Abstract
We have recently isolated a rice circadian clock-related mutant carrying a null mutation in Os-GIGANTEA(GI) gene, the solo ortholog of Arabidopsis GI. Time-course global transcriptome analyses of leaves from wild-type and osgi mutant grown in the field have revealed that Os-GI affects gene expression of more than half of genes on rice 44k microarray. To better understand the biological significance of circadian clock function in growth and development of rice, we here investigated the gene expression involved in phytohormone biosynthesis. Here we found that mRNA levels of a few major genes encoding GA2-oxidase which can inactivate bioactive gibberellins (GAs) were remarkably increased in osgi-1 plants. This suggests that Os-GI functions to maintain bioactive GA level through the regulation of the GA-deactivating enzyme genes in rice. Consistently, osgi-1 plants showed semi-dwarf phenotype with reduced internode and leaf sheath elongation.
Keywords: Circadian clocks, gibberellin 2-oxidase, microarray, phytohormone, rice
Molecular genetics using Arabidopsis have revealed that circadian clocks control variety of physiological responses including flowering, hypocotyl elongation, and guard cell opening.1 We have recently identified a rice circadian-clock related mutant, os-gigantea (osgi) mutant.2 Os-GI is the solo ortholog of GIGANTEA (GI) in the Arabidopsis circadian system.1,3,4 Time-course extensive transcriptome analyses of osgi-1 leaves grown in the field revealed that Os-GI affects gene expression of approximately 75% of genes among 27,201 genes and is required for strong amplitudes and proper phase-setting of global gene expression under natural field conditions.2 These field-transcriptome data imply that circadian clock in rice affects various physiological traits during growth and development of rice. In fact, a critical deficiency of photoperiodic flowering of osgi-1 plants has been shown due to a great reduction of mRNA levels of Hd3a, a rice florigen gene, and Ehd1, an inducer of Hd3a under laboratory short-day conditions.2,5 To deeper understand the molecular function of circadian clock in rice, we are comparing the field-transcriptome data between wild-type (the cultivar Norin 8) and osgi-1 plants and trying to find out key gene controlled by circadian clock systems. In this study, we analyzed the expression of phytohormone biosynthetic genes since circadian clock modulates hormonal action through the transcriptional regulation of phytohormone-related genes in Arabidopsis.6
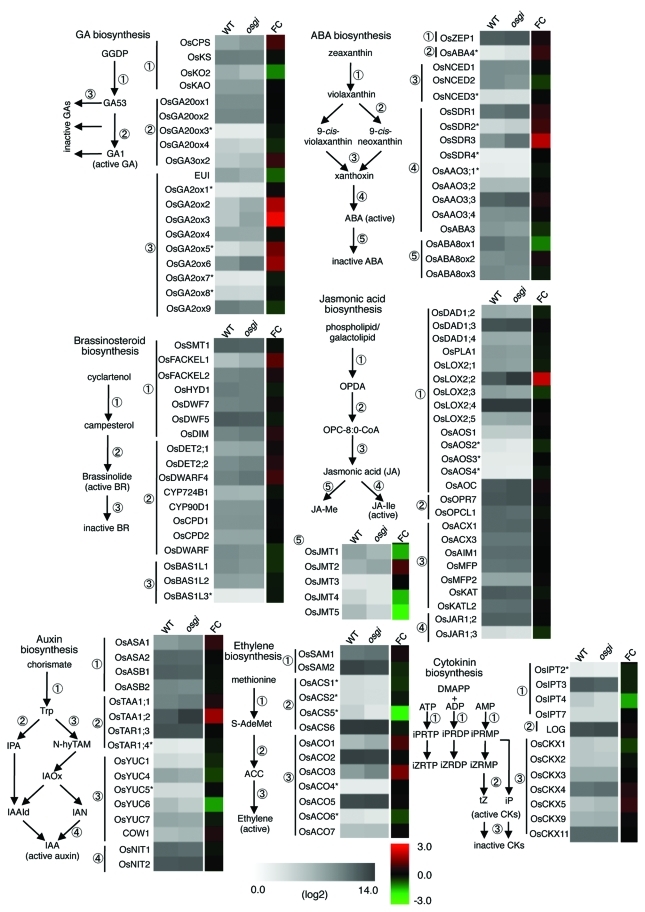
The transcriptome analyses were performed by using leaves of wild-type and osgi-1 plants sampled for 1 d (24 h) at 2 h intervals (total 13 time points).2 At each of time point, eight arrays were used with two replicates at four developmental stages. Because each microarray raw data converted to a logarithm fits to normal distribution, we first normalized all Wild-type and osgi-1 microarray data converted to log2 using the qspline method, which was implemented in R and included in the Bioconductor package (http://www.r-project.org/).7 Mean of all eight normalized array data sets per time point was used for further analysis. In this case, we did not focus on the difference of developmental stages and instead used as eight replicated samples per time point to highlight the Os-GI in the diurnal rhythms. We then listed up genes involved in seven phytohormone biosynthetic pathways, gibberellin (GA), abscisic acid (ABA), auxin, brassinosteroid, cytokinin (CK), jasmonic acid (JA), and ethylene for this study (Table S1) based on recent publication on the comprehensive transcriptome analysis of phytohormone biosynthesis and signaling genes during rice anther development.8 Hirano et al. also showed that bioactive GA, free IAA, bioactive CK, and ABA were detected in leaf blades, indicating that phytohormone biosynthesis works in rice leaves.8 To address whether Os-GI regulates the expression of phytohormone biosynthetic genes, the mean of the gene expression levels for 24 h and their ratios between wild-type and osgi-1 plants were calculated (Fig. 1). Although most tested genes showed only slight differences between wild-type and osgi-1 plants (less than 2-fold), however, some were remarkably affected in osgi-1 (blight red or green boxes indicated in Figure 1). Among them, we found that some members in gene families are consistently changed in osgi-1. In GA biosynthesis, four of nine OsGA2ox genes were upregulated in osgi-1 (3 to 5.8–fold increase). Three of five OsJMT genes in JA biosynthesis were downregulated in osgi-1 (2.6 to 4.5-fold reduction). In ABA biosynthesis, two OsSDR genes in osgi-1 were increased with lesser extent (1.5 to 3.9-fold increase). In addition, the expression of OsKO2 gene in early step of GA biosynthesis was reduced in osgi-1 plants. In auxin biosynthesis, OsTAA1;2 is highly expressed in osgi-1. Although genetic studies in Arabidopsis have demonstrated that TAA1 is essential for a local auxin production in elongating hypocotyl and growing root,9,10 biological action of TAA in leaves remains unknown. Furthermore, Os-GI affected the gene expression of OsABA8ox1, OsYUC6, OsLOX2;2, OsACS5, OsACO3, and OsIPT4 genes (Fig. 1).
Figure 1.
Comparison of gene expression levels of phytohormone biosynthetic genes. Average mRNA levels during 24 h in the wild-type (WT) and osgi-1 plants are indicated as single color gradient. Their ratios (FC; fold change) between WT and osgi-1 are indicated as different color gradient. Red and green colors indicate higher and lower expression in osgi-1 mutant, respectively. A simplified view of each phytohormone biosynthetic pathway and genes involved in the indicated step are described in left sides of the expression data. Asterisks on right shoulder of gene name indicate that peaked expression level was lower than 26. Color gradient display was drawn by means of software, MeV_4_7.25
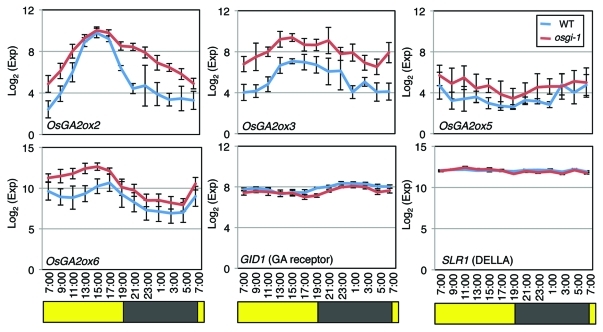
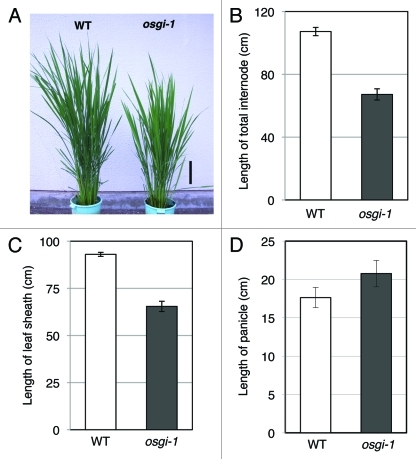
Considering the fact in which equal or much higher expression of their palalogous genes in both wild-type and osgi-1 plants suggests minor effects by Os-GI for these steps in each phytohormone biosynthesis, we focused on expression of OsGA2ox genes. GA 2-oxidase encoded by OsGA2ox genes can deactivate bioactive GAs or its precursors irreversibly to reduce bioactive GA levels.11,12 Both quantitative analyses of endogenous GA levels and gene expression analyses of GA biosynthetic genes have indicated circadian regulation of GA biosynthesis in other plant species.13-16 In RiceXPro, a public database of rice gene expression profiling,17 spatio-temporal gene expression patterns of OsGA20ox, OsGA3ox2, and OsGA2ox genes (especially OsGA20ox2 and OsGA2ox6), revealed that leaf organs are one of the major expression sites during rice life cycle. The expression patterns of OsGA2ox2, OsGA2ox3, OsGA2ox5, and OsGA2ox6 in our data generally showed diurnal oscillation that become high during daytime in wild-type leaves (blue lines in Figure 2). Interestingly, this expression pattern is highly similar to the diurnal fluctuation pattern of endogenous GA8, an inactive GA, in sorghum, another monocotyledonous plant.13 In contrast, in osgi-1, the diurnal expression of these OsGA2ox genes were consistently elevated than those of wild-type in all time points tested (red lines in Figure 2). The results suggest that Os-GI represses major OsGA2ox genes in order to maintain a proper bioactive GA level. Consistently with their elevated gene expression, osgi-1 plants showed a semi-dwarf phenotype (Fig. 3A). We also revealed that the semi-dwarfism in osgi-1 plants is mainly derived from reduced length of internode and leaf sheath, respectively (Fig. 3B, C), corresponding to organs that GA induces elongation to control the rice plant stature.11 On the other hand, panicle length in osgi-1 slightly longer than that of wild-type (Fig. 3D), suggesting that elevated expression of GA2ox in leaves does not affect the development of reproductive organ.8
Figure 2.
Diurnal expression profiles of the selected OsGA2ox genes, GID1, and SLR1. Average calibrated data were expressed as log2 scale. Data are represented as means ± SE (n = 8). Yellow and black boxes below sampling time indicate day and night periods are referred from the estimated photoperiods described in the recently published paper.2
Figure 3.
Semi-dwarf phenotype of osgi-1 plant. (A) Photograph of 90-d-old WT and osgi-1 plants grown under natural field conditions. Bar indicates 20 cm. (B) Total internode length of WT and osgi-1 plants. (C) Leaf sheath length at same developmental stage. (D) Panicle length. Bars are expressed as the mean ± SE (n = 9 in (B) and (D), n = 3 in (C).
In Arabidopsis, circadian clock regulates rhythmic hypocotyl elongation through transcriptional regulation of the GA receptor genes.18 However, there were no obvious differences of gene expression of rice GA receptor, GID1, and the DELLA protein, SLR1 (Fig. 2).19 Although the expression level of OsGA2ox9 was also high in both wild-type and osgi-1 plants (Fig. 1), we could exclude this due to a weak GA deactivation activity of OsGA2ox9.12
In ABA actions, there is a consistency of higher OsSDR expression with the accumulation of more sucrose and starch in osgi-1 (Fig. 1),2 since Arabidopsis ABA2 gene encoding a SDR is induced by sugar treatment and ABA can promote transcription of starch biosynthetic gene in sugar storage processes.20,21 In addition, osgi-1 might have more opened guard cells than the wild-type,2 possibly due to changes in ABA actions. In JA action, whereas JA-Ile, not JA-Me, is the biologically active compound,22,23 the effect of reduction of JMT expression on JA-Ile production remains unknown. Some secondary metabolites accumulated in osgi-1 are known to be phytoalexin- related flavonoids and stress-inducible,2,24 suggesting an elevated active JA synthesis in osgi-1 plants.
In addition to the reduced body size during vegetative growth and longer size of panicle (Fig. 3), osgi-1 plants have an increased numbers of both panicle and spikelet, and with some reduced fertility under stressed conditions.2 Although quantitative measurements of phytohormones are necessary to confirm the changes of genes, the differences of gene expression reported in this study give us some hints to understand how Os-GI modulates growth and development of rice. (1186 words)
Supplementary Material
Supplementary PDF file supplied by authors.
Acknowledgments
This work was supported by two grants from the Ministry of Agriculture, Forestry, and Fisheries of Japan (Genomics for Agricultural Innovation GPN0001 and RTR0004 to T.I.).
Glossary
Abbreviations:
- Os
Oryza sativa
- GI
GIGANTEA
- GA
gibberellin
- ABA
abscisic acid
- JA
jasmonic acid
- CK
cytokinin
- IAA
indole-3-acetic acid
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/18207
References
- 1.Harmer SL. The circadian system in higher plants. Annu Rev Plant Biol. 2009;60:357–77. doi: 10.1146/annurev.arplant.043008.092054. [DOI] [PubMed] [Google Scholar]
- 2.Izawa T, Mihara M, Suzuki Y, Gupta M, Itoh H, Nagano AJ, et al. Os-GIGANTEA confers robust diurnal rhythms on the global transcriptome of rice in the field. Plant Cell. 2011;23:1741–55. doi: 10.1105/tpc.111.083238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fowler S, Lee K, Onouchi H, Samach A, Richardson K, Morris B, et al. GIGANTEA: a circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J. 1999;18:4679–88. doi: 10.1093/emboj/18.17.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Park DH, Somers DE, Kim YS, Choy YH, Lim HK, Soh MS, et al. Control of circadian rhythms and photoperiodic flowering by the Arabidopsis GIGANTEA gene. Science. 1999;285:1579–82. doi: 10.1126/science.285.5433.1579. [DOI] [PubMed] [Google Scholar]
- 5.Itoh H, Nonoue Y, Yano M, Izawa T. A pair of floral regulators sets critical day length for Hd3a florigen expression in rice. Nat Genet. 2010;42:635–8. doi: 10.1038/ng.606. [DOI] [PubMed] [Google Scholar]
- 6.Thines B, Harmon FG. Four easy pieces: mechanisms underlying circadian regulation of growth and development. Curr Opin Plant Biol. 2011;14:31–7. doi: 10.1016/j.pbi.2010.09.009. [DOI] [PubMed] [Google Scholar]
- 7.Workman C, Jensen LJ, Jarmer H, Berka R, Gautier L, Nielser HB, et al. A new non-linear normalization method for reducing variability in DNA microarray experiments. Genome Biol 2002; 3:0048 [DOI] [PMC free article] [PubMed]
- 8.Hirano K, Aya K, Hobo T, Sakakibara H, Kojima M, Shim RA, et al. Comprehensive transcriptome analysis of phytohormone biosynthesis and signaling genes in microspore/pollen and tapetum of rice. Plant Cell Physiol. 2008;49:1429–50. doi: 10.1093/pcp/pcn123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stepanova AN, Robertson-Hoyt J, Yun J, Benavente LM, Xie DY, Dolezal K, et al. TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell. 2008;133:177–91. doi: 10.1016/j.cell.2008.01.047. [DOI] [PubMed] [Google Scholar]
- 10.Tao Y, Ferrer JL, Ljung K, Pojer F, Hong F, Long JA, et al. Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell. 2008;133:164–76. doi: 10.1016/j.cell.2008.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sakamoto T, Miura K, Itoh H, Tatsumi T, Ueguchi-Tanaka M, Ishiyama K, et al. An overview of gibberellin metabolism enzyme genes and their related mutants in rice. Plant Physiol. 2004;134:1642–53. doi: 10.1104/pp.103.033696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lo S-F, Yang S-Y, Chen K-T, Hsing Y-I, Zeevaart JAD, Chen L-J, et al. A novel class of gibberellin 2-oxidases control semidwarfism, tillering, and root development in rice. Plant Cell. 2008;20:2603–18. doi: 10.1105/tpc.108.060913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foster KR, Morgan PW. Genetic regulation of development in Sorghum bicolor. IX. The ma3 R allele disrupts diurnal control of gibberellin biosynthesis. Plant Physiol. 1995;108:337–43. doi: 10.1104/pp.108.1.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hisamatsu T, King RW, Helliwell CA, Koshioka M. The involvement of gibberellin 20-oxidase genes in phytochrome-regulated petiole elongation of Arabidopsis. Plant Physiol. 2005;138:1106–16. doi: 10.1104/pp.104.059055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carrera E, Jackson SD, Prat S. Feedback control and diurnal regulation of gibberellin 20-oxidase transcript levels in potato. Plant Physiol. 1999;119:765–74. doi: 10.1104/pp.119.2.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao X, Yu X, Foo E, Symons GM, Lopez J, Bendehakkalu KT, et al. A study of gibberellin homeostasis and cryptochrome-mediated blue light inhibition of hypocotyl elongation. Plant Physiol. 2007;145:106–18. doi: 10.1104/pp.107.099838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sato Y, Antonio BA, Namiki N, Takehisa H, Minami H, Kamatsuki K, et al. RiceXPro: a platform for monitoring gene expression in japonica rice grown under natural field conditions. Nucleic Acids Res. 2011;39(Database issue):D1141–8. doi: 10.1093/nar/gkq1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arana MV, Marín-de la Rosa N, Maloof JN, Blázquez MA, Alabadí D. Circadian oscillation of gibberellin signaling in Arabidopsis. Proc Natl Acad Sci U S A. 2011;108:9292–7. doi: 10.1073/pnas.1101050108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ueguchi-Tanaka M, Nakajima M, Motoyuki A, Matsuoka M. Gibberellin receptor and its role in gibberellin signaling in plants. Annu Rev Plant Biol. 2007;58:183–98. doi: 10.1146/annurev.arplant.58.032806.103830. [DOI] [PubMed] [Google Scholar]
- 20.Seo M, Koshiba T. Complex regulation of ABA biosynthesis in plants. Trends Plant Sci. 2002;7:41–8. doi: 10.1016/S1360-1385(01)02187-2. [DOI] [PubMed] [Google Scholar]
- 21.Rook F, Corke F, Card R, Munz G, Smith C, Bevan MW. Impaired sucrose-induction mutants reveal the modulation of sugar-induced starch biosynthetic gene expression by abscisic acid signalling. Plant J. 2001;26:421–33. doi: 10.1046/j.1365-313X.2001.2641043.x. [DOI] [PubMed] [Google Scholar]
- 22.Chini A, Fonseca S, Fernandez G, Adie B, Chico JM, Lorenzo O, et al. The JAZ family of repressor is the missing link in jasmonate signaling. Nature. 2008;448:666–71. doi: 10.1038/nature06006. [DOI] [PubMed] [Google Scholar]
- 23.Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, Liu G, et al. JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature. 2007;448:661–5. doi: 10.1038/nature05960. [DOI] [PubMed] [Google Scholar]
- 24.Grace SC, Logan BA. Energy dissipation and radical scavenging by the plant phenylpropanoid pathway. Philos Trans R Soc Lond B Biol Sci. 2000;355:1499–510. doi: 10.1098/rstb.2000.0710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mar JC, Wells CA, Quackenbush J. Defining an informativeness metric for clustalling gene expression data. Bioinformatics. 2011;15:1094–100. doi: 10.1093/bioinformatics/btr074. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary PDF file supplied by authors.