Risk assignment in breast cancer patients using the PAM50 Breast Cancer Intrinsic Classifier™ and the Oncotype DX® Recurrence Score in the same population was compared. There is good agreement between the two assays for high and low prognostic risk assignment but PAM50 assigns more patients to the low risk category. About half of the intermediate risk RS group was reclassified as low risk luminal A by PAM50, which suggests a potential complementary use for the assays.
Keywords: Oncotype DX®, PAM50 assay, Gene expression profiles, Breast cancer, Prognosis
Abstract
Purpose.
To compare risk assignment by PAM50 Breast Cancer Intrinsic Classifier™ and Oncotype DX_Recurrence Score (RS) in the same population.
Methods.
RNA was extracted from 151 estrogen receptor (ER)+ stage I–II breast cancers and gene expression profiled using PAM50 “intrinsic” subtyping test.
Results.
One hundred eight cases had complete molecular information; 103 (95%) were classified as luminal A (n = 76) or luminal B (n = 27). Ninety-two percent (n = 98) had a low (n = 59) or intermediate (n = 39) RS. Among luminal A cancers, 70% had low (n = 53) and the remainder (n = 23) had an intermediate RS. Among luminal B cancers, nine were high (33%) and 13 were intermediate (48%) by the RS. Almost all cancers with a high RS were classified as luminal B (90%, n = 9). One high RS cancer was identified as basal-like and had low ER/ESR1 and low human epidermal growth factor receptor 2 (HER2) expression by quantitative polymerase chain reaction in both assays. The majority of low RS cases were luminal A (83%, n = 53). Importantly, half of the intermediate RS cancers were re-categorized as low risk luminal A subtype by PAM50.
Conclusion.
There is good agreement between the two assays for high (i.e., luminal B or RS > 31) and low (i.e., luminal B or RS < 18) prognostic risk assignment but PAM50 assigns more patients to the low risk category. About half of the intermediate RS group was reclassified as luminal A by PAM50.
Introduction
Several multigene prognostic assays have been developed to aid adjuvant treatment selection for patient with early-stage breast cancer [1]. These assays can provide prognostic information that is independent of standard clinical and pathological information, and may be particularly helpful in cases for which measures of clinical risk are equivocal (i.e., small, node-negative, intermediate grade tumors). Practice guidelines issued by the American Society of Clinical Oncology (ASCO), the National Comprehensive Cancer Network, and the St. Gallen Expert Panel each acknowledge that genomic prognostic tests can provide additional prognostic and predictive information beyond anatomic staging, and such information may be considered when adjuvant chemotherapy is recommended for patients with early-stage breast cancer [2–4]. The most widely used prognostic assay in the U.S. is the 21-gene recurrence score (RS) (Oncotype DX®; Genomic Health, Inc., Redwood City, CA). This test is used to risk stratify estrogen receptor estrogen receptor-positive (ER)+ lymph node—negative breast cancer and to identify patients who can avoid adjuvant chemotherapy [5–7].
Several other genomic tests, including the MammaPrint® (Agendia, Inc., Amsterdam, The Netherlands), the Molecular Grade Index (bioTheranostics, Inc., San Diego, CA), and the immunohistochemistry (IHC)-based Mammostrat® (Clarient Inc., Aliso Viejo, CA) assays, have been introduced into the market to address similar prognostic questions [8–16]. The recently developed PAM50 Breast Cancer Intrinsic Classifier™ assay (ARUP Laboratories, Salt Lake City, UT) is a standardized test measuring 50 classifier genes and five control genes to identify the intrinsic subtypes commonly known as luminal A, luminal B, human epidermal growth factor (HER)-2 enriched, and basal-like [17, 18]. Research microarray studies have often considered the normal-like subtype as a tumor subtype, but it has been found that this subtype is often caused by having too much “normal” breast tissue in the cancer specimen [17, 18] and it is thus considered as “insufficient” for clinical purposes. Contaminating normal tissue has been shown to interfere with the subtype call, but in a systematic and predictable fashion [19], whereas a shift in risk score using Oncotype DX® or MammaPrint® can result in an unpredictable change. The normal-like subtype is not reported in the commercial PAM50 assay and we removed it from the main analyses but included it in supplemental online data.
Each of these tests can distinguish better prognosis breast cancers form worse prognosis cases, particularly among ER+ cancers. However, each of the tests relies on a different set of genes, with only a modest overlap between assays, and therefore may assign a different risk category to the same patient. Although several studies examined classification overlap among various genomic prognostic tests using proxy models, such as gene expression array–based unofficial versions of Oncotype DX® and modified versions of published assays [20, 21], no direct comparison of any two commercially available assays in the same patient population has been reported to date. An imminent clinical challenge for practicing physicians is to understand how different assays relate to each other and what to expect when more than one assay is used for the same cancer.
The objective of this study was to compare the PAM50 Breast Cancer Intrinsic Classifier™ and the Oncotype DX®, two conceptually different, commercially available, standardized prognostic risk predictors in the same patient population. Both tests were performed in the reference laboratories where they were developed. Results were generated blindly. We examined how often these two tests assigned discrepant risk to the same individual. Histologic grade and Ki-67 protein expression, two common prognostic markers in breast cancer, were also evaluated along with the molecular signatures.
Methods
Patients
We identified 304 consecutive patients who underwent Oncotype DX® testing at the Nellie B. Connally Breast Center of the University of Texas, MD Anderson Cancer Center (MDACC) as part of routine care (n = 254) or in the context of the Trial Assigning IndividuaLized Options for Treatment (Rx) (n = 55) until November 20, 2008. Demographic and clinicopathological data were extracted from a prospectively maintained clinical database (database search, May 2011). We recently published the characteristics of this patient cohort [22]. Archived, formalin-fixed, paraffin embedded (FFPE) tissue blocks were available for 151 (49%) patients from the institutional tumor bank, whereas the rest of the cases had no stored tissue. All cases were reviewed by a breast pathologist (S.K.) to centrally assess tumor grade (using the Nottingham histological three-tier grading system), tumor size, nodal status, and ER, progesterone receptor (PR), HER-2, and Ki-67 expression. The expression of Ki-67 (antibody MIB1; Dako, Glostrup, Denmark), ER (antibody 6F11; Novacastra/Vector Laboratories, Burlingame, CA), and PR (antibody 1A6; Novacastra/Vector Laboratories, Burlingame, CA) were determined during routine pathologic examination and were reported as percent positive nuclei in the invasive neoplastic compartment of the tissue. HER-2 status was assessed by either IHC (c-erbB/HER-2/neu AB-8 [Clone E2–4001]; Thermo Fisher Scientific, Fremont, CA) or fluorescence in situ hybridization (FISH) (PathVysion HER-2/neu DNA probe kit [LSI HER-2/neu Spectrum Orange/CEP17 spectrum green]; Abbott Molecular, Des Plaines, IL). Sixty nuclei were counted and results were categorized according to ASCO–College of American Pathologists guidelines. HER-2 positivity was defined as either HER-2 gene amplification by FISH or an IHC score of 3+. This biomarker study was approved by the MDACC Institutional Review Board.
Genomic Assays
The Oncotype DX® assay was performed as part of routine care on sections mailed to Genomic Health, Inc. and RS values were retrieved from medical records. Low, intermediate, and high risk categories were defined as RS values <18, 18–31, and >31, respectively. For the PAM50 assay, we extracted RNA from FFPE tissue blocks from four to six tumor-directed cores (1 mm) or 5-μm full-face cuts using the High Pure RNA Paraffin Kit (Roche Diagnostics, Indianapolis, IN). Total RNA (600 ng) was sent to the Department of Pathology, University of Utah Health Sciences Center for PAM50 testing by ARUP Laboratories, Inc. Researchers at the University of Utah were blinded to the RS values and demographic, clinical, and pathological details of the cases and received only RNA samples marked with a unique study identifier. The PAM50 tumor classifications and the individual gene scores that contribute to the PAM50 were returned to MDACC where the comparison with RS values was performed.
Statistical Analysis
For continuous variables, we compared the median among groups using the Wilcoxon rank test. For categorical variables, we used the χ2 test. All significance tests were two tailed. We constructed multivariate linear models to identify components of the PAM50 assay that independently predict the Oncotype DX® RS and the significance of models was assessed using the Wald test, with two-sided p-values <.05 considered significant. Models included IHC expression of Ki-67, mRNA levels of the estrogen receptor 1 gene (ESR1), progesterone receptor gene (PGR), and HER-2 and the PAM50 class as well as the proliferation and luminal scores of the PAM50 assay. The F-test was used to compare individual gene scores alone with subtypes. Receiver operating characteristic (ROC) curves were used to assess the ability of Ki-67 (IHC) to discriminate between RS categories and PAM50 intrinsic subtypes. All calculations and statistical tests were performed using SAS, version 9.2 (SAS Institute Inc., Cary, NC). The median follow-up time, calculated from the time of diagnosis and censored at the last follow-up visit for this patient cohort was short, 18 months; all patients were alive and only three recurrences occurred (one distant metastasis, one local chest wall recurrence, and one contralateral breast cancer).
Results
Samples missing any control genes or more than two classifier genes (n = 19) or classified as normal-like (n = 11) were excluded from further analyses, as per the standard operating procedure for the PAM50. Patients without Ki-67 IHC evaluation (n = 6) were also excluded from subsequent analyses (supplemental online data). The final cohort consisted of 108 patients, the median age was 57 years (25th and 75th percentiles, 50 years and 64 years, respectively), and most patients were postmenopausal (n = 79) (Table 1). All had ER+ HER-2− breast cancer; 96% had lymph node–negative disease (n = 102). The median tumor size was 1.5 cm (25th and 75th percentiles, 1.2 cm and 2.0 cm, respectively). Only 6% (n = 6) of the patients had histologic grade III breast cancer; 55% (n = 59) had grade II and the remaining 40% (n = 43) had grade I breast cancer. These patient characteristics are consistent with an intermediate clinical risk population for which Oncotype DX® is commonly used. Most patients (71%) did not receive chemotherapy, but almost all (94%) received adjuvant endocrine therapy.
Table 1.
Characteristics of the final cohort (n = 108)
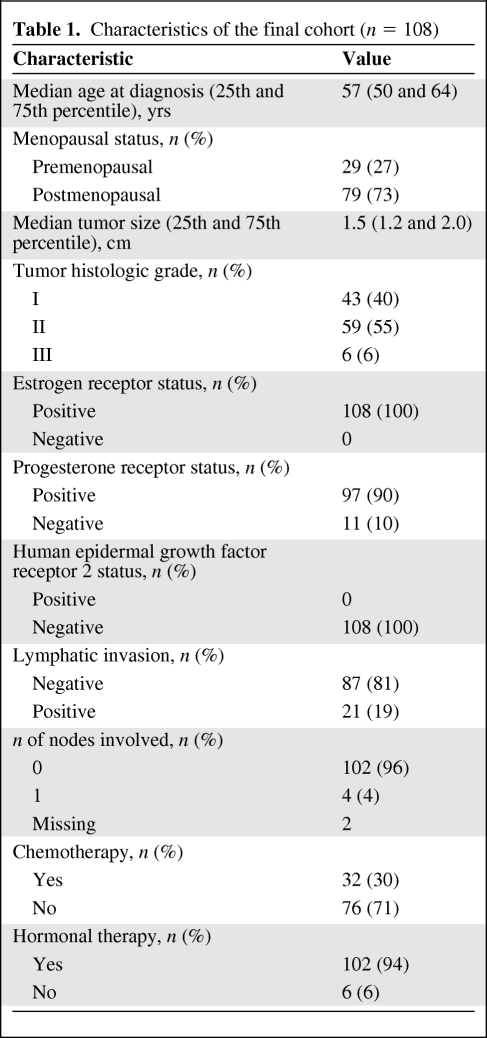
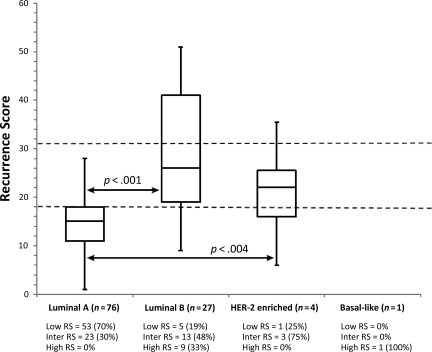
First, we assessed the distribution of RS values and risk groups across the intrinsic subtypes (Fig. 1). Luminal A cancers had a significantly lower median RS value (RS, 15; 25th and 75th percentiles, 11 and 18, respectively) than luminal B cancers (RS, 25; 25th and 75th percentiles, 19 and 40, respectively) (p < .0001) or any other subtype. The highest RS value was observed in the single basal-like cancer (RS, 57). We also examined the distribution of histologic tumor grade across RS and PAM50 risk groups. Luminal A cancers and the low and intermediate risk RS groups included approximately equal numbers of grade I and grade II cancers. We also looked specifically at how the 61 grade II tumors were classified by both tests and found that both Oncotype DX® and the PAM50 approximately split this group in half when classified as low risk RS (56%) and luminal A (63%), respectively. These results indicate that histologic grade is a suboptimal surrogate for genomic risk class as determined by either of these two assays.
Figure 1.
Distribution of recurrence score over PAM50 intrinsic subtypes. Dashed lines mark recurrence score cut points (<18 and >31). Abbreviations: HER-2, human epidermal growth factor receptor 2; RS, recurrence score.
Next, we examined the distribution of PAM50 intrinsic subtypes across the three RS groups (Fig. 2). Of 10 patients (8%) classified as high risk by Oncotype DX® (RS > 31), nine were assigned to the luminal B and one was assigned to the basal-like subtype. Among the 39 (36%) intermediate risk patients (RS, 18–31), 23 (59%) were classified as luminal A, 13 (33%) were classified as luminal B, and three (8%) were classified as HER-2 enriched. There were 59 (54%) patients who had a low risk RS (<18), and PAM50 classified 53 (89%) of those as luminal A, five (8%) as luminal B, and one (2%) as HER-2 enriched.
Figure 2.
Classification of each tumor by RS and PAM50 and distribution by histologic grade. (A): Classification of 108 individual breast cancer cases by RS and PAM50 intrinsic classifier. (B): RS. (C): PAM50 intrinsic classifier.
Abbreviations: HER-2, human epidermal growth factor receptor 2; RS, recurrence score.
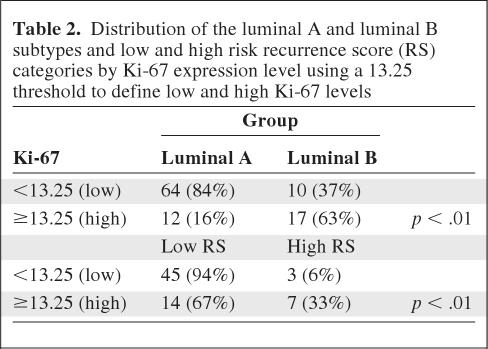
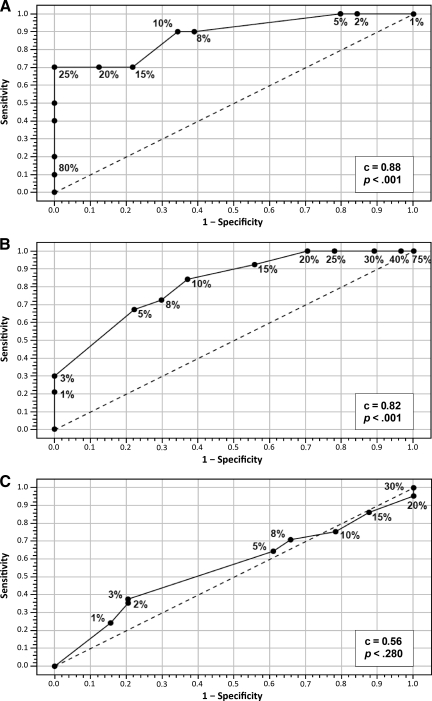
Ki-67 expression as measured by IHC was significantly lower in luminal A cancers (median Ki-67, 5%; 25th and 75th percentiles, 2% and 10%, respectively) than in luminal B cancers (median, 15%; 25th and 75th percentiles, 8% and 25%, respectively) (p < .01) and in low risk RS cancers (median, 5%; 25th and 75th percentiles, 5% and 10%, respectively) and intermediate risk RS cancers (median, 5%; 25th and 75th percentiles, 2% and 15%, respectively) than in high risk RS cancers (median, 27.5%; 25th and 75th percentiles, 10% and 40%, respectively) (p < .01). Table 2 shows Ki-67 classified as low or high risk using the 13.25 cutoff as found by Cheang et al. [23] to be optimal for discriminating between luminal A and luminal B cancers. Almost 80% of low risk RS and luminal A cancers had a low Ki-67 expression level, and 70% and 63% of luminal B and high risk RS cancers had a high Ki-67 expression level, respectively. We constructed ROC curves for Ki-67 values as defined by IHC (Fig. 3). Ki-67 performed well, with an area under the curve of 0.88 for the luminal A and luminal B subtypes (p < .0001) and 0.82 for low and high risk RS values (p = .0001). However, the ability of Ki-67 to distinguish between low and intermediate risk RS values was poor (c = 0.56; p > .28).
Table 2.
Distribution of the luminal A and luminal B subtypes and low and high risk recurrence score (RS) categories by Ki-67 expression level using a 13.25 threshold to define low and high Ki-67 levels
Figure 3.
Examination of the discriminatory ability of Ki-67 for the luminal A and luminal B subtypes and RS categories. Shown are receiver operating characteristic curves examining the ability of Ki-67 to discriminate between high and low risk RS values (A), the luminal A and luminal B subtypes (B), and low and intermediate risk RS values (C).
In order to further explore the relationship between the RS and individual components of the PAM50 assay, we built a number of multivariate linear regression models using subtype and metagene scores (proliferation and luminal) and individual gene scores (ESR1, PGR, ERBB2) reported by the PAM50 test (http://www.aruplab.com/Lab-Tests/General-Oncology/PAM50/index.jsp). Approximately 66% of the variation in the RS could be explained by components of the PAM50 (adjusted R2, 0.66). In addition to subtype (p = .038), PGR score (p < .001), luminal score (p = .004), and proliferation score (p = 2.50 × 10−5) each remained significant predictors of the RS. Ki-67 expression by IHC lost significance in the presence of the proliferation score (which includes MKI67).
Discussion
Genomic predictors are increasingly being used in the clinic as ancillary tests to assist in the assessment of prognosis and to select the proper adjuvant treatment for patient with early-stage ER+ breast cancer. Already, several multianalyte assays have been incorporated into clinical practice for breast cancer, so it is increasingly important to understand the degree of concordance between these tests and how they perform in relation to more conventional IHC testing.
When discordant risk results are obtained, it is currently unknown which assay will predict outcome more accurately. In ER+ breast cancer, these answers come particularly slow because relapses often do not occur until many years after the initial diagnosis. In our cohort, there were only three recurrences and all were luminal B cancers, including one contralateral breast cancer (RS, 45, high risk), one distant metastasis (RS, 31, intermediate risk), and one local chest wall recurrence (RS, 7, low risk).
The clinical PAM50 test classifies four tumor types: luminal A, luminal B, HER-2 enriched, and basal-like. As previously, discussed we excluded the normal-like tumor subtype from our analyses. In this study, there were 11 samples (13%) classified as normal-like (or insufficient), and these ranged from a low to an intermediate RS. This was an expected result because the luminal A subtype is most susceptible to switching to normal-like with normal tissue contamination and this was the most prevalent tumor subtype in this cohort.
Ninety-four percent of the samples were either luminal A (n = 76) or luminal B (n = 27), which is consistent with samples required to be ER+ for Oncotype DX® testing. Of the nonluminal subtypes, one was diagnosed as basal-like (0.9%) and four were HER-2 enriched (4%). Nielsen et al. [17] previously reported that 0.6% and 8% of clinically ER+ tumors were basal-like or HER-2 enriched, respectively. The identification of these subtypes in clinically ER+ tumors is important because these patients have a poor prognosis when given adjuvant tamoxifen therapy alone [17].
Because two reported routine variables—histologic grade and Ki-67 expression level by IHC—are commonly suggested as surrogates for molecular class or Oncotype DX® risk group [23], we also examined these variables as predictors of genomic risk group. With the caveat that our study population was biased toward an indeterminate clinical risk group for whom the genomic classification would be most beneficial, we observed that the high risk groups by genomics (luminal B and intermediate to high risk RS) often contained grade 2 tumors and not uncommonly contained grade 1 breast cancers. The intermediate risk RS group had an even split of grade I and grade II cancers, but the luminal B subtype included mostly (81%) grade II and grade III tumors. These data show that low grade does not reliably identify all low-risk patients using these genomic assays. Ki-67 IHC evaluation has also been used to better risk stratify ER+ tumors, although it has been shown to be less accurate than quantitative polymerase chain reaction using MKI67 alone or the proliferation score [17]. We found that the Ki-67 IHC count was significantly lower in luminal A and low risk RS cancers than in other groups, and that it had predictive value in distinguishing luminal A from luminal B cancers and low risk RS from high risk RS cancers. However, Ki-67 staining did not prove to be helpful in trying to discriminate between the intermediate and low risk RS categories, suggesting that more quantitative methods and multianalyte tests more accurately assess risk in ER+ tumors [17].
In conclusion, these results indicate that there is reasonably good agreement between the Oncotype DX® and PAM50 assays for high (i.e., luminal B and RS > 31) and low (i.e., luminal A and RS < 18) prognostic risk assignment. Overall, more patients were assigned to the low risk category by the PAM50 assay than by the RS. About half of the intermediate risk RS group was reclassified as the luminal A low risk subtype by the PAM50 assay, suggesting a potential complementary use for the assays. A significantly larger study with long follow-up and homogeneous adjuvant therapy is required to establish the true prognostic, and therefore clinical, value in patients considered to have a clinically indeterminate risk by current histopathological standards.
Supplementary Material
Acknowledgments
Dr. Catherine M. Kelly was funded by a Susan G. Komen Fellowship award. Prof. Lajos Pusztai received funding from the Breast Cancer Research Foundation of New York, NY.
Presented at the IMPAKT Breast Cancer Conference, Brussels, Belgium, May 5–7, 2011.
Presented at the 47th Annual Meeting of the American Society of Clinical Oncology, Chicago, Illinois, June 3–7, 2011.
Footnotes
- (C/A)
- Consulting/advisory relationship
- (RF)
- Research funding
- (E)
- Employment
- (H)
- Honoraria received
- (OI)
- Ownership interests
- (IP)
- Intellectual property rights/inventor/patent holder
- (SAB)
- Scientific advisory board
Author Contributions
Conception/Design: Catherine M. Kelly, Philip S. Bernard, Savitri Krishnamurthy, Lajos Pusztai
Provision of study material or patients: Catherine M. Kelly, Philip S. Bernard, Savitri Krishnamurthy, Lajos Pusztai
Collection and/or assembly of data: Catherine M. Kelly, Philip S. Bernard, Savitri Krishnamurthy, Bailiang Wang, Mark T.W. Ebbert, Roy R.L. Bastien, Elliana Young, Takayuki Iwamoto, Lajos Pusztai
Data analysis and interpretation: Catherine M. Kelly, Philip S. Bernard, Savitri Krishnamurthy, Mark T.W. Ebbert, Roy R.L. Bastien, Kenneth M. Boucher, Elliana Young, Takayuki Iwamoto, Lajos Pusztai
Manuscript writing: Catherine M. Kelly, Philip S. Bernard, Savitri Krishnamurthy, Bailiang Wang, Mark T.W. Ebbert, Roy R.L. Bastien, Kenneth M. Boucher, Elliana Young, Takayuki Iwamoto, Lajos Pusztai
Final approval of manuscript: Catherine M. Kelly, Philip S. Bernard, Savitri Krishnamurthy, Bailiang Wang, Mark T.W. Ebbert, Roy R.L. Bastien, Kenneth M. Boucher, Elliana Young, Takayuki Iwamoto, Lajos Pusztai
References
- 1.Sotiriou C, Pusztai L. Gene-expression signatures in breast cancer. The N Engl J Med. 2009;360:790–800. doi: 10.1056/NEJMra0801289. [DOI] [PubMed] [Google Scholar]
- 2.Gnant M, Harbeck N, Thomssen C. St. Gallen: Summary of the Consensus Discussion. Breast Care (Basel) 2011;6:136–141. doi: 10.1159/000328054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carlson RW, Brown E, Burstein HJ, et al. NCCN Task Force Report: Adjuvant therapy for breast cancer. J Natl Compr Canc Netw. 2006;4(suppl 1):S1–S26. [PubMed] [Google Scholar]
- 4.Khatcheressian JL, Wolff AC, Smith TJ, et al. American Society of Clinical Oncology 2006 update of the breast cancer follow-up and management guidelines in the adjuvant setting. J Clin Oncol. 2006;24:5091–5097. doi: 10.1200/JCO.2006.08.8575. [DOI] [PubMed] [Google Scholar]
- 5.Paik S, Shak S, Tang G, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 6.Paik S, Tang G, Shak S, et al. Gene expression and benefit of chemotherapy in women with node-negative, estrogen receptor-positive breast cancer. J Clin Oncol. 2006;24:3726–3734. doi: 10.1200/JCO.2005.04.7985. [DOI] [PubMed] [Google Scholar]
- 7.Paik S. Development and clinical utility of a 21-gene recurrence score prognostic assay in patients with early breast cancer treated with tamoxifen. The Oncologist. 2007;12:631–635. doi: 10.1634/theoncologist.12-6-631. [DOI] [PubMed] [Google Scholar]
- 8.Mook S, Van't Veer LJ, Rutgers EJ, et al. Individualization of therapy using MammaPrint: From development to the MINDACT trial. Cancer Genomics Proteomics. 2007;4:147–155. [PubMed] [Google Scholar]
- 9.Slodkowska EA, Ross JS. MammaPrint 70-gene signature: Another milestone in personalized medical care for breast cancer patients. Expert Rev Mol Diagn. 2009;9:417–422. doi: 10.1586/erm.09.32. [DOI] [PubMed] [Google Scholar]
- 10.Wittner BS, Sgroi DC, Ryan PD, et al. Analysis of the MammaPrint breast cancer assay in a predominantly postmenopausal cohort. Clin Cancer Res. 2008;14:2988–2993. doi: 10.1158/1078-0432.CCR-07-4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cuadros M, Llanos A. [Validation and clinical application of MammaPrint® in patients with breast cancer] Med Clin (Barc) 2011;136:627–632. doi: 10.1016/j.medcli.2010.02.009. In Spanish. [DOI] [PubMed] [Google Scholar]
- 12.Loi S, Haibe-Kains B, Desmedt C, et al. Definition of clinically distinct molecular subtypes in estrogen receptor-positive breast carcinomas through genomic grade. J Clin Oncol. 2007;25:1239–1246. doi: 10.1200/JCO.2006.07.1522. [DOI] [PubMed] [Google Scholar]
- 13.Ma XJ, Salunga R, Dahiya S, et al. A five-gene molecular grade index and HOXB13:IL17BR are complementary prognostic factors in early stage breast cancer. Clin Cancer Res. 2008;14:2601–2608. doi: 10.1158/1078-0432.CCR-07-5026. [DOI] [PubMed] [Google Scholar]
- 14.Filho OM, Ignatiadis M, Sotiriou C. Genomic Grade Index: An important tool for assessing breast cancer tumor grade and prognosis. Crit Rev Oncol Hematol. 2011;77:20–29. doi: 10.1016/j.critrevonc.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 15.Mathieu MC, KNC, Li H, et al. The role of the genomic breast cancer index in predicting pathologic complete response in breast cancer patients treated with neoadjuvant anthracycline plus taxane. J Clin Oncol. 2011;29(suppl):573. [Google Scholar]
- 16.Bartlett JM, Thomas J, Ross DT, et al. Mammostrat as a tool to stratify breast cancer patients at risk of recurrence during endocrine therapy. Breast Cancer Res. 2010;12:R47. doi: 10.1186/bcr2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nielsen TO, Parker JS, Leung S, et al. A comparison of PAM50 intrinsic subtyping with immunohistochemistry and clinical prognostic factors in tamoxifen-treated estrogen receptor-positive breast cancer. Clin Cancer Res. 2010;16:5222–5232. doi: 10.1158/1078-0432.CCR-10-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parker JS, Mullins M, Cheang MC, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27:1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elloumi F, Hu Z, Li Y, et al. Systematic bias in genomic classification due to contaminating non-neoplastic tissue in breast tumor samples. BMC Med Genomics. 2011;4:54. doi: 10.1186/1755-8794-4-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iwamoto T, Lee JS, Bianchini G, et al. First generation prognostic gene signatures for breast cancer predict both survival and chemotherapy sensitivity and identify overlapping patient populations. Breast Cancer Res Treat. 2011;130:155–164. doi: 10.1007/s10549-011-1706-9. [DOI] [PubMed] [Google Scholar]
- 21.Fan C, Oh DS, Wessels L, et al. Concordance among gene-expression-based predictors for breast cancer. N Engl J Med. 2006;355:560–569. doi: 10.1056/NEJMoa052933. [DOI] [PubMed] [Google Scholar]
- 22.Kelly CM, Krishnamurthy S, Bianchini G, et al. Utility of Oncotype DX risk estimates in clinically intermediate risk hormone receptor-positive, HER2-normal, grade II, lymph node-negative breast cancers. Cancer. 2010;116:5161–5167. doi: 10.1002/cncr.25269. [DOI] [PubMed] [Google Scholar]
- 23.Cheang MC, Chia SK, Voduc D, et al. Ki67 index, HER2 status, and prognosis of patients with luminal B breast cancer. J Natl Cancer Inst. 2009;101:736–750. doi: 10.1093/jnci/djp082. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.