Abstract
Dysregulation of canonical Wnt signaling is thought to play a role in colon carcinogenesis. β-Catenin, a key mediator of the pathway, is stabilized upon Wnt activation and accumulates in the nucleus, where it can interact with the transcription factor T cell factor (TCF) to transactivate gene expression. Normal colonic epithelia express a truncated TCF-1 form, called dnTCF-1, that lacks the critical β-catenin–binding domain and behaves as a transcriptional suppressor. How the cell maintains a balance between the two forms of TCF-1 is unclear. Here, we show that ERM-binding phosphoprotein 50 (EBP50) modulates the interaction between β-catenin and TCF-1. We observed EBP50 localization to the nucleus of human colorectal carcinoma cell lines at low cell culture densities and human primary colorectal tumors that manifested a poor clinical outcome. In contrast, EBP50 was primarily membranous in confluent cell lines. Aberrantly located EBP50 stabilized conventional β-catenin/TCF-1 complexes and connected β-catenin to dnTCF-1 to form a ternary molecular complex that enhanced Wnt/β-catenin signaling events, including the transcription of downstream oncogenes such as c-Myc and cyclin D1. Genome-wide analysis of the EBP50 occupancy pattern revealed consensus binding motifs bearing similarity to Wnt-responsive element. Conventional chromatin immunoprecipitation assays confirmed that EBP50 bound to genomic regions highly enriched with TCF/LEF binding motifs. Knockdown of EBP50 in human colorectal carcinoma cell lines compromised cell cycle progression, anchorage-independent growth, and tumorigenesis in nude mice. We therefore suggest that nuclear EBP50 facilitates colon tumorigenesis by modulating the interaction between β-catenin and TCF-1.
Introduction
The canonical Wnt signaling pathway, also called the β-catenin/TCF pathway, controls various cellular events such as migration, proliferation, and differentiation throughout development. Dysregulation of Wnt signaling is thought to underlie the mechanisms of colon tumorigenesis and other carcinogenesis such as melanoma and hepatocellular carcinoma (HCC) (1–3). β-Catenin is a key mediator of the Wnt pathway and was originally identified as a component of the adherens junctions, where β-catenin links E-cadherin to α-catenin, and thus the actin cytoskeleton (4). In the absence of Wnt, the β-catenin level is regulated by a “destruction complex,” which consists of adenomatous polyposis coli (APC), axin, and glycogen synthase kinase 3β (GSK3β). APC and axin assemble a structural scaffold that allows casein kinase 1 and GSK3β to subsequently phosphorylate β-catenin at the consensus serine and threonine motif in its N-terminal region. Phosphorylated β-catenin is then ubiquitinated and degraded by proteosome (5). Upon Wnt activation, the destruction complex is inhibited to accumulate unphosphorylated stabilized β-catenin in the cytoplasm and nucleus. β-Catenin itself does not bind to DNA. It has to interact with T cell factor (TCF) or lymphoid enhancer factor (LEF) transcription factors to execute its transactivating function in the nucleus (6, 7). Without nuclear β-catenin, TCF/LEF alone acts as a repressor of Wnt target genes by forming a complex with Groucho (8, 9), which can recruit histone deacetylase (HDAC) to condense chromatin. Upon Wnt activation, stabilized β-catenin converts TCF/LEF into a transcriptional activator by directly replacing those repressors (10).
In mammals, the TCF/LEF protein family has four members: TCF-1, TCF-3, TCF-4, and LEF-1 (11–13). The diversity of TCF/LEF protein is created by alternative splicing to generate multiple C-terminal tails and different promoter usage. TCF-1 and LEF-1 loci contain two different promoters for transcription. The first promoter produces mRNA encoding full-length TCF-1/LEF-1, whereas the second promoter produces mRNA encoding truncated proteins without an N-terminal β-catenin–binding domain (14–16). The most abundant TCF-1 isoforms in normal tissue lack a β-catenin–binding domain (14, 17). Those natural dominant negative dnTCF-1s can occupy promoter regions and recruit repressors to negatively regulate Wnt signaling. Full-length TCF1 is a target gene of β-catenin/TCF-4, and Tcf1–/– mice develop adenomas in the intestine and mammary glands, indicating that TCF-1 is a tumor suppressor (18). Full-length LEF-1 is also found to be induced by β-catenin/TCF complex and selectively expressed in colon cancer but not in normal colon tissues (15). The symmetry of dominant negative and full-length TCF-1/LEF-1 conceivably plays an important role in normal differentiation and tumor progression; however, the underlying molecular mechanism adopted by cells to maintain such a delicate balance remains obscure.
ERM-binding phosphoprotein 50 (EBP50), which contains two tandem PDZ domains and a C-terminal ERM–binding (ezrin-radixin-moesin–binding) domain, functions as an adaptor linking membrane protein to the underlying actin. EBP50 cellular targets include ion channels, GPCRs, signaling proteins, and nuclear proteins. The diversity of EBP50-interacting proteins indicates that EBP50 can orchestrate various signaling complexes in different subcellular compartments (19). EBP50 is a β-catenin–associated protein, and overexpressed EBP50 can enhance β-catenin transcriptional activity in cell lines in which β-catenin is already stabilized. Moreover, overexpression and nuclear localization of EBP50 are found in HCC (20), suggesting that nuclear EBP50 can promote cell growth. Controversially, EBP50 has also been proposed to be a tumor suppressor because it is required for β-catenin localization at the cell-cell junction to stabilize E-cadherin–mediated complex (21). Furthermore, EBP50 can interact with the tumor suppressor PTEN and form an EBP50/PTEN/PDGFR ternary complex, which can attenuate PDGF receptor signaling by restricting PI3K activation (22). Nevertheless, like β-catenin, the role of EBP50 in tumor development might be determined by its localization and associated partners (23).
Overexpression of the second PDZ domain of EBP50 can suppress β-catenin transcriptional activity (20), suggesting that EBP50, with two tandem repeat PDZ domains, might facilitate the interaction between β-catenin and other critical factors to transduce Wnt signaling. However, the molecular target of nuclear EBP50 remains unknown.
In this study, we demonstrated what we believe to be a novel function of EBP50: reinforcement of the TCF-1/β-catenin interaction and, potentially, switching of the dominantly negative dnTCF-1 into a tumor facilitator. Notably, the pathophysiological significance was validated by the finding that there was a significant correlation between aberrant nuclear EBP50 expression and poor clinical manifestation in patients with colorectal carcinoma (CRC).
Results
Nuclear EBP50 facilitates β-catenin/TCF–mediated signaling.
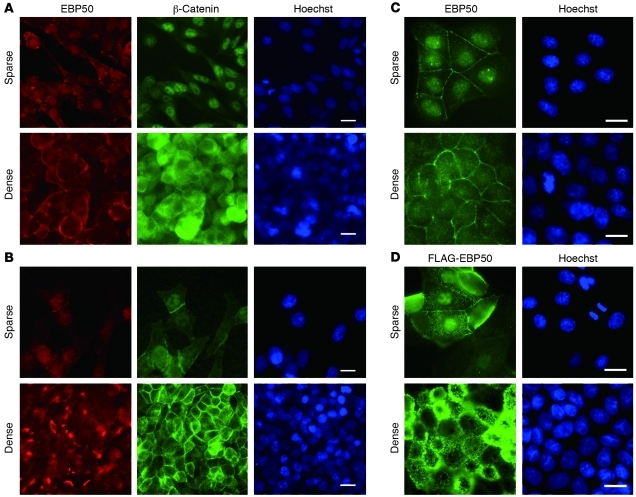
In investigating the subcellular localization of EBP50, we detected both membranous and nuclear distribution in the colon carcinoma cell lines SW480 and HT29. Interestingly, this nuclear localization apparently increased in isolated, unpolarized cells in contrast to the mainly membranous pattern in confluent polarized monolayers (Figure 1, A and B). We observed similar results when we extended our study from colon carcinoma cell lines to non-transformed differentiated MDCK cells (Figure 1C). This phenomenon was observed not only for endogenous EBP50 detected by commercial polyclonal antibody, but also in cells stably expressing a FLAG-tagged EBP50 construct employing an epitope tag–specific antibody (Figure 1D), further confirming the reliability of the staining results shown in Figure 1, A–C. This cell density–dependent EBP50 nuclear location was reminiscent of the β-catenin behavior and Wnt-dependent signaling activity in cells undergoing EMT (24–26) and prompted us to explore the potential role of EBP50 nuclear localization in β-catenin function. To examine whether EBP50 is involved in β-catenin–mediated signaling, we transfected siRNA targeted for EBP50 in SW480 cells, which express a truncated APC mutation and accumulate stabilized β-catenin in the nucleus (27). The reporter assay showed that β-catenin transcriptional activity was significantly inhibited after EBP50 knockdown (Figure 2A). The result was reproduced when another siRNA targeted for an independent EBP50 cDNA region was applied (Figure 2A), further confirming the role of EBP50 in modulating β-catenin transcriptional activity.
Figure 1. EBP50 displays nuclear translocation under low-density culture.
SW480 (A) and HT29 (B) colon cancer and MDCK (C) cell lines were plated in either sparse or dense conditions for 2 days and processed for EBP50 and β-catenin immunofluorescence study as well as Hoechst 33342 nuclear staining. (D) MDCK cells stably expressing FLAG-tagged EBP50 were processed for FLAG and Hoechst 33342 nuclear staining. Scale bars: 10 μm.
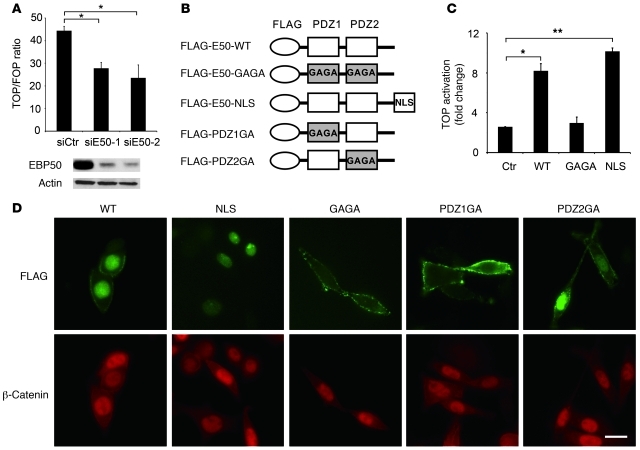
Figure 2. Nuclear EBP50 modulates Wnt/β-catenin signaling.
(A) SW480 cells were independently transfected with 40 μM siE50-1 or siE50-2, two siRNAs targeted at discrete regions of EBP50 cDNA, for 16 hours. Then, cells were trypsinized and replated into 12-well plates for 8 hours before being transfected with Top/Fop-flash reporter plasmids plus 40 μM of each EBP50 siRNA. After an additional 24 hours, cells were processed for luciferase activity assays. Western blotting demonstrated that both siRNAs efficiently suppressed the expression of EBP50. (B) The molecular configuration of the plasmids used in C and D. (C) HEK293 cells were transfected with the indicated plasmids, using empty vector pcDNA3 as a control (Ctr), and 1.5 μg of Top/Fop-flash reporter plasmid. Luciferase activity was assessed 24 hours later in triplicate. The data shown in A and C are mean ± SEM of 3 independent assays, each performed in triplicate. *P < 0.05, **P < 0.001. (D) EBP50-knockdown SW480 cells were transfected with the indicated FLAG-tagged EBP50 plasmids for 6 hours and stained with rabbit anti-FLAG (green) and mouse anti–β-catenin (red) antibodies. Scale bars: 10 μm.
EBP50 has two tandem PDZ domains that mediate the interaction between EBP50 and its binding partners (19). Although wild-type EBP50 localized in both the membrane and nucleus, a GAGA mutant, with alanine substitutions for the critical glycine residues in the binding pocket of the PDZ domains (Figure 2B), lost the nuclear entry potential of wild-type EBP50 (Figure 2D). Given the result of the above loss-of-function experiment, which implicated EBP50 in β-catenin signaling (Figure 2A), we overexpressed the GAGA mutant and an nuclear localization signal (NLS)–tagged EBP50 mutant in HEK293 cells whose β-catenin signaling pathway had been activated by LiCl treatment to clarify whether nuclear localization of EBP50 was required for its function in β-catenin signaling. Although expression of either wild-type EBP50 or the nucleus-exclusive EBP50-NLS mutant could enhance β-catenin transactivating ability, the GAGA mutant could not (Figure 2C). These data imply that the PDZ domains of EBP50 are important for its nuclear localization as well as its function in the β-catenin pathway. To further clarify which PDZ domain is responsible for this characteristic, we generated two mutant constructs, PDZ1GA and PDZ2GA, which have a glycine-to-alanine mutation at the PDZ motif binding pocket of the first and second PDZ domain in EBP50, respectively (Figure 2B). While the PDZ2GA mutant displayed both membranous and nuclear localization similar to that of wild-type EBP50, PDZ1GA mutant was membrane exclusive, implying that the first PDZ domain is responsible for nuclear localization of EBP50 (Figure 2D). The effect of PDZ domain mutations on the subcellular localization of EBP50 was also observed in HEK293 cells (Supplemental Figure 1; supplemental material available online with this article; doi: 10.1172/JCI45661DS1). Intriguingly, the nuclear β-catenin localization was not affected by the subcellular distribution of these EBP50 mutants (Figure 2D). In line with this result, EBP50 knockdown had no apparent influence on the nuclear distribution of β-catenin in SW480 cells (Supplemental Figure 2). Given these results and a previous report that EBP50 displayed marked upregulation upon Wnt protein treatment as well as stabilized β-catenin accumulation (28), we propose that a ligand protein of the first PDZ domain interacts with EBP50 in the nucleus and this interaction modulates Wnt/β-catenin signaling activity.
EBP50 interacts with TCF-1B through its first PDZ domain.
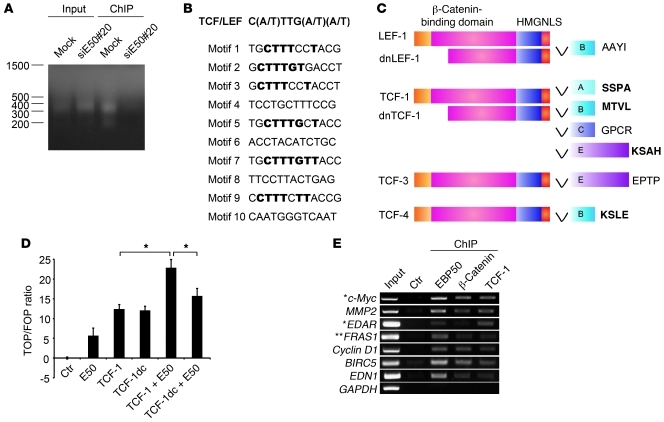
To further investigate whether EBP50 plays a role in gene transactivation in the nucleus, we performed genome-wide localization analysis of EBP50 occupancy in SW480 cells by ChIP-on-chip analysis (Figure 3A). Among the 816 bound events (Supplemental Table 1), we selected the 57 most significant (Supplemental Table 2), defined as having at least three consecutive probes at the same genomic locus, for consensus motif prediction analysis. Interestingly, 6 of the 10 highest-ranking motifs were identical or similar to the TCF/LEF binding motif (Figure 3B).
Figure 3. EBP50 binds to the TCF/LEF consensus motif.
(A) Mock- and an EBP50 siRNA–transfected SW480 clone (siE50#20) were processed for ChIP assay using anti-EBP50 antibody. The ligation-mediated PCR (LM-PCR) products from genomic (input) and anti-EBP50 antibody immunoprecipitated (ChIP) DNA are shown. (B) Ten motifs with the highest scores, compiled from the 57 most significantly EBP50-bound regions, as predicted by a BioProspector Search, are shown. Among them, 6 motifs identical or similar to the TCF/LEF consensus motif are shown in bold. (C) Molecular structure of TCF family proteins. HMG, high-mobility group proteins; NLS, nucleus localization signal. The last 4 amino acids at the C-terminal ends are shown in bold if those sequences match to a potential PDZ-binding motif. (D) SW480 cells were transfected with 2 μg of the indicated plasmids, using empty vector as a control, and 0.4 μg of Top/Fop-flash reporter plasmids. TCF-1dc is a C-terminally deleted TCF-1 construct lacking the EBP50 binding motif. Luciferase activity was assessed and presented as in Figure 2C. *P < 0.05. (E) SW480 cells were processed for ChIP assay using the indicated antibodies or a rabbit anti-podocalyxin antibody as a control. Selective Wnt/β-catenin–regulated gene sequences were amplified by PCR. *Genes reproducibly identified as Wnt/β-catenin targets by two previous systemic ChIP analyses (39, 40) as well as EBP50 binding sites by our ChIP-on-chip assay. **This gene, although not demonstrated to be Wnt/β-catenin regulated with vigorous experimental evidence, was revealed by previous systemic analysis (39, 40) and this study.
One important physiological function of EBP50 is to serve as a scaffold protein to facilitate assembly of multiprotein complexes at the membrane through its interaction with different ligand proteins by its two PDZ domains. Given this scenario and our previous finding that nuclear EBP50 facilitates Wnt/β-catenin signaling, we hypothesized that nuclear EBP50 serves as an adapter to assist the interaction between β-catenin and a transcription factor that is also critical in Wnt signaling. Based on this assumption and identification of the TCF/LEF motif as a strong candidate for the nuclear EBP50 binding site after ChIP-on-chip screening, we focused our attention on the TCF transcription factor family, which plays an essential role in transducing Wnt/β-catenin signaling (16, 29). Through bioinformatics search, we found that the C-terminal sequence (MTVL) of TCF-1B, the most abundant TCF-1 isoform, matched to a consensus PDZ-binding motif (Ser/Thr-Xxx-Val/Leu) for the PDZ domain of EBP50 (Figure 3C).
In a study complementary to our earlier gene knockdown approach (Figure 2A), EBP50 overexpression augmented β-catenin transcriptional activity and EBP50 worked synergistically with TCF-1 to activate the β-catenin–responsive promoter (Figure 3D). In contrast to full-length TCF-1, which worked synergistically with EBP50 to activate the β-catenin–responsive promoter (Figure 3D), TCF-1dc, a mutant lacking the EBP50 binding motif, failed to demonstrate a degree of synergism comparable to that of FLAG-EBP50 in activating β-catenin signaling, although it was still able to turn on the promoter by itself (Figure 3D), implying the importance of the C-terminal PDZ-binding motif of TCF-1 in interacting with EBP50. Furthermore, conventional ChIP analysis confirmed that EBP50 bound to the promoter regions of a battery of genes that are well-characterized Wnt-regulated targets (Figure 3E). These data together imply that EBP50 interacts with the Wnt regulatory elements on genomic DNA through the TCF/LEF family transcription factors and mediates gene expression.
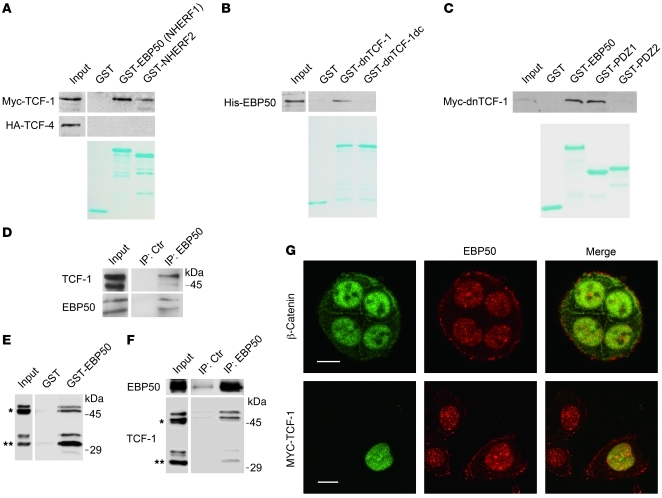
To examine whether EBP50 interacts with TCF-1B, we utilized a purified GST-EBP50 fusion protein to pull down Myc-tagged TCF-1 and HA-tagged TCF-4 from HEK293 lysates (Figure 4A). The results indicated that EBP50 interacted only with TCF-1 but not with its analog, TCF-4. In the meantime, EBP50 (also known as sodium hydrogen exchanger regulatory factor 1 [NHERF1]) showed a greater affinity to TCF-1 than its homolog NHERF2 (Figure 4A). Although GST-dnTCF-1, a recombinant protein of TCF-1 lacking its N-terminal β-catenin–binding domain, could bind purified His-EBP50 recombinant protein in vitro, GST–dnTCF-1dc, a PDZ-binding motif–deleted mutant, could no longer interact with His-EBP50 recombinant protein (Figure 4B). This result indicates that TCF-1 binds to EBP50 through its C-terminal PDZ-binding motif. To identify which PDZ domain of EBP50 is required for the binding, we generated recombinant GST fusion proteins expressing full-length EBP50 and the first and second PDZ domains of EBP50 for pull-down analysis. As shown in Figure 4C, only full-length EBP50 and its first PDZ domain, but not its second PDZ domain, interacted with dnTCF-1.
Figure 4. EBP50 interacts with TCF-1 and β-catenin through its first and second PDZ domains, respectively.
(A) Cell lysates from HEK293 cells overexpressing Myc–TCF-1 or HA–TCF-4 were incubated with glutathione beads loaded with GST, GST-EBP50 (NHERF1), or GST-NHERF2. After washing, bound materials were analyzed by immunoblotting. (B) In vitro GST pull-down assays employing purified His-EBP50 and GST, GST–dnTCF-1, or GST–dnTCF-1dc, a PDZ motif deletion mutant. (C) GST-fused full-length EBP50 (GST-EBP50) and the first (PDZ1) and second PDZ domains (PDZ2) of EBP50 were mixed with equal amounts of lysates from MDCK cells stably expressing Myc–dnTCF-1. After washing, bound materials were examined by immunoblotting. Lower panels of A–C: Coomassie blue–stained gels demonstrated the amount of each purified GST fusion protein used in the assays. (D) NCI-H28 lysates were immunoprecipitated by either control or anti-EBP50 antibody, followed by immunoblotting using TCF-1 or EBP50 antibodies. (E) Cellular lysates from Colo205 cells, which expressed both full-length (*) and N-terminally truncated TCF-1 (**), were incubated with GST or GST-EBP50 beads, and the bound material was analyzed by immunoblotting using anti–TCF-1 antibody. (F) Colo205 cell lysates were incubated with mouse anti-EBP50 or control antibody conjugated on NHS-activated beads and then processed for immunoblotting using rabbit anti-EBP50 and TCF-1 antibodies. (G) Top row: Double immunofluorescence and confocal microscopy revealed the localization of endogenous β-catenin (green) and EBP50 (red) in SW480 cells. Bottom: SW480 cells were transfected with Myc–TCF-1. Anti-Myc staining showed the colocalization of TCF-1 with endogenous EBP50 in the nucleus. Scale bars: 10 μm.
Having demonstrated the interaction between EBP50 and TCF-1 in vitro, we next analyzed the association of EBP50 and TCF-1 in vivo using the NCI-H28 mesothelioma cell line. Co-immunoprecipitation assays confirmed a direct interaction between EBP50 and TCF-1 independent of β-catenin, since NCI-H28 cells lacks endogenous β-catenin due to a genomic deletion in the β-catenin gene (Figure 4D). Using the colon carcinoma cell line Colo205, which expresses both full-length and N-terminally truncated TCF-1, we further demonstrated that EBP50 interacts with both forms of TCF-1 by in vitro pull-down analysis and co-immunoprecipitation study (Figure 4, E and F). As in a previous study showing that nuclear EBP50 is present in HCC tissue specimens and the human liver cancer cell line HepG2 (20), we also discovered that EBP50 was localized in the nucleus of the SW480 colonic cancer cell line as well as β-catenin and TCF-1 (Figure 4G). These findings are consistent with a potential role of EBP50 in promoting tumor growth through its interaction with β-catenin and TCF-1B and motivated us to pursue further evidence to support the presence of a nuclear β-catenin/EBP50/TCF-1 complex.
EBP50 rescues the repression effect of dnTCF-1.
Although TCF-1 is pivotal in enhancing malignant growth (30), a study in knockout mice surprisingly revealed its role as a tumor suppressor (18). In normal tissue, an N-terminally truncated form of TCF-1, dnTCF-1, plays a dominant negative role by constitutively binding to the promoter of Wnt/β-catenin–responsive genes with the silencer protein Groucho and suppressing the activation of downstream genes (9, 10). Nevertheless, the molecular mechanism by which dnTCF-1, lacking the critical β-catenin–binding domain, could revive its interaction with β-catenin remains unknown.
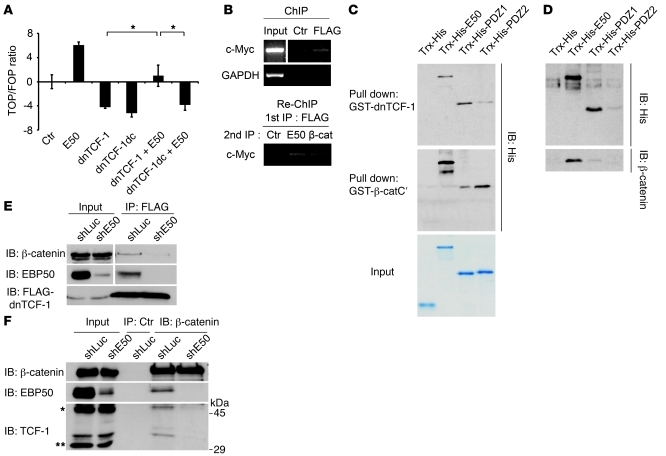
While overexpression of dnTCF-1, a negative regulator of Wnt signaling, indeed suppressed β-catenin activation, as proposed previously (31), cotransfection of EBP50 converted dnTCF-1 from a suppressor to an activator of β-catenin signaling (Figure 5A). By contrast, the PDZ-binding motif lacking mutant dnTCF-1dc failed to restore β-catenin signaling in the presence of EBP50 (Figure 5A), further underscoring the importance of the C-terminal PDZ-binding motif of TCF-1 in binding to EBP50 (Figure 4B). To exclude the possibility that overexpressed EBP50 drove away dnTCF-1, allowing endogenous TCF-1 to take over the promoter, which effected Wnt promoter transactivation, we constructed a SW480 clone stably expressing FLAG-tagged dnTCF-1. ChIP analysis showed that EBP50 overexpression stabilized dnTCF-1 retention on the target promoter instead of driving it away (Supplemental Figure 3), implying that EBP50 could help dnTCF-1 tether β-catenin on the target promoter region.
Figure 5. EBP50 promotes interaction of β-catenin and dnTCF-1.
(A) SW480 cells were transfected with 2 μg of the indicated plasmids and 0.4 μg of Top/Fop-flash reporter plasmids before luciferase activity was assessed as in Figure 2C. *P < 0.05. (B) SW480 cells stably expressing FLAG–dnTCF-1 were immunoprecipitated by either control or anti-FLAG antibody, followed by FLAG peptide elution. The eluate was then processed for Re-ChIP assay using EBP50 and β-catenin antibodies, respectively, and using rabbit anti-podocalyxin antibody as a control. Input represented 10% of total chromatin used for each experiment. (C) Purified Trx-His–fused full-length EBP50 (E50) and the first (PDZ1) and second PDZ domains (PDZ2) of EBP50 were incubated with glutathione beads conjugated with GST–dnTCF-1 (top panel) and GST–β-catenin C-terminal (middle panel) recombinant proteins. After incubation and washing, bound materials were assessed by immunoblotting. The amount and purity of the recombinant proteins are shown by a Coomassie blue–stained gel in the bottom panel. (D) Glutathione beads conjugated with GST–dnTCF-1 were incubated with the indicated Trx-His fusion proteins for 1 hour. Then untagged β-catenin, which was released from the GST–β-catenin by thrombin digestion, was added in the mixture for 1 more hour before the pulled-down materials were examined by immunoblotting. (E) SW480 cells stably expressing FLAG–dnTCF-1 were infected with lentivirus expressing siRNA against EBP50 (shE50) or luciferase control (shLuc). The cellular lysates were immunoprecipitated by mouse anti-FLAG antibody and processed for immunoblotting using the indicated antibodies. (F) Colo205 cells were infected with lentivirus expressing siRNA against EBP50 or luciferase control. The cellular lysates were immunoprecipitated by rabbit anti–β-catenin antibody and processed for immunoblotting using mouse anti–β-catenin, anti-EBP50, and anti–TCF-1 antibodies.
A β-catenin/EBP50/TCF-1 ternary complex is recruited to the promoter of Wnt target genes.
While an interaction of EBP50 and β-catenin has been previously suggested (20), the molecular mechanism by which EBP50 could help transduce Wnt/β-catenin signaling remains elusive. Results of reporter assays (Figure 2C and Figure 3D) implied that the localization of EBP50, a normally cortical membrane protein, at the promoter region of β-catenin–responsive genes is critical for transducing Wnt/β-catenin signaling. ChIP assays also demonstrated that EBP50 was recruited to the promoter regions of several well-characterized Wnt/β-catenin targets (Figure 3E). Furthermore, Re-ChIP experiments using eluted materials from TCF-1 immunoprecipitates implied the possible existence of a β-catenin/EBP50/TCF-1 complex at the c-Myc promoter (Figure 5B). This result is consistent with our ChIP-on-chip data indicating an EBP50 binding event in the Myc gene promoter (Supplemental Table 1).
An in vitro pull-own assay demonstrated that dnTCF-1 interacted with full-length EBP50 and the first PDZ domain of EBP50 and that β-catenin potentially interacted with EBP50 through the second PDZ domain (Figure 5C). To confirm the existence of a ternary protein complex as suggested by the Re-ChIP assay result (Figure 5B), we first purified recombinant GST-fused dnTCF-1 protein and immobilized it on beads. Then, we added full-length EBP50 or its individual PDZ domains and recombinant β-catenin sequentially to dissect the molecular interaction order. The result showed that N-terminally deleted dnTCF-1 could restore its interaction with β-catenin only when full-length EBP50 recombinant protein was there to provide a scaffold (Figure 5D). By contrast, neither the fusion protein tag thioredoxin-6His (Trx-His) nor the individual PDZ domains alone could provide such a linking function, although dnTCF-1 could pull down both the full-length EBP50 and its first PDZ domain (Figure 5D). This result confirms the existence of a ternary complex formed by β-catenin/EBP50/TCF-1 with EBP50 serving as the link for the other two proteins. It has been obscure how dnTCF-1, lacking a β-catenin–binding domain, as a major TCF-1 isoform in normal colonic tissue could promote the expression of Wnt/β-catenin target genes to promote carcinogenesis. Our model suggests that EBP50 acts as a mediator to restore this missing linkage. Compatible with this model, EBP50 knockdown indeed diminished the interaction between overexpressed dnTCF-1 and endogenous β-catenin (Figure 5E). To demonstrate that this effect could also be observed in endogenous dnTCF-1, we used Colo205 cells, which expressed both nuclear EBP50 and dnTCF-1 (Supplemental Figure 4), in which EBP50 was stably knocked down via a lentivirus system. Similar to the overexpressed dnTCF-1 in SW480 cells, endogenous dnTCF-1 showed a significant decrease in β-catenin binding after EBP50 downregulation (Figure 5F).
EBP50 participates in β-catenin–mediated cell growth.
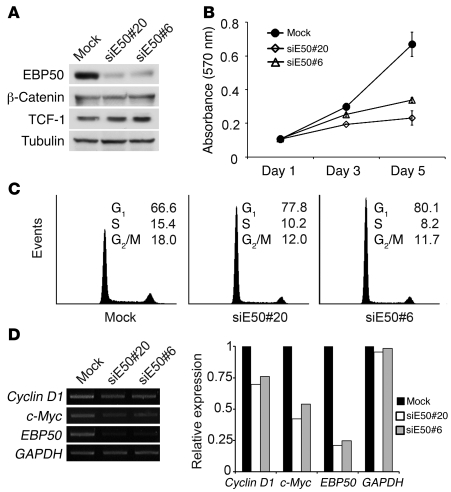
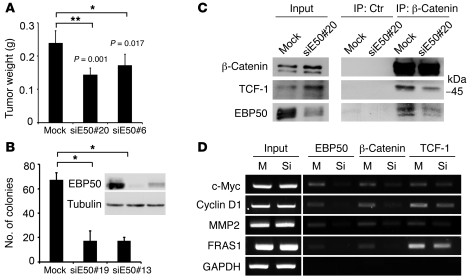
To demonstrate the functional impact of EBP50 interaction with β-catenin and TCF-1 in cell proliferation, we generated EBP50-knockdown stable clones by attenuating EBP50 expression using an siRNA approach and examined how cell growth was affected. Western blot analysis confirmed that EBP50 expression levels of two independent knockdown clones were less than 20% of the mock clone (Figure 6A). Cellular growth was significantly reduced in EBP50-knockdown clones (Figure 6B). Flow cytometry analysis showed that EBP50-knockdown cells had an increased proportion of cells at G0/G1 phase and a decreased distribution at the replicative S phase and mitotic phase compared with mock-transfected cells (Figure 6C). Semiquantitative RT-PCR analysis further confirmed that the gene transcripts of c-Myc and cyclin D1 were reduced in EBP50-depleted cells (Figure 6D). These results are consistent with our hypothesis that EBP50 modulates β-catenin signaling, since downregulation of β-catenin has been reported to induce G0/G1 arrest and cell growth inhibition by disrupting c-Myc and cyclin D1 expression (32, 33). Furthermore, to examine whether EBP50 affects tumorigenicity in vivo, we applied a tumor xenograft model using EBP50 knockdown SW480 cells. Consistent with our in vitro findings, the tumor growth of EBP50-knockdown cells was significantly diminished, demonstrating the importance of EBP50 in promoting growth in colon cancer cells both in vitro and in vivo (Figure 7A). Furthermore, EBP50 knockdown compromised the anchorage-independent growth of HT29 colon carcinoma cells in soft agar (Figure 7B), an essential feature of tumor transformation. These results indicated that EBP50 plays a significant role in cell growth and transformation. Given that SW480 cells only expressed full-length TCF-1 but not dnTCF-1, the effect of EBP50 knockdown on Wnt/β-catenin signaling (Figure 2A) and tumor cell growth (Figure 6B and Figure 7, A and B) implies that EBP50 is involved in the interaction between full-length TCF-1 and β-catenin. To clarify this issue, we evaluated TCF-1 and β-catenin association by co-immunoprecipitation in SW480 cells after EBP50 was knocked down. Although EBP50 knockdown seemed to have an unclear effect on upregulating the β-catenin expression level, the interaction between TCF-1 and β-catenin was diminished when EBP50 expression was downregulated (Figure 7C). The diminished association between TCF-1 and β-catenin after EBP50 knockdown is in agreement with our model whereby EBP50 bridges the connection between TCF-1 and β-catenin. To assess the importance of EBP50’s role as a scaffold allowing TCF to tether β-catenin at the target promoter sites, we inspected β-catenin occupancy at multiple sites when EBP50 was downregulated. Consistent with our hypothesis, ChIP assays demonstrated that β-catenin occupancy was diminished greatly at all the Wnt targets we examined, including c-Myc, cyclin D1, and MMP2 after EBP50 was knocked down (Figure 7D). Interestingly, TCF-1 binding seemed to decrease, although not to the same extent as β-catenin, with EBP50 downregulation (Figure 7D). This might explain why we consistently observed an increase in β-catenin and TCF-1 extractability when EBP50 expression was knocked down (Figure 7C).
Figure 6. EBP50 knockdown blocks cell cycle progression and inhibits cell proliferation.
(A) Western blot analysis for EBP50, β-catenin, TCF-1, and α-tubulin in mock transfectant and two independent SW480 clones in which EBP50 had been stably knocked down. (B) Proliferation (MTS) assays of control and EBP50-knockdown SW480 cells. The results represent mean ± SEM from 3 independent assays. (C) EBP50 knockdown induced G1 arrest in colon cancer SW480 cells. Cell cycle stage distribution of mock and EBP50-knockdown clones was analyzed by flow cytometry. The values represent average percentage of cells in each stage of the cell cycle from 3 independent experiments. (D) mRNA expression of c-Myc and cyclin D1 was quantified by semiquantitative PCR using GAPDH as an internal control. Results of one experiment representative of three are shown.
Figure 7. Tumorigenesis of the colon cancer cells is reduced by EBP50 knockdown, which disrupted β-catenin and TCF-1 interaction.
(A) Mock and EBP50 siRNA stably transfected (siE50) SW480 clones were injected beneath the dorsal skin of 6-week-old NOD-SCID mice. After 3 weeks, the mice were sacrificed. The injected tumors (n = 5 for each group) were dissected from the mice and weighed, and the results are shown (mean ± SEM). (B) Mock and EBP50 siRNA stably transfected HT29 clones were processed for soft agar assay to assess their anchorage-independent growth ability. The histogram shows results (mean ± SEM) from 3 different fields for each clone. The inset shows the result from Western blot analysis of EBP50 in mock and siRNA-expressing clones. *P < 0.05, **P < 0.001. (C) Cell lysates from mock and siE50 SW480 clone were immunoprecipitated with equal amounts of mouse normal serum (Ctr) and anti–β-catenin monoclonal antibody and processed for Western blot analysis using the indicated antibodies. (D) Mock (M) or siEBP50 SW480 (Si) cells were processed for ChIP assay using rabbit anti-EBP50, anti–β-catenin, or anti–TCF-1 antibody. Five percent of the immunoprecipitated DNA served as the templates for amplifying all the target sequences by PCRs, except those for MMP2, which used 0.5% of the immunoprecipitated materials as the templates for PCRs.
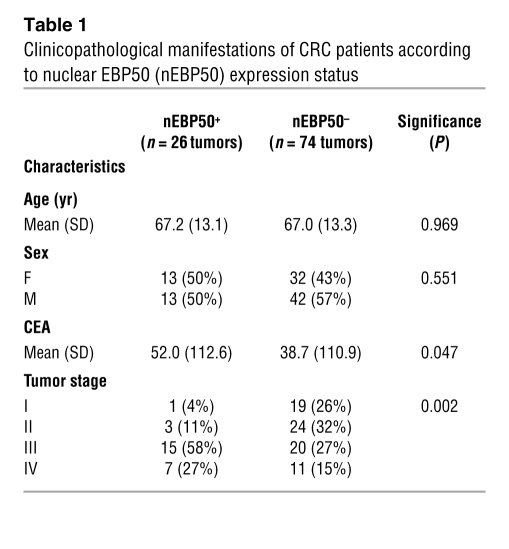
EBP50 is expressed in the nuclei of tumors in a subset of CRC patients who manifest a poor clinical outcome.
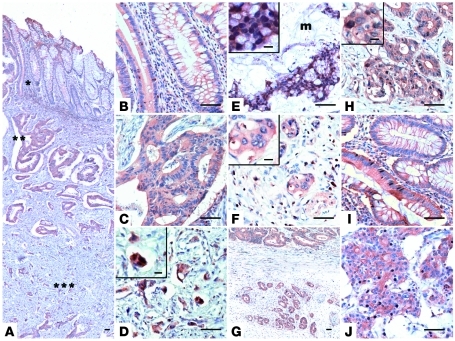
Because we had demonstrated that nuclear EBP50 can modulate cell growth by working together with TCF-1 and β-catenin, it is interesting to know whether this finding could be of any pathophysiological significance in human tumorigenesis. To definitively determine the EBP50 expression pattern in human CRC specimens, we performed immunohistochemistry (IHC) in archived CRC formalin-fixed, paraffin-embedded sections and analyzed the EBP50 expression pattern, with an emphasis on its nuclear expression. One hundred CRC patients were included in this study, and their relevant clinicopathological characteristics are provided in Table 1. Although nuclear EBP50 expression was observed in intraepithelial and the infiltrating intratumoral lymphocytes, EBP50 expression was limited to the membrane in normal colonic epithelia (Figure 8B, right portion, and Figure 8I, upper portion). Even within the neoplastic epithelia, nuclear EBP50 expression was detected in only one exceptional case, in which scattered EBP50 nuclear staining could be seen in adenomatous epithelia as well as the poorly differentiated adenocarcinoma (Figure 8, I and J, lower portion). EBP50 overexpression could be found in most adenomatous epithelial cells and differentiated neoplastic glands, yet the increased staining was limited in the cytosol and was not present in the nuclei (Figure 8, B and C). Moreover, we observed nuclear EBP50 staining in a subset of tumor cells in the deep invasive front away from the main tumor mass (Figure 8, D and H). In a group of CRC cases we studied, an increase in the staining intensity of EBP50 was observed from the mucosal portion to the deeper invasive front (Figure 8, B–D). This expression pattern resembled that of β-catenin in the same group (Supplemental Figure 5, B–D). Note that nuclear EBP50 expression was not uniformly observed in the invasive fronts of all CRC cases (Figure 8F and Supplemental Figure 5H); therefore, nuclear EBP50 location is not an autonomous finding for the invasive tumor cells. Nevertheless, we consistently observed robust nuclear EBP50 expression in floating tumor nests in the mucin pools (Figure 8E), which confirms that mucin coexpressed with nuclear β-catenin in colon cancer and could stabilize β-catenin/Wnt signaling (34, 35).
Table 1 .
Clinicopathological manifestations of CRC patients according to nuclear EBP50 (nEBP50) expression status
Figure 8. EBP50 expression in colorectal cancer specimens.
A representative CRC sample showed global EBP50 staining (A) in the mucosa (*), submucosa (**), and deeper invasive portions of the tumor (***); these regions are presented as magnified images in B–D, respectively. (E) EBP50 expression in CRC specimen with extracellular mucin pooling (m). (F) A representative sample showing negative nuclear EBP50 staining in the invasive front. Low- (G) and high-magnification (H) images of another CRC sample, which demonstrates aberrant nuclear EBP50 expression in the peripheral tumor nests (lower portion in G) outside the main tumor mass (upper portion in G). EBP50 expression was infrequently found concomitantly in the nuclei of the well-differentiated epithelial portion (I) and the poorly differentiated cancer part (J) in only a single case among our CRC patients. Scale bars: 50 μm; 10 μm, insets in D–F and H.
Differences in clinical and pathological features of the CRC patients by EBP50 nuclear expression are shown in Table 1. About 25% of the patients we studied were assessed to have nuclear EBP50 expression. Most significantly, these patients were associated with higher carcinoembryonic antigen (CEA) values at the initial presentation (P = 0.047) and more advanced tumor involvement (P = 0.002), two well-established prognostic factors for CRC cancer survival (36).
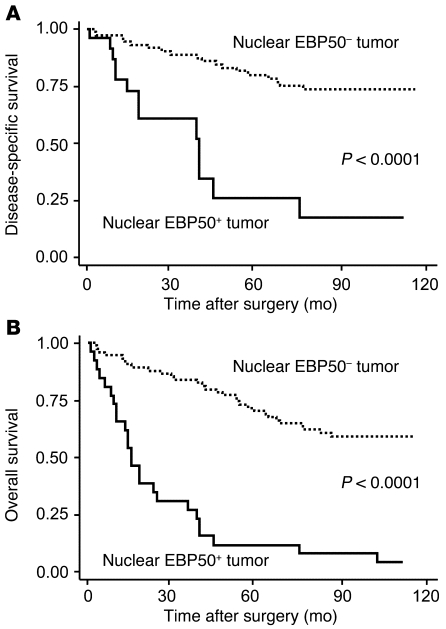
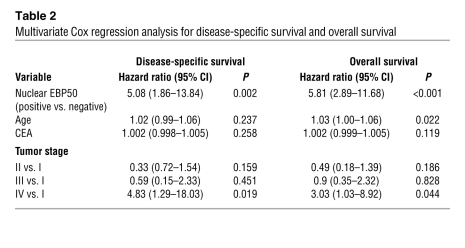
Aberrant nuclear EBP50 expression in primary CRC tumors was associated with poorer disease-specific and overall survival (Figure 9). Forty-five percent of patients were censored before the cut-off date of December 31, 2008. The median follow-up time was 70 months, and the median time to disease-specific and overall death was 29 and 26 months, respectively. In the univariate analysis (Supplemental Table 3), the hazard ratio of EBP50 nuclear expression was 5.49 (95% CI, 2.59–11.66; P < 0.001) for disease-specific survival and 6.13 (95% CI, 3.49–10.76; P < 0.001) for overall survival. To investigate whether nuclear EBP50 expression in primary CRC is an independent prognostic factor for clinical outcome, we performed multivariate analysis incorporating EBP50 nuclear expression status, age, initial CEA value, and tumor stage in the model as potential risk factors. The results showed that aberrant nuclear EBP50 expression was an independent and strong predictor, with a hazard ratio of 5.08 (95% CI, 1.86–13.84; P = 0.002) for disease-specific survival and 5.81 (95% CI, 2.89–11.68; P < 0.001) for overall survival, after controlling for all other prognostic factors (Table 2).
Figure 9. Kaplan-Meier survival analysis in CRC patients according to nuclear EBP50 expression status.
(A) Disease-specific survival. (B) Overall survival.
Table 2 .
Multivariate Cox regression analysis for disease-specific survival and overall survival
Discussion
In the present study, we established the role of EBP50 as a facilitator of the interaction between TCF-1 and β-catenin. We demonstrated that EBP50, a PDZ protein conventionally regarded as a membrane cytoskeleton linker protein, displayed nuclear localization when the cells were sparse, but retained a membrane expression pattern when cells were dense. In addition, we also revealed a PDZ1-mediated interaction between EBP50 and TCF-1. Through its two tandem PDZ domains, EBP50 can bridge the TCF-1/β-catenin complex on the promoter regions of Wnt/β-catenin–responsive genes to enhance target gene transcription. Overexpression of dnTCF-1 has been shown in the colon carcinoma cell lines SW480 and HT29 to induce CEACAM5/6 and mesothelin, which are typically expressed in the invasive front of colon cancers (37). Coincidentally, most colon cancer cell lines only express full-length TCF-1, but semi-adherent and highly dedifferentiated cancer cells such as Colo201 and Colo205 also express dnTCF-1 (38). Our findings further explain how a dominant negative dnTCF-1 could transform from playing a transcriptionally suppressive role to serving as an active partner of β-catenin to turn on Wnt signaling–responsive promoters and activate malignant growth. In addition to the effect of EBP50 on the interaction between dnTCF-1 and β-catenin, we found that EBP50 also stabilized the binding between β-catenin and full-length TCF-1, which retained the β-catenin–binding region (Figure 5F and Figure 7C). The results of our functional studies in SW480 cells also confirmed that knockdown EBP50 could compromise the relevant β-catenin/TCF-1 functions.
Two previous studies have adopted systemic approaches to collect genome-wide information for Wnt pathway targets. One study exploited TCF-4 antibody for ChIP-on-chip analysis (39), and the other used β-catenin antibody for ChIP followed by sequencing (40) to identify candidate genes upon Wnt signaling activation. Interestingly, there is only a limited degree of overlap between the gene lists of their studies and ours (Supplemental Table 1). Comparison of the EBP50 binding consensus sequence (Figure 3B) disclosed nucleotide differences at key positions of the Wnt response element (WRE), while the other two sets of genome-wide ChIP data revealed a near perfect match to the WRE consensus. This result raises the intriguing possibility that EBP50 directs TCF-1 to an alternative set of target genes, not the core set of Wnt targets identified by previous groups, such that EBP50 might act as a modifier rather than as a core oncogenic component in the Wnt signaling pathway.
Previously, EBP50 had been shown to act as a tumor suppressor or promoter. This controversy could be at least partly explained by our finding that the role of EBP50 in cell growth depended on the localization of EBP50 and the activation status of β-catenin. In addition to its membrane localization, EBP50 has been reported to be cytosolic and nucleic, especially in cancer cell lines and tissues. The molecular mechanism underlining such a mobile subcellular localization of EBP50 may depend on environmental signaling and also its post-translational modifications and interacting partners. According to our study, EBP50 showed a nuclear localization pattern when the cells were sparse, but was retained at the plasma membrane when cells reached confluence. Interestingly, we found that EBP50 was distributed in the nucleus at the invasive front of colon cancer specimens, while it was detected at the membranous and cytosolic portions in the central mass of tumors. These distinct distribution patterns are exactly the same as those of β-catenin as reported in colon cancer specimens (24). Cells at the invasive front of tumors usually display an unpolarized morphology and herald an epithelial-mesenchymal transition (EMT), suggesting that EBP50 may subject to regulatory mechanisms dominant during EMT that facilitate its entry into the nucleus.
Previous studies have shown that EBP50 stabilizes the β-catenin/E-cadherin complex at the plasma membrane and controls the cellular distribution of β-catenin in mouse embryonic fibroblasts (MEFs) (21). However, we were not able to detect visible changes in the localization of β-catenin upon overexpression or knockdown of EBP50 in SW480 and HT29 cells. In addition, the additive effect of EBP50 on β-catenin–mediated transcription of EBP50 is restricted to cells that express stabilized active β-catenin, implying that EBP50 has different effects on membrane- and nucleus-associated β-catenin.
An increasing number of PDZ proteins have been reported to switch their localization between the cytosol and the nucleus, thus modulating transcriptional activity. In addition to EBP50, β-catenin is associated with at least three class I PDZ proteins, LIN-7, MAGI-1, and Tax-interacting protein (TIP), through its C-terminal PDZ-binding motif (41–43). Notably, overexpression of either TIP-1, which contains a single PDZ domain, or the isolated PDZ domain of EBP50 can inhibit β-catenin transcriptional activity in colorectal cancer cells (43). It is reported that TIP-1 function as a negative regulator to disrupt PDZ-based scaffolding of membrane complexes (44). Therefore, whether TIP-1 suppresses Wnt/β-catenin signaling by interrupting the EBP50-mediated β-catenin/TCF-1 complex is worthy of further examination.
Several nuclear proteins and nuclear-cytoplasm shuttling proteins, including yes-associated protein (YAP) (45) and transcriptional coactivator with PDZ-binding motif (TAZ) (46), have been shown to be associated with EBP50 via PDZ domain interaction. Recently, YAP and TAZ have been reported to play a vital role in the Hippo pathway, which controls organ size and tumorigenesis through TEAD family transcription factors as well as EMT (47–49). Our ChIP-on-chip analysis identified significant numbers of TEAD binding motifs in EBP50 binding regions (Supplemental Table 2), and this result implies that nuclear EBP50 may be implicated in transcriptional activity other than the β-catenin/TCF-1 pathway. Therefore, whether nuclear EBP50 modulates EMT or tumorigenesis events through interaction with these two transcriptional modulators should be an intriguing issue to follow.
Methods
Plasmids.
FLAG-EBP50, its mutant derivatives, Myc-tagged full-length TCF-1, and FLAG-tagged dominant negative TCF-1 were cloned into pcDNA3.1 (Invitrogen) by a standard molecular cloning procedure. The core binding pocket sequence GYGF was replaced with GAGA singly or doubly in the tandem PDZ domains of a wild-type FLAG-EBP50 construct to create PDZ domain–defective mutant FLAG-PDZ1GA, FLAG-PDZ2GA, or FLAG-E50-GAGA. DNAs for full-length, PDZ1 domain (residues 1–148), and PDZ2 domain (residues 149–297) EBP50 were subcloned into pET32a for bacterial expression. The C terminus of β-catenin (residues 518–781) was subcloned into pGEX2T to create GST–β-catenin C′. All these constructs had been verified by automatic DNA nucleotide sequencing, and the sequence information is shown in Supplemental Table 4.
Cell culture, transfection, and viral infection.
The colon carcinoma cell lines SW480, HT29, and the kidney epithelial cell line HEK293T were cultured in DMEM supplemented with 10% fetal calf serum at 37°C in a humidified incubator containing 5% CO2. Colo205 and NCI-H28 mesothelioma cells were cultured in RPMI 1640 medium supplemented with 10% fetal calf serum. Transfection was performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Transient knockdown experiments were conducted with an siRNA duplex targeting two independent sites of human EBP50 cDNA (GATACAGAAGGAGAACAGTCGTGAA and TGACTTTAGGGAAGGTGAATGTGTT) and a low GC matched negative universal control (catalog 12935-200, Invitrogen). Alternatively, cells were transfected with pSuper-EGFP-neo-siE50 plasmid expressing the interfering RNA targeted against the EBP50 sequence (CCCATCCTAGACTTCAACA), and then stable clones were selected and maintained in 800 μg/ml G418 (Sigma-Aldrich). Lentiviral vector expressing siRNA against human EBP50 (pLKO.1-shEBP50) and its control (pLKO.1-Luc) were obtained from the RNAi consortium at Academia Sinica. Colo205 and SW480 cells were infected with virus plus 8 μg/ml Polybrene overnight and then selected by 1 μg/ml (Colo205) or 2 μg/ml (SW480) puromycin for 5 days.
Antibodies.
Mouse anti-EBP50 and anti–β-catenin antibodies were from BD Transduction Laboratories. Mouse anti–TCF-1 antibody was from Millipore. Rabbit anti–β-catenin antibody was a gift from W.J. Nelson (Stanford University, Stanford, California, USA). Rabbit anti-EBP50 polyclonal and anti–TCF-1 monoclonal antibodies were from Abcam and Cell Signaling Technology, respectively. Rabbit and mouse anti-FLAG (M2) antibodies were from Sigma-Aldrich. Anti-Myc hybridoma cells (9E10) were from ATCC.
Immunofluorescence.
Cells were fixed with 3.7% paraformaldehyde for 20 minutes and permeabilized with 50 mM NaCl, 300 mM sucrose, 10 mM PIPES (pH 6.8), 3 mM MgCl2, and 0.5% Triton X-100 for 5 minutes. Cells were blocked with PBS plus 10% goat serum, 1% BSA, and 50 mM NH4Cl for 1 hour and then incubated with primary antibodies for 1 hour. Cells were then incubated with FITC- and rhodamine-conjugated secondary antibodies plus Hoechst dye for 1 hour.
Reporter assay.
SW480 cells (2 × 105) were transfected with 0.4 μg Top-flash (50) Wnt/β-catenin reporter plasmid, which harbors 3 repeated TCF-binding sites, or Fop-flash with mutated TCF-binding sites and 0.02 μg pTK-RL for normalization of transfection efficiency. HEK293 cells (5 × 105) were transfected with 1.5 μg Top-flash reporter plasmids and 3 μg of the indicated Flag-tagged EBP50 plasmids. After transfection, cells were seeded to 12-well plates in regular medium with or without 20 mM LiCl for 24 hours and then assessed with a Firelite Luciferase Assay Kit (PerkinElmer).
Expression and purification of fusion proteins.
The recombinant GST and Trx-His fusion proteins were induced in DE3pLys bacterial strain with 0.5 mM IPTG for 30 minutes and then purified with glutathione beads and nickel-chelating resins, respectively, according to standard protocols. Trx-His proteins were eluted by 500 mM imidazole, and recombinant β-catenin was separated from the GST moiety using thrombin.
GST pull-down assay.
GST pull-down assays were used to examine the interaction between NHERF and TCF-1. GST-NHERF1 and GST-NHERF2 were induced by IPTG and purified with glutathione-Sepharose beads. HEK293 cells were transfected with Myc–TCF-1 or HA–TCF-4, and cell lysates were mixed with purified GST-NHERF1 or GST-NHERF2 immobilized on the beads. Pull-down assays were performed at 4°C for 1 hour. The beads were then washed thoroughly with PBS buffer. Bound proteins were eluted by boiling in SDS sample buffer, separated by SDS-PAGE, and detected by immunoblotting.
Immunoprecipitation and immunoblotting.
Cells were lysed in 20 mM Tris (pH 7.5), 150 mM NaCl, and 0.1% Triton X-100 plus 25 U/ml Benzonase endonuclease (Merck), and an aliquot of cell lysate was incubated with NHS (N-hydroxysuccinimide) beads, which were covalently conjugated with the appropriate antibody overnight at 4°C. Beads were then collected and washed 3 times in PBS buffer and eluted by boiling in SDS sample buffer. Protein eluates were separated by SDS-PAGE and processed for immunoblot analysis using the relevant primary antibodies, horseradish peroxidase–conjugated secondary antibody, and SuperSignal reagents (Pierce).
ChIP and Re-ChIP assays.
A total of 5 × 105 wild-type (ChIP) or FLAG–TCF-1 stably expressing (Re-ChIP) SW480 cells were cross-linked in culture media with 0.1% formaldehyde and quenched by 0.125 M glycine. After being washed with cold PBS, cells were scraped and lysed in 5 mM PIPES (pH 8), 85 mM KCl, and 0.5% NP40. The nuclei were pelleted by centrifugation and lysed in 50 mM Tris, 10 mM EDTA, and 1% SDS. The lysates were sonicated and incubated with salmon sperm DNA/protein A agarose. These precleaned lysates were diluted with a 4× volume of 0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl (pH 8.1), and 167 mM NaCl, and then mixed with rabbit anti-EBP50, TCF-1, β-catenin (ChIP), or mouse anti-FLAG (Re-ChIP) antibodies and salmon sperm DNA/protein A agarose overnight at 4°C. Complexes were pelleted and washed once with 2 mM EDTA and 50 mM Tris and three times each with 100 mM Tris, 500 mM LiCl, 1% deoxycholate, and 1% NP-40. The washed complexes were eluted with freshly prepared 1% SDS and 50 mM NaHCO3 with Na+ concentration adjusted to 300 mM, followed by incubation at 65°C for 4 hours to reverse formaldehyde cross-links. The DNA/protein solution was treated with proteinase K and extracted by phenol/chloroform. For Re-ChIP experiments, the anti-FLAG antibody–immunoprecipitated complexes were eluted by incubation with FLAG peptide for 30 minutes at 37°C and subjected to a repeated ChIP procedure with rabbit anti-EBP50 or anti–β-catenin antibody. The purified DNA was amplified by PCR using gene-specific primers (Supplemental Table 5).
ChIP-on-chip and data analysis.
Immunoaffinity-enriched DNA and input DNA were amplified and then fluorescently labeled with Cy5 and Cy3, respectively. The labeled DNA was hybridized to the Human Promoter Microarray (G4489A; Agilent) according to the manufacturer’s instructions. Genomic Workbench software (Agilent) was used to identify binding peak events. Data were normalized using the normalized output from the Agilent Feature Extraction program. An error model was applied to assign P values to each probe representing the likelihood of the probe being a bound probe. Bound peaks were detected by the Whitehead per-array neighborhood model (51). The software default setting was used, except for 500 bp on the maximum distance of two neighbor probes. Finally, extra bound probes were added if the probe adjacent to the bound peak had a P value less than 0.01. Bound regions were calculated according to the following functions: bound region start = (P(m-1,start) + P(m,end))/2 and bound region end = (P(n,start) + P(n+1,end))/2; where P(m) to P(n) belong to a bound peak, P(m,end) denotes the last genomic position of a probe m, and P(n,start) denotes the first genomic position of a probe n. DNA sequences of the bound region were extracted from the NCBI human genome database (build 36; March 2006; hg18). Sequences were analyzed with the motif-finding program BioProspector (52). These ChIP-on-chip data have been deposited in the Gene Expression Omnibus (GEO GSE33775).
In vitro binding.
Eluted Trx-His fusion proteins (1 μg) were purified, eluted from Ni+-chelating resins, and then added to immobilized GST–dnTCF-1 or GST–β-catenin C′ in a total volume of 1 ml and incubated for more than 2 hours in 4°C. For ternary complex formation, eluted Trx-His fusion proteins and β-catenin were incubated with GST–dnTCF-1. All in vitro binding assays were incubated and washed in 10 mM Tris, 150 mM NaCl, and 0.5% TX-100.
Semiquantitative RT-PCR.
The RNA of SW480 was extracted with TRIzol reagent (Invitrogen) according to the manufacturer’s instructions, and then mRNA was reverse transcribed to cDNA using an oligo-dT18 primer by Superscript III reverse transcriptase (Invitrogen) at 50°C for 60 minutes. PCR analysis was performed using c-Myc–, cyclin D1–, EBP50-, and GAPDH-specific primers (Supplemental Table 4).
Proliferation assay.
Cell growth was measured using the CellTiter 96 assay (Promega) as recommended by the manufacturer. Briefly, 1,500 cells were plated in 96-well plates and allowed to proliferate for 1, 3, and 5 days. Absorbance at 490 nm was evaluated 4 hours later after MTS assay reagent was added.
Cell cycle assay.
Subconfluent mock and siEBP50 cells were collected and fixed with 75% ethanol. After permeation with 0.1 M citric acid (pH 7.8) and 0.2 M Na2HPO4, cells were stained with propidium iodide, and then stained cells were analyzed by flow cytometry using FACSCalibur (BD Biosciences).
Soft agar assay.
Six-well plates were plated with 2 ml semisolid DMEM containing 0.5% agar and 10% FBS. A total of 4 × 104 HT29 cells were then plated in 1 ml semisolid medium consisting of 0.3% agar and 10% FBS. Additional regular medium was added to the wells to prevent the agar from drying. After incubation for 7 days, colonies were stained with cresyl violet (Sigma-Aldrich) and photographed under a microscope at ×40 magnification. Colonies larger than 50 μm in diameter were scored.
Tumor xenograft growth assay.
SW480 mock and knockdown stable cells in the log phase were trypsinized and washed twice with 137 mM NaCl, 5 mM KCl, 4 mM NaHCO3, 0.5 mM EDTA, 0.1% (w/v) glucose. A total of 5 × 106 cells in 100 μl 1:1 (v/v) BD Matrigel matrix/DMEM were injected subcutaneously into the back of 8-week-old NOD/SCID mice. After 4 weeks, the mice were sacrificed for tumor size estimation.
Patient selection and tumor samples.
Formalin-fixed, paraffin-embedded samples were obtained from 100 patients with primary CRC who underwent tumor resection between January 1, 1999, and December 31, 2003, at National Taiwan University Hospital, Taipei, Taiwan. The clinical information and the outcome status of the patients were obtained through chart reviewing, questionnaire recording, telephone calling, and linking the patients’ unique national identification numbers to the national death index profile. Staging was based on the 2009 TNM classification guidelines.
Immunohistochemistry.
Five-micrometer-thick sections of the paraffin-embedded tissues were cut and deparaffinized according to standard histological techniques. Subsequently, immunohistochemical staining was performed by applying a biotin-free detection system (SuperSensitive Polymer-HRP Detection Kit/DAB from BioGenex) according to the manufacturer’s instructions. In brief, antigen retrieval was performed using citrate buffer (pH 6.0) for 30 minutes at 95°C, and endogenous peroxidase activity was blocked by 3% hydrogen peroxide for 10 minutes. After blocking in 2.5% normal goat serum in PBS, the sections were incubated with monoclonal antibody against EBP50 or β-catenin diluted 1:100 and 1:1,000, respectively, at 4°C overnight, followed by incubating labeled polymer horseradish peroxidase (30 minutes) and detecting by the liquid chromogen. Finally, the nuclei were counterstained with hematoxylin.
Statistics.
Information on age, sex, tumor size, stage, and EBP50 status was obtained as baseline variables. Disease-specific or overall survival was measured from the date of operation to the date of death due to CRC or all causes, or censored on December 31, 2008. The baseline characteristics of all 100 patients were compared in nuclear EBP50–positive and –negative cases using the Wilcoxon rank sum test for continuous variables and Fisher’s exact test for categorical variables. Disease-specific and overall survival curves were estimated by the Kaplan-Meier method, and the difference between survival curves was tested by the log-rank test. Hazard ratios and 95% CIs were estimated using the Cox proportional hazards regression model to assess the role of nuclear EBP50 expression as an independent CRC prognostic factor. Statistical analyses were performed with STATA 8.2. A P value less than 0.05 was considered significant. All values were 2-sided.
Study approval.
All animal procedures were performed in accordance with protocols approved by the IACUC of the medical school of National Taiwan University. The informed consent requirement in this study was waived due to use of archived tissue sections collected prior to 2008 by a guideline of the Institutional Ethic Committee Review Board (National Taiwan University Hospital).
Supplementary Material
Acknowledgments
This study is supported by National Science Council grants (NSC100-2325-B-002-029 and NSC97-3112-B-002-041) and National Taiwan University Hospital grants (aNTUH99P21-1 and 98P26-1) to T.-S. Jou. We thank Winston W. Shen for valuable editing comments.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2012;122(5):1881–1894. doi:10.1172/JCI45661.
References
- 1.Gregorieff A, Clevers H. Wnt signaling in the intestinal epithelium: from endoderm to cancer. Genes Dev. 2005;19(8):877–890. doi: 10.1101/gad.1295405. [DOI] [PubMed] [Google Scholar]
- 2.Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127(3):469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 3.Hoppler S, Kavanagh CL. Wnt signalling: variety at the core. J Cell Sci. 2007;120(pt 3):385–393. doi: 10.1242/jcs.03363. [DOI] [PubMed] [Google Scholar]
- 4.Nelson WJ, Nusse R. Convergence of Wnt, beta-catenin, and cadherin pathways. Science. 2004;303(5663):1483–1487. doi: 10.1126/science.1094291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu C, et al. Control of beta-catenin phosphorylation/degradation by a dual-kinase mechanism. Cell. 2002;108(6):837–847. doi: 10.1016/S0092-8674(02)00685-2. [DOI] [PubMed] [Google Scholar]
- 6.Molenaar M, et al. XTcf-3 transcription factor mediates beta-catenin-induced axis formation in Xenopus embryos. Cell. 1996;86(3):391–399. doi: 10.1016/S0092-8674(00)80112-9. [DOI] [PubMed] [Google Scholar]
- 7.Behrens J, et al. Functional interaction of beta-catenin with the transcription factor LEF-1. Nature. 1996;382(6592):638–642. doi: 10.1038/382638a0. [DOI] [PubMed] [Google Scholar]
- 8.Roose J, et al. The Xenopus Wnt effector XTcf-3 interacts with Groucho-related transcriptional repressors. Nature. 1998;395(6702):608–612. doi: 10.1038/26989. [DOI] [PubMed] [Google Scholar]
- 9.Brantjes H, Roose J, van De Wetering M, Clevers H. All Tcf HMG box transcription factors interact with Groucho-related co-repressors. Nucleic Acids Res. 2001;29(7):1410–1419. doi: 10.1093/nar/29.7.1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daniels DL, Weis WI. β-catenin directly displaces Groucho/TLE repressors from Tcf/Lef in Wnt-mediated transcription activation. Nat Struct Mol Biol. 2005;12(4):364–371. doi: 10.1038/nsmb912. [DOI] [PubMed] [Google Scholar]
- 11.Hurlstone A, Clevers H. T-cell factors: turn-ons and turn-offs. EMBO J. 2002;21(10):2303–2311. doi: 10.1093/emboj/21.10.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Noort M, Clevers H. TCF transcription factors, mediators of Wnt-signaling in development and cancer. Dev Biol. 2002;244(1):1–8. doi: 10.1006/dbio.2001.0566. [DOI] [PubMed] [Google Scholar]
- 13.Arce L, Yokoyama NN, Waterman ML. Diversity of LEF/TCF action in development and disease. Oncogene. 2006;25(57):7492–7504. doi: 10.1038/sj.onc.1210056. [DOI] [PubMed] [Google Scholar]
- 14.Van de Wetering M, Castrop J, Korinek V, Clevers H. Extensive alternative splicing and dual promoter usage generate Tcf-1 protein isoforms with differential transcription control properties. Mol Cell Biol. 1996;16(3):745–752. doi: 10.1128/mcb.16.3.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hovanes K, et al. Beta-catenin-sensitive isoforms of lymphoid enhancer factor-1 are selectively expressed in colon cancer. Nat Genet. 2001;28(1):53–57. doi: 10.1038/ng0501-53. [DOI] [PubMed] [Google Scholar]
- 16.Waterman ML. Lymphoid enhancer factor/T cell factor expression in colorectal cancer. Cancer Metastasis Rev. 2004;23(1–2):41–52. doi: 10.1023/A:1025858928620. [DOI] [PubMed] [Google Scholar]
- 17.Castrop J, et al. The human TCF-1 gene encodes a nuclear DNA-binding protein uniquely expressed in normal and neoplastic T-lineage lymphocytes. Blood. 1995;86(8):3050–3059. [PubMed] [Google Scholar]
- 18.Roose J, et al. Synergy between tumor suppressor APC and the beta-catenin-Tcf4 target Tcf1. Science. 1999;285(5435):1923–1926. doi: 10.1126/science.285.5435.1923. [DOI] [PubMed] [Google Scholar]
- 19.Shenolikar S, Voltz JW, Cunningham R, Weinman EJ. Regulation of ion transport by the NHERF family of PDZ proteins. Physiology (Bethesda). 2004;19:362–369. doi: 10.1152/physiol.00020.2004. [DOI] [PubMed] [Google Scholar]
- 20.Shibata T, Chuma M, Kokubu A, Sakamoto M, Hirohashi S. EBP50, a beta-catenin-associating protein, enhances Wnt signaling and is over-expressed in hepatocellular carcinoma. Hepatology. 2003;38(1):178–186. doi: 10.1053/jhep.2003.50270. [DOI] [PubMed] [Google Scholar]
- 21.Kreimann EL, et al. Cortical stabilization of beta-catenin contributes to NHERF1/EBP50 tumor suppressor function. Oncogene. 2007;26(36):5290–5299. doi: 10.1038/sj.onc.1210336. [DOI] [PubMed] [Google Scholar]
- 22.Takahashi Y, Morales FC, Kreimann EL, Georgescu MM. PTEN tumor suppressor associates with NHERF proteins to attenuate PDGF receptor signaling. EMBO J. 2006;25(4):910–920. doi: 10.1038/sj.emboj.7600979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Georgescu MM, Morales FC, Molina JR, Hayashi Y. Roles of NHERF1/EBP50 in cancer. Curr Mol Med. 2008;8(6):459–468. doi: 10.2174/156652408785748031. [DOI] [PubMed] [Google Scholar]
- 24.Brabletz T, et al. Variable beta-catenin expression in colorectal cancers indicates tumor progression driven by the tumor environment. Proc Natl Acad Sci U S A. 2001;98(18):10356–10361. doi: 10.1073/pnas.171610498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davies ML, Roberts GT, Spiller DG, Wakeman JA. Density-dependent location and interactions of truncated APC and beta-catenin. Oncogene. 2004;23(7):1412–1419. doi: 10.1038/sj.onc.1207266. [DOI] [PubMed] [Google Scholar]
- 26.Conacci-Sorrell M, Simcha I, Ben-Yedidia T, Blechman J, Savagner P, Ben-Ze’ev A. Autoregulation of E-cadherin expression by cadherin-cadherin interactions: the roles of beta-catenin signaling, Slug, and MAPK. J Cell Biol. 2003;163(4):847–857. doi: 10.1083/jcb.200308162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Munemitsu S, Albert I, Souza B, Rubinfeld B, Polakis P. Regulation of intracellular beta-catenin levels by the adenomatous polyposis coli (APC) tumor-suppressor protein. Proc Natl Acad Sci U S A. 1995;92(7):3046–3050. doi: 10.1073/pnas.92.7.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tang LY, et al. Quantitative phosphoproteome profiling of Wnt3a-mediated signaling network: indicating the involvement of ribonucleoside-diphosphate reductase M2 subunit phosphorylation at residue serine 20 in canonical Wnt signal transduction. Mol Cell Proteomics. 2007;6(11):1952–1967. doi: 10.1074/mcp.M700120-MCP200. [DOI] [PubMed] [Google Scholar]
- 29.Clevers H, van de Wetering M. TCF/LEF factor earn their wings. Trends Genet. 1997;13(12):485–489. doi: 10.1016/S0168-9525(97)01305-X. [DOI] [PubMed] [Google Scholar]
- 30.Tang W, Dodge M, Gundapaneni D, Michnoff C, Roth M, Lum L. A genome-wide RNAi screen for Wnt/β-catenin pathway components identifies unexpected roles for TCF transcription factors in cancer. Proc Natl Acad Sci U S A. 2008;105(28):9697–9702. doi: 10.1073/pnas.0804709105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van de Wetering M, et al. The beta-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell. 2002;111(2):241–250. doi: 10.1016/S0092-8674(02)01014-0. [DOI] [PubMed] [Google Scholar]
- 32.Huang WS, Wang JP, Wang T, Fang JY, Lan P, Ma JP. ShRNA-mediated gene silencing of beta-catenin inhibits growth of human colon cancer cells. World J Gastroenterol. 2007;13(48):6581–6587. doi: 10.3748/wjg.13.6581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Verma UN, Surabhi RM, Schmaltieg A, Becerra C, Gaynor RB. Small interfering RNAs directed against beta-catenin inhibit the in vitro and in vivo growth of colon cancer cells. Clin Cancer Res. 2003;9(4):1291–1300. [PubMed] [Google Scholar]
- 34.Baldus SE, et al. MUC1 and nuclear beta-catenin are coexpressed at the invasion front of colorectal carcinomas and are both correlated with tumor prognosis. Clin Cancer Res. 2004;10(8):2790–2796. doi: 10.1158/1078-0432.CCR-03-0163. [DOI] [PubMed] [Google Scholar]
- 35.Schroeder JA, Adriance MC, Thompson MC, Camenisch TD, Gendler SJ. MUC1 alters beta-catenin-dependent tumor formation and promotes cellular invasion. Oncogene. 2003;22(9):1324–1332. doi: 10.1038/sj.onc.1206291. [DOI] [PubMed] [Google Scholar]
- 36.Bendardaf R, Lamlum H, Pyrhonen S. Prognostic and predictive molecular markers in colorectal carcinoma. Anticancer Res. 2004;24(4):2519–2530. [PubMed] [Google Scholar]
- 37.Liebig B, et al. Forced expression of deltaN-TCF-1B in colon cancer derived cell lines is accompanied by the induction of CEACAM5/6 and mesothelin. Cancer Lett. 2005;223(1):159–167. doi: 10.1016/j.canlet.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 38.Mayer K, Hieronymus T, Castrop J, Clevers H, Ballhausen WG. Ectopic activation of lymphoid high mobility group-box transcription factor TCF-1 and overexpression in colorectal cancer cells. Int J Cancer. 1997;72(4):625–630. doi: 10.1002/(SICI)1097-0215(19970807)72:4<625::AID-IJC13>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 39.Hatzis P, et al. Genome-wide pattern of TCF7L2/TCF4 chromatin occupancy in colorectal cancer cells. Mol Cell Biol. 2008;28(8):2732–2744. doi: 10.1128/MCB.02175-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bottomly D, Kyler SL, McWeeney SK, Yochum GS. Identification of {beta}-catenin binding regions in colon cancer cells using ChIP-Seq. Nucleic Acids Res. 2010;38(17):5735–5745. doi: 10.1093/nar/gkq363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perego C, Vanoni C, Massari S, Longhi R, Pietrini G. Mammalian LIN-7 PDZ proteins associate with beta-catenin at the cell-cell junctions of epithelia and neurons. EMBO J. 2000;19(15):3978–3989. doi: 10.1093/emboj/19.15.3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dobrosotskaya IY, James GL. MAGI-1 interacts with beta-catenin and is associated with cell-cell adhesion structures. Biochem Biophys Res Commun. 2000;270(3):903–909. doi: 10.1006/bbrc.2000.2471. [DOI] [PubMed] [Google Scholar]
- 43.Kanamori M, et al. The PDZ protein tax-interacting protein-1 inhibits beta-catenin transcriptional activity and growth of colorectal cancer cells. J Biol Chem. 2003;278(40):38758–38764. doi: 10.1074/jbc.M306324200. [DOI] [PubMed] [Google Scholar]
- 44.Alewine C, Olsen O, Wade JB, Welling PA. TIP-1 has PDZ scaffold antagonist activity. Mol Biol Cell. 2006;17(10):4200–4211. doi: 10.1091/mbc.E06-02-0129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mohler PJ, Kreda SM, Boucher RC, Sudol M, Stutts MJ, Milgram SL. Yes-associated protein 65 localizes p62(c-Yes) to the apical compartment of airway epithelia by association with EBP50. J Cell Biol. 1999;147(4):879–890. doi: 10.1083/jcb.147.4.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xing W, Kim J, Wergedal J, Chen ST, Mohan S. Ephrin B1 regulates bone marrow stromal cell differentiation and bone formation by influencing TAZ transactivation via complex formation with NHERF1. Mol Cell Biol. 2010;30(3):711–721. doi: 10.1128/MCB.00610-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao B, et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008;22(14):1962–1971. doi: 10.1101/gad.1664408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chan SW, Lim CJ, Loo LS, Chong YF, Huang C, Hong W. TEADs mediate nuclear retention of TAZ to promote oncogenic transformation. J Biol Chem. 2009;284(21):14347–14358. doi: 10.1074/jbc.M901568200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang H, et al. TEAD transcription factors mediate the function of TAZ in cell growth and epithelial-mesenchymal transition. J Biol Chem. 2009;284(20):13355–13362. doi: 10.1074/jbc.M900843200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Korinek V, et al. Two members of the Tcf family implicated in Wnt/β-catenin signaling during embryogenesis in the mouse. Mol Cell Biol. 1998;18(13):1248–1256. doi: 10.1128/mcb.18.3.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Boyer LA, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122(6):947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu X, Brutlag DL, Liu JS. BioProspector: discovering conserved DNA motifs in upstream regulatory regions of co-expressed genes. Pac Symp Biocomput. 2001;2001:127–138. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.