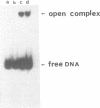
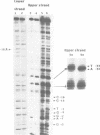
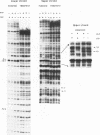
Abstract
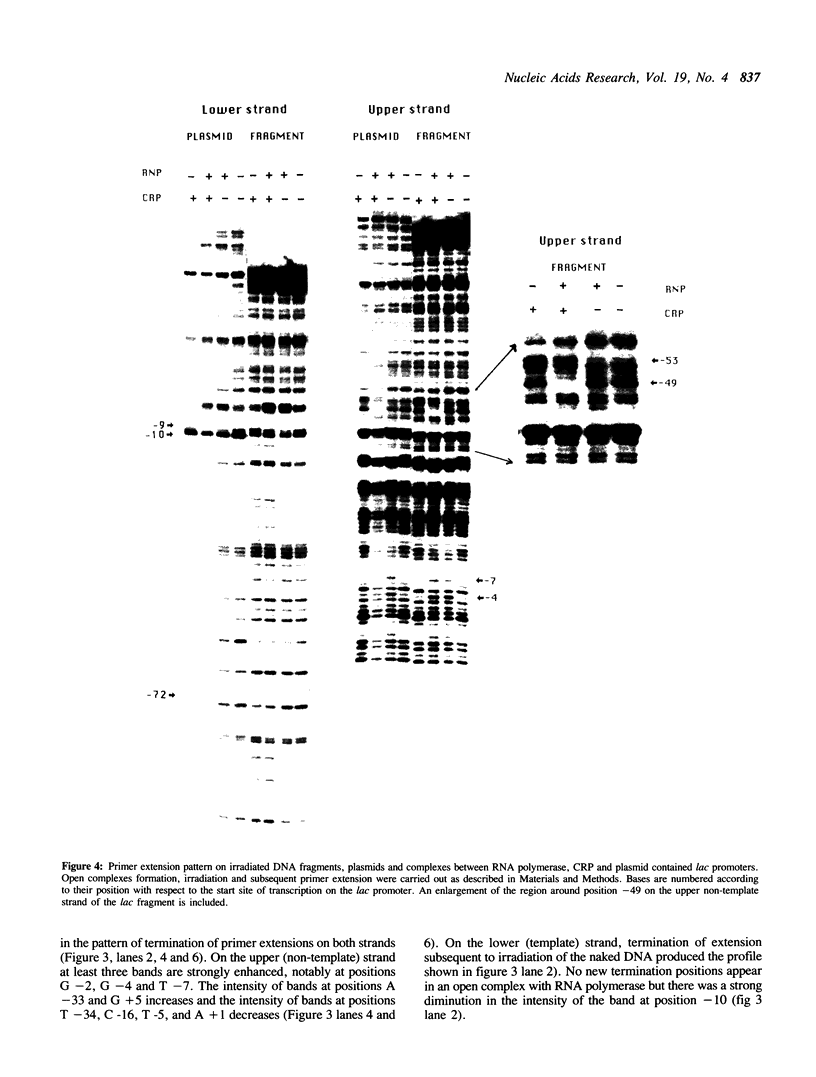
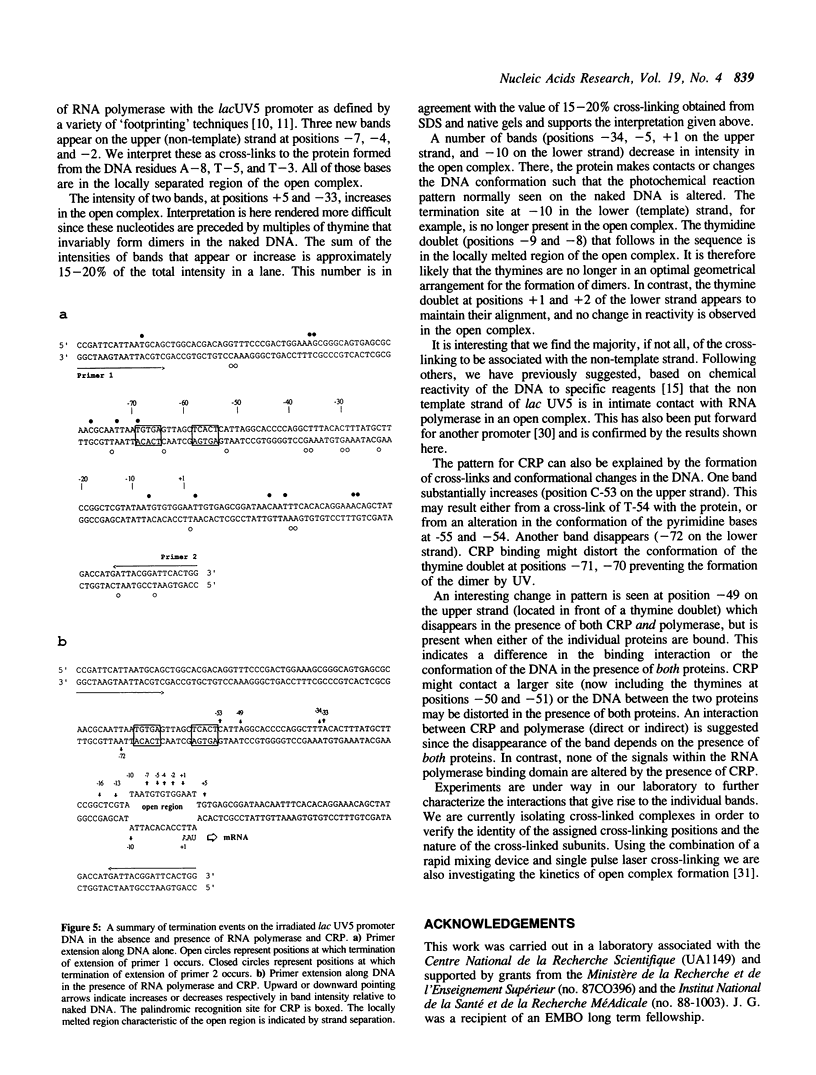
We report the results of photo-cross-linking of RNA polymerase and the cyclic AMP receptor protein (CRP) to the lac UV5 promoter region carried on either a linear fragment or a supercoiled plasmid. We have devised a protocol that allows the localisation of bases in contact with the protein. RNA polymerase makes contacts within the -10 and -35 regions of the promoter, essentially on the non-template strand. The CRP contact points found in a binary complex are affected by the formation of the ternary complex containing RNA polymerase. Supercoiling has no effect on the position of contacts in any of the complexes. These conclusions were derived from experiments performed using a generally applicable, non-interfering technique that reveals direct contacts between proteins and nucleic acids in nucleoprotein complexes.
Full text
PDF


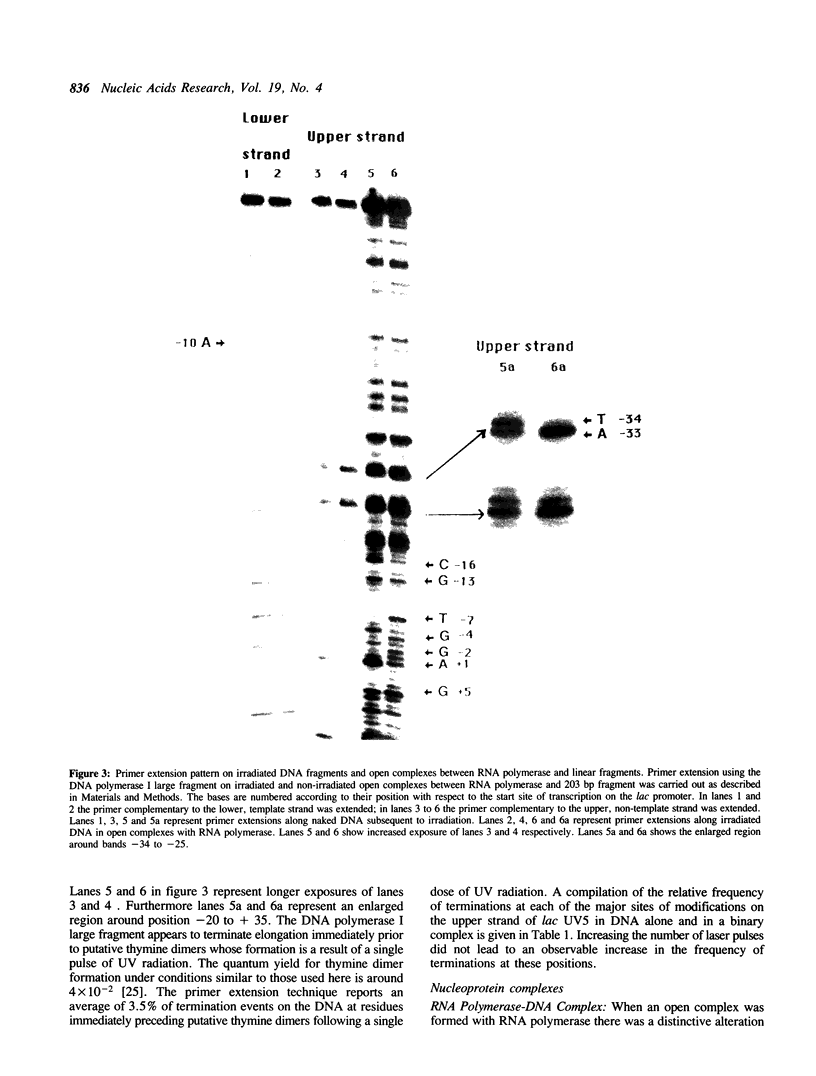


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Becker M. M., Wang J. C. Use of light for footprinting DNA in vivo. Nature. 1984 Jun 21;309(5970):682–687. doi: 10.1038/309682a0. [DOI] [PubMed] [Google Scholar]
- Buckle M., Buc H. Fine mapping of DNA single-stranded regions using base-specific chemical probes: study of an open complex formed between RNA polymerase and the lac UV5 promoter. Biochemistry. 1989 May 16;28(10):4388–4396. doi: 10.1021/bi00436a040. [DOI] [PubMed] [Google Scholar]
- Burgess R. R., Jendrisak J. J. A procedure for the rapid, large-scall purification of Escherichia coli DNA-dependent RNA polymerase involving Polymin P precipitation and DNA-cellulose chromatography. Biochemistry. 1975 Oct 21;14(21):4634–4638. doi: 10.1021/bi00692a011. [DOI] [PubMed] [Google Scholar]
- Chan B., Minchin S., Busby S. Unwinding of duplex DNA during transcription initiation at the Escherichia coli galactose operon overlapping promoters. FEBS Lett. 1990 Jul 2;267(1):46–50. doi: 10.1016/0014-5793(90)80284-p. [DOI] [PubMed] [Google Scholar]
- Chenchick A., Beabealashvilli R., Mirzabekov A. Topography of interaction of Escherichia coli RNA polymerase subunits with lac UV5 promoter. FEBS Lett. 1981 Jun 1;128(1):46–50. doi: 10.1016/0014-5793(81)81076-9. [DOI] [PubMed] [Google Scholar]
- Franklin W. A., Doetsch P. W., Haseltine W. A. Structural determination of the ultraviolet light-induced thymine-cytosine pyrimidine-pyrimidone (6-4) photoproduct. Nucleic Acids Res. 1985 Jul 25;13(14):5317–5325. doi: 10.1093/nar/13.14.5317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosaini L. R., Brown A. M., Sturtevant J. M. Scanning calorimetric study of the thermal unfolding of catabolite activator protein from Escherichia coli in the absence and presence of cyclic mononucleotides. Biochemistry. 1988 Jul 12;27(14):5257–5261. doi: 10.1021/bi00414a046. [DOI] [PubMed] [Google Scholar]
- Harrison C. A., Turner D. H., Hinkle D. C. Laser crosslinking of E. coli RNA polymerase and T7 DNA. Nucleic Acids Res. 1982 Apr 10;10(7):2399–2414. doi: 10.1093/nar/10.7.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heumann H., Lederer H., Baer G., May R. P., Kjems J. K., Crespi H. L. Spatial arrangement of DNA-dependent RNA polymerase of Escherichia coli and DNA in the specific complex. A neutron small angle scattering study. J Mol Biol. 1988 May 5;201(1):115–125. doi: 10.1016/0022-2836(88)90443-3. [DOI] [PubMed] [Google Scholar]
- Hillel Z., Wu C. W. Photochemical cross-linking studies on the interaction of Escherichia coli RNA polymerase with T7 DNA. Biochemistry. 1978 Jul 25;17(15):2954–2961. doi: 10.1021/bi00608a003. [DOI] [PubMed] [Google Scholar]
- Hockensmith J. W., Kubasek W. L., Vorachek W. R., von Hippel P. H. Laser cross-linking of nucleic acids to proteins. Methodology and first applications to the phage T4 DNA replication system. J Biol Chem. 1986 Mar 15;261(8):3512–3518. [PubMed] [Google Scholar]
- Jeppesen C., Jensen K. F., Nielsen P. E. A specific and efficient photoreaction between E. coli RNA polymerase and T+1 in the lacUV5 or deoP1 promoter. Nucleic Acids Res. 1988 Oct 25;16(20):9545–9555. doi: 10.1093/nar/16.20.9545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsrud L. Contacts between Escherichia coli RNA polymerase and a lac operon promoter. Proc Natl Acad Sci U S A. 1978 Nov;75(11):5314–5318. doi: 10.1073/pnas.75.11.5314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkegaard K., Buc H., Spassky A., Wang J. C. Mapping of single-stranded regions in duplex DNA at the sequence level: single-strand-specific cytosine methylation in RNA polymerase-promoter complexes. Proc Natl Acad Sci U S A. 1983 May;80(9):2544–2548. doi: 10.1073/pnas.80.9.2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe P. A., Hager D. A., Burgess R. R. Purification and properties of the sigma subunit of Escherichia coli DNA-dependent RNA polymerase. Biochemistry. 1979 Apr 3;18(7):1344–1352. doi: 10.1021/bi00574a034. [DOI] [PubMed] [Google Scholar]
- Malan T. P., Kolb A., Buc H., McClure W. R. Mechanism of CRP-cAMP activation of lac operon transcription initiation activation of the P1 promoter. J Mol Biol. 1984 Dec 25;180(4):881–909. doi: 10.1016/0022-2836(84)90262-6. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Panyutin I. G., Kovalsky O. I., Budowsky E. I. Irradiation of the template with high-intensity (pulse-laser) ultraviolet light results in DNA-polymerase termination events at deoxyguanosine residues. FEBS Lett. 1989 Dec 4;258(2):274–276. doi: 10.1016/0014-5793(89)81672-2. [DOI] [PubMed] [Google Scholar]
- Park C. S., Hillel Z., Wu C. W. Molecular mechanism of promoter selection in gene transcription. I. Development of a rapid mixing-photocrosslinking technique to study the kinetics of Escherichia coli RNA polymerase binding to T7 DNA. J Biol Chem. 1982 Jun 25;257(12):6944–6949. [PubMed] [Google Scholar]
- Pehrson J. R. Thymine dimer formation as a probe of the path of DNA in and between nucleosomes in intact chromatin. Proc Natl Acad Sci U S A. 1989 Dec;86(23):9149–9153. doi: 10.1073/pnas.86.23.9149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasse-Dwight S., Gralla J. D. KMnO4 as a probe for lac promoter DNA melting and mechanism in vivo. J Biol Chem. 1989 May 15;264(14):8074–8081. [PubMed] [Google Scholar]
- Schaeffer F., Kolb A., Buc H. Point mutations change the thermal denaturation profile of a short DNA fragment containing the lactose control elements. Comparison between experiment and theory. EMBO J. 1982;1(1):99–105. doi: 10.1002/j.1460-2075.1982.tb01131.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebenlist U., Gilbert W. Contacts between Escherichia coli RNA polymerase and an early promoter of phage T7. Proc Natl Acad Sci U S A. 1980 Jan;77(1):122–126. doi: 10.1073/pnas.77.1.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebenlist U., Simpson R. B., Gilbert W. E. coli RNA polymerase interacts homologously with two different promoters. Cell. 1980 Jun;20(2):269–281. doi: 10.1016/0092-8674(80)90613-3. [DOI] [PubMed] [Google Scholar]
- Simpson R. B. The molecular topography of RNA polymerase-promoter interaction. Cell. 1979 Oct;18(2):277–285. doi: 10.1016/0092-8674(79)90047-3. [DOI] [PubMed] [Google Scholar]
- Spassky A., Kirkegaard K., Buc H. Changes in the DNA structure of the lac UV5 promoter during formation of an open complex with Escherichia coli RNA polymerase. Biochemistry. 1985 May 21;24(11):2723–2731. doi: 10.1021/bi00332a019. [DOI] [PubMed] [Google Scholar]