Abstract
Autophagy is the process by which cells degrade their own components in lysosomes or vacuoles. Autophagy in tobacco BY-2 cells cultured in sucrose-free medium takes place in formed, autolysosomes in the presence of a cysteine protease inhibitor. The autolysosomes in BY-2 cells are located in the endocytotic pathway and thus can be stained with fluorescent endocytosis marker FM4–64. In the present study, in order to detect autophagy in the root cells of Arabidopsis, we incubated root tips from Arabidopsis seedlings in culture medium containing the membrane-permeable cysteine protease inhibitor E-64d and FM4–64, and examined whether autolysosomes stained with FM4–64 are accumulated. The results suggest that autophagy accompanying the formation of autolysosomes also occurs in Arabidopsis root cells. Such autophagy appeared to occur constitutively in the root cells in nutrient-sufficient culture medium. Even in atg5 mutants in which an autophagy-related gene is disrupted, accumulation of the structures stained with FM4–64, which likely correspond to autolysosomes, was seen although at lower level than in wild type roots.
Keywords: Arabidopsis, autolysosome/autophagosome, autophagy, cysteine protease, endocytosis, protease inhibitor, vacuole
Introduction
Autophagy is the process by which cells degrade their own components in lysosomes or vacuoles. In macroautophagy, parts of the cytoplasm are first enclosed in double-membrane-bounded structures called autophagosomes, which subsequently fuse with pre-existing lysosomes and/or vacuoles.1,2 This sequestration of a portion of the cytoplasm occurs non-selectively in some autophagic processes and by targeting selected organelles in other processes. In microautophagy, lysosomes and/or vacuoles directly incorporate portions of the cytoplasm without forming autophagosomes.3-5 In both types of autophagy, the cytoplasm taken up into lysosomes/vacuoles is degraded by lysosomal/vacuolar proteases. Autophagy contributes to the recycling of cellular components and the supply of respiratory substrates. In addition to such universal roles, autophagy has been suggested to play a role in various developmental processes.6-13
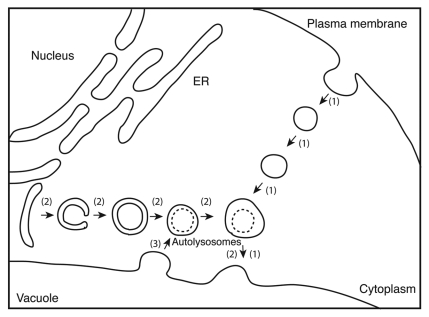
Inhibition of lysosomal/vacuolar proteases with inhibitors that block the degradation of sequestered parts of the cytoplasm causes the accumulation of cytoplasmic materials in lysosomes and/or vacuoles. The protease inhibitor leupeptin inhibits lysosomal proteases, cathepsins B and L, and leads to the accumulation of cytoplasmic inclusions in lysosomes in mammalian cells.14 In contrast, the serine protease inhibitor phenylmethylsulfonyl fluoride inhibits proteinase B, resulting in the accumulation of cytoplasmic drops called autophagic bodies in the vacuole of yeast cells.15 A similar phenomenon is found in the cultured tobacco cell line BY-2. When BY-2 cells at logarithmic growth phase are transferred to sucrose-free culture medium and further cultured, there is a net degradation of cellular proteins.16 Supplementation of culture medium with protease inhibitors, such as E-64c, E-64, leupeptin and antipain, inhibits intracellular cysteine protease and protein degradation in BY-2 cells, and causes the accumulation of lysosomes containing cytoplasmic inclusions, which we termed “autolysosomes” in plant cells (Fig. 1).16-18 This result suggests that protein degradation occurs in “autolysosomes,” which are newly formed upon autophagy in BY-2 cells.
Figure 1.
Convergence of autophagy and endocytosis. (1) the endocytotic pathway; (2) the macroautophagic pathway; (3) a retrograde membrane transport from the central vacuole to autolysosomes. A cysteine protease inhibitor blocks the degradation of cytoplasmic inclusions in autolysosomes and their fusion with the central vacuole, causing their accumulation.
The autophagic pathway (route 2 in Figure 1) merges with the endocytotic pathway (route 1 in Figure 1) in BY-2 cells19,20 as in mammalian cells.21 In BY-2 cells, the fluorescent tracer of endocytosis FM4–64 predominantly migrates from the plasma membranes to the membranes of the central vacuole by endocytosis, while FM4–64 fluorescence migrates from the plasma membrane but stops at the autolysosomes without arriving at the central vacuole in the cells shown to accumulate “autolysosomes.” This shows that the autolysosomes are located on the endocytotic pathway, downstream from the plasma membrane and upstream from the central vacuole.19
Retrograde transport of the membrane from the central vacuole to “autolysosomes” is also seen in BY-2 cells (Fig. 1, [3]). When the cells with vacuolar membranes stained with FM4–64 are placed in sucrose-free culture medium containing E-64c, there is predominant FM4–64 fluorescence in the emerging autolysosomes.19 This shows that when autolysosomes are formed, a membrane flow occurs from the central vacuole to the autolysosomes.
Taken together, FM4–64 added to the culture medium is incorporated into cells and finally associates with autolysosomes in the presence of E-64c. Therefore, the combination of FM4–64 and E-64c with fluorescence microscopy allows the detection of autolysosomes in BY-2 cells.
In the present study, we examined whether this staining method is suitable for detecting autolysosomes in the autophagy of Arabidopsis root cells.
Results
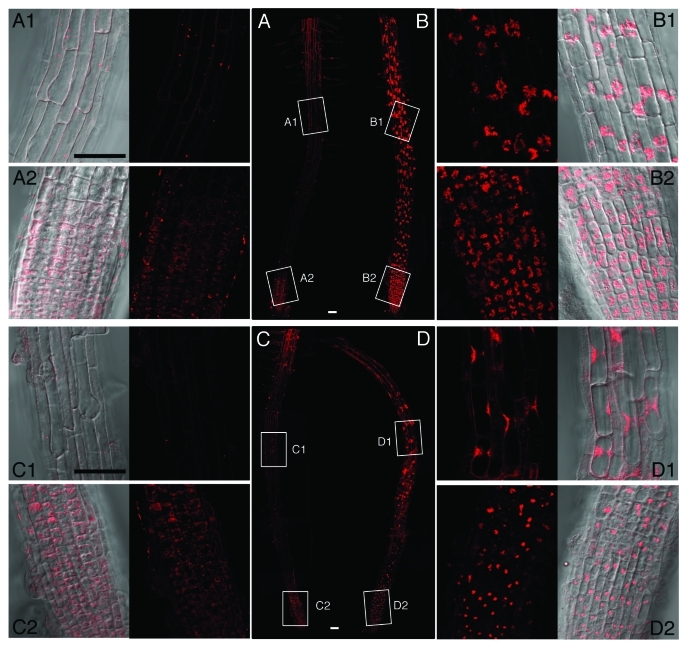
The FM4–64 staining protocol used in the present study visualized the plasma membrane immediately after staining and dotted structures in the cytoplasm, some of which are considered to be early endosomes (data not shown). After 18 h, FM4–64 fluorescence migrated to the membrane of the vacuoles for the root tips incubated in the absence of E-64d, and we could visualize vacuolar membrane together with some particulate structures, which are likely to be “bulbs” (Fig. 2A).22 FM4–64 fluorescence near the meristematic zone appeared stronger than that in the elongation zone (Fig. 2A, A1 vs. A2), probably because cells near the meristematic zone are smaller and the structures of the vacuolar membranes in these small cells are more convoluted. In contrast to root tips incubated without E-64d, root tips incubated in the presence of E-64d showed fluorescence primarily on the autolysosomes with negligible fluorescence associated with vacuolar membranes (Fig. 2B), suggesting that the endocytotic pathway from the plasma membrane to the central vacuole (route [1] in Figure 1) converges with the autophagic pathway leading to the central vacuole (route [2] in Figure 1) upstream from the step of autolysosome formation in Arabidopsis root cells in the same manner as in BY-2 cells. Thus, endocytosis appears to also contribute to the formation of “autolysosomes” in Arabidospsis root cells.
Figure 2.
Accumulation of autolysosomes in Arabidopsis root cells. Root tips were cut from Arabidopsis (wild type and atg5–2) seedlings and incubated in culture medium containing 1% methanol as a solvent control (A and C) or 100 µM E-64d (B and D) for 3 h, followed by staining with 10 µM FM4–64 for 30 min. After FM4–64 was removed, the root tips were placed in fresh media and incubated under the same culture conditions for 18 h. (A) wild type in the absence of E-64d; B, wild type in the presence of E-64d; (C) atg5–2 in the absence of E-64d; (D) atg5–2 in the presence of E-64d. A1, A2, B1, B2, C1, C2, D1, and D2 are the enlargements of portions enclosed by rectangles in (A, B, C, and D). In A1, A2, C1, and C2, the fluorescent image is at right, and the merged image of the fluorescent image and the Nomarski image is at left; in B1, B2, D1, and D2, the fluorescent and merged images are placed at the left and right, respectively. Scale bar, 50 µm.
The staining of root tips from atg5–1 and atg5–2 mutants was the same as that from the wild type plants for incubation in the absence of E-64d (Fig. 2, C vs. A), suggesting that the disruption of the ATG5 gene per se does not perturb the endocytotic pathway in Arabidopsis root cells. In contrast, a dramatic change in staining patterns was observed for root tips incubated in the presence of E-64d (Fig. 2, D); there was accumulation of some special structures stained with FM4–64, although the amount of such accumulation was less than that in wild type root tips both in the elongation zone and near the meristematic zone (Fig. 2, D vs. B). These structures appeared to take on a more highly condensed formation compared with autolysosome accumulation in wild type root tips (Fig. 2, D vs. B). Most of these structures were localized around the nucleus, similar to the patterns observed for the accumulation of autolysosomes; however, we occasionally found that some structures appeared to be located in the central vacuole. We suspect that the same structures accumulate in the cells of wild type roots, but we could not find any. This suggests that the emergence of these structures in the presence of E-64d is ATG-dependent and that these structures correspond to “autolysosomes” in wild type cells.
Discussion
Staining with FM4–64 appears to be more sensitive for the detection of autolysosomes in Arabidopsis root cells compared with our previous method using neutral red. In the previous study, root tips from Arabidopsis seedlings were cultured in a similar way as in the present study, and stained with neutral red.23 Neutral red is a lysosomotrophic dye, which tends to accumulate in acidic organelles such as vacuoles and autolysosomes, and cytoplasmic inclusions in these organelles were preferentially stained. In contrast, the staining method in the present study is based on the assumption that the endocytosis marker FM4–64 finally becomes associated with autolysosomes, if they emerge. Thus, this method, in principal, specifically stains autolysosomal membrane and not cytoplasmic inclusions in autolysosomes and the central vacuole. This staining allows the sensitive detection of autolysosomes in a wide range of Arabidopsis root cells.
Autophagy seems to occur constitutively in Arabidopsis root cells irrespective of nutritional conditions (see also ref. 24). Even when root tips were incubated in nutrient-sufficient culture medium, autolysosomes accumulated in all root cells in the presence of E-64d. In contrast, autolysosomes do not accumulate significantly in BY-2 cells cultured in nutrient-sufficient medium.25 Root tip cells are differ from BY-2 cells in that they are growing and the vacuoles are enlarging. This evidence leads us to suppose that constitutive autophagy in root cells is somehow involved in the genesis of vacuoles in root cells. There have been several reports that plant vacuoles are generated through autophagic processes.9,26-28
The present study clearly shows that autolysosomes are formed in Arabidopsis root cells, whereas experiments using concanamycin A have shown that autophagosomes directly fuse with the central vacuole without transforming to autolysosomes in Arabidopsis root cells,29 as occurs in yeast cells.30 Concanamycin A is an inhibitor of vacuolar H+-ATPase, and is thought to prevent the acidification of the vacuolar/lysosomal interior and thus inhibits vacuolar/lysosomal protease. Therefore although the two inhibitors E-64d and concanamycin are supposed to have the same effects, there is a conflict between the two. It has been reported that concanamycin A has effects other than increasing pH in the central vacuole, and it is likely that concanamycin A distorts the normal autophagic pathway in root cells. However, we need experimental evidence to prove this.
E-64d caused some structures stained with FM4–64 to accumulate, even in the atg5–1 and atg5–2 mutants. Although the pattern of accumulation of these structures appeared to be different from that of autolysosomes in wild type plants, most of these structures were located around the nucleus, like autolysosomes. We are now analyzing these structures by electron microscopy. If some of the accumulated structures are proved to be real autolysosomes, it would follow that unlike in yeast cells, the ATG5 gene is not essential for the progression of autophagy in Arabidopsis root cells.
Materials and methods
Seedlings of wild type and atg5–1 and atg5–2 mutant plants of Arabidopsis were grown for 1 week at 23 ± 1°C under continuous light from fluorescent lamps (Inoue et al. 2006). Root tips were cut 5 mm from the apex and cultured in the medium consisting of ½-strength Murashige and Skoog medium, 3% (w/v) sucrose, and either 100 µM E-64d (an ester of E-64c, and thus more membrane-permeable; Peptide Institute) or 1% (v/v) methanol as a solvent control at 23 ± 1°C with shaking at 50 rpm in the dark. After 3 h, FM4–64 (Molecular Probes) was added to each culture to a final concentration of 10 µM, and the cultures were kept at 23 ± 1°C for 30 min to stain the root cells, and unbound FM4–64 was washed out. The root tips were then transferred to fresh culture medium containing 100 µM E-64d or 1% (v/v) methanol and further cultured for 18 h at 23 ± 1°C with shaking at 50rpm in the dark. Root tips were observed by confocal laser microscopy (FV1000-D, Olympus).
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/18297
References
- 1.Dunn WA., Jr. Autophagy and related mechanisms of lysosome-mediated protein degradation. Trends Cell Biol. 1994;4:139–43. doi: 10.1016/0962-8924(94)90069-8. [DOI] [PubMed] [Google Scholar]
- 2.Klionsky DJ, Ohsumi Y. Vacuolar import of proteins and organelles from the cytoplasm. Annu Rev Cell Dev Biol. 1999;15:1–32. doi: 10.1146/annurev.cellbio.15.1.1. [DOI] [PubMed] [Google Scholar]
- 3.Ahlberg J, Glaumann H. Uptake--microautophagy--and degradation of exogenous proteins by isolated rat liver lysosomes. Effects of pH, ATP, and inhibitors of proteolysis. Exp Mol Pathol. 1985;42:78–88. doi: 10.1016/0014-4800(85)90020-6. [DOI] [PubMed] [Google Scholar]
- 4.Tuttle DL, Dunn WA., Jr. Divergent modes of autophagy in the methylotrophic yeast Pichia pastoris. J Cell Sci. 1995;108:25–35. doi: 10.1242/jcs.108.1.25. [DOI] [PubMed] [Google Scholar]
- 5.Sakai Y, Koller A, Rangell LK, Keller GA, Subramani S. Peroxisome degradation by microautophagy in Pichia pastoris: identification of specific steps and morphological intermediates. J Cell Biol. 1998;141:625–36. doi: 10.1083/jcb.141.3.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290:1717–21. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robinson DG, Hinz G. Vacuole biogenesis and protein transport to the plant vacuole: a comparison with the yeast vacuole and the mammlian lysosome. Protoplasma. 1997;197:1–25. doi: 10.1007/BF01279880. [DOI] [Google Scholar]
- 8.Herman EM, Larkins BA. Protein storage bodies and vacuoles. Plant Cell. 1999;11:601–14. doi: 10.1105/tpc.11.4.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marty F. Plant vacuoles. Plant Cell. 1999;11:587–600. doi: 10.1105/tpc.11.4.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moriyasu Y, Hillmer S. Autophagy and vacuole formation. In: Robinson DG, Rogers JC, eds. Vacuolar Compartments, Annu. Plant Reviews, vol. 5. Sheffield Academic Press, 2000: 71-89. [Google Scholar]
- 11.Moriyasu Y, Klionsky DJ. Autophagy in plants. In: Klionsky DJ, ed. Autophagy. Georgetown, TX: Landes Bioscience, 2004:208-215. [Google Scholar]
- 12.Bassham DC, Laporte M, Marty F, Moriyasu Y, Ohsumi Y, Olsen LJ, et al. Autophagy in development and stress responses of plants. Autophagy. 2006;2:2–11. doi: 10.4161/auto.2092. [DOI] [PubMed] [Google Scholar]
- 13.Bassham DC. Plant autophagy--more than a starvation response. Curr Opin Plant Biol. 2007;10:587–93. doi: 10.1016/j.pbi.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 14.Kominami E, Hashida S, Khairallah EA, Katunuma N. Sequestration of cytoplasmic enzymes in an autophagic vacuole-lysosomal system induced by injection of leupeptin. J Biol Chem. 1983;258:6093–100. [PubMed] [Google Scholar]
- 15.Takeshige K, Baba M, Tsuboi S, Noda T, Ohsumi Y. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J Cell Biol. 1992;119:301–11. doi: 10.1083/jcb.119.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moriyasu Y, Ohsumi Y. Autophagy in tobacco suspension-cultured cells in response to sucrose starvation. Plant Physiol. 1996;111:1233–41. doi: 10.1104/pp.111.4.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moriyasu Y, Inoue Y. Use of protease inhibitors for detecting autophagy. In: Klionsky DJ, Ed. Methods in Enzymology, vol 451. Elsevier Inc. Academic Press 2008:557-580. [DOI] [PubMed] [Google Scholar]
- 18.Takatsuka C, Inoue Y, Higuchi T, Hillmer S, Robinson DG, Moriyasu Y. Autophagy in tobacco BY-2 cells cultured under sucrose starvation conditions: isolation of the autolysosome and its characterization. Plant Cell Physiol. 2011;52:2074–87. doi: 10.1093/pcp/pcr137. [DOI] [PubMed] [Google Scholar]
- 19.Yano K, Matsui S, Tsuchiya T, Maeshima M, Kutsuna N, Hasezawa S, et al. Contribution of the plasma membrane and central vacuole in the formation of autolysosomes in cultured tobacco cells. Plant Cell Physiol. 2004;45:951–7. doi: 10.1093/pcp/pch105. [DOI] [PubMed] [Google Scholar]
- 20.Yamada K, Fuji K, Shimada T, Nishimura M, Hara-Nishimura I. Endosomal proteases facilitate the fusion of endosomes with vacuoles at the final step of the endocytotic pathway. Plant J. 2005;41:888–98. doi: 10.1111/j.1365-313X.2005.02349.x. [DOI] [PubMed] [Google Scholar]
- 21.Liou W, Geuze HJ, Geelen MJH, Slot JW. The autophagic and endocytic pathways converge at the nascent autophagic vacuoles. J Cell Biol. 1997;136:61–70. doi: 10.1083/jcb.136.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saito C, Ueda T, Abe H, Wada Y, Kuroiwa T, Hisada A, et al. A complex and mobile structure forms a distinct subregion within the continuous vacuolar membrane in young cotyledons of Arabidopsis. Plant J. 2002;29:245–55. doi: 10.1046/j.0960-7412.2001.01189.x. [DOI] [PubMed] [Google Scholar]
- 23.Inoue Y, Suzuki T, Hattori M, Yoshimoto K, Ohsumi Y, Moriyasu Y. AtATG genes, homologs of yeast autophagy genes, are involved in constitutive autophagy in Arabidopsis root tip cells. Plant Cell Physiol. 2006;47:1641–52. doi: 10.1093/pcp/pcl031. [DOI] [PubMed] [Google Scholar]
- 24.Yano K, Suzuki T, Moriyasu Y. Constitutive autophagy in plant root cells. Autophagy. 2007;3:360–2. doi: 10.4161/auto.4158. [DOI] [PubMed] [Google Scholar]
- 25.Inoue Y, Moriyasu Y. Autophagy is not a main contributor to the degradation of phospholipids in tobacco cells cultured under sucrose starvation conditions. Plant Cell Physiol. 2006;47:471–80. doi: 10.1093/pcp/pcj013. [DOI] [PubMed] [Google Scholar]
- 26.Marty F. Cytochemical studies on GERL, provacuoles, and vacuoles in root meristematic cells of Euphorbia. Proc Natl Acad Sci U S A. 1978;75:852–6. doi: 10.1073/pnas.75.2.852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Amelunxen F, Heinze U. Zur Entwicklung der Vacuole in Testa-Zellun des Leinsamens. Eur J Cell Biol. 1984;35:343–54. [On the development of the vacuole in the testa cells on Linum seeds] [Google Scholar]
- 28.Yano K, Hattori M, Moriyasu Y. A novel type of autophagy occurs together with vacuole genesis in miniprotoplasts prepared from tobacco culture cells. Autophagy. 2007;3:215–21. doi: 10.4161/auto.3739. [DOI] [PubMed] [Google Scholar]
- 29.Yoshimoto K, Hanaoka H, Sato S, Kato T, Tabata S, Noda T, et al. Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell. 2004;16:2967–83. doi: 10.1105/tpc.104.025395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kirisako T, Baba M, Ishihara N, Miyazawa K, Ohsumi M, Yoshimori T, et al. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J Cell Biol. 1999;147:435–46. doi: 10.1083/jcb.147.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]