Abstract
Keeping a delicate balance in the immune system by eliminating invading pathogens, while still maintaining self-tolerance to avoid autoimmunity, is critical for the body’s health. The gut microbiota that resides in the gastrointestinal tract provides essential health benefits to its host, particularly by regulating immune homeostasis. Moreover, it has recently become obvious that alterations of these gut microbial communities can cause immune dysregulation, leading to autoimmune disorders. Here we review the advances in our understanding of how the gut microbiota regulates innate and adaptive immune homeostasis, which in turn can affect the development of not only intestinal but also systemic autoimmune diseases. Exploring the interaction of gut microbes and the host immune system will not only allow us to understand the pathogenesis of autoimmune diseases but will also provide us new foundations for the design of novel immuno- or microbe-based therapies.
Keywords: animal model, antibiotics, autoimmune, commensal, cytokine, germ free, microbiota, mucosal immunity
Introduction
The mammalian gastrointestinal (GI) tract is home to an enormous and complex community of commensal bacteria.1-3 This gut microbial community (microbiota) has co-evolved with its host over millennia and provides benefits to its host in many ways, including, but not limited to, digestion, production of nutrients, detoxification, protection against pathogens and regulation of immune system.1-5 The immune system plays a vital role in keeping the body healthy by providing a fine balance between the elimination of invading pathogens and the maintenance of tolerance to healthy self-tissue. However, in the case of patients with autoimmune disorders, the mechanism to maintain self-tolerance fails and the result is that the immune system mistakenly attacks and destroys healthy self-tissue.6,7
Given the intimate interplay between gut microbiota and the host immune system, it is not surprising that some members of the gut microbiota have been linked to autoimmune diseases. However, only recently has the study of the gut microbiota and autoimmunity become a more navigable field, owing to the ground-breaking advances in “next-generation” sequencing technology, which have now provided culture-independent microbial analysis that greatly facilitates the characterization of these complex commensal communities.8-11 Additionally, extensive progress has been made as investigators have begun to reveal the cellular and molecular interactions between commensals and the mucosal immune system, particularly with the help of animal autoimmune models. This review will discuss the rapidly advancing field of host-microbiota interaction, with particular focus on the role of gut microbiota in immune homeostasis and autoimmune diseases both within and outside the intestine.
Gut Microbiota and Immune Homeostasis
Several approaches have been used to demonstrate that signals derived from gut microbiota are critical for the development of the immune system. Among them, germ-free (GF) models, where animals are reared in a sterile environment and thus have never been exposed to any microorganisms, are a powerful approach that reveals the importance of the microbiota in shaping both innate and adaptive immunity.12 Alternatively, the manipulation of microbiota, either with antibiotic treatment or microbiota reconstitution, also provides key evidence for the role of the microbiota in immune homeostasis.13-18 These approaches are also useful in determining the role of the microbiota in autoimmunity, which will be discussed in a later section. One critical note is that the gut microbiota can regulate not only the local intestinal immune system but also can have a profound influence on systemic immune responses. In this section, we will review how gut microbiota shapes the innate and adaptive immunity to achieve immune homeostasis.
Microbiota and innate immune homeostasis
Antigen presenting cells (APCs)
Having co-evolved with microbiota, a key feature of intestinal APCs is their ability to protect the body against infection while still maintaining immune tolerance to the normal gut microbiota. For example, dendritic cells (DCs) of Peyer's patches (lymphoid nodules embedded in the gut wall) produce high levels of interleukin-10 (IL-10), compared with splenic DCs activated under similar conditions.19 Similar to DCs, gut macrophages are located in close proximity to the intestinal microbiota, and they develop a unique phenotype, so called “inflammation anergy,” referring to the noninflammatory profile of intestinal macrophages when they encounter microbial stimuli in homeostatic conditions.20 For example, intestinal macrophages do not produce pro-inflammatory cytokines in response to microbial stimuli such as Toll-like receptor (TLR) ligands, a set of microbe-associated molecular patterns.21
Several reports provide direct evidence that demonstrates the pivotal role of the gut microbiota in regulating the development of APCs. A reduced number of intestinal but not systemic DCs was observed in GF animals and the monocolonization of GF animals with Escherichia coli was sufficient to recruit DCs to the intestines.22,23 Moreover, microbe-derived ATP has recently been shown to stimulate a subset of DCs that express CD70 and CX3CR1 on their surface, which then induce the differentiation of Th17 cells.24 Intestinal macrophages represent the largest population of tissue macrophages in the body.25 While the numbers of gut macrophages were either normal in GF mice23 or decreased in GF pigs,26 systemic macrophages were reduced in GF pigs.26 Additionally, macrophage functions such as chemotaxis, phagocytosis and microbicidal activities have been shown to be compromised in peritoneal macrophages of GF mice.27,28 GF mice were also devoid of macrophage activation markers such as major histocompatibility complex class II.29
Neutrophils
Neutrophils are a crucial component of the innate immune system and a systemic influence of microbiota in the regulation of neutrophils has been demonstrated. One particularly stark phenotype of GF rats is that they are neutropenic.30 Furthermore, impaired superoxide anion and nitric oxide generation and decreased phagocytic function were also observed in the peripheral blood neutrophils of GF rats.31 Interestingly, the transfer of GF rats back to the conventional or specific pathogen-free (SPF) environment could not restore a normal superoxide anion phenotype. A recent mechanistic study showed that the recognition of peptidoglycan from the gut microbiota by the cytosolic receptor-nucleotide oligomerization domain 1 (NOD1), enhanced the killing activity of bone marrow neutrophils. This data elegantly demonstrated how systemic immunomodulation by intestinal microbiota could be achieved.32
Other innate cell types
Conventional natural killer (NK) cells are innate lymphocytes that can detect and eliminate transformed and infected target cells by producing interferon-γ (IFNγ) or perforin. Recently, studies have identified two types of NK cells that express the NK cell natural cytotoxicity receptor NKp46 in the gut mucosa.33 One type of gut NKp46+ cell closely resembles conventional NK cells; the other type differs from classical NK cells by its limited IFNγ production and absence of perforin. Additionally, these unusual gut NKp46+ cells differ from classical NK cells by their expression of the nuclear hormone receptor retinoic acid receptor-related orphan receptor gamma t (RORγt) and interleukin-22 (IL-22). As GF mice lack IL-22-producing NKp46+ cells, this suggests that the gut microbiota may play a crucial role in promoting IL-22+NKp46+ cell differentiation.34
Mast cells represent 2–3% of lamina propria (LP) cells in the GI tract. Intestinal mast cells have a number of regulatory functions, such as controlling blood flow and coagulation, smooth muscle peristalsis, and permeability and electrolyte exchange by intestinal epithelial cells (IECs).35 GF mice were observed to have lower intestinal mast cell densities and higher mast cell percentages in the blood than conventionally raised mice. Further mechanistic studies have suggested that the gut microbiota can promote the migration of mast cells into the intestine through the induction of CXCR2 ligands from IECs and this promotional effect was dependent on MyD88, an adaptor molecule in the TLR signaling pathway.36
The intestinal epithelium, consisting of a single layer of IECs, provides the primary physical barrier that separates the commensals harbored in the intestinal lumen from the underlying sterile tissue. Aside from their mechanical protective function, IECs, though typically not classified as immune cells, also have a number of immunoregulatory roles such as the secretion of antimicrobial peptides, cytokines and chemokines. A reduced proliferation rate and lower expression of antimicrobial genes of IECs was observed in GF and broad-spectrum antibiotic-treated mice.37,38 These data suggest that the gut microbiota can condition the immunoregulatory roles of IECs by regulating the expression of antimicrobial factors.
Microbiota and adaptive immune homeostasis
T cells
CD4+ T cells are a key component of the adaptive immune system. Intestinal CD4+ T cells are mostly located in the LP of the intestine. Upon stimulation, naive CD4+ T cells can differentiate into four major subtypes: T helper 1 (Th1), Th2, Th17, or regulatory T cell (Treg). These various CD4+ T cell subtypes are distinguished by their expression of various transcription factors and cytokines (Fig. 1). The proper regulation and balance of T-cell subtypes is a crucial factor in determining one’s health status. For example, Th1 cells are critical for the host defense against intracellular microbial infection, while Th2 cells play an important role in eliminating parasite infections. Uncontrolled Th responses can be pathological, as the Th1 and Th17 responses have been linked to autoimmune diseases while the Th2 response has been associated with allergic reactions. Treg is a key mediator of immune tolerance; its dysfunction can lead to autoimmune disorders.
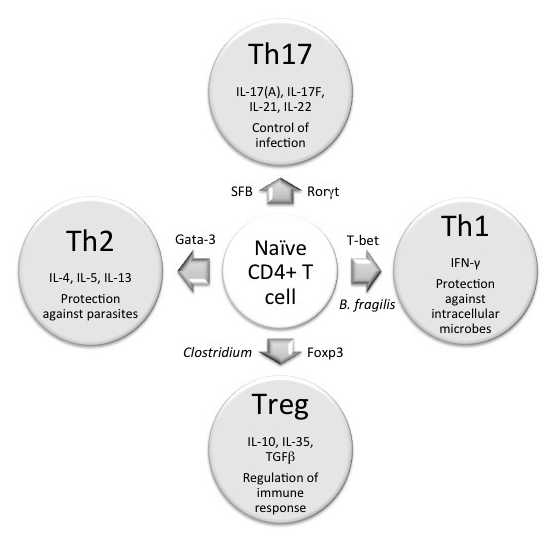
Figure 1. Commensal bacteria induce CD4+T cell differentiation. Naïve CD4+T cells can differentiate into four major cell types: Th1, Th2, Tregs and Th17. The differentiation of each lineage requires the induction of a transcription factor that is unique to each lineage. Once differentiated, each lineage secretes a special (set of) cytokine, as shown in the figure. Th1 cells play an important role in eliminating intracellular pathogens while Th2 function to control parasitic infection. The primary role of Th17 is to control infection and Tregs is to regulate immune response. The type of bacteria species that has been shown to induce a particular T cell differentiation pathway is indicated in the figure.
The gut microbiota plays an important role in the development of CD4+ T cells, both within and outside the intestine. Thus, there is a marked decrease in the number of LP CD4+ cells in GF mice.39 Systemically, the spleens and mesenteric lymph nodes of GF animals also exhibit defects, as lymphocyte zones are absent in these animals.18 GF mice were also observed to have a Th1/Th2 imbalance: their immune response is biased toward the Th2 response. Recent studies even revealed the association of specific bacterial species with the development of particular T-cell subtypes. Bacteroides fragilis was shown to induce the development of a systemic Th1 response through its polysaccharide A (PSA) molecules.18 In contrast, segmented filamentous bacteria (SFB) were found to be potent inducers of LP Th17 cells.17,40
As IL-17 is a crucial pro-inflammatory cytokine, it will be of interest to see if there are other intestinal bacteria that can also induce Th17 cells. Identifying other microbes as Th17 inducers is especially important in humans because a recent report indicated that only a small number of reads corresponding to 0.31% of the mouse SFB genome were identified in the data set of human gut metagenome sequences, suggesting other bacteria may function as the main inducers of human Th17 cells.41,42 Alternatively, it is possible that no detection of SFB in human is a result of the human metagenome data set not including samples from children at weaning periods. In mice, SFB colonization becomes prevalent at the weaning age. If mouse SFB colonize humans at the same time period, fecal samples of weaning children would be required in order to detect SFB in human.41
Recently, Clostridia, particularly those of cluster IV and XIVa, were shown to be capable of promoting the induction of colonic Tregs.43 In another study, TLR9 signaling induced by DNA from the gut microbiota was shown to maintain immune homeostasis by limiting Treg cell conversion in the intestinal sites.44 Interestingly, the PSA of B. fragilis can signal through TLR2 on Tregs to subsequently suppress a Th17 response.45 Lathrop et al. has recently demonstrated that colonic Tregs have a unique TCR repertoire that mostly recognizes the bacteria of colonic contents.46 Moreover, colonic Tregs express low levels of the transcription factor Helios, a putative marker for thymus-derived Treg. If T cells with colonic-specific TCR fail to undergo Treg development and instead become T effector cells, they have the potential to induce colitis. Together, their findings suggest that most colonic Tregs are of peripheral origin and are educated by the gut microbiota to be tolerant to commensal-derived foreign antigen. In summary, a shift in the composition of the gut microbiota can cause either a pathological or beneficial outcome mediated by the regulation of particular CD4+ T cell subtypes induced by the gut microbiota.
Intestinal CD8+ T cells are mostly found in the intraepithelial compartment of the gut. A reduced number and decreased cytotoxicity of intestinal CD8+ T cells in GF mice indicate that signals from the microbiota are critical in maintaining the population and function of intestinal CD8+ T cells.47-49 These defects might be due in part to the impaired clonal expansion of intraepithelial CD8+ cells in GF mice. Though not required for shaping the systemic CD8+ T-cell repertoire, the gut microbiota plays an important role in conditioning CD8+ T cells to modulate other peripheral immune cells, such as marginal zone B cells, plasmacytoid DCs, and invariant natural killer T cells.50-52
Gamma delta (γδ) T cells are often considered to be the bridge between the innate and adaptive immunity. The percentage of γδ T cells among intestinal intraepithelial lymphocytes is quite high, compared with their percentage in the lymph nodes or spleen (50% vs. 1–5%).53 Intestinal intraepithelial γδ T cells express CD8 and have lytic activity.48 Although the absence of commensal microbiota had little effect on the pool size and characteristics of γδ T cells, the cytolytic activity of γδ T cells was reduced in GF mice, suggesting a key role of microbiota in maintaining the function of γδ T cells.
B cells
Gut-associated B cells can mostly be found in the Peyer’s patches, most of which are immunoglobulin (Ig) A-secreting plasma cells. An estimated 0.8 g of IgA per meter of intestine is secreted each day, considerably exceeding the combined production of all other Ig classes.54 The number and cellularity of the Peyer’s patches were significantly reduced in GF animals and as a result, a decreased level of IgA and reduced number of plasma cells were observed in the intestine of GF animals.55 Thus, the gut microbiota is a major driving force for mucosal IgA production; a large dose (109 colony-forming unit or CFU) of live bacteria was required to induce a high titer of secretory IgA in GF mice.56 Mucosal IgA induction lacks a memory response, which explains the recognition of intestinal IgA to mostly the current existing microbiota. Systemically, the spleens of the GF mice also contain fewer and smaller germinal centers, where the differentiation and affinity maturation of B cells occur.57 Accordingly, serum natural IgG level was severely reduced while serum natural IgM level was normal in GF animals.58,59 Interestingly, the allergy-associated Ig isotype, IgE was found to be increased locally in the intestine as well as systemically in the serum of GF rats.60 This observation is consistent with the Th2-predisposed phenotype of GF animals, which can promote natural IgE induction, a Th2 humoral immune response.
Gut Microbiota and Autoimmunity
As the gut microbiota has such profound effects on both the innate and adaptive immune system, it is not surprising that some members of the gut microbiota have been linked to autoimmune diseases. Significant attention has been focused on the role of gut microbiota in GI-related autoimmune diseases. Remarkably, as discussed earlier, the gut microbiota has a role beyond the local gut immune system and impacts many systemic immune components. Accordingly, recent studies have also unraveled the effects of gut microbiota in extraintestinal diseases. Here, the roles of intestinal microbiota in autoimmune disorders both within and outside the gut will be discussed (summarized in Table 1). In particular, we focus on studies that show how changing in a single microbial species and/or global commensal communities can alter the outcome of autoimmune diseases by tipping the balance between a pathological or protective immune response.
Table 1. Effect of gut microbiota on autoimmune diseases.
| Disease | Animal model | Manipulation method of microbiota (signal) | Effects in GF animal | Reference |
|---|---|---|---|---|
| IBD |
IL-2−/− |
GF |
less severity |
64 |
| |
TCRαβ−/− |
GF |
no disease |
66 |
| |
IL-10−/− |
GF |
no disease |
65 |
| |
Helicobacter hepaticus-induced colitis in Rag−/− |
introducing B. fragilis |
less severity |
73 |
| |
DSS-induced colitis |
introducing Clostridium |
less severity |
43 |
| RA |
IL-1Rn−/− |
GF |
no disease |
84 |
| |
IL-1Rn−/− |
introducing Lactobacillus bifidus to ex-GF IL-1Rn−/− mice |
restores disease |
84 |
| |
TLR2−/−IL-1Rn−/− |
n/a |
increased severity |
84 |
| |
TLR4−/−-IL-1Rn−/− |
n/a |
less severity |
84 |
| |
K/BxN |
GF |
less severity |
16 |
| |
K/BxN |
introducing SFB to ex-GF K/BxN |
restores disease |
16 |
| T1D |
NOD |
GF |
severe disease to no difference* |
88–90,92,93 |
| |
NOD female |
SFB natural colonization |
protection by SFB |
91 |
| |
NOD male |
SFB natural colonization |
no difference |
91 |
| |
MyD88−/−NOD |
GF |
severe disease |
90 |
| |
MyD88−/−NOD |
n/a |
no disease |
90 |
| Multiple sclerosis |
EAE |
GF |
less severity |
95 |
| |
EAE |
introducing SFB to GF EAE |
restores disease |
95 |
| |
EAE |
introducing B. fragilis |
less severity |
96 |
| |
Established EAE |
introducing 3 strains of Lactobacillus |
therapeutic effect |
99 |
| APECED |
AIRE−/− |
GF |
no difference |
101 |
| |
MyD88−/−Aire−/− |
n/a |
no difference |
101 |
| Systemic lupus erythematosus |
MRL/1pr |
GF |
no difference |
102 |
| Autoimmune gastritis | AID−/− | GF | no difference | 103 |
The discrepancy of difference in disease severity between GF and SPF NOD mice is likely due to the variability of microbiota composition among different SPF facilities.
Gut microbiota and GI-associated autoimmune disease
Inflammatory bowel disease (IBD)
An autoimmune disorder that affects the GI tract, IBD consists of two main forms: Crohn disease and ulcerative colitis. Several lines of compelling evidence indicate that bacteria play a critical role in the pathogenesis of IBD. For example, patients with IBD, as well as animal IBD models, often benefit from antibiotic treatment.61,62 In addition, the phyla of gut microbiota differ greatly in patients with IBD when compared with normal adults.63 Importantly, many IBD animal models show either a milder form of disease (such as in the IL-2−/− IBD model) or are protected against disease (such as in the IL-10−/− or T-cell receptor α/β−/− IBD models) after GF rederivation, which indicates that the normal gut microbiota contributes to the inflammatory state of IBD.64-66 Recently, progress has been made in identifying the dysbiosis of specific microbiota in IBD patients. A reduction in Firmicutes and Bacteroides species and an overgrowth of proteobacteria has been characterized in IBD patients.67,68 Interestingly, similar changes in the microbial communities were found in a mouse model of acute colitis, where inflammation was induced by the adoptive transfer of transgenic CD8+ T cells that attacked the intestinal epithelium.69
Despite compelling evidence that demonstrates dysbiosis in patients and animals with IBD, it is difficult to assign host-predisposing factors that cause dysbiosis. However, two animal studies have elegantly demonstrated that intestinal inflammation can be the major cause of dysbiosis, leading to the selection of microbiota species with a colitogenic phenotype. T-bet is a member of the T-box transcription factor family that plays a crucial role in the regulation of immune cells. T-bet−/−Rag−/− ulcerative colitis (TRUC) mice have colonic inflammation that resembles ulcerative colitis in humans.70 The colitis in TRUC mice is driven by the overproduction of the pro-inflammatory cytokine TNF-α by colonic DCs and transfer of the microbiota from TRUC mice into wild type recipients transmits colitis. A later study discovered that the presence of Klebsiella pneumoniae and Proteus mirabilis in TRUC mice can elicit colitis in SPF but not GF wildtype mice.71 This suggested that Klebsiella pneumoniae and Proteus mirabilis worked in concert with other members of the endogenous microbial community to induce inflammation.
Another study using mice deficient in the inflammasome pathway also highlights the importance of inflammation as a major cause of dysbiosis and disease. Inflammasomes are cytoplasmic multiprotein complexes that are composed of one of several nucleotide-binding oligomerization domain-like receptor proteins (NLRP), which function as sensors for stress stimuli. In NLRP−/− mice, a defect in the inflammasome pathway resulted in an alteration of the gut microbiota—specifically, an increase of Prevotella and TM7 species, rendering the NLRP−/− animal susceptible to colitis.72 As in the TRUC mice, the gut microbiota in NLRP−/− mice can also cause the disease in wildtype animals. Importantly, both animal studies suggest that once certain colitis-associated microbiota is created in IBD-susceptible animals, they can transmit colitis horizontally to even cause disease in wild-type animals that are not genetically predisposed to IBD. These findings highlight that aggressive microbiota species can be the real cause rather than just the result of a disease.
Not surprisingly, there are also “beneficial” commensal bacteria that can ameliorate disease. For example, B. fragilis can reduce the colitis induced by Helicobacter hepaticus in immunocompromised mice through its production of PSA, which suppresses disease by both stimulating the anti-inflammatory IL-10 production from CD4+ T cells and downregulating the pro-inflammatory IL-17 production in the colonic tissue.73 Bacteroides thetaiotaomicron was also demonstrated to attenuate Salmonella enterica-induced inflammation by enhancing the nuclear export of peroxisome proliferator activated receptor-γ (PPAR-γ), a transcription factor that plays key roles in the regulation of lipid metabolism and inflammation.74,75 Short-chain fatty acids (SCFAs) produced by the gut microbiota have also been shown to reduce inflammation in the dextran sulfate sodium (DSS)-induced colitis model. This anti-inflammatory effect required the interaction of SCFAs with G-protein-coupled receptor 43 expressed on immune cells.76 The introduction of Clostridium upregulated the colon Treg population and coincided with the reduction of DSS-induced colitis, suggesting that Tregs might be responsible for the anti-inflammatory effects mediated by Clostridium.43
An elegant study by Feng et al. demonstrated that microbiota-derived innate and TCR specific signals are both required for the induction of disease using a murine model of IBD.77 Accordingly, it was shown that homeostatic proliferation of transferred T cells were only observed in SPF but not in GF Rag−/− mice, indicating that the presence of the gut microbiota is required for the T cell proliferation. This microbiota-mediated T cell proliferation requires a MyD88-dependant IL-6 induction in DCs. Additionally, transfer of CD4+ T cells from CBir1 TCR transgenic mice that have a TCR specific for the microbiota flagellin, CBir1, induces colitis in SPF Rag−/− mice. This disease induction is driven by an antigen specific response as co-transfer of CBir1 T cells with an abundance of OT-II transgenic T cells that recognizes ovalbumin (which does not exist in the gut lumen) ameliorates the colitis development due to a lack of cognate antigen recognition in the gut. These data indicate that microbiota-mediated T cell spontaneous proliferation and antigen-specific T-cell activation both contribute to the disease pathogenesis.
Gut microbiota and extraintestinal autoimmune disorders
Rheumatoid arthritis (RA)
RA is an autoimmune disease that causes chronic inflammation of the joints and affects approximately 1% of the world’s population. The low concordance rate of RA in monozygotic twins (15%) compared with other autoimmune diseases such as type I diabetes (~50%) suggests that environmental factors must play a crucial role in the etiopathogenesis of RA.78,79 Among the possible environmental triggers of RA, the microbes we encounter in our surroundings are a likely candidate.80 Attention has mostly been devoted to disease correlations with infectious microbes81 until recently; a dysbiosis of gut microbial communities have been reported in patients with early (< 6 mo duration) RA when compared with patients of fibromyalgia, as assessed from the 16S rRNA composition of fecal samples.82 Additionally, the therapeutic effect of some antibiotics (i.e., sulfasalazine and minocycline) for some RA patients may be related to the bactericidal activity of these molecules, as they are likely modulating gut microbiota.
Early GF studies using different RA models showed a discrepant role of the microbiota on disease severity, ranging from inhibition to augmentation.83 However, the significance of these studies is difficult to assess because in general, they relied on the administration of bacterial products (often Complete Freund’s Adjuvant, or CFA) for the induction of disease, which could complicate the effect of commensal bacteria on RA. More recently, a spontaneous T cell-mediated arthritis model, the IL-1 receptor antagonist deficient (IL-1Rn−/−) mouse model, was used to examine the importance of microbiota in autoimmune arthritis.84 The gut microbiota was required for the arthritis development in IL-1Rn−/− mice as GF IL-1Rn−/− mice did not develop disease. Additionally, monocolonization of GF IL-1Rn−/− mice with Lactobacillus bifidus restored the disease. The reduction of Tregs and Th17 cells in the spleens and lymph nodes were found to be associated with disease enhancement in non-GF TLR2−/− IL-1Rn−/− mice and disease amelioration in non-GF TLR4−/− IL-1Rn−/− mice. Together, these observations suggest that the gut microbiota or microbial signals such as TLR stimuli can affect a non-gut disease by regulating systemic immune components. However, it is not clear how the microbiota located in the gut can modulate systemic immune cells, which in turn regulate a non-gut disease.
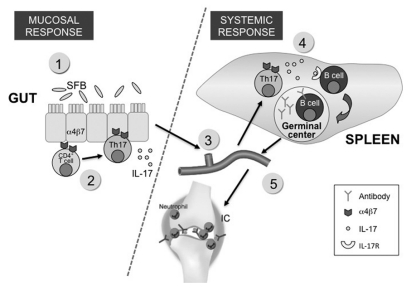
Recently, the K/BxN mouse, another spontaneous arthritis model, was used to provide a mechanism to explain the missing links among gut microbiota, systemic immunity and autoimmune arthritis.16 At first, arthritis was shown to be reduced in K/BxN mice housed under GF conditions, which indicated a pathological role for microbiota. Mechanistic studies then revealed that a distal augmentation of disease by gut microbiota on the joints was made possible through the gut microbiota-mediated induction of LP Th17 cells of small intestine, which subsequently migrate into the peripheral lymphoid tissue and secrete IL-17 (Fig. 2). IL-17, in turn, acts directly on B cells to provide help in systemic B cell differentiation and autoantibody production that ultimately lead to the development of disease. Importantly, the introduction of a single gut microbiota species, SFB, into GF K/BxN mice was able to trigger disease development. The tie lies within the unique ability of SFB to induce the robust differentiation of LP Th17 cells. The mechanism of how the gut microbiota exerts its effects at systemic sites remains largely unknown. While it was postulated that the effect of the microbiota on the systemic immune response is mediated by the circulation of microbiota-derived soluble factors from the gut into the periphery,32 the K/BxN study clearly provides an alternative mechanism where microbiota-derived products can affect the immune system at a distal sites without leaving the gut.
Figure 2. An autoimmune arthritis model that demonstrates the link between gut microbiota and an extraintestinal disease. The K/BxN arthritis model was used to demonstrate how the gut microbiota can influence a non-gut-associated disease. K/BxN mice express a transgene-encoded T-cell receptor that reacts to a self-peptide. Colonization of SFB on the gut induces the differentiation of Th17 cells (step 1 and 2), which subsequently exit the gut and migrate into the peripheral lymphoid tissue. The gut-origin of Th17 cells can be identified by their expression of the α4β7 receptor, imprinted on these T cells by intestinal-mucosa-associated DCs (step 3). IL-17, in turn, acts directly on B cells to provide help in the differentiation of germinal center B cells and the production of autoantibody in spleen (step 4). The autoantibody then circulates into its target organ joints, which ultimately leads to the development of disease.
Type 1 diabetes (T1D)
T1D is an autoimmune disease that results from T cell-mediated destruction of insulin-producing β-cells in the pancreas. A significant reduction of intestinal Tregs was observed in T1D patients, suggesting the possible involvement of the gut microbiota in T1D.85 While many of the autoimmune models mentioned above display a weaker disease phenotype in the GF environment, T1D—especially in the prototypic spontaneous NOD mouse model—provides a clear exception to this “rule.” The diabetic incidence of NOD mice in the GF facility is often significantly higher when compared with their SPF counterparts, an observation that is consistent with the finding that T1D is more prevalent in countries with stringent hygiene practices.86-89 In non-GF conditions, disease incidence can vary by facility but is still generally higher in the NOD females than males.88 Consistent with the above results, another study indicated that the gut microbiota might have a protective role, as MyD88−/− NOD mice have been shown to be protected from diabetes onset in an SPF environment.90 Interestingly, the protective effect of MyD88 deficiency required the presence of gut microbiota, since MyD88−/− NOD mice readily developed diabetes in the GF facility. These results indicate that the protective effect of MyD88 deficiency is not due to the prevention of MyD88 signaling from detrimental bacteria but rather the induction of MyD88-independent signaling from the expansion of beneficial bacteria, which would have been otherwise kept in check by MyD88. Moreover, an attenuation of diabetes was observed in GF NOD mice colonized with the microbiota from SPF MyD88−/− NOD mice. An increase in Bacteroidetes was identified in SPF MyD88−/− NOD mice, which suggests a role of immunoregulation by these gut commensals.
Despite the higher disease incidence observed in female NOD mice, some of them never develop diabetes under SPF housing conditions. A more recent study reported that a single commensal species, SFB, can protect female NOD mice against diabetes.91 The authors observed a strong cosegregation of SFB-colonization and diabetes protection in NOD females where males were highly protected regardless of their SFB status. As in other experimental contexts, SFB also promoted a robust induction of SI-LP Th17 cells in NOD mice. While Th17 cells appeared to be likely mediators for SFB-associated protection in female NOD mice, male NOD mice that lacked SFB and Th17 cells still displayed a low disease incidence when compared with GF male NOD mice, suggesting that other microbial species and immunoregulatory pathways are responsible for the protection of NOD males from diabetes. In addition, the role of IL-17 in diabetes is still a topic of debate. The effect of IL-17-producing cells on diabetes ranges from inhibition to even exacerbation of disease. Importantly, this study also offered an explanation for the discrepancy in disease severity between GF and SPF NOD mice observed by several groups.88-90,92,93 Because of a near complete penetrance of diabetes in SFB-negative NOD females and an almost full protection from diabetes onset in SFB-positive NOD females, one can expect that disease exacerbation in GF mice will be more obvious when comparing a GF to a SPF-housed SFB-positive but not SFB-negative NOD colony.
Experimental autoimmune encephalomyelitis (EAE)
EAE is a mouse model of multiple sclerosis (MS), where an autoimmune response causes demyelination in the central nervous system (CNS). Although EAE is generally accepted as a murine model for human MS, the pathological mechanism of EAE might differ significantly from human MS, as EAE is not a spontaneous model and disease induction requires the bacterial adjuvant CFA. With these points in mind, there is still valuable information that has been obtained using the EAE model and several studies have indicated a role for microbiota in EAE. Antibiotic-mediated modification of the gut microbiota can significantly dampen the disease severity.94 GF mice induced for EAE had an attenuated disease phenotype, which is consistent with their lower production of pro-inflammatory cytokines, such as IL-17.95 Finally, monocolonization of GF mice with SFB increased the number of Th17 cells in the CNS and restored their development and progression of EAE, suggesting a pathological role for the SFB in EAE.
In contrast, some commensals can have a beneficial role in EAE development. The introduction of the human commensal B. fragilis can ameliorate disease through its expression of PSA.96,97 This protection was associated with an enhanced number of Treg cells and CD5+ B cells in the B. fragilis-treated group.96,98 Excitingly, it was found that treatment with a combination of three Lactobacillus strains, L. paracasei DSM 13434, L. plantarum DSM 15312 and DSM 15313, suppressed and reversed the clinical symptoms of established EAE, and IL-10-producing Tregs were found to be involved in this Lactobacillus-mediated therapeutic effect.99
Microbiota-independent autoimmune disease
Although many autoimmune diseases result from the interaction of both genetic and environmental factors, there are some exceptions. Sometimes, only the genetic factor contributes to disease development. Accordingly, it is important to keep in mind that the severity of some autoimmune disorders does not differ depending on the presence or absence of commensal bacteria, such as in autoimmune regulator (Aire) deficient mice. The Aire−/− mouse is an animal model of human autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy (APECED), a polyendocrine autoimmune disease that occurs from mutations in the Aire protein, a transcriptional regulator that plays an important role in T cell tolerance induction in the thymus.100 Re-deriving Aire−/− mice into a GF condition or crossing these mice into a MyD88−/− background did not alter the disease phenotype of Aire−/− mice, suggesting that the breaking of central tolerance in the thymus alone can lead to autoimmunity that overrides peripheral tolerance mechanisms without the need for microbial stimulation.101
Additionally, the MRL/lpr mouse model of human systemic lupus erythematosus and activation-induced cytidine deaminase (AID) deficient mice, an autoimmune gastritis model, both exhibit comparable disease phenotypes in the GF condition.102,103 These findings demonstrate that live commensal organisms are not involved in the pathology observed in these models and it is primarily the genetic factors that play the major role in the development of some autoimmune diseases. However, these studies do not exclude the possibility of immune stimulation by low level of microbe-derived products in the diet, that may fill in the requirement of environment stimuli for the development of disease in GF mice.103
Human Practices that Influences the Composition of Microbiota Communities
There are many practices adapted during the development of human civilization that pose a dominant effect on the composition of gut microbiota. Serious attention needs to be paid to these practices because alterations in the gut microbiota cannot only impact the development of autoimmune diseases, as discussed in this review, but can also affect many other health-related issues, such as allergy and obesity. For example, dietary habit is one of the major factors influencing the diversity of gut microbiota. By using GF mice that were fecal-transplanted with human gut microbiota, one study demonstrated that switching from a low-fat, plant polysaccharide-rich diet to a high-fat, high-sugar diet can shift the configuration of the microbiome in one day.104 This diet-altered microbiome was able to rapidly promote obesity in the mice within two weeks.
In another study, the gut microbiota was compared in fecal samples of children from Europe and rural Africa.105 The diet of African children is rich in fiber, starch and plant polysaccharides and low in fat and animal protein, whereas the diet of European children is high in sugar, starch and fat and low in fiber. As compared with the European cohort, the microbiota of the African cohort showed a significant depletion in Firmicutes and an increase in Bacteroidetes. Interestingly, species of the bacteria Prevotella and Xylanibacter, which are known to encode genes required for metabolizing plant polysaccharide, were observed in the African cohort but completely absent in the European cohort. A significantly higher level of anti-inflammatory molecules, like SCFAs, was also found in the African cohort. Moreover, an animal study showed that NOD mice fed with a special soy-based diet had a significantly lower incidence of diabetes, which was associated with reduced pro-inflammatory cytokines, IL-17 and IL-23 in colon.106
Antibiotic treatments, vaccinations and hygiene practices all can alter gut microbiota composition. Antibiotic use was associated with the reduction of Bacteroides and Bifidobacterium and the outgrowth of Campylobacter, Streptococcus, Leuconostoc or yeasts such as Candida albicans in the intestinal microbial communities.14,107-109 As we are born sterile, bacterial colonization during and shortly after birth also plays an important role in shaping the communities of gut microbiota. Thus, the prematurity of an infant’s birth, the method of delivery, and the infant’s food source (e.g., breast milk, commercial formula, etc.) all have a major impact during the acquisition phase of the gut microbiota development. For example, vaginally-born infants were dominantly colonized by bacteria communities that resembled their mother’s vaginal microbiota which includes Lactobacillus, Prevotella, or Sneathia spp, while caesarean section (C-section)-born infants harbored bacteria mostly found on the skin surface such as Staphylococcus, Corynebacterium and Propionibacterium spp110 Furthermore, premature infants had a predominant colonization of C. difficile.109 Formula-fed infants were often associated with the colonization of Staphylococci, E. coli, C. difficile, Bacteroides, Atopobium and Lactobacilli and a delayed colonization of Bifidobacterium species.109,111-115 Some of the changes in microbial communities in early life due to certain practices such as C-section might increase one’s risk in developing asthma, allergy and autoimmune disease in the later childhood.116-118
Conclusion
The influence of commensals on health and disease through the regulation of immune function has emerged as an area of scientific and clinical importance. The recent advancements in “next-generation” sequencing have led to a revolution in developing a culture-independent and thorough method to characterize gut microbial communities. It is now evident that the gut microbiota has a profound effect on the host immune system and can affect autoimmune-related diseases both within and outside the gut. Aside from the genetic factors, environmental factors play an important role in shaping the microbiota as well. These factors should be treated with caution as inappropriate practices such as overuse of antibiotics might increase the risk of autoimmune disease by the microbiota-mediated immunomodulation.
The challenge lying ahead is to distinguish cause from effect, i.e., whether the gut microbiota is the cause of the disease or a result of the disease status. The use of animal models where the intestinal flora can be manipulated, such as in GF animals, provides a power tool for such mechanistic studies. Another daunting task is to consider the effect of the intestinal microbiota on the results of every animal experiment, as the composition of the microbiota can vary in different animal facilities: we know now that even a change in a single bacterial species within the gut can have a drastic impact on host immunity and pathology. Greater attention will be necessary in order to interpret results and compare published studies correctly. This practice has already been demonstrated to be critical when comparing the disease incidence of GF NOD mice with NOD mice from various SPF facilities.91 Understanding the interaction of gut microbes with the host immune system is a timely and important health topic as the rate of many diseases such as numerous immune disorders are rising at an alarmingly high speed and may result from dysbiosis of commensals.86
Acknowledgments
H.-J.W. is supported by the Arthritis National Research Foundation. We thank G. Yuen for her critical review and editorial assistance. We also wish to thank Dr L. Lybarger, Dr J. Nikolich-Zugich, Dr M. Kuhns, Dr V. Viswanathan, Dr M. Kriegel, Dr W. Fu, and Dr T. Feng for their critical reading of the manuscript.
Glossary
Abbreviations:
- GI
gastrointestinal
- GF
germ-free
- APC
antigen presenting cell
- DC
dendritic cell
- IL
interleukin
- TLR
Toll-like receptor
- SPF
specific pathogen-free
- NOD1
nucleotide oligomerization domain 1
- NK
natural killer
- IFNγ
interferon-gamma
- RORγt
retinoic acid receptor-related orphan receptor gamma t
- LP
lamina propria
- IEC
intestinal epithelial cell
- Th
T helper
- Treg
regulatory T cell
- PSA
polysaccharide A
- SFB
segmented filamentous bacteria
- γδ
gamma delta
- Ig
immunoglobulin
- IBD
inflammatory bowel disease
- TRUC
T-bet−/−Rag−/− ulcerative colitis
- NLRP
nucleotide-binding oligomerization domain-like receptor proteins
- PPARγ
peroxisome proliferator activated receptor-gamma
- SCFA
short-chain fatty acid
- DSS
dextran sulphate sodium
- RA
rheumatoid arthritis
- CFA
complete Freund’s adjuvant
- T1D
type-1 diabetes
- EAE
experimental autoimmune encephalomyelitis
- MS
multiple sclerosis
- CNS
central nervous system
- APECED
autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy
- AID
activation-induced cytidine deaminase
- C-section
caesarean section
Footnotes
Previously published online: www.landesbioscience.com/journals/gutmicrobes/article/19320
References
- 1.Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124:837–48. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 2.Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI. Worlds within worlds: evolution of the vertebrate gut microbiota. Nat Rev Microbiol. 2008;6:776–88. doi: 10.1038/nrmicro1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bäckhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–20. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 4.Finke D. Induction of intestinal lymphoid tissue formation by intrinsic and extrinsic signals. Semin Immunopathol. 2009;31:151–69. doi: 10.1007/s00281-009-0163-6. [DOI] [PubMed] [Google Scholar]
- 5.Hill DA, Artis D. Intestinal bacteria and the regulation of immune cell homeostasis. Annu Rev Immunol. 2010;28:623–67. doi: 10.1146/annurev-immunol-030409-101330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goodnow CC, Sprent J, Fazekas de St Groth B, Vinuesa CG. Cellular and genetic mechanisms of self tolerance and autoimmunity. Nature. 2005;435:590–7. doi: 10.1038/nature03724. [DOI] [PubMed] [Google Scholar]
- 7.Westerberg LS, Klein C, Snapper SB. Breakdown of T cell tolerance and autoimmunity in primary immunodeficiency--lessons learned from monogenic disorders in mice and men. Curr Opin Immunol. 2008;20:646–54. doi: 10.1016/j.coi.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature. 2007;449:804–10. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grice EA, Kong HH, Conlan S, Deming CB, Davis J, Young AC, et al. NISC Comparative Sequencing Program Topographical and temporal diversity of the human skin microbiome. Science. 2009;324:1190–2. doi: 10.1126/science.1171700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–4. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith K, McCoy KD, Macpherson AJ. Use of axenic animals in studying the adaptation of mammals to their commensal intestinal microbiota. Semin Immunol. 2007;19:59–69. doi: 10.1016/j.smim.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 13.Sekirov I, Tam NM, Jogova M, Robertson ML, Li Y, Lupp C, et al. Antibiotic-induced perturbations of the intestinal microbiota alter host susceptibility to enteric infection. Infect Immun. 2008;76:4726–36. doi: 10.1128/IAI.00319-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hill DA, Hoffmann C, Abt MC, Du Y, Kobuley D, Kirn TJ, et al. Metagenomic analyses reveal antibiotic-induced temporal and spatial changes in intestinal microbiota with associated alterations in immune cell homeostasis. Mucosal Immunol. 2010;3:148–58. doi: 10.1038/mi.2009.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–41. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 16.Wu HJ, Ivanov II, Darce J, Hattori K, Shima T, Umesaki Y, et al. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 2010;32:815–27. doi: 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–98. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–18. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 19.Iwasaki A, Kelsall BL. Freshly isolated Peyer’s patch, but not spleen, dendritic cells produce interleukin 10 and induce the differentiation of T helper type 2 cells. J Exp Med. 1999;190:229–39. doi: 10.1084/jem.190.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smythies LE, Sellers M, Clements RH, Mosteller-Barnum M, Meng G, Benjamin WH, et al. Human intestinal macrophages display profound inflammatory anergy despite avid phagocytic and bacteriocidal activity. J Clin Invest. 2005;115:66–75. doi: 10.1172/JCI19229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smythies LE, Shen R, Bimczok D, Novak L, Clements RH, Eckhoff DE, et al. Inflammation anergy in human intestinal macrophages is due to Smad-induced IkappaBalpha expression and NF-kappaB inactivation. J Biol Chem. 2010;285:19593–604. doi: 10.1074/jbc.M109.069955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haverson K, Rehakova Z, Sinkora J, Sver L, Bailey M. Immune development in jejunal mucosa after colonization with selected commensal gut bacteria: a study in germ-free pigs. Vet Immunol Immunopathol. 2007;119:243–53. doi: 10.1016/j.vetimm.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 23.Williams AM, Probert CS, Stepankova R, Tlaskalova-Hogenova H, Phillips A, Bland PW. Effects of microflora on the neonatal development of gut mucosal T cells and myeloid cells in the mouse. Immunology. 2006;119:470–8. doi: 10.1111/j.1365-2567.2006.02458.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Atarashi K, Nishimura J, Shima T, Umesaki Y, Yamamoto M, Onoue M, et al. ATP drives lamina propria T(H)17 cell differentiation. Nature. 2008;455:808–12. doi: 10.1038/nature07240. [DOI] [PubMed] [Google Scholar]
- 25.Lee SH, Starkey PM, Gordon S. Quantitative analysis of total macrophage content in adult mouse tissues. Immunochemical studies with monoclonal antibody F4/80. J Exp Med. 1985;161:475–89. doi: 10.1084/jem.161.3.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang W, Wen K, Azevedo MS, Gonzalez A, Saif LJ, Li G, et al. Lactic acid bacterial colonization and human rotavirus infection influence distribution and frequencies of monocytes/macrophages and dendritic cells in neonatal gnotobiotic pigs. Vet Immunol Immunopathol. 2008;121:222–31. doi: 10.1016/j.vetimm.2007.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mørland B, Midtvedt T. Phagocytosis, peritoneal influx, and enzyme activities in peritoneal macrophages from germfree, conventional, and ex-germfree mice. Infect Immun. 1984;44:750–2. doi: 10.1128/iai.44.3.750-752.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mitsuyama M, Ohara R, Amako K, Nomoto K, Yokokura T, Nomoto K. Ontogeny of macrophage function to release superoxide anion in conventional and germfree mice. Infect Immun. 1986;52:236–9. doi: 10.1128/iai.52.1.236-239.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mikkelsen HB, Garbarsch C, Tranum-Jensen J, Thuneberg L. Macrophages in the small intestinal muscularis externa of embryos, newborn and adult germ-free mice. J Mol Histol. 2004;35:377–87. doi: 10.1023/B:HIJO.0000039840.86420.b7. [DOI] [PubMed] [Google Scholar]
- 30.Ohkubo T, Tsuda M, Tamura M, Yamamura M. Impaired superoxide production in peripheral blood neutrophils of germ-free rats. Scand J Immunol. 1990;32:727–9. doi: 10.1111/j.1365-3083.1990.tb03216.x. [DOI] [PubMed] [Google Scholar]
- 31.Ohkubo T, Tsuda M, Suzuki S, El Borai N, Yamamura M. Peripheral blood neutrophils of germ-free rats modified by in vivo granulocyte-colony-stimulating factor and exposure to natural environment. Scand J Immunol. 1999;49:73–7. doi: 10.1046/j.1365-3083.1999.00456.x. [DOI] [PubMed] [Google Scholar]
- 32.Clarke TB, Davis KM, Lysenko ES, Zhou AY, Yu Y, Weiser JN. Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat Med. 2010;16:228–31. doi: 10.1038/nm.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Satoh-Takayama N, Vosshenrich CA, Lesjean-Pottier S, Sawa S, Lochner M, Rattis F, et al. Microbial flora drives interleukin 22 production in intestinal NKp46+ cells that provide innate mucosal immune defense. Immunity. 2008;29:958–70. doi: 10.1016/j.immuni.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 34.Sanos SL, Bui VL, Mortha A, Oberle K, Heners C, Johner C, et al. RORgammat and commensal microflora are required for the differentiation of mucosal interleukin 22-producing NKp46+ cells. Nat Immunol. 2009;10:83–91. doi: 10.1038/ni.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bischoff SC. Physiological and pathophysiological functions of intestinal mast cells. Semin Immunopathol. 2009;31:185–205. doi: 10.1007/s00281-009-0165-4. [DOI] [PubMed] [Google Scholar]
- 36.Kunii J, Takahashi K, Kasakura K, Tsuda M, Nakano K, Hosono A, et al. Commensal bacteria promote migration of mast cells into the intestine. Immunobiology. 2011;216:692–7. doi: 10.1016/j.imbio.2010.10.007. [DOI] [PubMed] [Google Scholar]
- 37.Khoury KA, Floch MH, Hersh T. Small intestinal mucosal cell proliferation and bacterial flora in the conventionalization of the germfree mouse. J Exp Med. 1969;130:659–70. doi: 10.1084/jem.130.3.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reikvam DH, Erofeev A, Sandvik A, Grcic V, Jahnsen FL, Gaustad P, et al. Depletion of murine intestinal microbiota: effects on gut mucosa and epithelial gene expression. PLoS One. 2011;6:e17996. doi: 10.1371/journal.pone.0017996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Macpherson AJ, Martinic MM, Harris N. The functions of mucosal T cells in containing the indigenous commensal flora of the intestine. Cell Mol Life Sci. 2002;59:2088–96. doi: 10.1007/s000180200009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gaboriau-Routhiau V, Rakotobe S, Lécuyer E, Mulder I, Lan A, Bridonneau C, et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31:677–89. doi: 10.1016/j.immuni.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 41.Kuwahara T, Ogura Y, Oshima K, Kurokawa K, Ooka T, Hirakawa H, et al. The lifestyle of the segmented filamentous bacterium: a non-culturable gut-associated immunostimulating microbe inferred by whole-genome sequencing. DNA Res. 2011;18:291–303. doi: 10.1093/dnares/dsr022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sczesnak A, Segata N, Qin X, Gevers D, Petrosino JF, Huttenhower C, et al. The genome of th17 cell-inducing segmented filamentous bacteria reveals extensive auxotrophy and adaptations to the intestinal environment. Cell Host Microbe. 2011;10:260–72. doi: 10.1016/j.chom.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–41. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hall JA, Bouladoux N, Sun CM, Wohlfert EA, Blank RB, Zhu Q, et al. Commensal DNA limits regulatory T cell conversion and is a natural adjuvant of intestinal immune responses. Immunity. 2008;29:637–49. doi: 10.1016/j.immuni.2008.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Round JL, Lee SM, Li J, Tran G, Jabri B, Chatila TA, et al. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science. 2011;332:974–7. doi: 10.1126/science.1206095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lathrop SK, Bloom SM, Rao SM, Nutsch K, Lio CW, Santacruz N, et al. Peripheral education of the immune system by colonic commensal microbiota. Nature. 2011;478:250–4. doi: 10.1038/nature10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Imaoka A, Matsumoto S, Setoyama H, Okada Y, Umesaki Y. Proliferative recruitment of intestinal intraepithelial lymphocytes after microbial colonization of germ-free mice. Eur J Immunol. 1996;26:945–8. doi: 10.1002/eji.1830260434. [DOI] [PubMed] [Google Scholar]
- 48.Kawaguchi-Miyashita M, Shimizu K, Nanno M, Shimada S, Watanabe T, Koga Y, et al. Development and cytolytic function of intestinal intraepithelial T lymphocytes in antigen-minimized mice. Immunology. 1996;89:268–73. doi: 10.1046/j.1365-2567.1996.d01-740.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Helgeland L, Dissen E, Dai KZ, Midtvedt T, Brandtzaeg P, Vaage JT. Microbial colonization induces oligoclonal expansions of intraepithelial CD8 T cells in the gut. Eur J Immunol. 2004;34:3389–400. doi: 10.1002/eji.200425122. [DOI] [PubMed] [Google Scholar]
- 50.Wei B, Su TT, Dalwadi H, Stephan RP, Fujiwara D, Huang TT, et al. Resident enteric microbiota and CD8+ T cells shape the abundance of marginal zone B cells. Eur J Immunol. 2008;38:3411–25. doi: 10.1002/eji.200838432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fujiwara D, Wei B, Presley LL, Brewer S, McPherson M, Lewinski MA, et al. Systemic control of plasmacytoid dendritic cells by CD8+ T cells and commensal microbiota. J Immunol. 2008;180:5843–52. doi: 10.4049/jimmunol.180.9.5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wei B, Wingender G, Fujiwara D, Chen DY, McPherson M, Brewer S, et al. Commensal microbiota and CD8+ T cells shape the formation of invariant NKT cells. J Immunol. 2010;184:1218–26. doi: 10.4049/jimmunol.0902620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Goodman T, Lefrancois L. Intraepithelial lymphocytes. Anatomical site, not T cell receptor form, dictates phenotype and function. J Exp Med. 1989;170:1569–81. doi: 10.1084/jem.170.5.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brandtzaeg P, Halstensen TS, Kett K, Krajci P, Kvale D, Rognum TO, et al. Immunobiology and immunopathology of human gut mucosa: humoral immunity and intraepithelial lymphocytes. Gastroenterology. 1989;97:1562–84. doi: 10.1016/0016-5085(89)90406-x. [DOI] [PubMed] [Google Scholar]
- 55.Crabbé PA, Bazin H, Eyssen H, Heremans JF. The normal microbial flora as a major stimulus for proliferation of plasma cells synthesizing IgA in the gut. The germ-free intestinal tract. Int Arch Allergy Appl Immunol. 1968;34:362–75. doi: 10.1159/000230130. [DOI] [PubMed] [Google Scholar]
- 56.Hapfelmeier S, Lawson MA, Slack E, Kirundi JK, Stoel M, Heikenwalder M, et al. Reversible microbial colonization of germ-free mice reveals the dynamics of IgA immune responses. Science. 2010;328:1705–9. doi: 10.1126/science.1188454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bauer H, Horowitz RE, Levenson SM, Popper H. The response of the lymphatic tissue to the microbial flora. Studies on germfree mice. Am J Pathol. 1963;42:471–83. [PMC free article] [PubMed] [Google Scholar]
- 58.Hooijkaas H, Benner R, Pleasants JR, Wostmann BS. Isotypes and specificities of immunoglobulins produced by germ-free mice fed chemically defined ultrafiltered “antigen-free” diet. Eur J Immunol. 1984;14:1127–30. doi: 10.1002/eji.1830141212. [DOI] [PubMed] [Google Scholar]
- 59.Pereira P, Forni L, Larsson EL, Cooper M, Heusser C, Coutinho A. Autonomous activation of B and T cells in antigen-free mice. Eur J Immunol. 1986;16:685–8. doi: 10.1002/eji.1830160616. [DOI] [PubMed] [Google Scholar]
- 60.Durkin HG, Chice SM, Gaetjens E, Bazin H, Tarcsay L, Dukor P. Origin and fate of IgE-bearing lymphocytes. II. Modulation of IgE isotype expression on Peyer’s patch cells by feeding with certain bacteria and bacterial cell wall components or by thymectomy. J Immunol. 1989;143:1777–83. [PubMed] [Google Scholar]
- 61.Khan KJ, Ullman TA, Ford AC, Abreu MT, Abadir A, Marshall JK, et al. Antibiotic therapy in inflammatory bowel disease: a systematic review and meta-analysis. Am J Gastroenterol. 2011;106:661–73. doi: 10.1038/ajg.2011.72. [DOI] [PubMed] [Google Scholar]
- 62.Gionchetti P, Rizzello F, Lammers KM, Morselli C, Sollazzi L, Davies S, et al. Antibiotics and probiotics in treatment of inflammatory bowel disease. World J Gastroenterol. 2006;12:3306–13. doi: 10.3748/wjg.v12.i21.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Manichanh C, Rigottier-Gois L, Bonnaud E, Gloux K, Pelletier E, Frangeul L, et al. Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut. 2006;55:205–11. doi: 10.1136/gut.2005.073817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schultz M, Tonkonogy SL, Sellon RK, Veltkamp C, Godfrey VL, Kwon J, et al. IL-2-deficient mice raised under germfree conditions develop delayed mild focal intestinal inflammation. Am J Physiol. 1999;276:G1461–72. doi: 10.1152/ajpgi.1999.276.6.G1461. [DOI] [PubMed] [Google Scholar]
- 65.Sellon RK, Tonkonogy S, Schultz M, Dieleman LA, Grenther W, Balish E, et al. Resident enteric bacteria are necessary for development of spontaneous colitis and immune system activation in interleukin-10-deficient mice. Infect Immun. 1998;66:5224–31. doi: 10.1128/iai.66.11.5224-5231.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dianda L, Hanby AM, Wright NA, Sebesteny A, Hayday AC, Owen MJ. T cell receptor-alpha beta-deficient mice fail to develop colitis in the absence of a microbial environment. Am J Pathol. 1997;150:91–7. [PMC free article] [PubMed] [Google Scholar]
- 67.Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A. 2007;104:13780–5. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sokol H, Seksik P, Rigottier-Gois L, Lay C, Lepage P, Podglajen I, et al. Specificities of the fecal microbiota in inflammatory bowel disease. Inflamm Bowel Dis. 2006;12:106–11. doi: 10.1097/01.MIB.0000200323.38139.c6. [DOI] [PubMed] [Google Scholar]
- 69.Stecher B, Robbiani R, Walker AW, Westendorf AM, Barthel M, Kremer M, et al. Salmonella enterica serovar typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol. 2007;5:2177–89. doi: 10.1371/journal.pbio.0050244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Garrett WS, Lord GM, Punit S, Lugo-Villarino G, Mazmanian SK, Ito S, et al. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell. 2007;131:33–45. doi: 10.1016/j.cell.2007.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Garrett WS, Gallini CA, Yatsunenko T, Michaud M, DuBois A, Delaney ML, et al. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe. 2010;8:292–300. doi: 10.1016/j.chom.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Elinav E, Strowig T, Kau AL, Henao-Mejia J, Thaiss CA, Booth CJ, et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145:745–57. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–5. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- 74.Kelly D, Campbell JI, King TP, Grant G, Jansson EA, Coutts AG, et al. Commensal anaerobic gut bacteria attenuate inflammation by regulating nuclear-cytoplasmic shuttling of PPAR-gamma and RelA. Nat Immunol. 2004;5:104–12. doi: 10.1038/ni1018. [DOI] [PubMed] [Google Scholar]
- 75.Dubuquoy L, Rousseaux C, Thuru X, Peyrin-Biroulet L, Romano O, Chavatte P, et al. PPARgamma as a new therapeutic target in inflammatory bowel diseases. Gut. 2006;55:1341–9. doi: 10.1136/gut.2006.093484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, Yu D, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461:1282–6. doi: 10.1038/nature08530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Feng T, Wang L, Schoeb TR, Elson CO, Cong Y. Microbiota innate stimulation is a prerequisite for T cell spontaneous proliferation and induction of experimental colitis. J Exp Med. 2010;207:1321–32. doi: 10.1084/jem.20092253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Serrano NC, Millan P, Páez MC. Non-HLA associations with autoimmune diseases. Autoimmun Rev. 2006;5:209–14. doi: 10.1016/j.autrev.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 79.Kyvik KO, Green A, Beck-Nielsen H. Concordance rates of insulin dependent diabetes mellitus: a population based study of young Danish twins. BMJ. 1995;311:913–7. doi: 10.1136/bmj.311.7010.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Edwards CJ. Commensal gut bacteria and the etiopathogenesis of rheumatoid arthritis. J Rheumatol. 2008;35:1477–14797. [PubMed] [Google Scholar]
- 81.Leirisalo-Repo M. Early arthritis and infection. Curr Opin Rheumatol. 2005;17:433–9. doi: 10.1097/01.bor.0000166388.47604.8b. [DOI] [PubMed] [Google Scholar]
- 82.Vaahtovuo J, Munukka E, Korkeamäki M, Luukkainen R, Toivanen P. Fecal microbiota in early rheumatoid arthritis. J Rheumatol. 2008;35:1500–5. [PubMed] [Google Scholar]
- 83.Björk J, Kleinau S, Midtvedt T, Klareskog L, Smedegård G. Role of the bowel flora for development of immunity to hsp 65 and arthritis in three experimental models. Scand J Immunol. 1994;40:648–52. doi: 10.1111/j.1365-3083.1994.tb03518.x. [DOI] [PubMed] [Google Scholar]
- 84.Abdollahi-Roodsaz S, Joosten LA, Koenders MI, Devesa I, Roelofs MF, Radstake TR, et al. Stimulation of TLR2 and TLR4 differentially skews the balance of T cells in a mouse model of arthritis. J Clin Invest. 2008;118:205–16. doi: 10.1172/JCI32639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Badami E, Sorini C, Coccia M, Usuelli V, Molteni L, Bolla AM, et al. Defective differentiation of regulatory FoxP3+ T cells by small-intestinal dendritic cells in patients with type 1 diabetes. Diabetes. 2011;60:2120–4. doi: 10.2337/db10-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bach JF. The effect of infections on susceptibility to autoimmune and allergic diseases. N Engl J Med. 2002;347:911–20. doi: 10.1056/NEJMra020100. [DOI] [PubMed] [Google Scholar]
- 87.Zipris D. Epidemiology of type 1 diabetes and what animal models teach us about the role of viruses in disease mechanisms. Clin Immunol. 2009;131:11–23. doi: 10.1016/j.clim.2008.12.011. [DOI] [PubMed] [Google Scholar]
- 88.Pozzilli P, Signore A, Williams AJ, Beales PE. NOD mouse colonies around the world--recent facts and figures. Immunol Today. 1993;14:193–6. doi: 10.1016/0167-5699(93)90160-M. [DOI] [PubMed] [Google Scholar]
- 89.Rygaard J. Immune-deficient animals in biomedical research. Basel; New York: Karger, 1987. [Google Scholar]
- 90.Wen L, Ley RE, Volchkov PY, Stranges PB, Avanesyan L, Stonebraker AC, et al. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature. 2008;455:1109–13. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kriegel MA, Sefik E, Hill JA, Wu HJ, Benoist C, Mathis D. Naturally transmitted segmented filamentous bacteria segregate with diabetes protection in nonobese diabetic mice. Proc Natl Acad Sci U S A. 2011;108:11548–53. doi: 10.1073/pnas.1108924108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Alam C, Bittoun E, Bhagwat D, Valkonen S, Saari A, Jaakkola U, et al. Effects of a germ-free environment on gut immune regulation and diabetes progression in non-obese diabetic (NOD) mice. Diabetologia. 2011;54:1398–406. doi: 10.1007/s00125-011-2097-5. [DOI] [PubMed] [Google Scholar]
- 93.King C, Sarvetnick N. The incidence of type-1 diabetes in NOD mice is modulated by restricted flora not germ-free conditions. PLoS One. 2011;6:e17049. doi: 10.1371/journal.pone.0017049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ochoa-Repáraz J, Mielcarz DW, Ditrio LE, Burroughs AR, Foureau DM, Haque-Begum S, et al. Role of gut commensal microflora in the development of experimental autoimmune encephalomyelitis. J Immunol. 2009;183:6041–50. doi: 10.4049/jimmunol.0900747. [DOI] [PubMed] [Google Scholar]
- 95.Lee YK, Menezes JS, Umesaki Y, Mazmanian SK. Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4615–22. doi: 10.1073/pnas.1000082107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ochoa-Repáraz J, Mielcarz DW, Ditrio LE, Burroughs AR, Begum-Haque S, Dasgupta S, et al. Central nervous system demyelinating disease protection by the human commensal Bacteroides fragilis depends on polysaccharide A expression. J Immunol. 2010;185:4101–8. doi: 10.4049/jimmunol.1001443. [DOI] [PubMed] [Google Scholar]
- 97.Ochoa-Repáraz J, Mielcarz DW, Wang Y, Begum-Haque S, Dasgupta S, Kasper DL, et al. A polysaccharide from the human commensal Bacteroides fragilis protects against CNS demyelinating disease. Mucosal Immunol. 2010;3:487–95. doi: 10.1038/mi.2010.29. [DOI] [PubMed] [Google Scholar]
- 98.Ochoa-Repáraz J, Mielcarz DW, Haque-Begum S, Kasper LH. Induction of a regulatory B cell population in experimental allergic encephalomyelitis by alteration of the gut commensal microflora. Gut Microbes. 2010;1:103–8. doi: 10.4161/gmic.1.2.11515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lavasani S, Dzhambazov B, Nouri M, Fåk F, Buske S, Molin G, et al. A novel probiotic mixture exerts a therapeutic effect on experimental autoimmune encephalomyelitis mediated by IL-10 producing regulatory T cells. PLoS One. 2010;5:e9009. doi: 10.1371/journal.pone.0009009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Anderson MS, Venanzi ES, Klein L, Chen Z, Berzins SP, Turley SJ, et al. Projection of an immunological self shadow within the thymus by the aire protein. Science. 2002;298:1395–401. doi: 10.1126/science.1075958. [DOI] [PubMed] [Google Scholar]
- 101.Gray DH, Gavanescu I, Benoist C, Mathis D. Danger-free autoimmune disease in Aire-deficient mice. Proc Natl Acad Sci U S A. 2007;104:18193–8. doi: 10.1073/pnas.0709160104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Maldonado MA, Kakkanaiah V, MacDonald GC, Chen F, Reap EA, Balish E, et al. The role of environmental antigens in the spontaneous development of autoimmunity in MRL-lpr mice. J Immunol. 1999;162:6322–30. [PubMed] [Google Scholar]
- 103.Hase K, Takahashi D, Ebisawa M, Kawano S, Itoh K, Ohno H. Activation-induced cytidine deaminase deficiency causes organ-specific autoimmune disease. PLoS One. 2008;3:e3033. doi: 10.1371/journal.pone.0003033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med. 2009;1:6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A. 2010;107:14691–6. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Alam C, Valkonen S, Palagani V, Jalava J, Eerola E, Hänninen A. Inflammatory tendencies and overproduction of IL-17 in the colon of young NOD mice are counteracted with diet change. Diabetes. 2010;59:2237–46. doi: 10.2337/db10-0147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Noverr MC, Falkowski NR, McDonald RA, McKenzie AN, Huffnagle GB. Development of allergic airway disease in mice following antibiotic therapy and fungal microbiota increase: role of host genetics, antigen, and interleukin-13. Infect Immun. 2005;73:30–8. doi: 10.1128/IAI.73.1.30-38.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.McKenna P, Hoffmann C, Minkah N, Aye PP, Lackner A, Liu Z, et al. The macaque gut microbiome in health, lentiviral infection, and chronic enterocolitis. PLoS Pathog. 2008;4:e20. doi: 10.1371/journal.ppat.0040020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Penders J, Thijs C, Vink C, Stelma FF, Snijders B, Kummeling I, et al. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics. 2006;118:511–21. doi: 10.1542/peds.2005-2824. [DOI] [PubMed] [Google Scholar]
- 110.Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A. 2010;107:11971–5. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bezirtzoglou E, Tsiotsias A, Welling GW. Microbiota profile in feces of breast- and formula-fed newborns by using fluorescence in situ hybridization (FISH) Anaerobe. 2011;17:478–82. doi: 10.1016/j.anaerobe.2011.03.009. [DOI] [PubMed] [Google Scholar]
- 112.Penders J, Vink C, Driessen C, London N, Thijs C, Stobberingh EE. Quantification of Bifidobacterium spp., Escherichia coli and Clostridium difficile in faecal samples of breast-fed and formula-fed infants by real-time PCR. FEMS Microbiol Lett. 2005;243:141–7. doi: 10.1016/j.femsle.2004.11.052. [DOI] [PubMed] [Google Scholar]
- 113.Stark PL, Lee A. The microbial ecology of the large bowel of breast-fed and formula-fed infants during the first year of life. J Med Microbiol. 1982;15:189–203. doi: 10.1099/00222615-15-2-189. [DOI] [PubMed] [Google Scholar]
- 114.Harmsen HJ, Wildeboer-Veloo AC, Raangs GC, Wagendorp AA, Klijn N, Bindels JG, et al. Analysis of intestinal flora development in breast-fed and formula-fed infants by using molecular identification and detection methods. J Pediatr Gastroenterol Nutr. 2000;30:61–7. doi: 10.1097/00005176-200001000-00019. [DOI] [PubMed] [Google Scholar]
- 115.Yoshioka H, Iseki K, Fujita K. Development and differences of intestinal flora in the neonatal period in breast-fed and bottle-fed infants. Pediatrics. 1983;72:317–21. [PubMed] [Google Scholar]
- 116.Renz-Polster H, David MR, Buist AS, Vollmer WM, O’Connor EA, Frazier EA, et al. Caesarean section delivery and the risk of allergic disorders in childhood. Clin Exp Allergy. 2005;35:1466–72. doi: 10.1111/j.1365-2222.2005.02356.x. [DOI] [PubMed] [Google Scholar]
- 117.Salam MT, Margolis HG, McConnell R, McGregor JA, Avol EL, Gilliland FD. Mode of delivery is associated with asthma and allergy occurrences in children. Ann Epidemiol. 2006;16:341–6. doi: 10.1016/j.annepidem.2005.06.054. [DOI] [PubMed] [Google Scholar]
- 118.Decker E, Hornef M, Stockinger S. Cesarean delivery is associated with celiac disease but not inflammatory bowel disease in children. Gut Microbes. 2011;2:91–8. doi: 10.4161/gmic.2.2.15414. [DOI] [PubMed] [Google Scholar]