Abstract
CYP2C9 is involved in metabolism of nearly 25% of clinically used drugs. Coding region polymorphisms CYP2C9*2 and *3 contribute to interperson variability in drug dosage and clinical outcomes, whereas the role of a regulatory polymorphism remains uncertain. Measuring allelic RNA expression in 87 human liver samples, combined with genotyping, sequencing, and reporter gene assays, we identified a promoter variable number tandem repeat polymorphism (pVNTR) that fully accounted for allelic CYP2C9 mRNA expression differences. Present in three different variant forms [short (pVNTR-S), medium (pVNTR-M), and long (pVNTR-L)], only the pVNTR-S allele reduced the CYP2C9 mRNA level compared with the pVNTR-M (reference) allele. pVNTR-S is in linkage disequilibrium with *3, with linkage disequilibrium r2 of 0.53 to 0.75 in different populations. In patients who were taking a maintenance dose of warfarin, the mean warfarin dose was associated with the copies of pVNTR-S (p = 0.0001). However, in multivariate regression models that included the CYP2C9*3, pVNTR-S was no longer a significant predictor of the warfarin dose (p = 0.60). These results indicate that although pVNTR-S reduced CYP2C9 mRNA expression, the in vivo effects of pVNTR-S on warfarin metabolism cannot be separated from the effects of *3. Therefore, it is not necessary to consider pVNTR-S as an additional biomarker for warfarin dosing. Larger clinical studies are needed to define whether the pVNTR-S has a minimal effect in vivo, or whether the effect attributed to *3 is really a combination of effects on expression by the pVNTR-S along with effects on catalytic activity from the nonsynonymous *3 variant.
Introduction
CYP2C9 metabolizes nearly 25% of clinically used drugs. Genetic variability in CYP2C9 can exert robust effects on treatment outcomes with drugs displaying a narrow therapeutic index, including the commonly prescribed anticonvulsant phenytoin, anticoagulant warfarin, antidiabetic tolbutamide and glipizide, antihypertensive losartan, and antidepressant fluoxetine and the nonsteroidal anti-inflammatory drugs ibuprofen, diclofenac, and celecoxib (Klose et al., 1998; Miners and Birkett, 1998; Davies et al., 2000). The human gene encoding the CYP2C9 protein was mapped to chromosome 10q24.2 and spans over 55 kilobases (kb). Coding region polymorphisms in CYP2C9 have been studied extensively, with more than 30 alleles identified (http://www.cypalleles.ki.se). The two clinically most important alleles, CYP2C9*2 and CYP2C9*3, occur at a minor allele frequency of ∼10 and 6%, respectively, in white populations but are less abundant or undetectable in individuals of African and Asian descent (Lee et al., 2002). In contrast, other potentially important variants, e.g., CYP2C9*8, *9, and *11, occur in the African population with frequencies of 3.6, 13, and 5.6%, respectively, whereas these alleles, in turn, are low or undetectable in other racial groups (Allabi et al., 2004; Blaisdell et al., 2004). Convincing evidence suggests that CYP2C9*2 and CYP2C9*3 alleles convey reduced enzyme activity and have been associated with drug dosage requirements and treatment outcomes (Aithal et al., 1999; Higashi et al., 2002; Lee et al., 2002). Consequently, CYP2C9 variants are listed as a candidate biomarker test in the U.S. Food and Drug Administration, Table of Pharmacogenomics Biomarkers in Drug Labels for celecoxib and warfarin.
Genetic variability in CYP2C9 is not fully accounted for by the known coding region polymorphisms (Shintani et al., 2001; Takahashi et al., 2004). The clearance of warfarin varies 12-fold (Scordo et al., 2002), and the level of CYP2C9 protein expression varies 6-fold in human liver microsomes in homozygous CYP2C9*1/*1 carriers (Yasar et al., 2001). Moreover, warfarin metabolism varied among individuals carrying different promoter haplotypes that do not contain *2 and *3 (Veenstra et al., 2005). For example, haplotype *1D carriers required a significantly lower warfarin dose than reference *1/*1 allele carriers in a small subject group (Veenstra et al., 2005), suggesting that regulatory polymorphisms exist in CYP2C9. To take full advantage of CYP2C9 as a genetic biomarker for drug therapy, it is important to consider the full complement of relevant polymorphisms.
Studies on regulatory polymorphisms in the CYP2C9 promoter region affecting transcription (Shintani et al., 2001; Takahashi et al., 2004; Kramer et al., 2008) have yielded inconsistent results (King et al., 2004; Veenstra et al., 2005). In particular, reporter gene assays showed promoter SNP −4302C>T and haplotypes H3A and H3B (or pattern 6, containing −981G>A, −1537C>T, −1885C>G, and −1911T>A) reduced constitutive promoter activity (Takahashi et al., 2004; Kramer et al., 2008), whereas −2663delTG and/or −3089G>A reduced pregnane X receptor (PXR) or phenytoin-mediated induction of CYP2C9 promoter activity (Kramer et al., 2008; Chaudhry et al., 2010). However, it is unclear whether these polymorphisms or haplotypes affect CYP2C9 mRNA expression in human livers, because conclusions derived from reporter gene assays in transfected cells are not always consistent with in vivo gene expression and regulation. The purpose of this study was to determine the presence of any regulatory CYP2C9 polymorphisms that would change the constitutive mRNA expression in human livers. Because the total mRNA level is strongly influenced by trans-acting factors confounding the effect of cis-acting polymorphisms, we measured allelic mRNA expression, i.e., the relative amount of mRNA derived from each of the two alleles in the same individual. This cancels out trans-acting factors that would have the same effects on both alleles. Allelic RNA ratios significantly deviating from 1 (normalized by measured DNA ratios) demonstrate allelic expression imbalance (AEI), a strong indicator of a cis-acting regulatory polymorphism(s). Then, we used AEI as a phenotypic trait to scan for functional polymorphisms that are responsible for observed AEI, as demonstrated previously (Wang et al., 2005, 2011a). Using this approach, we reevaluated the effects of previously identified polymorphisms on constitutive liver CYP2C9 expression and searched for novel regulatory variants. We identified a promoter variable number tandem repeat polymorphism (pVNTR) located 4 kb upstream of the translation start site to be functional that affects constitutive CYP2C9 mRNA expression in human livers. We then assessed the clinical effect of this pVNTR polymorphism by testing the association of the pVNTR genotype with warfarin dose requirements in patients undergoing warfarin therapy.
Materials and Methods
Tissue and DNA Samples.
One hundred sixty-eight human liver biopsy or autopsy samples were obtained from the Cooperative Human Network Midwestern and Western Division, under the approval of The Ohio State University Institutional Review Board. Because CYP2C9 promoter polymorphisms can affect CYP2C9 inducibility (Kramer et al., 2008; Chaudhry et al., 2010), we excluded livers from individuals with known usage of CYP2C9 inducers (phenytoin, phenobarbital, ethanol, carbamazepine, etc.). Four hundred thirty DNA samples from patients who were taking sulfomethoxazole [(SMX) cohort], collected for another study (Wang et al., 2011b), were also used in this study to determine the distribution of CYP2C9*2, CYP2C9*3, and the newly identified pVNTR-S. The study protocol was approved by The Ohio State University Institutional Review Board.
DNA and RNA Preparation.
Preparation of genomic DNA and RNA was performed as described previously (Pinsonneault et al., 2004). cDNA was prepared using gene-specific primers and oligo(dT) as described previously (Wang et al., 2005, 2011a).
Genotyping.
Twenty-eight SNPs were genotyped in liver genomic DNA using multiplex SNapShot assays or gene-specific real-time PCR. The CYP2C9 pVNTR polymorphism was genotyped using PCR with fluorescently labeled primer, yielding three main amplicons of different lengths: long (pVNTR-L), medium (pVNTR-M reference sequence), and short (pVNTR-S). PCR conditions and sequence of primers are provided in Supplemental Table 1.
Promoter Region Sequencing.
CYP2C9 promoter region [6343 base pairs (bp) upstream of the translation start site, reference sequence NT_030059] was PCR amplified from two samples with allelic RNA ratios deviating from 1, showing significant AEI (L012 and L052), and two samples without AEI (L50 and L71). PCR products were purified and sequenced using primers shown in Supplemental Table 1. To sequence the pVNTR polymorphism region, a fragment of ∼1000 bp surrounding the pVNTR was PCR amplified from 10 samples having different genotypes for pVNTR polymorphisms and cloned into pCR 2.1-TOPO vector, using the TOPO cloning kit (Invitrogen, Carlsbad, CA). Clones were sequenced using PCR primers shown in Supplemental Table 1.
Quantitative Analysis of Allelic Ratios in Genomic DNA and mRNA Using SNapShot.
SNapShot assays were performed as described (Wang et al., 2005). We used three marker SNPs, two in the coding region (rs1057911, A>T and rs9332242, C>G) and one in the intron region (rs2298037, C>T), to detect allelic RNA expression of mRNA or heteronuclear RNA (hnRNA) as described for CYP3A4 (Wang et al., 2011a). All 168 livers were first genotyped for these marker SNPs, and then 87 samples heterozygous for at least one marker SNP were selected for measuring allelic RNA expression. The allelic RNA ratios were normalized to DNA ratios set to 1. Copy number variants (e.g., gene duplication) were not detectable in genomic DNA.
Reporter Gene Constructs.
Luciferase reporter gene constructs containing a 4465-bp promoter region (from 388355 to 392820 in NT_030059) were generated from two PCR-amplified fragments. Overlapping fragments of 1487 bp (from 388,355 to 389,842 in NT_030059, 5P construct) and 3598 bp (from 389,221 to 392,819 in NT_030059, 3P construct) were PCR amplified and cloned into pCR 2.1-TOPO vector, respectively. Both forward primers were tagged with the XhoI site, and reverse primers were tagged with NcoI sites. Then, the 3P construct was cut with KpnI (located at pCR 2.1-TOPO vector) and NdeI (located in an overlapping segment of 3P and 5P) and joined with the 5P construct cut with the same enzymes. The resulting 4465-bp promoter fragment was cut with XhoI and NcoI and cloned into pGL3 vector. Constructs containing different alleles were generated by PCR amplification from different individuals with certain haplotypes and exchanging the fragments representing the 5P or 3P portions. The inserted SNPs/haplotypes were confirmed by DNA sequencing and shown to be free of spurious in vitro mutations.
Reporter Gene Assay.
Reporter gene constructs (1 μg) were transfected into HepG2 cells together with a transcription factor expression cocktail (50 ng of HNF1α + 25 ng of pCDB1 + 50 ng of HNF4α, cotransfected in all experiments) known to be essential for CYP2C9 promoter activity (Kramer et al., 2008). As an internal transfection control, a thymidine kinase promoter-driven Renilla luciferase construct (TK-pRL, 50 ng) was cotransfected. Cells were harvested after 48 h, and luciferase activity was measured with Dual-Glo luciferase assay (Promega, Madison, WI) on a luminescence plate reader (PerkinElmer Life and Analytical Sciences, Waltham, MA). In some experiments, in addition to the transcription factor expression cocktail, we cotransfected 75 ng of other transcription factors [constitutive androstane receptor (CAR), PXR, CCAAT/enhancer binding protein α (CEBPA), or GATA4], or as a control, pcDNA3 empty vector, together with pGL3-fused reporter plasmids to test the effects of transcription factors on promoter activity. To test the effects of inducers, 50 μM phenytoin or 10 μM rifampicin was added to cells 24 h after transfection, and luciferase activity was measured another 24 h after treatment. For each construct, we selected three clones for plasmid DNA preparation. Each experiment was repeated three times, each with triplicates.
Cell Culture and Transfection.
HepG2 cells were cultured in DMEM supplemented with 10% fetal bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin. Cells were plated into 24-well plates 1 day before transfection. Transfection was performed by using LipoJet reagent (SignaGen, Ijamsville, MD) according to the manufacturer's protocol.
Transcription Factor Constructs.
Full-length cDNA of human hepatocyte nuclear factor 1α (HNF1α), HNF4α, CAR splice variant 1, CEBPA, PXR splice variant 1, and GATA4 were PCR amplified from human liver cDNA using primers shown in Supplemental Table 1. PCR products were cloned into pCR 2.1-TOPO vector using the TOPO cloning kit and confirmed by DNA sequencing. Then, the cDNA fragments were subcloned into pCDNA3 vector for transfection. HNF1α cofactor pterin-4-alpha-carbinolamine dehydratase was purchased from Thermo Fisher Scientific (Waltham, MA) and subcloned into pcDNA3 vector.
Clinical Warfarin Dosing Study.
To test the clinical effect of the promoter-variable number tandem repeat (pVNTR) polymorphism, we assessed the association of the pVNTR genotype with the maintenance warfarin dose required in patients undergoing stable warfarin anticoagulation therapy from the University of Florida (n = 348) (Aquilante et al., 2006) and Cairo, Egypt (n = 207) (Shahin et al., 2011). The relevant protocols were approved by the local institutional review boards.
Data Analysis.
Data are expressed as mean ± S.D. Statistical analysis of allelic DNA and RNA ratios was performed using Prism (GraphPad Software Inc., San Diego, CA). Correlations between AEI status (displaying AEI, allelic RNA ratio >1.25 or <0.8, or no AEI, allelic RNA ratio <1.25 and >0.8) and SNP heterozygosity were determined by calculating κ coefficients using SPSS (SPSS Inc., Chicago, IL). A κ coefficient of 1.0 indicates a perfect agreement between heterozygosity and displaying AEI (all heterozygotes for the SNP show AEI and all homozygotes show no AEI), which would indicate that the SNP itself or other polymorphisms are in complete LD with it, is responsible for the AEI. The significance of κ coefficients was calculated using Z-score with a one-sided p value (κ coefficient, >0).
For analysis of the clinical study, the weekly warfarin maintenance dose was transformed using square root transformation to improve model fit. Different pVNTR genotypes (L/L, M/L, M/M, S/L, S/M, S/S) were combined into zero, one, or two copies of the short allele (pVNTR-S). The contribution of pVNTR-S to the model predicting the weekly warfarin dose was assessed using linear regression. The stepwise selection method was used in multiple linear regression to evaluate the contribution of pVNTR-S, CYP2C9*2, and CYP2C9*3, to the warfarin dose requirement, using p < 0.2 and p < 0.05 as criteria for entering and staying in the model. Independent studies in whites (Aquilante et al., 2006), African Americans (Gage et al., 2004), Asians (Zhao et al., 2004), and Egyptians (Shahin et al., 2011) have confirmed that both CYP2C9*2 and *3 are functional variants that influence the warfarin dose required to maintain the therapeutic international normalized ratio. Therefore, we performed combined analysis in all the warfarin patients. This is the approach that the International Warfarin Pharmacogenomics Consortium (Klein et al., 2009) used to create the warfarin dosing algorithm. The linkage disequilibrium between pVNTR-S and CYP2C9*2 and *3 alleles was analyzed in Haploview software (Barrett et al., 2005). All statistical analyses for the clinical association were performed in SAS (version 9.2; SAS Institute, Cary, NC).
Results
Allelic Expression Imbalance of CYP2C9 in Human Livers.
Allelic RNA expression of CYP2C9 was measured in 87 livers heterozygous for at least one of the three SNP markers, including two exonic (SNP26 rs1057911 and SNP28 rs9332242) to measure mature mRNA and one intronic (SNP25 rs2298037) to measure heteronuclear RNA, as reported previously for CYP3A4 (Wang et al., 2011a). Thirteen samples showed allelic RNA ratios significantly different from 1 and were considered displaying AEI, with the major allele consistently associated with higher levels of mRNA than the minor allele [allelic RNA ratio of 1.38 ± 0.12 (mean ± S.D.), ranging from 25 to 60% in different samples]. This result suggests the presence of a cis-acting regulatory polymorphism(s) that affects the mRNA level by influencing either RNA transcription or RNA processing.
Searching for Polymorphisms Responsible for Observed AEI.
We genotyped 28 SNPs in samples with allelic RNA expression data, including promoter SNPs, which have been reported to affect promoter activity, and coding region SNPs (Table 1), which could also affect mRNA stability and change RNA level as reported for multidrug resistance polypeptide 1 (Wang et al., 2005) and mu opioid receptor (Zhang et al., 2005), or modulate early hnRNA processing. Six SNPs were monomorphic in this population, due to low allele frequency in whites (83 of 87 liver samples tested). The remaining 22 SNPs formed four haplotype blocks with high LD between blocks 1, 2, and 4 (Supplemental Fig. 1). Two haplotypes containing completely linked SNPs (LD r2 = 100%) were identified. Haplotype 1 (H1) contains nine SNPs (SNP2, 3, 4, 13, 14, 16, 18, 23, and 26), whereas haplotype 2 (H2) contains three SNPs (SNP7, 9, and 10). H1 and H2 are also in LD with each other, with r2 = 0.79. We then correlated AEI status (displaying AEI or not displaying AEI) with heterozygosity of SNPs/haplotypes by calculating a κ coefficient. SNP7, SNP9, and SNP10 in haplotype 2 yielded the highest K coefficient (k = 0.953; Table 2), with all 12 samples heterozygous for these three SNPs showing AEI (Supplemental Fig. 2). However, one sample with AEI (L114) was homozygous for the major allele, suggesting that SNP7, SNP9, or SNP10 cannot fully account for the AEI and might not be responsible. H1 encompasses a previously identified haplotype (*3A and 3B, with SNP13, 14, 16, and 18) that has been associated with reduced CYP2C9 promoter activity in reporter gene assays (Shintani et al., 2001; Kramer et al., 2008). However, the AEI data again failed to fully agree with heterozygosity of this haplotype (k = 0.915). Specifically, two samples heterozygous for H1 SNPs did not show AEI (L17 and L163; Supplemental Fig. 2), arguing against H1 haplotype or any SNPs in H1 as causative factors. SNP6 previously was shown to reduce promoter activity in a reporter gene assay (Kramer et al., 2008) but did not correlate with AEI status (k = −0.136) (Supplemental Fig. 3A). SNP11 and SNP12 were shown to reduce rifampicin- or phenytoin-induced promoter activity in reporter gene assays (Kramer et al., 2008; Chaudhry et al., 2010) and were associated with the phenytoin maintenance dose (Chaudhry et al., 2010), but they did not associate with AEI status (k coefficient of 0.066 and 0.096, respectively) (Supplemental Fig. 3, B and C), indicating these two SNPs did not affect constitutive CYP2C9 expression in livers, consistent with the previous studies by Kramer et al. (2008) and Chaudhry et al. (2010) that these variants do not affect constitutive CYP2C9 promoter activity but only inducible activity.
TABLE 1.
CYP2C9 polymorphisms genotyped in liver samples
The translation start site A (in ATG) is taken as position 1.
| SNP Number | rs Number | Position | Gene Region | Nucleotide or Amino Acid Changes | Note |
|---|---|---|---|---|---|
| 1 | −8897 | Promoter | T>C | ||
| 2 | −7419 | Promoter | A>G | Link to *3, H1 | |
| 3 | −5813 | Promoter | A>G | Link to *3, H1 | |
| 4 | −5661 | Promoter | C>A | Link to *3, H1 | |
| 5 | rs74150723 | −4877 | Promoter | G>A | |
| 6 | rs12251841 | −4302 | Promoter | C>T | |
| 7 | −3849 | Promoter | G>A | H2 | |
| 8 | rs61886768 | −3597 | Promoter | A>G | |
| 9 | −3579 | Promoter | G>A | H2 | |
| 10 | rs61886769 | −3360 | Promoter | T>C | H2 |
| 11 | rs12782374 | −3089 | Promoter | G>A | |
| 12 | rs71486745 | −2663 | Promoter | del GT | |
| 13 | rs9332092 | −1911 | Promoter | T>C | Link to *3, H1 |
| 14 | rs9332093 | −1885 | Promoter | C>G | Link to *3, H1 |
| 15 | rs9332096 | −1565 | Promoter | C>T | Monomorphic |
| 16 | rs61604699 | −1537 | Promoter | G>A | Link to *3, H1 |
| 17 | rs4918758 | −1188 | Promoter | T>C | |
| 18 | rs9332098 | −981 | Promoter | G>A | Link to *3, H1 |
| 19 | rs1799853 | cDNA 430 | Exon 3 | C>T, R144C, *2 | |
| 20 | rs7900194 | cDNA 449 | Exon 3 | G>A, R150H, *8 | Monomorphic |
| 21 | rs9332131 | cDNA 818 | Exon 5 | del A, frame shift | Monomorphic |
| 22 | rs28371685 | cDNA1003 | Exon 7 | C>T, R335W, *11 | Monomorphic |
| 23 | rs1057910 | cDNA1075 | Exon 7 | A>C, I359L, *3 | H1 |
| 24 | rs28371686 | cDNA1080 | Exon 7 | C>G, D360E, *5 | Monomorphic |
| 25 | rs2298037 | Intron 8 | C>T | Marker SNP | |
| 26 | rs1057911 | cDNA1425 | Exon 9 | A>T, G457G | Marker SNP, link to *3, H1 |
| 27 | rs9332239 | cDNA1465 | Exon 9 | C>T, P489S, *12 | Monomorphic |
| 28 | rs9332242 | cDNA1581 | Exon 9 | C>G, 3′UTR | Marker SNP |
UTR, untranslated region.
TABLE 2.
Correlation between SNP heterozygosity and AEI positive status (allelic RNA ratio, ≥1.25)
A K coefficient of 1 indicates a perfect agreement.
| SNP Number | K Coefficient | P Value | Haplotype |
|---|---|---|---|
| SNP1 | 0.869 | <0.0001 | |
| SNP2, SNP3, SNP4, SNP13, SNP14, SNP16, SNP18, SNP23, SNP26 | 0.915 | <0.0001 | H1 |
| SNP5 | 0.876 | <0.0001 | |
| SNP6 | −0.136 | 0.9719 | |
| SNP7, SNP9, SNP10 | 0.953 | <0.0001 | H2 |
| SNP8 | −0.291 | 0.999 | |
| SNP11 | 0.066 | 0.549 | |
| SNP12 | 0.096 | 0.366 | |
| SNP17 | −0.067 | 0.972 | |
| SNP19 | −0.296 | 0.999 | |
| SNP25 | −0.270 | 0.998 | |
| SNP28 | −0.314 | 0.999 | |
| pVNTR-S | 1 | <0.0001 |
To search for the causative polymorphism, we sequenced >6 kb of the promoter region in two samples with AEI and two samples without AEI. Besides known SNPs genotyped in this study and identified by Kramer et al. (2008), a region between nucleotide positions 388,753 to 388,890 in reference sequence NT0030059.13 (−3979 bp upstream of the translation start site) was found to contain a pVNTR. To measure the length and distribution of pVNTR in liver samples, we PCR amplified genomic DNA using a fluorescently labeled primer (Supplemental Table 1) followed by analysis on an ABI 3730 sequencer. This yielded a pattern of three fragment types varying in length: a short allele ranging from 417 to 438 bp in length, a medium allele with 446 to 488 bp, and a long allele with 512 to 522 bp. Of 159 liver DNA samples tested, 62% of samples were homozygous for pVNTR-M, and 23% were heterozygous for pVNTR-M/pVNTR-L. Therefore, pVNTR-M was considered as a reference allele.
Linkage disequilibrium analysis showed pVNTR-S is in high LD with H1 and H2, with LD r2 of 0.87 and 0.93, respectively. All 13 samples displaying AEI were heterozygous for pVNTR-S, whereas all samples lacking AEI either did not carry the pVNTR-S or were homozygous for pVNTR-S (Supplemental Fig. 2). K coefficient analysis showed a complete agreement between heterozygosity of the pVNTR-S and displaying AEI (k = 1; Table 2). This result strongly suggests that pVNTR-S is the causative polymorphism and responsible for allelic CYP2C9 mRNA expression imbalance. In contrast, pVNTR-M and pVNTR-L alleles showed a similar RNA level and did not display AEI (Supplemental Fig. 3D).
Structure of the pVNTR in CYP2C9.
After PCR amplification of the pVNTR region from 10 individuals and TOPO cloning the PCR products into pCR 2.1-TOPO vector, 16 clones representing variable lengths of pVNTR-S, pVNTR-M, and pVNTR-L alleles were sequenced. As shown in Fig. 1, pVNTR-S, pVNTR-M, and pVNTR-L alleles not only have different lengths but have distinct motif patterns [nTGnTAnTG(or CA)nTA(+/−CG)]. pVNTR-L contains four motif copies, pVNTR-M has two copies (the first and third motifs are deleted), and pVNTR-S contains only the most 3′ motif common to all three alleles (Fig. 1). All four motifs have variable lengths in different individuals because of variable numbers of dinucleotide repeats (TG 1–4 and TA 5–9, CG being always 1). Moreover, there is a variable number of poly A and TA stretch preceding this repeat region (ranging from 11 to 31 As and from 9 to 13 TAs in the clones sequenced). Because of the variable length of the preceding poly A and TA stretch and variable lengths of the motifs, all three alleles show variable lengths in different individuals, with pVNTR-L ranging from 168 to 178 bp, pVNTR-M ranging from 107 to 138 bp, and pVNTR-S ranging from 79 to 90 bp.
Fig. 1.
Representative DNA sequence (A) and the motif structure (B) of the long, medium, and short alleles of pVNTR in CYP2C9.
Promoter Activity of pVNTR Polymorphism with Reporter Gene Assays.
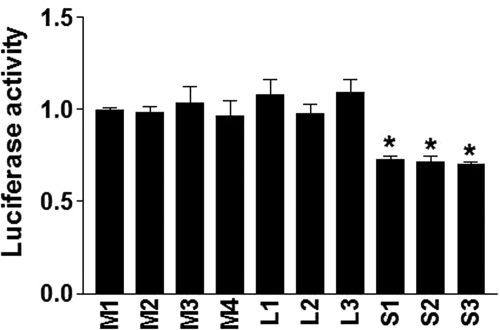
Because pVNTR-S allele completely accounts for the observed AEI results (reducing allelic mRNA levels 38% compared with the pVNTR-M or pVNTR-L alleles), we tested all three pVNTR alleles on CYP2C9 promoter activity using reporter gene assays. A CYP2C9 fragment of 4465 bp upstream of the translation start site was PCR amplified and cloned into pGL3 reporter gene vector upstream of fly luciferase cDNA. To test for interactions between the pVNTR-S and previously reported SNPs or haplotypes, different reporter gene plasmids were constructed containing pVNTR-S, pVNTR-M, or pVNTR-L alleles in combination with other SNPs or haplotypes as they occur in the 4465-bp region of the test subjects (Table 3). As shown in Fig. 2, in the presence of a transcription factor expression cocktail (HNF1α 50 ng + pCDB1 25 ng + HNF4α 50 ng), reporter plasmids containing pVNTR-S in any combination with other SNPs or haplotypes all displayed lower basal promoter activity compared with the reference construct (M1), whereas there were no differences between pVNTR-M and pVNTR-L alleles in any combination with other SNPs or haplotypes.
TABLE 3.
Reporter gene constructs
| Construct | Nucleotide Changes |
|---|---|
| M1 | pVNTR-M, reference sequence |
| M2 | pVNTR-M, −1911T>C, −1885C>G, −1537C>T, −1188T>C, −981G>A |
| M3 | pVNTR-M, −3089G>A, −2663delTG |
| M4 | pVNTR-M, −4302C>T |
| L1 | pVNTR-L |
| L2 | pVNTR-L, −1911T>C, −1885C>G, −1537C>T, −1188T>C, −981G>A |
| L3 | pVNTR-L, −3089G>A, −2663delTG |
| S1 | pVNTR-S |
| S2 | pVNTR-S, −1911T>C, −1885C>G, −1537C>T, −1188T>C, −981G>A |
| S3 | pVNTR-S, −3089G>A, −2663delTG |
Fig. 2.
Effects of pVNTR-S, pVNTR-M, and pVNTR-L alleles on constitutive CYP2C9 promoter activity. HepG2 cells were transfected with reporter gene constructs containing pVNTR-M, pVNTR-M, or pVNTR-S in combination with other SNPs/haplotypes in naturally occurring combinations, as shown in Table 3. Luciferase activities were measured 48 h after transfection. Data are mean ± S.D., n = 3. Compared with reference construct M1; *, P < 0.05 (analysis of variance with Dunnett's post hoc test).
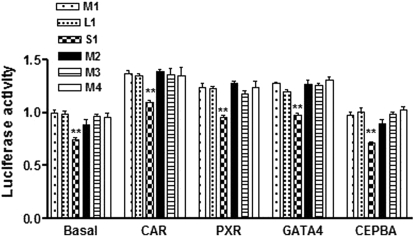
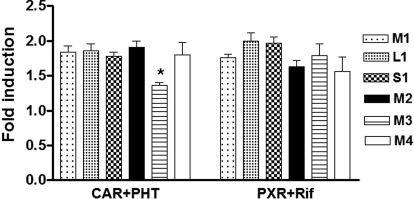
Testing the effect of enzyme induction, in addition to the transcription factor expression cocktail, cotransfection of CAR, PXR, and GATA4, but not CEPBA, increased reporter gene activity across all constructs, consistent with a previous study by Chen et al. (2005) with HNF4α, CAR, and PXR. However, only the pVNTR-S allele maintained lower promoter activity, compared with the pVNTR-M allele (Fig. 3), to the same extent as observed with basal promoter activity (in the presence of the transcription factor expression cocktail). This result indicates that the reduced promoter activity of pVNTR-S is not caused by the lack of these transcription factors known to regulate CYP2C expression in the cell transfection system. We next tested whether the pVNTR-S allele affects promoter activity after drug induction. CAR- or PXR-cotransfected cells were treated with phenytoin or rifampicin for 24 h before luciferase activity was measured. Phenytoin and rifampicin generally increased promoter activity 1.7- to 2-fold over basal activity, but the induction ratio did not differ between pVNTR-S and pVNTR-M alleles after treatment with either phenytoin or rifampicin (Fig. 4). In contrast, the reporter plasmid containing minor alleles of SNP11 and SNP 12 displayed lower induction after CAR/phenytoin but not PXR/rifampicin treatment (Fig. 4).
Fig. 3.
Cotransfection of transcription factors did not alter pVNTR-S allele effects on CYP2C9 promoter activity. HepG2 cells were cotransfected with reporter gene constructs containing the pVNTR-M, pVNTR-L, or pVNTR-S allele, in combination with other SNPs/haplotypes as shown in Table 3, together with different transcription factors (CAR, PXR, GATA4, or CEPBA). Luciferase activities were measured 48 h after transcription. Data are mean ± S.D., n = 3. Compared with reference construct M1; **, P < 0.01 (analysis of variance with Dunnett's post hoc test).
Fig. 4.
Effects of CAR-phenytoin- or PXR-rifampicin-mediated induction on pVNTR-S modulation of CYP2C9 promoter activity. HepG2 cells were cotransfected with reporter gene constructs containing the pVNTR-M, pVNTR-L, or pVNTR-S allele, in the presence or absence of other SNPs/haplotypes, together with transcription factors CAR or PXR. Cells were then treated with either 50 μM phenytoin (PHT) or 10 μM rifampicin (Rif), respectively, at 24 h after transfection, and luciferase activities were measured another 24 h after treatment. Data are mean ± S.D., n = 3. Compared with reference construct M1, *, P < 0.05 (analysis of variance with Dunnett's post hoc test).
Allele Frequency of pVNTR-S.
Because pVNTR-S is highly linked to H1, which contains the known loss-of-function allele *3, we tested the allele frequency of pVNTR-S allele, *3 and *2, in DNA samples from a total of 804 whites (146 from the liver cohort, 334 from the SMX cohort, 324 from the University of Florida warfarin study), 120 African Americans (96 from the SMX cohort and 24 from the University of Florida warfarin study), and 207 Egyptians (Egyptian warfarin study). Of 72 pVNTR-S carriers, 59 subjects were similarly heterozygous or homozygous for *3, indicating that most of the pVNTR-S carriers also carry *3. However, the pVNTR-S is not always present concordantly with *3, and in African Americans, pVNTR-S (5.1%) occurs at a slightly higher allele frequency than *3 (2.5%) (Table 4). Two homozygous pVNTR-S alleles and nine heterozygous carriers did not carry *3, whereas 11 pVNTR-S allele homozygotes were heterozygous for *3 and six *3 heterozygotes and two *3 homozygotes did not carry the pVNTR-S allele, indicating incomplete concordance between *3 and pVNTR-S. The LD (r2) between *3 and pVNTR-S is 0.75 for whites, 0.53 for African Americans, and 0.59 for Egyptians. In contrast, there is no LD between *2 and pVNTR-S in all cohorts (LD r2 = 0). Because *3 reduces CYP2C9 enzyme activity and pVNTR-S reduces mRNA expression, it is possible that pVNTR-S may contribute to the in vivo effects attributed to *3 when they coexist.
TABLE 4.
Minor allele frequency of CYP2C9 *3, *2 and promoter pVNTR alleles
The LD r2 refers to the pVNTR-S and *3, determined in different cohorts. The LD r2 between *2 and pVNTR-S is zero in all cohorts. The SMX cohort was used in another study in which patients were taking SMX as described under Materials and Methods.
| Individuals | Minor Allele Frequency |
Number | LD r2 | ||||
|---|---|---|---|---|---|---|---|
| *3 | *2 | pVNTR-S | pVNTR-M | pVNTR-L | |||
| Caucasian | |||||||
| Liver cohort | 0.048 | 0.154 | 0.062 | 0.778 | 0.160 | 146 | 0.76 |
| SMX cohort | 0.052 | 0.145 | 0.054 | 0.799 | 0.147 | 334 | 0.69 |
| Warfarin cohort | 0.048 | 0.120 | 0.062 | 0.777 | 0.161 | 324 | 0.79 |
| Combined | 0.050 | 0.137 | 0.058 | 0.789 | 0.152 | 804 | 0.75 |
| Black | |||||||
| SMX cohort | 0.031 | 0.036 | 0.052 | 0.784 | 0.164 | 96 | 0.58 |
| Warfarin cohort | 0 | 0.042 | 0.045 | 0.954 | 0 | 24 | N.A. |
| Combined | 0.025 | 0.038 | 0.051 | 0.883 | 0.065 | 120 | 0.53 |
| Egyptian cohort | 0.092 | 0.112 | 0.115 | 0.780 | 0.100 | 207 | 0.59 |
N.A., not applicable.
Clinical Significance of pVNTR-S.
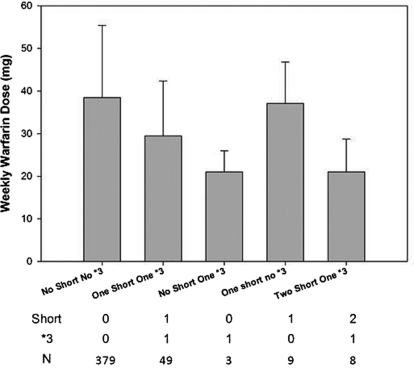
The clinical significance of the pVNTR-S was tested by association of pVNTR-S with the weekly warfarin maintenance dose in patients undergoing stable warfarin anticoagulation therapy (Aquilante et al., 2006; Shahin et al., 2011). The pVNTR genotype was determined in 453 patients. The mean weekly warfarin dose was 38.2 ± 16.9, 30.7 ± 12.7, and 21.0 ± 7.7 mg/week for zero, one, and two copies of pVNTR-S, respectively (p = 0.0001). Univariate linear regression showed that the pVNTR-S was a significant predictor for the warfarin dose (p = 3.1 × 10−6, r2 = 0.047), as was the CYP2C9*3 allele (p = 5.7 × 10−7, r2 = 0.054) and the CYP2C9*2 allele (p = 0.0027, r2 = 0.0199). However, in multivariable regression models that included the CYP2C9*3 allele, pVNTR-S was no longer a significant predictor of the warfarin dose (p = 0.60). When the genotypes of CYP2C9*3 and pVNTR-S were considered together, the effect of pVNTR-S alone was not significant in the absence of CYP2C9*3 (Fig. 5). On the other hand, the CYP2C9*2 allele did not affect the association of the warfarin dose with pVNTR-S. When all three variants were evaluated together in the multiple regression model, the only significant predictors were CYP2C9*3, which explained 5.4% of the variance, and CYP2C9*2, which explained 1.3% of variance, in the warfarin dose required, whereas pVNTR-S was not an independent predictor.
Fig. 5.
Weekly warfarin dose (milligrams per week) for each combination of pVNTR-S and CYP2C9*3 allele.
Discussion
In this study, we have identified and characterized a regulatory promoter enhancer pVNTR, located nearly 4 kb upstream of the translation start site, that fully accounts for differences in the allelic CYP2C9 mRNA constitutive expression in human livers. This pVNTR consists of tandem-repeat motifs grouped into three alleles according to length and structure: short (pVNTR-S), medium (pVNTR-M), and long (pVNTR-L) alleles. pVNTR-S is associated with 25 to 60% (allelic RNA ratio of 1.38 ± 0.12, mean ± S.D.) reduced CYP2C9 mRNA expression, compared with pVNTR-M or pVNTR-L. However, because pVNTR-S is in high LD with the known loss-of-function allele CYP2C9*3, the in vivo effects of pVNTR-S on warfarin metabolism cannot be fully assessed in this study.
Several CYP2C9 promoter SNPs/haplotypes were identified previously (Shintani et al., 2001; Takahashi et al., 2004; Kramer et al., 2008), but none of them fully accounts for the observed allelic RNA expression in this study. This finding argues against these variants directly modulating constitutive CYP2C9 mRNA expression in human livers. In support of the allelic RNA expression results, reporter gene assays confirmed that only the pVNTR-S allele significantly reduces basal promoter activity, consistent with an ∼38% allelic expression imbalance observed in human livers. Sequencing of 6 kb upstream CYP2C9 promoter region in two samples showing AEI and two samples lacking AEI did not reveal any other polymorphisms that associate with AEI. Because the pVNTR-S allele is in high LD with the promoter haplotype containing 981G>A, −1537C>T, −1885C>G, and −1911T>A (previously identified as *3A/*3B), it is possible that previously used reporter gene constructs inadvertently contained the pVNTR-S allele (Kramer et al., 2008). To determine whether the pVNTR-S allele interacts with other SNPs/haplotypes, we tested the promoter activity of pVNTR-S in the context of other naturally occurring SNPs/haplotypes. The pVNTR-S allele consistently reduced promoter activity regardless of the presence or absence of other SNPs, indicating that the pVNTR-S allele is active independently. Overall, the pVNTR-S allele seems to be the only functional polymorphism present within the 6 kb upstream promoter region that reduces constitutive CYP2C9 mRNA expression, at least in a white population, because a majority of our liver samples were from whites. Because of ethnic differences in the frequency of CYP2C9 variants and haplotypes, we cannot rule out the possibility that other regulatory polymorphisms exist in Asian and African populations, whereas none was detected in whites (Lee et al., 2010; Perera et al., 2011).
The molecular mechanisms underlying pVNTR-S regulation of CYP2C9 mRNA expression remains to be resolved. Promoter variable tandem repeat polymorphisms can regulate gene expression via different ways: 1) altering transcription factors or other protein binding sites and inhibiting or promoting gene expression; and 2) changing DNA structure, such as varying the spacing between functional motif or altering the structure and melting properties of DNA in their proximity (Lesch et al., 1994; Contente et al., 2002). VNTRs have been shown frequently to affect gene expression (Albanese et al., 2001; Borrmann et al., 2003; Amador et al., 2004; Donninger et al., 2004; Okada et al., 2006) and have been associated with disease risk (Heshmati et al., 2009; Zarif Yeganeh et al., 2010). Using Mfold calculations, we found that the pVNTR-S allele has a secondary structure of reduced complexity than pVNTR-M and pVNTR-L alleles, indicating that pVNTR-S may alter the DNA structure necessary for transcription factors or other regulatory protein binding. Alternatively, use of the Matinspector program (Genomatix software suite) to search for transcription factor binding sites revealed that the pVNTR-M and pVNTR-L alleles contain a putative hypoxia-inducible factor binding site (tatatataCGTGtgtg) located at the junction of the second and third motifs, which is deleted in the pVNTR-S allele. This requires further investigation.
Because of the high LD between *3 and pVNTR-S, the independent in vivo effect of pVNTR-S may be limited and cannot be fully evaluated in this study, at least in the ethnic groups studied. Because the occurrence of pVNTR-S and *3 is not completely concordant, pVNTR-S may reduce in vivo drug metabolism independent of *3, especially in populations where *3 and pVNTR-S are in relatively low LD (African Americans). Detecting an independent effect of pVNTR-S from *3 will require testing in a larger cohort. In 2005, Veenstra et al. (2005) reported that promoter haplotype CYP2C9 *1D was significantly associated with a decreased warfarin dose requirement, time to stable dosing, and time to bleeding event. Comparing the SNPs in *1D with the pVNTR-S allele haplotype lacking the*3 allele, both contain the minor allele of SNP rs61886769 (SNP10), which is a part of H2 we have determined here to be in high LD with pVNTR-S. Therefore, *1D may have affected the warfarin dosage requirements by inclusion of pVNTR-S. Moreover, CYP2C9*3 reduces enzyme activity in a substrate-specific fashion, with over 10-fold differences in either Km or Vmax for different substrates, tested in a CYP2C9 yeast expression system (Takanashi et al., 2000), suggesting variable effects of *3 with different substrates. Any independent effect of pVNTR-S would become more apparent with substrates that have less reduction of intrinsic clearance caused by coding SNP *3.
In summary, the pVNTR identified here regulates CYP2C9 mRNA expression. Because the reduction in mRNA level is moderate and pVNTR-S is in high LD with the loss-of-function CYP2C9*3 allele, the independent in vivo effect of pVNTR-S on warfarin metabolism seems to be limited. Thus, pVNTR-S is not considered as an additional biomarker for warfarin dosing. The independent in vivo effects of pVNTR-S on metabolism of other substrates requires further investigation. We also confirm the previously proposed reduction of CYP2C9 inducibility caused by the minor alleles of SNP11 and SNP 12 (Kramer et al., 2008; Chaudhry et al., 2010). Together, this study provides a comprehensive analysis of regulatory variants in the CYP2C9 promoter region that can serve as a guide for assessing regulatory genetic factors in CYP2C9 with potential impact on drug therapy.
Supplementary Material
Acknowledgments
We acknowledge Dr. Wolfgang Sadee for critical reading, editing, and comments.
This work was supported by the National Institutes of Health National Institute of Allergy and Infectious Diseases [Grant R21-AI074399] (to D.W.); and the National Institutes of Health National Institute of General Medical Sciences [Grants U01-GM092655, U01-GM074492] (to D.W. and J.A.J., respectively).
Article, publication date, and citation information can be found at http://dmd.aspetjournals.org.
 The online version of this article (available at http://dmd.aspetjournals.org) contains supplemental material.
The online version of this article (available at http://dmd.aspetjournals.org) contains supplemental material.
- kb
- kilobase
- pVNTR
- promoter variable number tandem repeat
- pVNTR-S
- short pVNTR
- pVNTR-M
- medium pVNTR
- pVNTR-L
- long pVNTR
- AEI
- allelic expression imbalance
- LD
- linkage disequilibrium
- H1
- haplotype 1
- H2
- haplotype 2
- HNF1α
- human hepatocyte nuclear factor 1α
- HNF4α
- human hepatocyte nuclear factor 4α
- CAR
- constitutive androstane receptor
- CEBPA
- CCAAT/enhancer binding protein α
- PXR
- pregnane X receptor
- SNP
- single-nucleotide polymorphism
- PCR
- polymerase chain reaction
- SMX
- sulfomethoxazole
- hnRNA
- heteronuclear RNA
- bp
- base pair
- GATA4
- GATA binding protein 4.
Authorship Contributions
Participated in research design: Wang, Shahin, Khalifa, and Johnson.
Conducted experiments: Sun, Gawronski, and Langaee.
Performed data analysis: Wang, Gong, and Gawronski.
Wrote or contributed to the writing of the manuscript: Wang, Gong, and Johnson.
References
- Aithal GP, Day CP, Kesteven PJ, Daly AK. (1999) Association of polymorphisms in the cytochrome P450 CYP2C9 with warfarin dose requirement and risk of bleeding complications. Lancet 353:717–719 [DOI] [PubMed] [Google Scholar]
- Albanèse V, Biguet NF, Kiefer H, Bayard E, Mallet J, Meloni R. (2001) Quantitative effects on gene silencing by allelic variation at a tetranucleotide microsatellite. Hum Mol Genet 10:1785–1792 [DOI] [PubMed] [Google Scholar]
- Allabi AC, Gala JL, Horsmans Y, Babaoglu MO, Bozkurt A, Heusterspreute M, Yasar U. (2004) Functional impact of CYP2C95, CYP2C96, CYP2C98, and CYP2C911 in vivo among black Africans. Clin Pharmacol Ther 76:113–118 [DOI] [PubMed] [Google Scholar]
- Amador ML, Oppenheimer D, Perea S, Maitra A, Cusatis G, Iacobuzio-Donahue C, Baker SD, Ashfaq R, Takimoto C, et al. (2004) An epidermal growth factor receptor intron 1 polymorphism mediates response to epidermal growth factor receptor inhibitors. Cancer Res 64:9139–9143 [DOI] [PubMed] [Google Scholar]
- Aquilante CL, Langaee TY, Lopez LM, Yarandi HN, Tromberg JS, Mohuczy D, Gaston KL, Waddell CD, Chirico MJ, Johnson JA. (2006) Influence of coagulation factor, vitamin K epoxide reductase complex subunit 1, and cytochrome P450 2C9 gene polymorphisms on warfarin dose requirements. Clin Pharmacol Ther 79:291–302 [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21:263–265 [DOI] [PubMed] [Google Scholar]
- Blaisdell J, Jorge-Nebert LF, Coulter S, Ferguson SS, Lee SJ, Chanas B, Xi T, Mohrenweiser H, Ghanayem B, Goldstein JA. (2004) Discovery of new potentially defective alleles of human CYP2C9. Pharmacogenetics 14:527–537 [DOI] [PubMed] [Google Scholar]
- Borrmann L, Seebeck B, Rogalla P, Bullerdiek J. (2003) Human HMGA2 promoter is coregulated by a polymorphic dinucleotide (TC)-repeat. Oncogene 22:756–760 [DOI] [PubMed] [Google Scholar]
- Chaudhry AS, Urban TJ, Lamba JK, Birnbaum AK, Remmel RP, Subramanian M, Strom S, You JH, Kasperaviciute D, Catarino CB, et al. (2010) CYP2C9*1B promoter polymorphisms, in linkage with CYP2C19*2, affect phenytoin autoinduction of clearance and maintenance dose. J Pharmacol Exp Ther 332:599–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Kissling G, Negishi M, Goldstein JA. (2005) The nuclear receptors constitutive androstane receptor and pregnane X receptor cross-talk with hepatic nuclear factor 4alpha to synergistically activate the human CYP2C9 promoter. J Pharmacol Exp Ther 314:1125–1133 [DOI] [PubMed] [Google Scholar]
- Contente A, Dittmer A, Koch MC, Roth J, Dobbelstein M. (2002) A polymorphic microsatellite that mediates induction of PIG3 by p53. Nat Genet 30:315–320 [DOI] [PubMed] [Google Scholar]
- Davies NM, McLachlan AJ, Day RO, Williams KM. (2000) Clinical pharmacokinetics and pharmacodynamics of celecoxib: a selective cyclo-oxygenase-2 inhibitor. Clin Pharmacokinet 38:225–242 [DOI] [PubMed] [Google Scholar]
- Donninger H, Cashmore TJ, Scriba T, Petersen DC, Janse van Rensburg E, Hayes VM. (2004) Functional analysis of novel SLC11A1 (NRAMP1) promoter variants in susceptibility to HIV-1. J Med Genet 41:e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gage BF, Eby C, Milligan PE, Banet GA, Duncan JR, McLeod HL. (2004) Use of pharmacogenetics and clinical factors to predict the maintenance dose of warfarin. Thromb Haemost 91:87–94 [DOI] [PubMed] [Google Scholar]
- Heshmati Y, Mirabzadeh A, Feizzade G, Gilanipour M, Etminan MR, Khoram Khorshid HR, Kamali K, Fakhri M, Moghimi N, Najmabadi H, et al. (2009). A novel polymorphic purine complex at the 1.5 kb upstream region of the human caveolin-1 gene and risk of Alzheimer's disease; extra-short alleles and accumulated allele homozygosity. Am J Med Genet B Neuropsychiatr Genet 150B:248–253 [DOI] [PubMed] [Google Scholar]
- Higashi MK, Veenstra DL, Kondo LM, Wittkowsky AK, Srinouanprachanh SL, Farin FM, Rettie AE. (2002) Association between CYP2C9 genetic variants and anticoagulation-related outcomes during warfarin therapy. JAMA 287:1690–1698 [DOI] [PubMed] [Google Scholar]
- King BP, Khan TI, Aithal GP, Kamali F, Daly AK. (2004) Upstream and coding region CYP2C9 polymorphisms: correlation with warfarin dose and metabolism. Pharmacogenetics 14:813–822 [DOI] [PubMed] [Google Scholar]
- International Warfarin Pharmacogenetics Consortium, Klein TE, Altman RB, Eriksson N, Gage BF, Kimmel SE, Lee MT, Limdi NA, Page D, Roden DM, et al. (2009) Estimation of the warfarin dose with clinical and pharmacogenetic data. N Engl J Med 360:753–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose TS, Ibeanu GC, Ghanayem BI, Pedersen LG, Li L, Hall SD, Goldstein JA. (1998) Identification of residues 286 and 289 as critical for conferring substrate specificity of human CYP2C9 for diclofenac and ibuprofen. Arch Biochem Biophys 357:240–248 [DOI] [PubMed] [Google Scholar]
- Kramer MA, Rettie AE, Rieder MJ, Cabacungan ET, Hines RN. (2008) Novel CYP2C9 promoter variants and assessment of their impact on gene expression. Mol Pharmacol 73:1751–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CR, Goldstein JA, Pieper JA. (2002) Cytochrome P450 2C9 polymorphisms: a comprehensive review of the in-vitro and human data. Pharmacogenetics 12:251–263 [DOI] [PubMed] [Google Scholar]
- Lee SJ, Jang YJ, Cha EY, Kim HS, Lee SS, Shin JG. (2010) A haplotype of CYP2C9 associated with warfarin sensitivity in mechanical heart valve replacement patients. Br J Clin Pharmacol 70:213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesch KP, Balling U, Gross J, Strauss K, Wolozin BL, Murphy DL, Riederer P. (1994) Organization of the human serotonin transporter gene. J Neural Transm Gen Sect 95:157–162 [DOI] [PubMed] [Google Scholar]
- Miners JO, Birkett DJ. (1998) Cytochrome P4502C9: an enzyme of major importance in human drug metabolism. Br J Clin Pharmacol 45:525–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada T, Hashimoto R, Numakawa T, Iijima Y, Kosuga A, Tatsumi M, Kamijima K, Kato T, Kunugi H. (2006) A complex polymorphic region in the brain-derived neurotrophic factor (BDNF) gene confers susceptibility to bipolar disorder and affects transcriptional activity. Mol Psychiatry 11:695–703 [DOI] [PubMed] [Google Scholar]
- Perera MA, Gamazon E, Cavallari LH, Patel SR, Poindexter S, Kittles RA, Nicolae D, Cox NJ. (2011) The missing association: sequencing-based discovery of novel SNPs in VKORC1 and CYP2C9 that affect warfarin dose in African Americans. Clin Pharmacol Ther 89:408–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinsonneault J, Nielsen CU, Sadée W. (2004) Genetic variants of the human H+/dipeptide transporter PEPT2: analysis of haplotype functions. J Pharmacol Exp Ther 311:1088–1096 [DOI] [PubMed] [Google Scholar]
- Scordo MG, Pengo V, Spina E, Dahl ML, Gusella M, Padrini R. (2002) Influence of CYP2C9 and CYP2C19 genetic polymorphisms on warfarin maintenance dose and metabolic clearance. Clin Pharmacol Ther 72:702–710 [DOI] [PubMed] [Google Scholar]
- Shahin MH, Khalifa SI, Gong Y, Hammad LN, Sallam MT, El Shafey M, Ali SS, Mohamed ME, Langaee T, Johnson JA. (2011) Genetic and nongenetic factors associated with warfarin dose requirements in Egyptian patients. Pharmacogenet Genomics 21:130–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani M, Ieiri I, Inoue K, Mamiya K, Ninomiya H, Tashiro N, Higuchi S, Otsubo K. (2001) Genetic polymorphisms and functional characterization of the 5′-flanking region of the human CYP2C9 gene: in vitro and in vivo studies. Clin Pharmacol Ther 70:175–182 [DOI] [PubMed] [Google Scholar]
- Takahashi H, Ieiri I, Wilkinson GR, Mayo G, Kashima T, Kimura S, Otsubo K, Echizen H. (2004) 5′-Flanking region polymorphisms of CYP2C9 and their relationship to S-warfarin metabolism in white and Japanese patients. Blood 103:3055–3057 [DOI] [PubMed] [Google Scholar]
- Takanashi K, Tainaka H, Kobayashi K, Yasumori T, Hosakawa M, Chiba K. (2000) CYP2C9 Ile359 and Leu359 variants: enzyme kinetic study with seven substrates. Pharmacogenetics 10:95–104 [DOI] [PubMed] [Google Scholar]
- Veenstra DL, Blough DK, Higashi MK, Farin FM, Srinouanprachan S, Rieder MJ, Rettie AE. (2005) CYP2C9 haplotype structure in European American warfarin patients and association with clinical outcomes. Clin Pharmacol Ther 77:353–364 [DOI] [PubMed] [Google Scholar]
- Wang D, Johnson AD, Papp AC, Kroetz DL, Sadée W. (2005) Multidrug resistance polypeptide 1 (MDR1, ABCB1) variant 3435C>T affects mRNA stability. Pharmacogenet Genomics 15:693–704 [PubMed] [Google Scholar]
- Wang D, Guo Y, Wrighton SA, Cooke GE, Sadee W. (2011a) Intronic polymorphism in CYP3A4 affects hepatic expression and response to statin drugs. Pharmacogenomics J 11: 274–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Para MF, Koletar SL, Sadee W. (2011b) Human N-acetyltransferase 1 *10 and *11 alleles increase protein expression through distinct mechanisms and associate with sulfamethoxazole-induced hypersensitivity. Pharmacogenet Genomics 21:652–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasar U, Eliasson E, Forslund-Bergengren C, Tybring G, Gadd M, Sjöqvist F, Dahl ML. (2001) The role of CYP2C9 genotype in the metabolism of diclofenac in vivo and in vitro. Eur J Clin Pharmacol 57:729–735 [DOI] [PubMed] [Google Scholar]
- Zarif Yeganeh M, Mirabzadeh A, Khorram Khorshid HR, Kamali K, Heshmati Y, Gozalpour E, Veissy K, Olad Nabi M, Najmabadi H, Ohadi M. (2010) Novel extreme homozygote haplotypes at the human caveolin 1 gene upstream purine complex in sporadic Alzheimer's disease. Am J Med Genet B Neuropsychiatr Genet 153B:347–349 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wang D, Johnson AD, Papp AC, Sadée W. (2005) Allelic expression imbalance of human mu opioid receptor (OPRM1) caused by variant A118G. J Biol Chem 280:32618–32624 [DOI] [PubMed] [Google Scholar]
- Zhao F, Loke C, Rankin SC, Guo JY, Lee HS, Wu TS, Tan T, Liu TC, Lu WL, Lim YT, et al. (2004) Novel CYP2C9 genetic variants in Asian subjects and their influence on maintenance warfarin dose. Clin Pharmacol Ther 76:210–219 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.