PDZ domains are part of a molecular scaffold that holds multiprotein signaling complexes together. It is generally believed that the role of PDZ domains is to position target ion channels, receptors, or other signaling molecules in correct spatial arrangement in relation to each other and to specialized regions of the cell (1–4). Two recent reports, one published in Cell (5) and another in this issue of PNAS (6) challenge this paradigm. These reports are focused on biochemical and functional analysis of cystic fibrosis transmembrane conductance regulator (CFTR) channel interactions with multi-PDZ domain adaptor proteins CAP70 (CFTR-associated protein 70 kDa) (5) and NHERF (Na+/H+ exchanger regulatory factor) (6). CFTR contains a classical type I PDZ-domain binding carboxyl-terminal consensus −T-K/R-L, conserved from Xenopus to humans (5). Biochemical association of CFTR with the NHERF PDZ1 domain has previously been reported by several groups (7–9), and the importance of this interaction for apical sorting of CFTR in polarized epithelia has been demonstrated (10, 11). The previous findings fit with the general paradigm regarding the role of the PDZ domain containing proteins in membrane protein targeting in polarized cells.
What makes the reports by Raghuram et al. (6) and Wang et al. (5) particularly interesting is the notion that the association with PDZ domains does more than simply position the CFTR in the appropriate location. Both groups concluded that PDZ domains in NHERF and CAP70 also play a modulatory function by directly affecting CFTR channel gating. Raghuram et al. found that an addition of the NHERF PDZ1–2 tandem protein to the cytosolic side of inside-out patches taken from Calu-3 (lung submucosal gland) cells resulted in an increase of endogenous CFTR channel activity. Remarkably, only the tandem construct of 1–2 NHERF PDZ domains activated CFTR, but not when first or second PDZ domains added independently, mixed together or when one of the PDZ domains in the tandem construct was mutated. Moreover, first and second PDZ domains of NHERF inhibit potentiation of CFTR activity by PDZ1–2 NHERF construct, and the tandem construct itself exerts a biphasic effect on CFTR activity. Wang et al. (5) performed similar experiments with CFTR channels transiently expressed in HEK-293 cells. Using inside-out patches or marcopatches, Wang et al. found that the activity of a CFTR channel potentiated in a biphasic manner by the tandem construct of 3–4 PDZ domains of CAP70, whereas CAP70 PDZ 1–2 and 2–3 tandem constructs had no effect. The PDZ 3 domain alone did not activate CFTR channels, but the PDZ 3–3 tandem construct was effective. Moreover, Wang et al. found that the monoclonal antibody directed against CFTR carboxyl termini had a potentiating effect on CFTR channel activity similar to that of the CAP70 PDZ 3–4 tandem construct.
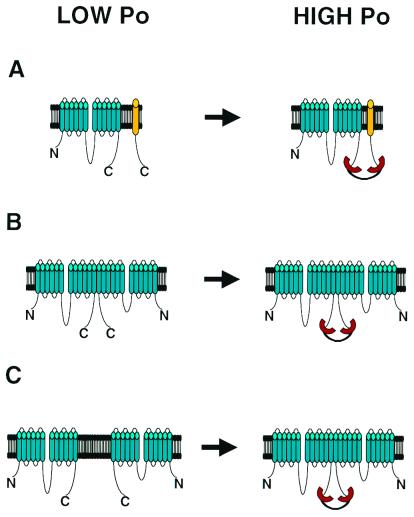
Three general models can explain the findings of Raghuram et al. (6) and Wang et al. (5). In the first model PDZ domain tandem links CFTR with an additional membrane or membrane-associated protein (Fig. 1A). In the second model PDZ domain tandem cross-links two CFTR subunits within the dimeric channel (Fig. 1B). In the third model PDZ domain tandem promotes an assembly of the dimeric CFTR channel from two monomeric subunits (Fig. 1C). For some channels recruitment of additional subunits (Fig. 1A) has been shown to have a functional effect (12). However, both groups conclude that this scenario is unlikely in the case of CFTR. Indeed, the effects of NHERF PDZ1–2 on CFTR activity were observed not only in Calu-3 cells, but also when CFTR was expressed in Chinese hamster ovary, NIH 3T3, and HEK-293 cells (6), suggesting that a putative regulatory subunit must be ubiquiously expressed. The ability of monoclonal carboxyl-terminal CFTR antibody to activate channels (5) makes this scenario even less likely.
Figure 1.
Bivalent PDZ domain activation of CFTR channel. (A) PDZ domains link CFTR channel with an additional regulatory subunit. (B) PDZ domains link two CFTR subunits within pre-existing dimeric channel complex. (C) PDZ domains shift equilibrium between monomeric and dimeric forms of CFTR channel.
The second possibility (Fig. 1B) is favored by Raghuram et al. who argued that the bivalent NHERF PDZ1–2 domain tandem cross-links carboxyl termini of CFTR subunits in a preexisting dimeric (or multimeric) CFTR channel complex (6). Oligomeric state of CFTR channel is controversial. Biochemical experiments suggested that CFTR channel is a monomer (13), but more recent freeze-fracture electron microscopyl (14) and functional (15) data are more consistent with the dimeric CFTR channel. Raghuram et al. explained their results by assuming that the dimeric CFTR channel has low open probability (Fig. 1B Left). By linking carboxyl termini of CFTR monomers within a dimer NHERF PDZ1–2 tandem construct causes additional elevation in open channel probability (Fig. 1B Right). The strongest argument in support of this model comes from the observation of the NHERF PDZ1–2 tandem construct effect on CFTR activity in patches containing just one channel (6).
Wang et al. (5) are in favor of the third model (Fig. 1C). They argue that the dimeric state of the CFTR channel is transient and present in equilibrium with the monomeric form, which may account for conflicting data regarding CFTR channel stochiometry as discussed above. The data by Wang et al. can then be explained assuming that both monomeric and dimeric CFTR channels are functional, but the open probability of a dimeric channel is higher. In this case a bivalent PDZ domain construct will increase an open probability of a CFTR channel by promoting its dimerization (Fig. 1C). More information regarding the oligomeric state of the CFTR channel should help to discriminate between the models depicted on Fig. 1 B and C.
Can the message from the papers by Raghuram et al. (6) and Wang et al. (5) be applicable to channels other than CFTR? On some level, the conclusion that PDZ domains play not only a structural but also a regulatory role is analogous to the discovery of a modulatory effect of synaptic t-SNARE syntaxin on neuronal Ca2+ channels. The interactions of syntaxin and Ca2+ channels was also initially assumed to play a purely structural role, important for positioning of synaptic vesicles close to the mouth of the channel. However, after syntaxin modulation of Ca2+ channel gating was demonstrated (16, 17), the ability of syntaxin to modulate CFTR (18) and EnaC (19) also has been shown, leading to a conclusion that syntaxin may play the general role of ion channel modulator in a variety of contexts. From the example of syntaxin we can expect that the role of PDZ domains in modulation of ion channels will not be limited to CFTR, and more examples of ion channel modulation by multivalent scaffold proteins are likely to be uncovered.
As a first step to this unchartered territory, we listed some examples of the ion channels that bind to multi-PDZ domain proteins (Table 1). With the exception of CFTR, the stochiometry of most known ion channels is fixed, and the mechanism that involves the shift in equilibrium between monomeric and oligomeric states of the channel (Fig. 1C) will not be appropriate for these channels. However, the general mechanisms depicted in Fig. 1 A and B can potentially work for any multi-PDZ domain-associated ion channel. If direct modulation of these channels by multivalent PDZ domain protein is demonstrated, it will establish a generality of phenomenon discovered by Raghuram et al. and Wang et al. in their studies of CFTR.
Table 1.
Ion channels associated with multi-PDZ domain proteins
| Ion channel | Scaffold protein (number of PDZ domains) | Reference |
|---|---|---|
| NMDA receptors | PSD-95 (3) | (20) |
| SAP-102 (3) | (21) | |
| AMPA receptors | GRIP1 (7) | (22) |
| PICK-1 (1*) | (23) | |
| SAP97 (3) | (24) | |
| Shaker K+ channels | PSD-95 (3) | (25) |
| Dlg-1 (3) | (26) | |
| TRP | INAD (5) | (27) |
| Ca2+ channels | Mint-1 (2) | (28) |
| CFTR | CAP70 (4) | (5) |
| NHERF (2) | (6) | |
| Kir4.1 K+ channels | CIPP1 (4) | (29) |
| ENaC | PICK1 (1*) | † |
NMDA, N-methyl-d-aspartate; AMPA, α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid. TRP, transient receptor potential.
PICK1 protein forms oligomers via PICK1–PICK1 interactions (23).
Hruska-Hageman, A. M., Price, M. P., Johnson, W. A. & Welsh, M. J. (2000) J. Neurosci. 26, 1906 (abst.).
Acknowledgments
We thank Drs. Shmuel Muallem and Phil Thomas for helpful discussions and Phyllis Foley for help with preparation of the manuscript. Work in our laboratory is supported by the Robert A. Welch Foundation and National Institutes of Health Grants R01 NS39552 and NS38082.
Footnotes
See companion article on page 1300.
References
- 1.Kornau H C, Seeburg P H, Kennedy M B. Curr Opin Neurobiol. 1997;7:368–373. doi: 10.1016/s0959-4388(97)80064-5. [DOI] [PubMed] [Google Scholar]
- 2.Ponting C P, Phillips C, Davies K E, Blake D J. BioEssays. 1997;19:469–479. doi: 10.1002/bies.950190606. [DOI] [PubMed] [Google Scholar]
- 3.Fanning A S, Anderson J M. Curr Biol. 1996;6:1385–1388. doi: 10.1016/s0960-9822(96)00737-3. [DOI] [PubMed] [Google Scholar]
- 4.Sheng M, Wyszynski M. BioEssays. 1997;19:847–853. doi: 10.1002/bies.950191004. [DOI] [PubMed] [Google Scholar]
- 5.Wang S, Yue H, Derin R B, Guggino W B, Li M. Cell. 2000;103:169–179. doi: 10.1016/s0092-8674(00)00096-9. [DOI] [PubMed] [Google Scholar]
- 6.Raghuram V, Mak D D, Foskett J K. Proc Natl Acad Sci USA. 2001;98:1300–1305. doi: 10.1073/pnas.031538898. . (First Published January 23, 2001; 10.1073/pnas.031538898) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hall R A, Ostedgaard L S, Premont R T, Blitzer J T, Rahman N, Welsh M J, Lefkowitz R J. Proc Natl Acad Sci USA. 1998;95:8496–8501. doi: 10.1073/pnas.95.15.8496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Short D B, Trotter K W, Reczek D, Kreda S M, Bretscher A, Boucher R C, Stutts M J, Milgram S L. J Biol Chem. 1998;273:19797–19801. doi: 10.1074/jbc.273.31.19797. [DOI] [PubMed] [Google Scholar]
- 9.Wang S, Raab R W, Schatz P J, Guggino W B, Li M. FEBS Lett. 1998;427:103–108. doi: 10.1016/s0014-5793(98)00402-5. [DOI] [PubMed] [Google Scholar]
- 10.Moyer B D, Denton J, Karlson K H, Reynolds D, Wang S, Mickle J E, Milewski M, Cutting G R, Guggino W B, Li M, Stanton B A. J Clin Invest. 1999;104:1353–1361. doi: 10.1172/JCI7453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moyer B D, Duhaime M, Shaw C, Denton J, Reynolds D, Karlson K H, Pfeiffer J, Wang S, Mickle J E, Milewski M, et al. J Biol Chem. 2000;275:27069–27074. doi: 10.1074/jbc.M004951200. [DOI] [PubMed] [Google Scholar]
- 12.Ammala C, Moorhouse A, Gribble F, Ashfield R, Proks P, Smith P A, Sakura H, Coles B, Ashcroft S J, Ashcroft F M. Nature (London) 1996;379:545–548. doi: 10.1038/379545a0. [DOI] [PubMed] [Google Scholar]
- 13.Marshall J, Fang S, Ostedgaard L S, O'Riordan C R, Ferrara D, Amara J F, Hoppe H T, Scheule R K, Welsh M J, Smith A E, et al. J Biol Chem. 1994;269:2987–2995. [PubMed] [Google Scholar]
- 14.Eskandari S, Wright E M, Kreman M, Starace D M, Zampighi G A. Proc Natl Acad Sci USA. 1998;95:11235–11240. doi: 10.1073/pnas.95.19.11235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zerhusen B, Zhao J, Xie J, Davis P B, Ma J. J Biol Chem. 1999;274:7627–7630. doi: 10.1074/jbc.274.12.7627. [DOI] [PubMed] [Google Scholar]
- 16.Bezprozvanny I, Scheller R H, Tsien R W. Nature (London) 1995;378:623–626. doi: 10.1038/378623a0. [DOI] [PubMed] [Google Scholar]
- 17.Wiser O, Bennett M K, Atlas D. EMBO J. 1996;15:4100–4110. [PMC free article] [PubMed] [Google Scholar]
- 18.Naren A P, Nelson D J, Xie W, Jovov B, Pevsner J, Bennett M K, Benos D J, Quick M W, Kirk K L. Nature (London) 1997;390:302–305. doi: 10.1038/36882. [DOI] [PubMed] [Google Scholar]
- 19.Qi J, Peters K W, Liu C, Wang J M, Edinger R S, Johnson J P, Watkins S C, Frizzell R A. J Biol Chem. 1999;274:30345–30348. doi: 10.1074/jbc.274.43.30345. [DOI] [PubMed] [Google Scholar]
- 20.Kornau H C, Schenker L T, Kennedy M B, Seeburg P H. Science. 1995;269:1737–1740. doi: 10.1126/science.7569905. [DOI] [PubMed] [Google Scholar]
- 21.Muller B M, Kistner U, Kindler S, Chung W J, Kuhlendahl S, Fenster S D, Lau L F, Veh R W, Huganir R L, Gundelfinger E D, Garner C C. Neuron. 1996;17:255–265. doi: 10.1016/s0896-6273(00)80157-9. [DOI] [PubMed] [Google Scholar]
- 22.Dong H, O'Brien R J, Fung E T, Lanahan A A, Worley P F, Huganir R L. Nature (London) 1997;386:279–284. doi: 10.1038/386279a0. [DOI] [PubMed] [Google Scholar]
- 23.Xia J, Zhang X, Staudinger J, Huganir R L. Neuron. 1999;22:179–187. doi: 10.1016/s0896-6273(00)80689-3. [DOI] [PubMed] [Google Scholar]
- 24.Leonard A S, Davare M A, Horne M C, Garner C C, Hell J W. J Biol Chem. 1998;273:19518–19524. doi: 10.1074/jbc.273.31.19518. [DOI] [PubMed] [Google Scholar]
- 25.Kim E, Niethammer M, Rothschild A, Jan Y N, Sheng M. Nature (London) 1995;378:85–88. doi: 10.1038/378085a0. [DOI] [PubMed] [Google Scholar]
- 26.Zito K, Fetter R D, Goodman C S, Isacoff E Y. Neuron. 1997;19:1007–1016. doi: 10.1016/s0896-6273(00)80393-1. [DOI] [PubMed] [Google Scholar]
- 27.Shieh B H, Zhu M Y. Neuron. 1996;16:991–998. doi: 10.1016/s0896-6273(00)80122-1. [DOI] [PubMed] [Google Scholar]
- 28.Maximov A, Sudhof T C, Bezprozvanny I. J Biol Chem. 1999;274:24453–24456. doi: 10.1074/jbc.274.35.24453. [DOI] [PubMed] [Google Scholar]
- 29.Kurschner C, Mermelstein P G, Holden W T, Surmeier D J. Mol Cell Neurosci. 1998;11:161–172. doi: 10.1006/mcne.1998.0679. [DOI] [PubMed] [Google Scholar]