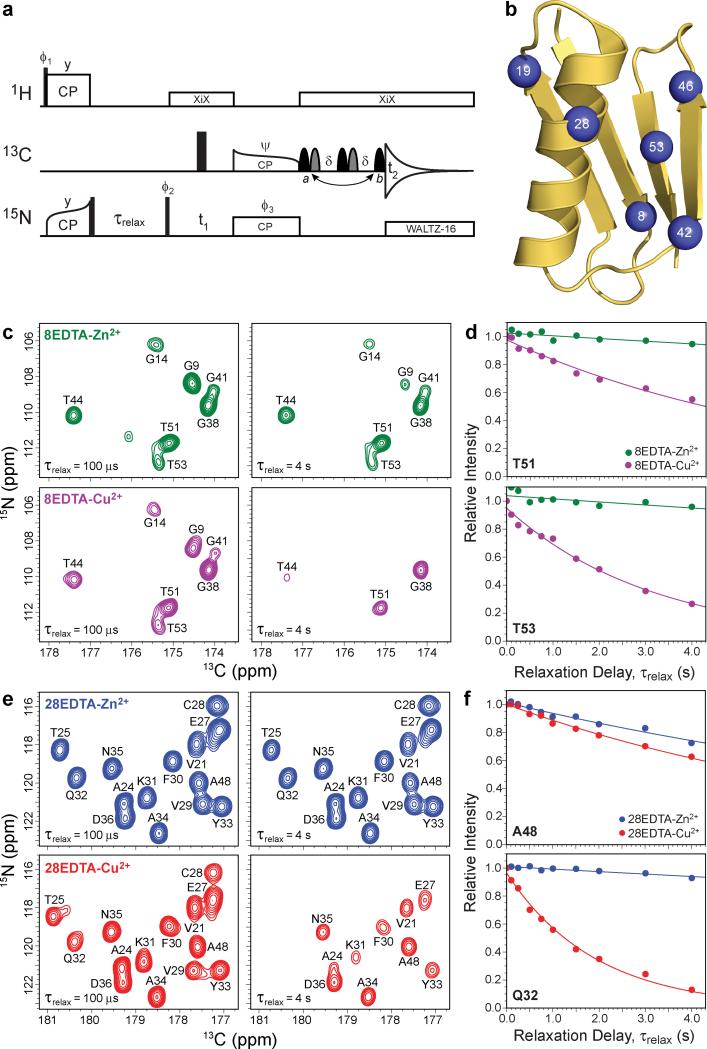
Figure 1. Determination of longitudinal backbone amide 15N paramagnetic relaxation enhancements in Cys-EDTA-Cu2+ GB1 mutants by solid-state NMR spectroscopy.
a, Pulse scheme used to determine residue-specific longitudinal 15N rate constants, R1, from a series of 2D 15N-13CO correlation spectra recorded with different values of the relaxation delay, τrelax. See Supplementary Fig. S1 for the complete pulse scheme description. b, Ribbon diagram of GB1 (PDB entry 2GI9) with the locations of the non-native Cys-EDTA-Cu2+/Zn2+ tags in point mutants investigated in this study (residues 8, 19, 28, 42, 46 or 53) indicated by blue spheres. c, Small regions of 2D 15N-13CO spectra recorded for 13C,15N-labeled 8EDTA-Zn2+ (green contours) and 8EDTA-Cu2+ (magenta contours) mutants with longitudinal 15N relaxation delays of 100 μs and 4 s, as indicated in the bottom left corner of each spectrum. The resonance assignments have been established previously,35 and cross-peaks are labeled according to the amide 15N frequency of the residue involved. d, Representative site-resolved 15N longitudinal relaxation trajectories for residues T51 and T53 in 8EDTA-Cu2+ and Zn2+. Experimental trajectories for 8EDTA-Zn2+ (green circles) and 8EDTA-Cu2+ (magenta circles) correspond to integrated cross-peak intensities in a series of 2D NCO spectra recorded as a function of the relaxation delay. Simulated best-fit trajectories to decaying single exponentials used to extract the longitudinal 15N relaxation rate constants are shown as solid lines of the corresponding color. The 15N PREs are calculated by taking the difference between the 15N R1 values obtained for the EDTA-Cu2+ and EDTA-Zn2+ proteins. e, Same as panel c for 28EDTA-Zn2+ (blue contours) and 28EDTA-Cu2+ (red contours). f, Same as panel d for residues Q32 and A48 in 28EDTA-Zn2+ (blue circles/lines) and 28EDTA-Cu2+ (red circles/lines).
In Fig. 1c and 1e we show representative small regions of 2D NCO spectra for 8EDTA-Cu2+/Zn2+ and 28EDTA-Cu2+/Zn2+ recorded with τrelax values of 100 μs (reference spectra) and 4 s. While the reference spectra for the corresponding EDTA-Cu2+ and Zn2+ proteins are effectively indistinguishable from one another and contain all of the expected intense one-bond 15N-13CO correlations, a number of cross-peaks are severely attenuated or altogether missing in spectra recorded for the EDTA-Cu2+ proteins with longer relaxation delays (τrelax ≈ 2-4 s). The most attenuated resonances (for example G9, G14 and T53 in 8EDTA-Cu2+ and T25, C28 and Q32 in 28EDTA-Cu2+) are invariably associated with residues located in the spatial proximity of the paramagnetic tag.34,35 This is to be expected based on the Solomon dipolar relaxation mechanism,24,25 and previously we have demonstrated that most experimental solid-state NMR 15N PREs and 15N-Cu2+ distances determined in this fashion are in quantitative agreement with the corresponding values predicted using protein structural models based on the WT GB1 fold.34,35 Fig. 1d and 1f show the complete experimental relaxation trajectories and best fits to decaying single exponentials (i.e., I = A·exp{–R1 × τrelax}, where I is the instantaneous cross-peak intensity, A is the peak intensity at τrelax = 0, and R1 the 15N longitudinal relaxation rate constant; the two fit parameters are A and R1) for representative residues in 8EDTA-Cu2+/Zn2+ and 28EDTA-Cu2+/Zn2+, respectively.
Collectively, these experiments yield 231 amide 15N longitudinal PRE restraints (Supplementary Table S1) for the six paramagnetic GB1 mutants (i.e., an average of ~38 out of 55 possible restraints per mutant or ~4-5 restraints per residue; the remaining PREs could not be determined reliably due to peak overlap in 2D NCO spectra). Approximately half of the PRE restraints (110 of 231) correspond to Γ1N values greater than 0.1 s-1, and were used as is in the course of the protein structure calculations discussed below. The remaining 121 restraints were converted to purely repulsive ‘NOE-type’ distance restraints preventing the associated atoms from approaching closer than 15.1 Å. This was done in order to avoid potential complications with the convergence of the calculated structures, associated with small but detectable effects on the extracted Γ1N values stemming from the presence of intermolecular 15N-Cu2+ couplings and secondary protein Cu2+ binding sites.36
Validation of the utility of solid-state NMR 15N PRE restraints for protein structure determination
In order to evaluate the ability of the longitudinal solid-state NMR 15N PREs to determine the correct protein fold in the absence of traditional dipolar coupling based distance restraints, we performed a set of preliminary, ‘ideal case’ scenario calculations for GB1 in Xplor-NIH39 using protocols where all native sidechains as well as the backbone atoms for regular secondary structure elements were frozen in rigid bodies corresponding to their conformations in the 1.14 Å GB1 X-ray structure40,41 (PDB entry 2GI9), and the Cys-EDTA-Cu2+ sidechain conformations (initially optimized using PRE data and 2GI9 atomic coordinates) were also fixed. The regular secondary structure elements included the α-helix (residues 22-37) and the four β-strands, β1 to β4, spanning amino acids (aa) 2-8, 13-19, 42-46 and 51-55, respectively. The backbone torsion angles for the remaining residues were randomized. Moreover, to enable the simultaneous use of all PRE restraints within the same structure calculation protocol, the Cys-EDTA-Cu2+ tags were placed on all six modified residues (even though separate NMR experiments were used to record PRE data for each of the GB1 mutants) with the concurrent disabling of interactions between atoms associated with the different tags.
In Fig. 2a and 2b we show representative sets of 10 lowest energy structures (out of a total of 1000 structures) calculated without and with the use of solid-state NMR longitudinal 15N PRE restraints, respectively, in addition to the more conventional potential energy terms described in the Methods section. Not surprisingly, when no experimental NMR data were incorporated into the calculations, the lowest energy protein structures obtained in Xplor-NIH are effectively random and show little resemblance to one another or to the GB1 X-ray structure (Fig. 2a). Within this set of 10 lowest energy structures, no structure, including the one displaying the smallest backbone atom (C′, Cα, N and O) coordinate root-mean-square deviation (RMSD) of 3.4 Å relative to 2GI9, correctly predicts the protein fold. Though occasionally (3 times out of 1000 for this dataset) the calculations with no experimental restraints included were able to yield protein structures with the correct GB1 fold (backbone RMSDs versus 2GI9 of ~1.5-2 Å), these ‘correct’ structures were associated with energies that were considerably higher than those obtained for numerous ‘incorrect’ structures having backbone atom RMSDs in the ~3-9 Å range. In stark contrast, analogous calculations which also include the 231 experimental 15N PRE restraints yield lowest energy protein structures that cluster tightly together, and, most importantly, correspond to the correct GB1 fold with backbone RMSDs of 0.9-1.8 Å relative to 2GI9 (Fig. 2b).
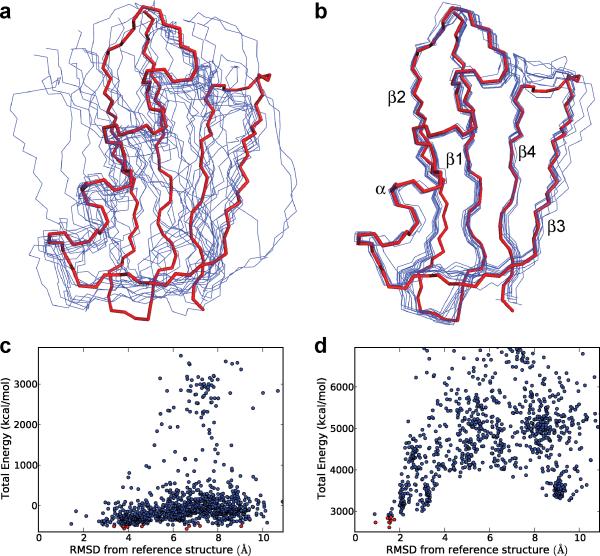
Figure 2. Results of idealized preliminary structure calculations for GB1 in the absence and presence of solid-state NMR longitudinal 15N paramagnetic relaxation enhancement restraints.
a, Backbone traces (blue) for the 10 lowest energy GB1 structures (out of 1000 total structures), calculated using Xplor-NIH39 in the absence of experimental 15N PRE restraints. Shown for comparison is the backbone trace (red) corresponding to the 1.14 Å GB1 X-ray structure40,41 (PDB entry 2GI9). Native sidechain torsion angles and backbone torsion angles for residues in regular secondary structure elements (β1: aa 2-8; β2: aa 13-19; α: aa 22-37; β3: aa 42-46; β4: aa 51-55) were fixed to their X-ray structure values during the calculations, and the remaining backbone torsion angles were randomized (a detailed description of the structure calculations is provided in the Methods section). The backbone coordinates of the 10 lowest energy structures show RMSDs ranging from 3.4 to 9.1 Å relative to the X-ray structure. b, Same as panel a except that 231 15N PRE restraints recorded for the six paramagnetic GB1 mutants were additionally included in the structure calculations. The backbone coordinates of the 10 lowest energy structures show RMSDs relative to 2GI9 ranging from 0.9 to 1.8 Å. c,d, Plots of total energy vs. backbone atom coordinate RMSD from the 2GI9 reference X-ray structure for each of the 1000 structures calculated without (c) and with (d) the use of the 15N PRE restraints. In each plot, the 10 lowest energy structures are indicated by red circles.
The complete results for both sets of calculations employing large ensembles of 1000 random starting protein structures are summarized in Fig. 2c and 2d. These data clearly illustrate that in the absence of 15N PRE restraints there is effectively no correlation between the total energy of a particular protein structure obtained in Xplor-NIH and that structure displaying the correct GB1 fold (Fig. 2c). Indeed, nearly all structures obtained in this manner bear little resemblance to the reference GB1 X-ray structure. On the other hand, the addition of 15N PRE restraints to the structure calculation protocol leads to a funnel-like effect where a relatively strong correlation emerges between the total energy of a calculated structure and its RMSD relative to the 2GI9 backbone atom coordinates (Fig. 2d). While the convergence properties of these calculations may not be ideal and the calculation procedure generates multiple incorrect protein structures, it is important to note that all of these incorrect structures have high energies and can be readily identified. Most significantly, however, the lowest energy structures (associated with total energies of ~3000 kcal/mol or less in this case; the 10 lowest energy structures are indicated by red circles in Fig. 2c and 2d) all possess similar backbone folds that also closely match the GB1 X-ray structure (Fig. 2b).
De novo determination of the GB1 backbone fold
Having established the utility of solid-state NMR 15N PRE restraints for protein structure calculations, we proceeded to investigate whether the fold of GB1 could be determined in a de novo manner by using only 15N PRE data (Supplementary Table S1) and standard solid-state NMR chemical shift based backbone dihedral angle restraints for WT GB1 (Supplementary Table S2) to define local secondary structure. As described in detail in the Methods section, these realistic de novo calculations consisted of a two stage protocol. In the initial stage, backbone atoms for residues comprising regular secondary structure elements (identified within the torsion angle likelihood obtained from shifts and sequence similarity, TALOS+, software42 as having α or β conformation with greater than 85% confidence) were frozen in rigid bodies with their dihedral angles fixed to the ideal TALOS+ predicted values, while all other backbone and sidechain torsion angles (including the Cys-EDTA-Cu2+ tags) were randomized. We found that fixing the TALOS+ identified secondary structure regions during this initial stage resulted in significantly improved convergence relative to the standard protocols which allow more degrees of freedom, as well as improved accuracy of the final calculated structures. The initial calculation stage was followed by a second stage, in which the 10 lowest energy structures from the initial calculations (indicated by red circles in Fig. 3a) were further refined using 10 different sets of randomized velocities in a protocol that allowed all sidechains to move as well as all backbone dihedral degrees of freedom constrained only by the TALOS+ dihedral angle potential with minimum dihedral angle uncertainties of ±20°, to yield a final set of 100 structures. From this set, the 20 lowest energy structures, shown in Fig. 3b and indicated by yellow circles in Fig. 3a, were used to obtain a regularized mean structure.
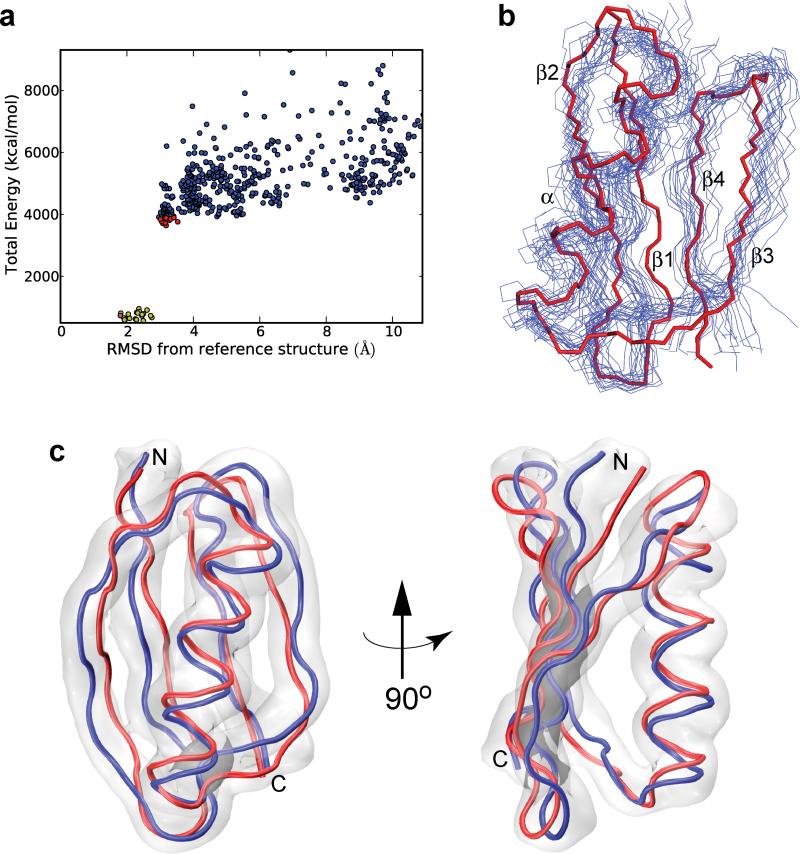
Figure 3. De novo calculation of the GB1 fold using solid-state NMR longitudinal 15N paramagnetic relaxation enhancement restraints.
a, Plot of total energy vs. backbone atom RMSD from the reference X-ray structure obtained during the course of the de novo calculations. The 10 lowest energy structures from the initial calculations are indicated by red circles. These 10 structures were further refined as described in the text to yield a set of 100 structures. From this final set, the 20 lowest energy structures indicated by yellow circles were used to obtain the regularized mean structure (magenta circle). b, Backbone traces for the 20 lowest energy GB1 structures (blue), corresponding to the yellow circles in panel a, and the reference X-ray structure (red). c, Comparison of the GB1 X-ray structure with the regularized mean solid-state NMR backbone fold determined using the realistic, de novo calculations. The coordinates of the mean NMR structure (blue) show a backbone atom RMSD of 1.8 Å and an all heavy atom RMSD of 2.7 Å relative to the X-ray structure (red). The gray cloud is a reweighted atomic probability map50 of the final ensemble of 20 lowest energy NMR structures shown in panel b plotted at 15% of maximum value, representing the conformational space occupied by the associated backbone atoms. Structure calculations were performed as described in the Methods section using 231 longitudinal 15N PRE restraints recorded for six paramagnetic GB1 analogs and solid-state NMR chemical shift-based dihedral angle restraints for WT GB1 obtained using TALOS+.42
Remarkably, using these largely unrestricted de novo calculations, which did not utilize any conventional dipolar coupling based interatomic distance restraints or structural information derived from X-ray diffraction data, we were able to determine a backbone fold for GB1 that is in good agreement with the 1.14 Å X-ray structure (Fig. 3c; structure indicated by the magenta circle in Fig. 3a). The mean solid-state NMR structure obtained in this manner (blue) displays a backbone atom RMSD of 1.8 Å and an all heavy atom RMSD (excluding sidechains of residues 8, 19, 28, 42, 46 and 53) of 2.7 Å relative to the 2GI9 reference structure (red), and the backbone atoms for the vast majority of residues in the X-ray structure (with exception of several aa located in the loop regions or near the N-terminus) fall within the conformational space occupied by the ensemble of the 20 lowest energy conformers (gray cloud). Finally, in the course of these calculations we have also considered the possible influence of Cys-EDTA-Cu2+ sidechain disorder on the quality of the derived structures. The PRE refinement facilities within Xplor-NIH allow such structural heterogeneity to be modeled using ensembles of several sidechains, by either representing the PRE as a sum of contributions from each ensemble member (corresponding to slow interconversion between ensemble members) or 1/r6 averaging of the different ensemble members (for fast interconversion), and both approaches were attempted using ensembles of two and three Cys-EDTA-Cu2+ tags. We found that the use of such ensembles did not improve the results relative to using a single Cys-EDTA-Cu2+ conformer, presumably due to the presence of additional degrees of freedom for which there are insufficient data in this study.
Discussion
Notwithstanding the tremendous current progress,5-19 the routine determination of protein structures by solid-state NMR remains a formidable task mainly due to challenges associated with the detection of large numbers of correlations that report on >5 Å internuclear distances. Measurements of such long-distance restraints in paramagnetic proteins, including PREs33,34 and pseudocontact shifts (PCS),30,31 have the potential to significantly accelerate and enhance solid-state NMR protein structure elucidation efforts. In fact, the utility of paramagnetic restraints of this type was recently illustrated by Bertini and co-workers for a 159-residue (~17.6 kDa) native metalloprotein, cobalt(II)-substituted matrix metalloproteinase 12,31,32 where over 300 13C PCS restraints were used in conjunction with ~30031 and ~80032 internuclear distances obtained from PDSD/DARR, CHHC and PAR experiments to arrive at 3D protein structures.
The approach toward global protein fold determination described here utilizes PRE restraints recorded for structural analogs of the protein of interest containing covalently-linked paramagnetic tags, and is readily applicable to a wide variety of protein molecules in the solid phase, including natively diamagnetic proteins. A major advantage of this approach, in addition to the fact that nuclear PREs yield almost exclusively structurally-relevant inter-residue restraints generally corresponding to >10 Å distances, is that these PRE restraints are very straightforward to extract by monitoring cross-peak intensities as a function of a relaxation delay in the simplest 2D or 3D solid-state NMR chemical shift correlation spectra that contain a minimum number of signals arising from adjacent, strongly dipolar coupled nuclei in the protein. As such, this methodology is pertinent to larger protein systems. While the implementation of this method requires the preparation of several point mutants of the protein under study, such biochemical manipulations are routine nowadays. It is also important to note that although the known 3D structure of GB1 was helpful in identifying suitable sites for introduction of the Cys-EDTA-Cu2+ tags in this study, the incorporation of such tags into proteins of unknown structure, with minimal influence on the backbone fold,38 should be readily implementable using an approach discussed by Battiste and Wagner27 that utilizes secondary structure information obtained from chemical shifts for the WT protein and the analysis of hydrophilicity patterns to identify the most probable solvent-accessible residues.43
The current study demonstrates that ~4-5 longitudinal amide 15N PRE restraints per residue, collected using a condensed NMR data acquisition approach35 for a set of six Cys-EDTA-Cu2+ mutants available in small quantities (~150-200 nanomoles of 13C,15N-labeled protein per sample), are sufficient to obtain a protein backbone fold for GB1 that is in close agreement with the X-ray structure without the use of any conventional internuclear distance restraints. Although the global protein fold determined in this manner does not correspond to the highest achievable resolution structure, it can be derived very rapidly and provides a reasonable structural model that is likely to be sufficient for many applications. Moreover, such a PRE-based model can serve as a major aid for resolving assignment ambiguities in conventional dipolar coupling based solid-state NMR spectra, and can readily be further refined to higher resolution, if desired, by incorporating additional interatomic distance restraints or by combining the PRE restraints with more advanced fragment-based chemical shift/molecular mechanics (CSMM) based structure calculation protocols such as CHESHIRE13 or CS-ROSETTA.14 It should be noted that the structure of GB1 has, in fact, been successfully solved (and to higher resolution) using the CHESHIRE approach with chemical shifts as the sole experimental input,13 whereas in this work we only extracted broad backbone torsion angle ranges from chemical shift data. While such CSMM methods, which utilize molecular mechanics force fields to obtain the proper protein fold, have chalked up an impressive record of correctly determined protein structures and we strongly advocate their use, they are also known to fail at times due to inadequacies of the MM force fields and the fact that chemical shift data contain essentially no long-distance information. In contrast, the PRE-based approach reported here uses direct distance restraints simply related to experimental observables, and thus serves as an independent means to solve or validate a protein fold.
Importantly, the number of paramagnetic-based structural restraints in these studies can be dramatically increased, by at least ~2-3-fold, with minimal effort by measuring other types of PREs, including longitudinal backbone and sidechain 13C PREs and transverse PREs for the different nuclei, in the same protein samples, as well as PCS restraints for 13C, 15N and potentially 1H nuclei in analogous Cys-EDTA-Co2+ samples. This is expected to substantially enhance the resolution of the derived structures, and bodes well for the extension of this paramagnetic solid-state NMR methodology to larger proteins for which a significant fraction of resonances can be assigned. Indeed, such sequential resonance assignments in larger systems may be facilitated by using spectral editing approaches that take advantage of the presence of the paramagnetic tags.
Methods
Sample preparation
Plasmid cDNAs encoding for N8C, E19C, K28C, E42C, D46C or T53C mutants were constructed as described previously,33 using T2Q-GB1 cDNA (referred to as GB1) and Stratagene QuikChange II site-directed mutagenesis protocol. Proteins were expressed in E. coli BL21(DE3) with Luria-Bertani medium or M9 minimal medium containing 1 g/L 15NH4Cl and 3 g/L 13C-glucose (Cambridge Isotope Laboratories) and purified by gel filtration chromatography.33,34 Cysteines were reacted with the sulfhydryl-specific reagent N-[S-(2-pyridylthio)cysteaminyl]EDTA37 (Toronto Research Chemicals), pre-loaded with 1.1 equivalents of Cu2+ or Zn2+ (diamagnetic control). Protein microcrystals for solid-state NMR were prepared by co-precipitating the 13C,15N-EDTA-Cu2+/Zn2+ proteins with natural abundance GB1 in a 1:3 molar ratio using microdialysis at 4 °C and a 2:1:1 (v/v) 2-methylpentane-2,4-diol/isopropanol/deionized water precipitant solution.34,36 Samples, each containing ~1.0-1.2 mg (~150-200 nanomoles) of 13C,15N-labeled protein, were packed into 1.6 mm zirconia rotors (Agilent Technologies) by centrifugation.
NMR spectroscopy
NMR spectra were recorded on a 500 MHz Varian spectrometer equipped with a 1.6 mm FastMAS™ probe. MAS rate and sample temperature were regulated at 40,000 ± 20 Hz and ~5 °C, respectively. Residue-specific amide 15N longitudinal relaxation rate constants, R1, for the EDTA-Cu2+/Zn2+ GB1 mutants were determined from series of 2D NCO spectra recorded using the pulse scheme in Fig. 1a with relaxation delays, τrelax, of 100 μs to 4 s. Spectra were collected with acquisition times of 25.6 ms (t1) and 30 ms (t2) and recycle delays of 0.4 s and 1.3 s for EDTA-Cu2+ and EDTA-Zn2+ proteins, respectively. The total experiment time required to record 15N R1 data for all EDTA-Cu2+/Zn2+ GB1 mutants was ~18 days. Spectra were processed using NMRPipe44 and analyzed with home-built software (available at http://code.google.com/p/nmrglue). 15N PREs, Γ1N, were calculated as Γ1N = R1(Cu2+) – R1(Zn2+), where 15N R1 values for Cu2+ and Zn2+ proteins were obtained by fitting residue-specific relaxation trajectories to decaying single exponentials.
Structure calculations
Structure calculations were carried out in Xplor-NIH39 with a protocol in which backbone atoms of regular secondary structure elements were frozen in rigid bodies corresponding to their X-ray (during initial calculations in Fig. 2) or ideal target conformations as predicted from TALOS+42 (during the realistic de novo calculations in Fig. 3). Regions of secondary structure were identified by inspection of the GB1 X-ray structure40,41 (PDB entry 2GI9) for the initial calculations and from TALOS+ hits predicted with >85% confidence in the α or β Ramachandran regions for the realistic calculations. The remaining backbone torsion angles were randomized. Sidechain torsion angles were fixed in their X-ray conformation for the initial calculations and randomized for the realistic calculations. The EDTA-Cu2+ tags were present on all six residues which were modified, but inter-residue interactions of the associated atoms were disabled. In the initial calculations, conformations of the EDTA-Cu2+ sidechains were first optimized using PRE data and the 2GI9 coordinates and held fixed during structure calculations. For the realistic calculations, EDTA-Cu2+ sidechains were treated as other sidechains with torsion angles randomized.
PRE restraints consisted of explicit restraints using the PREPot term45 with an electron correlation time of 3 ns25 for Γ N1 values above a cutoff of 0.1 s-1 (Supplementary Table S1). Smaller PREs were included using a purely repulsive ‘NOE-type’ term restraining the associated 15N-Cu2+ distances to values >15.1 Å. TALOS+ was used to derive 102 (of 110 possible) backbone torsion angle restraints from GB1 13C and 15N solid-state NMR chemical shifts (Supplementary Table S2). The radius of gyration term46 was used to achieve appropriate protein packing density. Additional knowledge-based energy terms included the torsion angle potential of mean force,47 the hydrogen bond potential of mean force,48 and the low resolution residue contact term.49 Standard bond, bond angle and improper torsion angle restraints were used to maintain proper covalent geometry, and a repulsive quartic van der Waals term used to prevent atomic overlap. During calculations in which EDTA-Cu2+ sidechains were allowed to move, the associated dihedral angles were restrained by a dihedral energy term.
The protocol consisted of initial conjugate gradient minimization without the PRE and repulsive distance terms, followed by minimization including all energy terms. This was followed by variable step-size dynamics for the smaller of 500 ps or 20,000 steps at 3000 K. Next, simulated annealing was performed from 3000 K to 25 K in 12.5 K increments, and at each temperature dynamics for the smaller of 1.6 ps or 400 steps was run. During simulated annealing the force constants of various energy terms were ramped to their final values as specified in Supplementary Table S3. For the realistic calculations, the initial structure calculation was followed by an additional calculation stage in which the 10 lowest energy structures from the initial calculation were refined using 10 different sets of randomized velocities (for a total of 100 structures) in a protocol which allowed all backbone dihedral degrees of freedom and omitted the initial minimization step. A regularized mean structure was calculated from the 20 lowest energy refined structures.
Supplementary Material
Acknowledgements
This research was supported by the National Science Foundation (CAREER Award MCB-0745754 to C.P.J.). C.D.S. was supported by the NIH Intramural Research Program of the Center for Information Technology. The GB1 plasmid was kindly provided by Dr. Angela M. Gronenborn.
Footnotes
Author contributions
C.P.J. designed research. I.S. and P.S.N. prepared samples. I.S., P.S.N. and J.J.H. recorded and analyzed the NMR data. C.D.S. and C.P.J. performed the structure calculations. C.D.S. and C.P.J. wrote the paper.
Additional information
The authors declare no competing financial interests. Supplementary information accompanies this paper. Correspondence and requests for materials should be addressed to C.P.J.
References
- 1.Wüthrich K. NMR studies of structure and function of biological macromolecules (Nobel lecture). Angew. Chem. Int. Ed. 2003;42:3340–3363. doi: 10.1002/anie.200300595. [DOI] [PubMed] [Google Scholar]
- 2.Lange A, et al. Toxin-induced conformational changes in a potassium channel revealed by solid-state NMR. Nature. 2006;440:959–962. doi: 10.1038/nature04649. [DOI] [PubMed] [Google Scholar]
- 3.Cady SD, et al. Structure of the amantadine binding site of influenza M2 proton channels in lipid bilayers. Nature. 2010;463:689–692. doi: 10.1038/nature08722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Petkova AT, et al. Self-propagating, molecular-level polymorphism in Alzheimer's β-amyloid fibrils. Science. 2005;307:262–265. doi: 10.1126/science.1105850. [DOI] [PubMed] [Google Scholar]
- 5.Wasmer C, et al. Amyloid fibrils of the HET-s(218-289) prion form a β solenoid with a triangular hydrophobic core. Science. 2008;319:1523–1526. doi: 10.1126/science.1151839. [DOI] [PubMed] [Google Scholar]
- 6.Castellani F, et al. Structure of a protein determined by solid-state magic-angle spinning NMR spectroscopy. Nature. 2002;420:98–102. doi: 10.1038/nature01070. [DOI] [PubMed] [Google Scholar]
- 7.Zech SG, Wand AJ, McDermott AE. Protein structure determination by high-resolution solid-state NMR spectroscopy: Application to microcrystalline ubiquitin. J. Am. Chem. Soc. 2005;127:8618–8626. doi: 10.1021/ja0503128. [DOI] [PubMed] [Google Scholar]
- 8.Loquet A, et al. 3D structure determination of the Crh protein from highly ambiguous solid-state NMR restraints. J. Am. Chem. Soc. 2008;130:3579–3589. doi: 10.1021/ja078014t. [DOI] [PubMed] [Google Scholar]
- 9.Manolikas T, Herrmann T, Meier BH. Protein structure determination from 13C spin-diffusion solid-state NMR spectroscopy. J. Am. Chem. Soc. 2008;130:3959–3966. doi: 10.1021/ja078039s. [DOI] [PubMed] [Google Scholar]
- 10.Korukottu J, et al. High-resolution 3D structure determination of kaliotoxin by solid-state NMR spectroscopy. PLoS ONE. 2008;3:e2359. doi: 10.1371/journal.pone.0002359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Franks WT, et al. Dipole tensor-based atomic-resolution structure determination of a nanocrystalline protein by solid-state NMR. Proc. Natl. Acad. Sci. USA. 2008;105:4621–4626. doi: 10.1073/pnas.0712393105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Paëpe G, Lewandowski JR, Loquet A, Böckmann A, Griffin RG. Proton assisted recoupling and protein structure determination. J. Chem. Phys. 2008;129:245101. doi: 10.1063/1.3036928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robustelli P, Cavalli A, Vendruscolo M. Determination of protein structures in the solid state from NMR chemical shifts. Structure. 2008;16:1764–1769. doi: 10.1016/j.str.2008.10.016. [DOI] [PubMed] [Google Scholar]
- 14.Shen Y, Vernon R, Baker D, Bax A. De novo protein structure generation from incomplete chemical shift assignments. J. Biomol. NMR. 2009;43:63–78. doi: 10.1007/s10858-008-9288-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nieuwkoop AJ, Wylie BJ, Franks WT, Shah GJ, Rienstra CM. Atomic resolution protein structure determination by three-dimensional transferred echo double resonance solid-state nuclear magnetic resonance spectroscopy. J. Chem. Phys. 2009;131:095101. doi: 10.1063/1.3211103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y, et al. Resonance assignment and three-dimensional structure determination of a human alpha-defensin, HNP-1, by solid-state NMR. J. Mol. Biol. 2010;397:408–422. doi: 10.1016/j.jmb.2010.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jehle S, et al. Solid-state NMR and SAXS studies provide a structural basis for the activation of αB-crystallin oligomers. Nat. Struct. Mol. Biol. 2010;17:1037–1042. doi: 10.1038/nsmb.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Linser R, Bardiaux B, Higman V, Fink U, Reif B. Structure calculation from unambiguous long-range amide and methyl 1H-1H distance restraints for a microcrystalline protein with MAS solid-state NMR spectroscopy. J. Am. Chem. Soc. 2011;133:5905–5912. doi: 10.1021/ja110222h. [DOI] [PubMed] [Google Scholar]
- 19.Huber M, et al. A proton-detected 4D solid-state NMR experiment for protein structure determination. Chemphyschem. 2011;12:915–918. doi: 10.1002/cphc.201100062. [DOI] [PubMed] [Google Scholar]
- 20.Helmus JJ, Nadaud PS, Höfer N, Jaroniec CP. Determination of methyl 13C-15N dipolar couplings in peptides and proteins by three-dimensional and four-dimensional magic-angle spinning solid-state NMR spectroscopy. J. Chem. Phys. 2008;128:052314. doi: 10.1063/1.2817638. [DOI] [PubMed] [Google Scholar]
- 21.Jaroniec CP, Filip C, Griffin RG. 3D TEDOR NMR experiments for the simultaneous measurement of multiple carbon-nitrogen distances in uniformly 13C,15N-labeled solids. J. Am. Chem. Soc. 2002;124:10728–10742. doi: 10.1021/ja026385y. [DOI] [PubMed] [Google Scholar]
- 22.Takegoshi K, Nakamura S, Terao T. 13C-1H dipolar-assisted rotational resonance in magic-angle spinning NMR. Chem. Phys. Lett. 2001;344:631–637. [Google Scholar]
- 23.Lange A, Luca S, Baldus M. Structural constraints from proton-mediated rare-spin correlation spectroscopy in rotating solids. J. Am. Chem. Soc. 2002;124:9704–9705. doi: 10.1021/ja026691b. [DOI] [PubMed] [Google Scholar]
- 24.Solomon I. Relaxation processes in a system of two spins. Phys. Rev. 1955;99:559–565. [Google Scholar]
- 25.Bertini I, Luchinat C, Parigi G. Solution NMR of Paramagnetic Molecules: Applications to Metallobiomolecules and Models. Elsevier; Amsterdam: 2001. [Google Scholar]
- 26.Gillespie JR, Shortle D. Characterization of long-range structure in the denatured state of staphylococcal nuclease. II. Distance restraints from paramagnetic relaxation and calculation of an ensemble of structures. J. Mol. Biol. 1997;268:170–184. doi: 10.1006/jmbi.1997.0953. [DOI] [PubMed] [Google Scholar]
- 27.Battiste JL, Wagner G. Utilization of site-directed spin labeling and high-resolution heteronuclear nuclear magnetic resonance for global fold determination of large proteins with limited nuclear overhauser effect data. Biochemistry. 2000;39:5355–5365. doi: 10.1021/bi000060h. [DOI] [PubMed] [Google Scholar]
- 28.Gaponenko V, et al. Protein global fold determination using site-directed spin and isotope labeling. Protein Sci. 2000;9:302–309. doi: 10.1110/ps.9.2.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pintacuda G, et al. Solid-state NMR spectroscopy of a paramagnetic protein: Assignment and study of human dimeric oxidized CuII-ZnII superoxide dismutase (SOD). Angew. Chem. Int. Ed. 2007;46:1079–1082. doi: 10.1002/anie.200603093. [DOI] [PubMed] [Google Scholar]
- 30.Balayssac S, Bertini I, Lelli M, Luchinat C, Maletta M. Paramagnetic ions provide structural restraints in solid-state NMR of proteins. J. Am. Chem. Soc. 2007;129:2218–2219. doi: 10.1021/ja068105a. [DOI] [PubMed] [Google Scholar]
- 31.Balayssac S, Bertini I, Bhaumik A, Lelli M, Luchinat C. Paramagnetic shifts in solid-state NMR of proteins to elicit structural information. Proc. Natl. Acad. Sci. USA. 2008;105:17284–17289. doi: 10.1073/pnas.0708460105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bertini I, et al. High-resolution solid-state NMR structure of a 17.6 kDa protein. J. Am. Chem. Soc. 2010;132:1032–1040. doi: 10.1021/ja906426p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nadaud PS, Helmus JJ, Höfer N, Jaroniec CP. Long-range structural restraints in spin-labeled proteins probed by solid-state nuclear magnetic resonance spectroscopy. J. Am. Chem. Soc. 2007;129:7502–7503. doi: 10.1021/ja072349t. [DOI] [PubMed] [Google Scholar]
- 34.Nadaud PS, Helmus JJ, Kall SL, Jaroniec CP. Paramagnetic ions enable tuning of nuclear relaxation rates and provide long-range structural restraints in solid-state NMR of proteins. J. Am. Chem. Soc. 2009;131:8108–8120. doi: 10.1021/ja900224z. [DOI] [PubMed] [Google Scholar]
- 35.Nadaud PS, Helmus JJ, Sengupta I, Jaroniec CP. Rapid acquisition of multidimensional solid-state NMR spectra of proteins facilitated by covalently bound paramagnetic tags. J. Am. Chem. Soc. 2010;132:9561–9563. doi: 10.1021/ja103545e. [DOI] [PubMed] [Google Scholar]
- 36.Nadaud PS, Sengupta I, Helmus JJ, Jaroniec CP. Evaluation of the influence of intermolecular electron-nucleus couplings and intrinsic metal binding sites on the measurement of 15N longitudinal paramagnetic relaxation enhancements in proteins by solid-state NMR. J. Biomol. NMR. 2011;51:293–302. doi: 10.1007/s10858-011-9536-y. [DOI] [PubMed] [Google Scholar]
- 37.Ermácora MR, Delfino JM, Cuenoud B, Schepartz A, Fox RO. Conformation-dependent cleavage of staphylococcal nuclease with a disulfide-linked iron chelate. Proc. Natl. Acad. Sci. USA. 1992;89:6383–6387. doi: 10.1073/pnas.89.14.6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hubbell WL, Altenbach C. Investigation of structure and dynamics in membrane proteins using site-directed spin labeling. Curr. Opin. Struct. Biol. 1994;4:566–573. [Google Scholar]
- 39.Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 40.Gallagher T, Alexander P, Bryan P, Gilliland GL. Two crystal structures of the B1 immunoglobulin-binding domain of streptococcal protein G and comparison with NMR. Biochemistry. 1994;33:4721–4729. [PubMed] [Google Scholar]
- 41.Franks WT, Wylie BJ, Stellfox SA, Rienstra CM. Backbone conformational constraints in a microcrystalline U-15N-labeled protein by 3D dipolar-shift solid-state NMR spectroscopy. J. Am. Chem. Soc. 2006;128:3154–3155. doi: 10.1021/ja058292x. [DOI] [PubMed] [Google Scholar]
- 42.Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: A hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rost B, Sander C. Conservation and prediction of solvent accessibility in protein families. Proteins. 1994;20:216–226. doi: 10.1002/prot.340200303. [DOI] [PubMed] [Google Scholar]
- 44.Delaglio F, et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 45.Iwahara J, Schwieters CD, Clore GM. Ensemble approach for NMR structure refinement against 1H paramagnetic relaxation enhancement data arising from a flexible paramagnetic group attached to a macromolecule. J. Am. Chem. Soc. 2004;126:5879–5896. doi: 10.1021/ja031580d. [DOI] [PubMed] [Google Scholar]
- 46.Kuszewski J, Gronenborn AM, Clore GM. Improving the packing and accuracy of NMR structures with a pseudopotential for the radius of gyration. J. Am. Chem. Soc. 1999;121:2337–2338. [Google Scholar]
- 47.Kuszewski J, Gronenborn AM, Clore GM. Improving the quality of NMR and crystallographic protein structures by means of a conformational database potential derived from structure databases. Protein Sci. 1996;5:1067–1080. doi: 10.1002/pro.5560050609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grishaev A, Bax A. An empirical backbone-backbone hydrogen-bonding potential in proteins and its applications to NMR structure refinement and validation. J. Am. Chem. Soc. 2004;126:7281–7292. doi: 10.1021/ja0319994. [DOI] [PubMed] [Google Scholar]
- 49.Ryabov Y, Suh J-Y, Grishaev A, Clore GM, Schwieters CD. Using the experimentally determined components of the overall rotational diffusion tensor to restrain molecular shape and size in NMR structure determination of globular proteins and protein-protein complexes. J. Am. Chem. Soc. 2009;131:9522–9531. doi: 10.1021/ja902336c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schwieters CD, Clore GM. Reweighted atomic densities to represent ensembles of NMR structures. J. Biomol. NMR. 2002;23:221–225. doi: 10.1023/a:1019875223132. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.