Abstract
Many DNA-hypermethylated cancer genes are occupied by the Polycomb (PcG) repressor complex in embryonic stem cells (ESCs). Their prevalence in the full spectrum of cancers, the exact context of chromatin involved, and their status in adult cell renewal systems are unknown. Using a genome-wide analysis, we demonstrate that ∼75% of hypermethylated genes are marked by PcG in the context of bivalent chromatin in both ESCs and adult stem/progenitor cells. A large number of these genes are key developmental regulators, and a subset, which we call the “DNA hypermethylation module,” comprises a portion of the PcG target genes that are down-regulated in cancer. Genes with bivalent chromatin have a low, poised gene transcription state that has been shown to maintain stemness and self-renewal in normal stem cells. However, when DNA-hypermethylated in tumors, we find that these genes are further repressed. We also show that the methylation status of these genes can cluster important subtypes of colon and breast cancers. By evaluating the subsets of genes that are methylated in different cancers with consideration of their chromatin status in ESCs, we provide evidence that DNA hypermethylation preferentially targets the subset of PcG genes that are developmental regulators, and this may contribute to the stem-like state of cancer. Additionally, the capacity for global methylation profiling to cluster tumors by phenotype may have important implications for further refining tumor behavior patterns that may ultimately aid therapeutic interventions.
It is now recognized that abnormal DNA hypermethylation at gene promoter CpG island contributes to tight transcriptional repression of many genes in cancer (Jones and Baylin 2007). For many well-defined tumor-suppressor genes, this epigenetic silencing constitutes an alternative to genetic mechanisms that mediate loss of function (Jones and Baylin 2007). Importantly, virtually every single tumor type harbors hundreds of epigenetically silenced coding genes or microRNAs (Jones and Baylin 2007; Lujambio and Esteller 2007). It is known that a subset of DNA-hypermethylated genes are important tumor suppressor genes. However, a more complete understanding of which additional subsets of genes are methylated in tumors is important for characterizing the role of DNA hypermethylation within tumor cells.
Our laboratory (Ohm et al. 2007) and others (Schlesinger et al. 2007; Widschwendter et al. 2007) provided a clue for the possibility of an instructive program for promoter DNA hypermethylation rather than random targeting. Schlesinger et al. showed that de novo DNA hypermethylation is mediated by the presence of H3K27Me3. Ohm et al. and Widschwendter et al. both demonstrate the strong association between genes with H3K27Me3 and de novo DNA hypermethylation. It was found that many genes with de novo promoter hypermethylation in colon cancer were among the subset of genes marked in embryonic cells by repressive Polycomb group proteins (PcG), in the context of “bivalent” chromatin. In the embryonic system, the bivalent chromatin occurs in non-DNA-methylated promoter CpG islands and consists of the simultaneous presence of the repressive PcG mark, H3K27Me3, and the active transcription marks, H3K4Me2/Me3 (Mikkelsen et al. 2007). Such chromatin is thought to maintain low, but poised, transcription of genes that otherwise upon active transcription would cause lineage commitment and disruption of stemness and the self-renewal status of ESCs (Squazzo et al. 2006; Mikkelsen et al. 2007; Ku et al. 2008).
Thus far, these relationships between abnormal DNA hypermethylation and PcG have emerged from comparing embryonic cells with cancer cells. Cancer cells possess hallmarks of embryonic stem cells, namely, the capacity for self-renewal and an undifferentiated cell state (Clarke and Fuller 2006; Ben-Porath et al. 2008; Kim et al. 2010), which are a fundamental property of the most tumorigenic, and often therapy-resistant, subpopulations of cells in human cancers (Trumpp and Wiestler 2008; Sharma et al. 2010). However, most human cancers are not derived from embryonic cells, and the relationship between cancer and adult cell renewal systems has been less clearly described. To understand the evolution of abnormal DNA hypermethylation in genes that display gene promoter PcG occupancy in embryonic cells, we have analyzed the nature of chromatin occupancy in adult stem and progenitor cells for genes hypermethylated in cancer. We have taken an integrated genomics approach using genome-wide chromatin analyses of adult mesenchymal stem cells (MSCs), their differentiated osteoblast progeny, and osteosarcoma cells (Fig. 1A), and cross-referenced these data with multiple databases. We compared gene expression, PcG marking, and DNA-hypermethylation status for genes that undergo abnormal, de novo promoter CpG-island DNA hypermethylation during human tumorigenesis.
Figure 1.
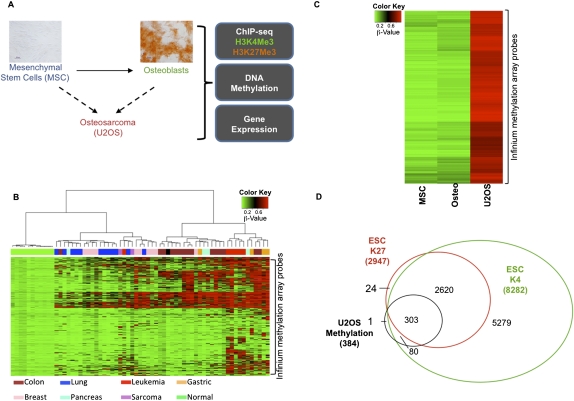
Genes with promoter-proximal CpG hypermethylation in osteosarcoma are greatly enriched for a bivalent chromatin history in ESCs and are down-regulated in osteosarcoma cells compared with ESCs. (A) Schematic of the experimental design. von Kossa staining shows differentiation of MSCs to osteoblasts (scale bar, 50 μm). (B) Heat map of β-values for 2489 Infinium probes (Supplemental Table 1) within CpG islands of 1891 genes for cell lines corresponding to different tumor types. These are probes that are methylated in at least one cell line and not methylated in any of the normal cells (see Methods). Different cell types are shown below the plot. (C) Heat map of the β-values for MSCs, osteoblasts, and U2OS cells. Genes with β-value >0.75 in U2OS and <0.25 in MSCs and osteoblasts were selected as hypermethylated genes in osteosarcoma. (D) Extent of overlap of the osteosarcoma-hypermethylated genes with genes marked by H3K4Me3 or H3K27Me3 in ESCs. Osteosarcoma-hypermethylated genes overlap significantly with ESC-bivalent genes (P-value < 0.001).
Results
Cancer-specific promoter CpG-island DNA hypermethylation in osteosarcoma occurs primarily at genes with PcG-marked chromatin in embryonic and adult stem cells
Previous studies have shown that approximately half of the genes with abnormal, promoter, CpG-island DNA hypermethylation in colon cancer tend to be PcG-marked in ESCs (Ohm et al. 2007; Schlesinger et al. 2007; Widschwendter et al. 2007). To understand this on a genome-wide level, we used the Illumina Infinium methylation array to identify a comprehensive list of genes that are specifically DNA-hypermethylated in tumor cell lines, including the osteosarcoma cell line U2OS, but not in normal cells (Fig. 1B). Infinium probes in CpG islands that had a β-value >0.75 were called “methylated,” while those with values <0.25 were called “unmethylated” (see Methods). The normal cells included ESCs, and the adult cell renewal system related to osteosarcoma, mesenchymal stem cells (MSCs), and osteoblasts (Fig. 1B). We then compared the 399 hypermethylated genes identified in U2OS (Fig. 1C; Supplemental Table 1) with a list of genes in ESCs that have H3K4Me3 and/or H3K27Me3 enrichment in a 5000-bp region upstream of and downstream from the transcription start site (TSS) by mining previously published ChIP-seq data (Ku et al. 2008) and from analyzing ChIP-seq data for ESCs from the NCBI Epigenome Roadmap project. A striking percentage, ∼80%, of the 384 methylated osteosarcoma genes annotated on the methylation arrays and the ChIP-seq data are marked by the PcG signature H3K27Me3 in ESCs, and this virtually always occurs in the setting of bivalent chromatin (Fig. 1D; gene examples in Fig. 2 A–D). In contrast, the set of unmethylated genes does not have a higher incidence of being PcG/bivalent-marked in ESCs; rather, they are H3K4Me3-marked in ESCs (Supplemental Fig. 1A).
Figure 2.
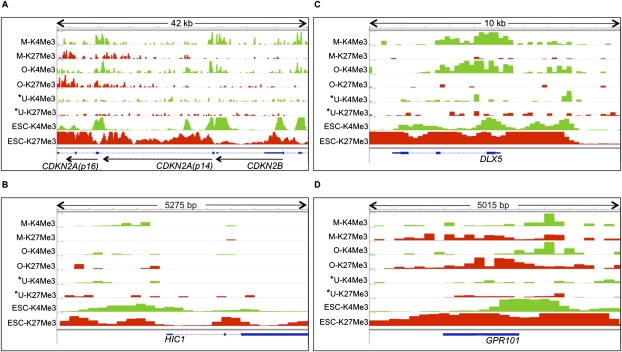
Plots of H3K4Me3 and H3K27Me3 enrichment in MSCs (M-K4Me3 and M-K27Me3), osteoblasts (O-K4Me3 and O-K27Me3), U2OS (U-K4Me3 and U-K27Me3), and ESCs (ESC-K4Me3 and ESC-K27Me3). (*) Cell type in which the gene is identified as methylated. (Green and red boxes) Peak height normalized to background enrichment for H3K4Me3 and H3K27Me3, respectively. Genes represented include CDKN2A (p14), CDKN2B (p15), and CDKN2A (p16) (A), DLX5 (B), HIC1 (C), and GPR101 (D).
The cancer-specific methylation of genes with bivalent marking in ESCs is highlighted by key aspects of the data. Very few genes (15) are differentially methylated in MSCs/osteoblasts compared with ESCs (data not shown). Of these, 13 are bivalently marked in ESCs as opposed to the 303 such genes that are hypermethylated in U2OS. This finding firmly illustrates that promoter, CpG-island methylation of ESC-bivalent genes is a highly specific abnormality in cancers and not a general phenomenon during lineage differentiation.
Having established this relationship between osteosarcoma hypermethylated genes and bivalent chromatin in ESCs, we queried the chromatin marking of these genes in the non-embryonic bone marrow compartment cells, MSCs, and osteoblasts, from which osteosarcomas are thought to arise (Fig. 1A; Cleton-Jansen et al. 2009; Levings et al. 2009; Siclari and Qin 2010). The epigenetic landscape of these cells was examined by ChIP-seq profiling for H3K4Me3 and H3K27Me3 in adult bone-marrow-derived MSCs and osteoblasts derived from these MSCs (Fig. 1A). Figure 2 demonstrates the characteristic patterns of H3K4Me3 and H3K27Me3 marks in the MSCs, osteoblasts, U2OS, and ESCs for selected U2OS-hypermethylated genes. These genes were selected to give a representation of the ChIP-enrichment patterns in MSCs/osteoblasts of U2OS-hypermethylated genes with no particular implied function (GPR101), based on their importance as cancer-hypermethylated genes with demonstrated tumor-suppressor functions, CDKN2A/CDKN2B locus (Merlo et al. 1995) and HIC1 (Wales et al. 1995; Chen et al. 2003), and a gene that should be activated during bone differentiation, DLX5 (Lee et al. 2006a). At the CDKN2A/CDKN2B locus in Chr 9p, the promoters of CDKN2A (p14), CDKN2B, and CDKN2A (p16) are hypermethylated in U2OS, as observed in the Infinium methylation platform and previous studies (McGarvey et al. 2007). Interestingly, all three gene promoters showed low H3K4Me3 and H3K27Me3 levels in the osteosarcoma cells (Fig. 2A). In MSCs, CDKN2A (p16) is bivalent, while CDKN2A (p14) and CDKN2B retain only the active H3K4Me3 mark (Fig. 2A). In contrast, all three genes have bivalent marks in the ESCs, suggesting that during normal development in the bone lineage, these genes resolve from a poised, bivalent state to a monovalent state. The other example genes show a range of markings similar to those observed in the CDKN2A (p16) locus, i.e., bivalency in ESCs that either remains bivalent or resolves to monovalency in the progenitor cells, with lack of both marks in the osteosarcoma cells (Fig. 2B–D). The distribution of chromatin patterns in U2OS for the majority of U2OS-hypermethylated genes is similar to the individual genes shown in Figure 2 (Supplemental Fig. 2A). Although ∼30% of the U2OS-hypermethylated promoters have low levels of H3K4Me3 enrichment, their ChIP enrichment and gene expression levels are comparable to those for promoters of low-expressing genes in U2OS (21st to 40th percentile gene expression group) (Supplemental Fig. 2B/C). At the global level, H3K4Me3 and H3K27Me3 are reduced in U2OS compared with MSCs/osteoblasts at the U2OS-hypermethylated gene promoters as observed in Figure 2 for the example hypermethylated genes (Supplemental Fig. 2D). The loss of H3K4Me3 is more pronounced than that of H3K27Me3, suggesting that, in their hypermethylated state in cancers, silencing at these promoters may be mediated more by the loss of H3K4Me3.
Global analysis of the chromatin patterns for the U2OS-hypermethylated genes in ESCs, MSCs, and osteoblasts is shown in Figure 3 (top panels). In MSCs and osteoblasts, U2OS-hypermethylated genes vary from being marked by H3K27Me3, H3K4Me3, bivalent, or none of the marks. In general, the MSC and osteoblast genomes have very few bivalent-marked genes compared with ESCs, with the majority of ESC-bivalent genes having resolved their chromatin to either H3K27Me3 or H3K4Me3 alone (Supplemental Fig. 1B). This could be due to the heterogeneity of the MSCs in culture (stromal stem cells, multipotent stromal cells, mesenchymal stromal cells, and multipotent adult progenitor cells; MAPC) (Wagner and Ho 2007). However, most significantly, in MSCs and osteoblasts, the PcG mark H3K27Me3 is more predictive of genes that undergo DNA hypermethylation in osteosarcoma (P-value = 8.1 × 10−73) than the H3K4Me3 mark. Thus, PcG-marked genes in both embryonic and adult progenitor systems have a higher chance of getting hypermethylated in cancers.
Figure 3.
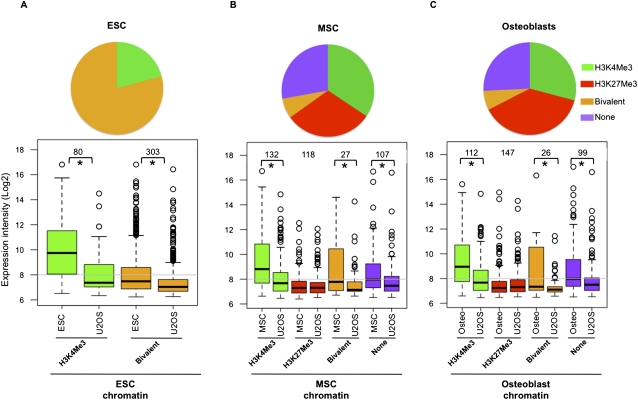
Enrichment of chromatin marks and levels of gene expression in ESCs, MSCs, and osteoblasts for genes methylated in U2OS. (Pie chart) Proportion of genes methylated in U2OS that have the different chromatin marks or none of the marks analyzed in this study in ESCs (A), MSCs (B), or osteoblasts (C). (Lower panels) Array expression intensities (log2) in ESCs, MSCs, or osteoblasts of gene probes constituting these subsets; in each case, they are compared to that in U2OS. (*) Significant gene expression changes (P-value < 0.005). (Gray line in the plot) Median log2 intensity of all genes in the corresponding cell type. See also Supplemental Figures 1 and 3.
In the context of the above chromatin relationships, we examined the expression of the U2OS-hypermethylated genes versus ESCs, MSCs, and osteoblasts using previous (Ohm et al. 2010) and present Agilent whole-genome expression arrays. Each bottom panel in Figure 3 compares the expression between U2OS and either ESCs, MSCs, or osteoblasts for U2OS-hypermethylated genes marked by the different chromatin marks in the latter cell types. In the case of ESCs, only the H3K4Me3 and bivalent-marked genes are shown because none of the U2OS-hypermethylated genes are marked only by H3K27Me3 as observed in Figure 1D. As would be predicted, ESC-bivalent genes display low expression in ESCs when compared with the genes marked only by the active transcription mark H3K4Me3, and these genes also have low expression in the U2OS cells (Fig. 3A). Importantly, these genes have lower expression in U2OS compared with ESCs, indicating that CpG methylation causes a tighter transcriptional repression (Fig. 3A).
Since ESC-bivalent genes have low expression, it is possible that, in general, genes that are silenced in ESCs are susceptible to hypermethylation in cancers. To test this, we stratified CpG-island genes into five groups in the order of increasing gene expression in ESCs (0–20, 21–40, 41–60, 61–80, 81–100 percentiles) (Supplemental Fig. 3A). Thus the 0th–20th and 81st–100th groups respectively represent the silenced/low- and high-expressing genes in ESCs. The 0–20th group is significantly enriched for U2OS-hypermethylated genes (odds ratio of 3.3) (Supplemental Fig. 3B) in contrast to the lack of enrichment in the 81st–100th group (odds ratio of 0.6) (Supplemental Fig. 3C). Most importantly, 303 of the 384 U2OS-hypermethylated genes are enriched for the ESC-bivalent genes (odds ratio of 7.6) (Supplemental Fig. 3D). Thus, the U2OS-hypermethylated genes are far more enriched in the ESC-bivalent genes than in the low-expressing/silenced gene group (compare odds ratio of 7.6 vs. 3.3). Furthermore, 91% of U2OS-hypermethylated genes that overlap with the 0th–20th group are bivalent in ESCs (Supplemental Fig. 3E). As would be expected, the expression in ESCs of U2OS-hypermethylated genes is similar to the ESC-bivalent genes (Supplemental Fig. 3F). Thus, although low-expressing genes do show a bias to get hypermethylated in cancer, it is the bivalent genes, which cluster within the low-expressing groups, that have a far higher probability of being hypermethylated.
As also would be expected, osteosarcoma-hypermethylated genes marked only by H3K4Me3 in MSCs and osteoblasts are silenced in U2OS, while those marked only by H3K27Me3 in MSCs and osteoblasts are similarly down-regulated in all cell types (Fig. 3B,C). Thus, this set of H3K27Me3-marked genes in MSCs and osteoblasts start off in a low-expression state and continue to maintain this state when DNA-methylated (Fig. 3B,C). These observations are consistent with a previous study that reported a change from a PcG-dependent silencing to methylation-dependent silencing in normal prostate cells and their cancer counterpart (Gal-Yam et al. 2008).
The substantial proportion of osteosarcoma hypermethylated genes with the active H3K4Me3 mark in MSCs and osteoblasts are of particular interest (Fig. 3B,C). These genes, which are expressed in the progenitor cells, have reduced expression when DNA-hypermethylated in the cancer cells (Fig. 3B,C). Therefore, this population of genes may constitute an example of true de novo silencing in association with tumor-specific DNA hypermethylation. This is in contrast to just a tightening of silencing seen for most of the other low-expressing genes and marked by bivalent chromatin or PcG alone in normal stem and/or progenitor cells. Interestingly, the set of U2OS-hypermethylated genes marked by H3K4Me3 in MSCs and osteoblasts generally showed lower expression in the respective latter cells compared with the whole set of H3K4Me3-marked genes (data not shown). This might indicate that the hypermethylation in cancer may partly be targeted to genes that initially have a low-expression state.
Taken together, these data clearly show that promoter hypermethylation in osteosarcoma has an extraordinarily high tendency to occur at genes marked by bivalent chromatin in ESCs. In the adult osteoblast self-renewal system, these genes resolve to monovalency, and those that resolve to H3K27Me3 are more likely to become methylated in osteosarcoma. Given the evidence that the PcG complex may recruit DNA methyltransferases and other silencing factors (Vire et al. 2006), the above data suggest that such a mechanism is involved in acquisition of methylation at a majority of cancer-specific hypermethylated genes and that this could possibly arise in adult stem and/or progenitor cells.
Tumor-specific promoter CpG-island methylation in various cancer cell lines/primary cancers traces to bivalently marked genes in non-embryonic stem cells
The above data appear to indicate that cancer-specific DNA-hypermethylated genes are marked by bivalent chromatin in ESCs or H3K27Me3 in adult MSCs/osteoblasts. Thus, we determined (1) if this relationship exists in other tumor types and its corresponding adult tissue stem and early progenitor cells and (2) how universal this relationship is when tumors are compared with unrelated adult stem progenitor cells. To this end, we compared the epigenetic landscape of the cancer-specific hypermethylated genes in the osteosarcoma cells and a spectrum of tumors to previously published ChIP-seq data from peripheral blood–derived adult hematopoietic stem cells (HSCs), as marked by a uniform population of CD34+/CD133+ cells, and erythroid progenitor cells derived from these HSCs and marked by CD34−/CD36+ (Cui et al. 2009). We thus extended our analyses to include a list of 1891 genes that, as determined by Infinium, are DNA-hypermethylated in our compendium of 54 tumor cell lines (Fig. 1B), reflecting most common human cancers. For this objective, it is important to make certain that our analyses are not biased to genes that are DNA-hypermethylated only in cancer cells in culture. This has been a controversial issue wherein it has been conjectured that cell culture introduces abnormal, promoter DNA hypermethylation of the type being studied here (Baylin and Bestor 2002). By comparing the cell culture methylation array data to that of the Cancer Genome Atlas (TCGA) patient data for hundreds of primary breast, colon, and lung cancers, we show that genes identified as hypermethylated in cancer cell lines are indeed hypermethylated in the primary cancers, and thus are fully relevant to tumorigenesis (Supplemental Fig. 4A–C). We find that 80%–99% of the hypermethylated genes from each specific tumor type in culture are also hypermethylated in 5% or more of the respective type of primary human cancers analyzed (Supplemental Fig. 4D). Importantly, a very low proportion (13%–18%) of genes unmethylated in the corresponding cell lines were observed to be hypermethylated in 5% or more of the respective type of primary human cancers, thus highlighting that hypermethylated genes identified in cell cultures indeed represent hypermethylated genes in primary tumors.
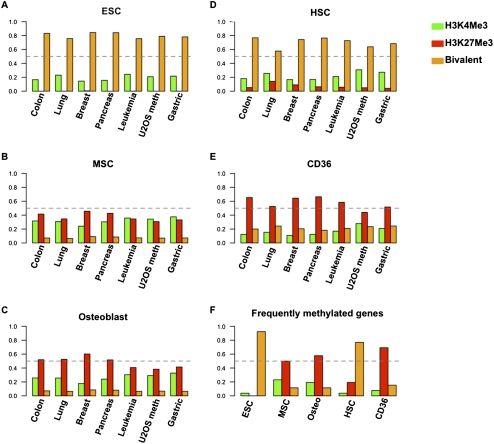
Interestingly, we find that a vast majority of the hypermethylated genes in the different tumor types bear bivalent chromatin in ESCs and the non-embryonic HSCs, ∼80% and 70% respectively (Fig. 4A,D) (P-values < 0.001). Also, the majority of DNA-hypermethylated genes in other cancer types, including the U2OS-hypermethylated genes, are PcG-marked in the CD36+ erythroid progenitor cells (Fig. 4E) (P-values < 0.001), and also have a much lower, but significant, overlap with PcG-marked genes in MSCs and osteoblasts (Fig. 4B,C) (P-values < 0.001). Also, the set of most frequently methylated genes in the different tumor types matches closely to the above-mentioned chromatin patterns in the embryonic/adult stem cells (Fig. 4F).
Figure 4.
Genes hypermethylated in a wide range of cancers are enriched for bivalent chromatin in ESCs and HSCs or H3K27Me3 in MSCs and CD36 erythroid progenitor cells or osteoblasts. Plots of the percentage of methylated genes tracing to each chromatin category (indicated by color for the chromatin type) of each of the cell types listed are indicated for each of the tumor types listed on the x-axis. (Dashed lines) The 50% mark for fraction of methylated genes. (A–E) Plots track the histone marks present for methylated genes based on the chromatin composition of ESCs, HSCs, MSCs, CD36, and osteoblasts, respectively. (F) Summary of the chromatin marks in each cell type for genes that are hypermethylated in at least 50% of cell lines in each tumor type.
In summary, the above data show that a vast majority of all genes with de novo, promoter CpG-island DNA hypermethylation in a full spectrum of primary human cancers and cancer cell lines are marked by bivalent chromatin and/or PcG (H3K27Me3) in adult stem cells or early progenitors, respectively. Furthermore, hypermethylated genes, regardless of the cancer tissue type in which they occur, tend to be Polycomb-marked in all of the adult stem/progenitor systems analyzed here.
Developmental regulator genes adopt abnormal DNA hypermethylation and have lower expression in cancer than in stem and progenitor cells
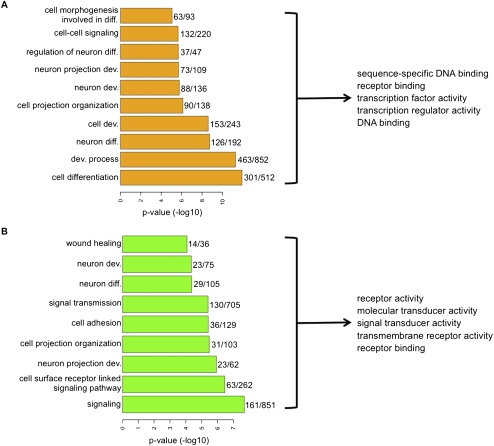
PcG-marked genes in ESCs, which nearly all reside in the context of bivalent chromatin, have previously been shown to be enriched for developmental regulators when compared with genes present in the whole genome (Lee et al. 2006b). To test if the cancer-specific hypermethylated genes that are bivalent in ESCs belong to defined biological functions in the Gene Ontology (GO) groups, we pooled the bivalent genes in ESCs that become DNA-hypermethylated across different tumor types and tested for enrichment of GO groups against the ESC-bivalent genes (see Methods). For these analyses, the enrichment of GO classes was tested against a background list of genes that have the same mark in ESCs, are represented in the ChIP-seq data, and have a CpG-island promoter probe in the methylation array data. We observe that bivalent genes in ESCs that become DNA-hypermethylated across the different tumor types are particularly enriched for genes important in development and differentiation (Fig. 5A; Supplemental Table 2). This finding suggests that within the ESC-bivalent genes, the subset of genes involved in developmental processes are targeted by DNA hypermethylation in cancers. Genes with a role in neuronal development are especially prominent in this category (Fig. 5A; Supplemental Table 2). Strikingly, among the developmental genes, most encode for proteins that interact with DNA and/or are involved in regulation of gene expression as transcription factors (Fig. 5A; Supplemental Table 2). In the cancer-hypermethylated genes that are only H3K4Me3-marked, while developmental genes involved in neuronal development are enriched, in contrast to the bivalent set, genes involved in cell signaling, such as kinases, and cell surface properties, are also enriched (Fig. 5B; Supplemental Tables 2, 3). GO analysis of genes unmethylated in the cancers that are either bivalent or H3K4Me3-marked in ESCs are not enriched for any of the developmental regulators or other gene categories enriched in the methylated gene sets (data not shown), thus supporting our observation that developmental regulators are targeted by DNA hypermethylation in cancer.
Figure 5.
Cancer-specific hypermethylated genes are enriched for functions associated with developmental regulation and comprise a list of genes that are tightly silenced in tumor cells compared with MSCs. Enrichment of biological processes in hypermethylated genes marked by bivalency (A) or H3K4Me3 (B) in ESCs against a background list of genes that, respectively, have the same mark and are present on the methylation array. (Right) Top five molecular function categories of the genes constituting the significantly enriched biological processes. The numbers adjacent to the bars are the total number of genes in the bivalent or H3K4Me3-marked genes in that category (denominator) and the number of genes in the category that are DNA hypermethylated in 50% or more of all the cancers examined (numerator). Supplemental Tables 2 and 3 give the gene lists used for GO analysis and all the top categories identified.
A “DNA hypermethylation module” within the ESC signature of cancer
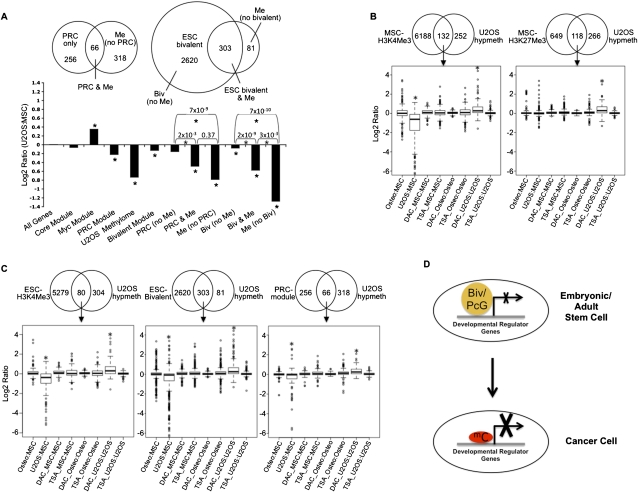
The relationships in the preceding sections regarding stem cell chromatin characteristics, gene expression, and cancer-hypermethylated genes invite the question of how modifications in tumor cells might relate to gene expression patterns in the ESC signature of cancer (Ben-Porath et al. 2008; Kim et al. 2010). In this concept, Ben-Porath et al. (2008) have proposed that particularly poorly differentiated tumors display an expression pattern reminiscent of ESCs that largely involves over-expression of genes but also repression of expression of PcG target genes. Recently, Kim et al. (2010) modified the concept to show that the highly expressed genes common to ESCs and cancer are not the targets of embryonic transcription factors, such as POU5F1, NANOG, and SOX2, termed the “core module” that directly programs ESCs, but rather are predominantly MYC complex genes, termed the “Myc module.” Importantly, Kim et al. also report that genes occupied by PcG in ESC cells, termed the “PRC module,” are down-regulated in ESCs and cancer and suggest that these genes emphasize the key role of PcG-complex proteins and their targets in cancer initiation and/or progression.
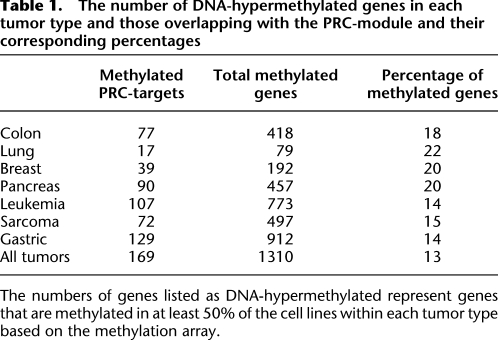
Considering that the PRC module is essentially a subset of the bivalent-marked genes in ESCs, we examined the role of DNA hypermethylation in silencing these genes in cancer by analyzing the relative expression in the osteosarcoma cell line to that of MSCs (Fig. 6A). In osteosarcoma cells, we find in agreement with Kim et al. (2010) that the core module is not significantly over-expressed in osteosarcoma (Fig. 6A). We also observe that the Myc-module genes are significantly over-expressed, and the PRC-module genes are significantly down-regulated (P-values < 0.01). As expected, a majority (93%) of the 322 PRC genes are also bivalent in ESCs. Among these genes, 66 are hypermethylated in the osteosarcoma cells (Fig. 6A). Strikingly, it is the subset of the PRC-module genes hypermethylated in osteosarcoma that drives the significantly silenced state of this module compared with the unmethylated PRC genes (Fig. 6A, PRC & Me and PRC [no Me], respectively). Furthermore, although the whole set of ESC-bivalent genes is only marginally down-regulated in U2OS compared with MSCs, the U2OS-hypermethylated subset is markedly down-regulated in U2OS, significantly more than the subset of ESC-bivalent genes that lack DNA hypermethylation (Fig. 6A, Bivalent module, Biv & Me, and Biv [no Me], respectively). Importantly, further examination of the DNA-methylated genes across all tumors revealed that PRC-module genes are frequently methylated in all of the tumor cell lines being examined, suggesting that silencing of the PRC module by DNA hypermethylation is a common feature of all cancers (Table 1).
Figure 6.
PRC-module and ESC-bivalent genes undergo significant repression of expression upon hypermethylation in cancer. (A, top panel) Venn plot of the overlap between the 384 genes hypermethylated in U2OS and the PRC module (Kim et al. 2010) (left) or the ESC-bivalent module (right). (Bottom panel) Expression changes between U2OS and MSCs for the Myc, Core, PRC-modules, ESC-bivalent genes, and the set of genes hypermethylated in osteosarcoma (termed “U2OS Methylome”). (*) Significant change from background based on Wilcoxon rank-sum test. The methylated genes that are not bivalent in ESCs (called “Me no bivalent”) are all marked by H3K4Me3. (B) Effect of 5-deoxy-aza-cytidine (DAC) and trichostatin (TSA) on re-expression of U2OS-hypermethylated genes marked by H3K4Me3 or H3K27Me3 in MSCs (B) or those marked by H3K4Me3, bivalent or PRC in ESCs (C). The log2 gene expression ratios for these genes from Agilent gene expression data summarizing the relative expression of osteoblasts versus MSCs, U2OS versus MSCs, and the relative expression of DAC- or TSA-treated MSCs, osteoblasts, and U2OS versus their control-treated counterparts are shown in B and C. (*) P-values < 0.001. (D) Model depicting the change of repressive mechanism from bivalent/PcG-marking to DNA hypermethylation. Developmental regulator genes are marked by bivalent (embryonic/adult stem cell) or PcG (adult stem/early progenitor cells) and have relatively low levels of expression. In tumors, DNA hypermethylation (mC) of bivalent/PcG-marked genes in stem cells leads to tight silencing at these genes. This epigenetic switch is responsible for the stable silencing seen at developmental regulators in cancer cells and may explain how cancer cells recapitulate aspects of the transcriptional and phenotypic state of stem cells.
Table 1.
The number of DNA-hypermethylated genes in each tumor type and those overlapping with the PRC-module and their corresponding percentages
A recent study in breast cancer suggests that methylation mainly targets genes that are already in a state of low expression in normal tissues, and that the majority of these genes frequently cannot be reactivated by using demethylating agents (Sproul et al. 2011). This has raised the controversy that DNA-hypermethylation status in tumors is not functionally relevant because the genes are transcriptionally incompetent. Thus, we examined the ability of the DNA demethylating agent, 5-deoxy-aza-cytidine (DAC), versus the histone deactylase inhibitor (HDACi), trichostatin (TSA), to reactivate methylated genes in osteosarcoma that are H3K4Me3-marked (active) or H3K27Me3 (PcG)–marked (silenced) in MSCs. DAC is well established to re-express genes with dense promoter DNA hypermethylation, while TSA does so poorly (Cameron et al. 1999; Suzuki et al. 2002). In agreement with these previous studies, in U2OS cells, hypermethylated genes in general are re-expressed upon DAC treatment and show little change upon TSA treatment, while in MSCs and osteoblasts, these same genes, when not DNA-methylated, are more expressed upon TSA treatment compared with DAC treatment (Supplemental Fig. 5). When these genes are categorized based on their chromatin nature in MSCs, we find that the H3K4Me3-marked genes in MSCs that are methylated in osteosarcoma undergo significant down-regulation (average 0.4-fold) as expected and, importantly, are reactivated on an average about 1.4-fold specifically by DAC and not TSA (P-value = 1.8 × 10−15) (Fig. 6B). Moreover, genes marked by H3K27Me3 in MSCs and that get methylated in osteosarcoma generally are not further down-regulated, but are significantly up-regulated on an avergae about 1.3-fold by DAC and not TSA (P-value = 2.2 × 10−16) (Fig. 6B). The U2OS-hypermethylated genes that are H3K4Me3-marked in MSCs are partially re-expressed toward the levels in normal MSCs/osteoblasts in response to DAC. Those marked by H3K27Me3 in MSCs actually attain higher levels in U2OS cells after DAC than in MSCs (data not shown). In addition, DNA-hypermethylated genes in cancers that are bivalent-marked in ESCs had a significant increase in expression after DAC treatment (average 1.4-, 1.35-, and 1.3-fold up-regulation, respectively, for genes belonging to the ESC-H3K4Me3, bivalent, or PRC module) but importantly little change with TSA treatment (P-value = 2.2 × 10−11 for all three sets of genes) (Fig. 6C). Furthermore, these genes showed little increase in DAC-treated MSCs or osteoblasts in which they are not DNA-hypermethylated (Fig. 6C). These results indicate that hypermethylated genes in cancers, even those that are otherwise maintained in a suppressed state by PcG marking in normal progenitor cells, are competent to reactivate upon removal of the methylation mark. However, the effects of such gene reactivation on phenotypic responses of cancer cells must be further explored in subsequent studies, and our data highlight the importance of such investigation.
We conclude from the above studies that a critical component of the ESC signature in cancer is a subset of genes from the PRC module that undergo de novo CpG-island methylation. The tighter repression of gene expression and the nature of these genes further strengthen the potentially important difference between bivalent marking and DNA hypermethylation for the cancer genes under study (Fig. 6D).
Matching DNA hypermethylation patterns with cancer subcategories
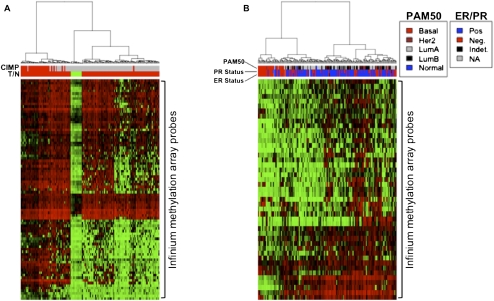
Previous studies have shown that tumors form distinct subtypes based on the degree of CpG-island methylation. A group of colon cancers, particularly those from the right colon, can have extensive promoter CpG-island hypermethylation, termed the CpG-island methylator phenotype (CIMP+) (Toyota et al. 1999; Weisenberger et al. 2006). These often have a better prognosis despite appearing less well differentiated, contain BRAF mutations, and contain a subset of tumors that have epigenetic silencing of mismatch repair genes and a resultant microsatellite hypermutation phenotype (Weisenberger et al. 2006). We find that PRC-module genes crisply separate the primary TCGA colon tumor samples into CIMP+ and CIMP− colon tumors (Fig. 7A). Thus, there is one group of hypermethylated genes common to both tumor types and another group methylated predominantly in the CIMP+ tumors as categorized by a previously defined marker panel (Weisenberger et al. 2006). Of note, the H3K4Me3- and bivalent-marked gene categories in ESCs also separate these tumors (Supplemental Fig. 6).
Figure 7.
Hierarchical cluster analysis of primary TCGA colon and breast cancer samples based on Infinium β-values of PRC-module genes identified as methylated in colon (A) or breast (B) cancer cell lines. (A) Colon tumors are classified as CIMP+ (top red bars in the heat map) if eight or more of a 12-CIMP marker panel (Weisenberger et al. 2006) are methylated in a tumor sample, and otherwise classified as CIMP− (top gray bars in the heat map). (Red) Tumor and (green) normal (T/N) samples. (B) Breast tumor samples were classified as estrogen (ER)/progesterone (PR) positive or negative and also classified according to their gene expression (PAM50) classes. The PRC-module genes sufficiently discern the CIMP+ and CIMP− tumors in both colon and breast samples and tightly cluster important breast cancers subtypes that are independently defined based on gene expression signatures.
In breast cancers, three groups have recently suggested a CIMP+ and CIMP−-like phenotype (Holm et al. 2010; Fackler et al. 2011; Fang et al. 2011). In this scenario, DNA hypermethylation of genes linked to a PcG lineage history in ESCs occurs more frequently in luminal-type, estrogen and progesterone expression–positive (ER+, PR+) cancers with a better survival than in basal-type cancers with a more aggressive phenotype (Holm et al. 2010; Fang et al. 2011). Holm et al. (2010) suggest that abnormal repression is primarily maintained by PcG factors in cancers arising from basal bipotential stem/progenitor cells in the mammary tissue, while DNA hypermethylation dominates abnormal silencing in cancers from the more restricted, epithelial stem/progenitor cells. We now show that the PRC-module genes identified as methylated in the breast cancer cell lines separate TCGA primary breast cancers into basal versus luminal tumors defined by the gene expression signatures for these tumor types (PAM50) (Perou et al. 1999), with much less frequent hypermethylation in the former (Fig. 7B). Again, similar clustering separating basal from the other subtypes is also seen with the hypermethylated genes from breast cell lines that are H3K4Me3- or bivalent-marked in ESCs (Supplemental Fig. 6). This latter observation is of interest because a recent study suggested that CIMP+ subtypes in breast cancer are mainly driven by the methylation of genes marked by PcG in ESCs (Fang et al. 2011). However, we find that CIMP+ appears to be driven by a pressure to hypermethylate genes belonging to all of the ESC chromatin lineage categories we have characterized in this study.
Thus, we show that the hypermethylated gene subsets that are either Polycomb- or H3K4Me3-marked in ESCs efficiently classify tumor subcategories in breast and colon cancers, and thus potentially provide a new category of hypermethylated genes within the ESC cancer signature. Furthermore, it demonstrates that the genes methylated in cell lines and with the different chromatin lineage histories are relevant to the biology of the cancer subtypes.
Discussion
Previous work with small numbers of candidate genes have suggested that ∼50% of the DNA-hypermethylated genes in colon cancer are PcG-marked in ESCs (Ohm et al. 2007). However, there is an insufficient understanding of the relationships between cancer-specific promoter CpG-island DNA hypermethylation and their history for PcG marking in adult stem/progenitor cells. Our current findings not only strengthen the relationships between promoter, CpG-island, DNA hypermethylation in cancer, and their history for PcG marking in ESCs, but extend this concept in several novel ways. Firstly, we find that, compared with previous reports, a much higher percentage of hypermethylated genes in cancer harbor bivalent chromatin in ESCs. Second, these hypermethylated genes, which predominantly consist of developmental genes, have lower expression in cancer cells prior to drug-induced re-expression (Figs. 3, 6B,C). Thus, DNA hypermethylation may constitute a tighter mode of gene silencing than bivalency or H3K27Me3 occupation, making induction of key genes or groups of genes potentially more difficult in terms of preventing abnormal growth or self-renewal. This observation is compatible with our previous chromosome conformational studies for the GATA4 gene (Tiwari et al. 2008). When DNA-hypermethylated in colon cancer cells, GATA4 is silenced and assumes a much tighter repressive chromatin state than in embryonic carcinoma cells, where it is bivalent-marked and not DNA-hypermethylated. Our current findings strongly suggest that such tightening of silencing occurs at multiple developmental regulators, which may provide a selective advantage to cancers.
We observe that developmental regulator genes that are PcG-marked in ESCs (bivalent) or adult stem/progenitor cells have the highest probability to be methylated in cancers, thus causing these promoters to be permanently silenced and unavailable to normal differentiation cues, and hence potentially supporting tumorigenesis (Fig. 6D). However, this is true only for a small proportion of PcG-occupied genes; the majority remain unmethylated in cancer. Thus, other features of these genes, in addition to these genes being PcG-marked at various stages of development, might play a role in their susceptibility to methylation. A recent study showed that age-related methylation of specific CpG islands, which also gets methylated in cancers, targets genes marked by PcG in stem cells (Teschendorff et al. 2010). Thus, the mechanisms underlying the selective targeting of a subset of PcG-marked genes seems to operate during normal physiology and disease. However, more studies are needed in order to determine how just a subset of PcG-marked genes is methylated in cancer.
Studies so far have focused on silencing of individual genes by hypermethylation. We observe that within the set of bivalently marked genes in ESCs, multiple developmental regulators are significantly biased toward silencing by hypermethylation in cancers. This observation comes to the fore only when all hypermethylated genes across different tumor types are pooled together and analyzed for enrichment of GO categories within the group of ESC-bivalent genes. Thus, the pressure to deregulate key developmental pathways is a common principle among different tumor types, although the individual genes that get silenced might differ. Furthermore, the selective advantage to tumors may arise from cumulative silencing of a group of developmental regulators rather than just individual genes. By laying out the hypermethylated gene sets across various tumors and by tracing their chromatin nature in ESCs and adult stem/progenitor cells, we provide here gene sets that should be analyzed for their role in deregulating developmental pathways that might potentially result in tumorigenesis.
Recent studies have shown that cancers maintain a stem cell–like gene expression program, significantly advancing our understanding of how cancers fundamentally differ from normal differentiated cells. A critical subset of genes in the ESC signature are PcG-regulated in ESCs and abnormally down-regulated in cancers. We show here that the silencing of a significant portion of these genes is correlated with hypermethylation in different tumor types. Thus, relative to the concepts of the ESC signature of cancers proposed by Weinberg and colleagues (Ben-Porath et al. 2008) and further refined by Orkin and colleagues (Kim et al. 2010), our studies here suggest that an important component of the ESC signature in cancers is attributable to DNA hypermethylation, which we term the “DNA hypermethylation module.” This module defines subsets of genes within the PRC module of the ESC cancer signature and among genes marked by bivalency in early cell compartments, which have undergone DNA hypermethylation during tumor initiation and/or progression. Further studies on this subset of genes, whose silencing is associated with tumorigenesis, will further elucidate the mechanisms by which cancer cells adopt traits of stem cells, namely, poor capacity to differentiate and unlimited self-renewal.
Methods
Cell culture and differentiation
Adult bone marrow–derived MSCs were cultured and differentiated as previously described (Jaiswal et al. 1997). Staining for mineral deposition was used to monitor differentiation. A list of the tumor cell lines used are provided in Supplemental Table 4. Normal cells used for Infinium methylation analyses are: human embryonic stem cell (H1ES), MSC, osteoblasts, human mammary epithelial (X184D, X240L, X96R, X122L, X153L), prostate epithelial passage 6 (PRECP6), normal colon (NC5A), human bronchial epithelial passage 8 (HBECP8) (Supplemental Table 1). Standard culturing conditions described in the American Type Culture Collection (ATCC) were used for culturing the various tumor cells.
Genome-wide DNA-hypermethylation analysis
The Infinium methylation array (Illumina) (Bibikova and Fan 2009) was used to analyze bisulfite-treated DNA (EZ DNA-Hypermethylation Kit, Zymo Research) as previously described (Easwaran et al. 2010). β-Values were computed as the signal of the methylation-specific probe over the sum of the signals of the methylation- and unmethylated-specific probes. Probes with poor signals (P-value > 0.05) were not considered. In vitro DNA-methylated genomic DNA (IVD) and DNA from DKO cells genetically deleted for DNMT1 and DNMT3B (Rhee et al. 2002) served as methylated and unmethylated controls, respectively. All probes were mapped to the human genome Build 36.3 (National Center for Biotechnology Information, NCBI) using the bowtie algorithm and ultrafast and memory-efficient alignment of short DNA sequences (Langmead et al. 2009) with genome annotation via the matching release of the Ensembl database. X-linked genes were removed from analyses, and only probes with β-values that are high in IVD and low in the DKO DNA were considered for analysis. Furthermore, only probes positioned within CpG islands from −1000 to +200 bp around transcription start sites (TSS) were analyzed. Probes that have β-value ≥ 0.75 in the cancer cell lines and ≤0.25 in all the normal cell lines/tissues were called “methylated.” Genes methylated in at least 50% of the tumor cell lines of a specific tissue type were defined as “methylated genes” for that tumor type. Heat maps are based on hierarchical clustering of β-values using Euclidean distance and Ward's algorithm (R package gplots).
Infinium methylation array data for primary lung, colon, and breast tumors were used to test if genes identified as methylated in cell lines are also methylated in primary tumors. Data were downloaded from the Cancer Genome Atlas (TCGA; http://tcga.cancer.gov/dataportal; file names are provided in Supplemental Table 5). β-Values were calculated as above followed by quantile normalization of samples within the same tumor type. A conservative cut-off of β-values > 0.5 in the TCGA data was used to call samples as methylated.
For CIMP classification of the TCGA primary colon cancer samples, the β-values for a set of 14 genes (BCL2, BDNF, CACNA1G, CALCA, CRABP1, DLEC1, GATA3, IGF2, KL, NEUROG1, RUNX3, SOCS1) (Weisenberger et al. 2006) were obtained, and only genes that were variable across the tumor samples (standard deviation > 0.15), and therefore capable of discerning methylation differences, were used for CIMP classification (two of the genes, HOXA1, NR3C1, were dropped in this process). CIMP+ tumors were classified as tumors that have eight or more of these genes methylated.
Chromatin immunoprecipitation-sequencing (ChIP-seq) and analysis
ChIP was performed as described previously (Bernstein et al. 2005). Briefly, cells were cross-linked with formaldehyde, lysed, and sonicated to get chromatin fragments ranging from 200 to 600 bp. Chromatin was captured by antibodies to H3K4Me3 (Millipore) or H3K27Me3 (kind gift from Thomas Jenuwein/ Nicholas Shukeir) and ProteinA/G magnetic beads (DynaBeads). Captured chromatin was washed in low-salt, high-salt buffers and TE (McGarvey et al. 2008) and subjected to simultaneous elution, de-cross-linking, and Proteinase-K treatment at 65°C in elution buffer followed by phenol:chloroform:isoamyl alcohol extraction and ethanol precipitation. ChIPed DNA was quantified using PicoGreen (Invitrogen).
Purified DNA was then used to prepare a sequencing library and sequenced on Applied Biosystems SOLiD (V3). Sequencing reads were aligned to hg18 (NCBI36) using Bioscope 1.2.1, which guarantees finding all alignments between the first 25 bp of the read (seed) and the reference sequence with up to two mismatches. Each match is extended to the full length of the read, scoring 1 point for matching and −2 points for mismatching bases. The read is trimmed to the length with the highest score. If there is only one alignment or if an alignment scores significantly higher than the others for the same read, it is considered unique and reported.
Aligned reads were analyzed using Model-based Analysis of ChIP-seq (MACS) (Zhang et al. 2008) to detect regions enriched for the histone marks (called peaks) with default settings. Peaks were detected from the raw aligned reads and from reads that were extended 300 bp (average size of the sonicated fragments) as described before (Mikkelsen et al. 2007). Although the two approaches identified peaks that were mostly similar, the latter allowed detection of regions with low but broader enrichment, especially for H3K27Me3. The peaks identified are provided in Supplemental Table 6 (labeled as Peaks1/2). Genes that had peak(s) within 5000 bp upstream/downstream from their transcription start sites were called “enriched.” The Integrative Genomics Viewer (IGV) browser was used to display ChIP-seq data (Robinson et al. 2011).
The following H3K4Me3 and H3K27Me3 ChIP-seq data from previously published sources were included in our analysis (using MACS): (a) ChIP-seq data for human ESCs from the following two sources: (1) GEO data sets GSM327662 and GSM327663 (Ku et al. 2008); (2) NIH Roadmap Epigenomics project (http://www.roadmapepigenomics.org/) data sets GSM433170 and GSM433167. (b) ChIP-seq data sets for HSC (GSM317587 and GSM317584) and CD36-positive erythroid cells (GSM317597 and GSM317594) (Cui et al. 2009). To get an exhaustive list of H3K4Me3- and H3K27Me3-marked genes in ESCs, we pooled the peak calls from our analysis using MACS and that described by Ku et al. (2008).
The list of H3K4Me3 and H3K27Me3 peaks detected in the different cell types is provided in Supplemental Table 6. The list of genes that have peaks within 5000 bp from the TSS is provided in Supplemental Table 7.
Gene expression analysis
RNA was extracted from MSCs, osteoblasts, U2OS, and ESCs (H9) and processed for hybridization on an Agilent 4×44K array as previously described (Schuebel et al. 2007). For comparing the gene expression intensities for the different groups of genes in Figure 3, the data sets were derived from cells that were either mock-treated or 5-aza-deoxycytidine-treated. The mock channels were extracted and quantile-normalized using the R statistical computing platform and limma package from the Bioconductor bioinformatics software project (http://bioinf.wehi.edu.au/limma/). The log2 intensities of probes are plotted in Figure 3. In addition to using our ESC (H9 cells) data, we compared previously published Affymetrix data (GEO data set GSE22246) for the same cells (Tchieu et al. 2010). To directly compare the expression of the different gene sets between U2OS and MSCs in Figure 6B, RNA from the two cell types were paired and hybridized followed by lowess normalization. Log2 ratios of probe intensities are plotted in Figure 6B.
The gene expression subtypes of the TCGA breast cancer data were identified using the gene expression signature described previously (Perou et al. 1999).
Gene class enrichment analysis
Gene Ontology (GO) analyses for biological processes (BP) and molecular function (MF) were performed using the GOSTATS package in Bioconductor (Falcon and Gentleman 2007). The set of genes that were used as universe for a query list of genes was chosen based on the following two criteria: (1) all genes marked by the same mark as the query list; (2) only genes present in the ChIP-seq data and the Illumina methylation array data (with annotated CpG-island probes). Only categories that are below a false discovery rate (Benjamini and Hochberg) of 0.01 are reported (Benjamini and Hochberg 1995). MF categories of the genes that constitute the significant BP categories were analyzed by testing if the genes that constitute each of the significant BP categories are enriched for MF categories (universe defined as above).
Data access
The ChIP-seq and microarray data used in this study have been submitted to the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) under accession no. GSE27900.
Acknowledgments
We thank Leslie Cope and Ludmila Danilova for help with analyzing TCGA data. We acknowledge the support of the Johns Hopkins Sidney Kimmel Comprehensive Cancer Center's Next Generation Sequencing Center (http://nextgenseq.onc.jhmi.edu/nextgenseq/) and its personnel, including Jennifer Meyers and Sarah Wheelan, for assisting with all stages of the ChIP-seq experiments. This work was supported by a State of MD TEDCO grant, the National Institutes of Health (NIH) HL099775, National Institute of Environmental Health Sciences (NIEHS) ES015226, ES011858, and a Hodson Junior Scholarship Award.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.131169.111.
Freely available online through the Genome Research Open Access option.
References
- Baylin S, Bestor TH 2002. Altered methylation patterns in cancer cell genomes: Cause or consequence? Cancer Cell 1: 299–305 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y 1995. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol 57: 289–300 [Google Scholar]
- Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, Weinberg RA 2008. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet 40: 499–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ III, Gingeras TR, et al. 2005. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell 120: 169–181 [DOI] [PubMed] [Google Scholar]
- Bibikova M, Fan JB 2009. GoldenGate assay for DNA methylation profiling. Methods Mol Biol 507: 149–163 [DOI] [PubMed] [Google Scholar]
- Cameron EE, Bachman KE, Myohanen S, Herman JG, Baylin SB 1999. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat Genet 21: 103–107 [DOI] [PubMed] [Google Scholar]
- Chen WY, Zeng X, Carter MG, Morrell CN, Chiu Yen RW, Esteller M, Watkins DN, Herman JG, Mankowski JL, Baylin SB 2003. Heterozygous disruption of Hic1 predisposes mice to a gender-dependent spectrum of malignant tumors. Nat Genet 33: 197–202 [DOI] [PubMed] [Google Scholar]
- Clarke MF, Fuller M 2006. Stem cells and cancer: Two faces of Eve. Cell 124: 1111–1115 [DOI] [PubMed] [Google Scholar]
- Cleton-Jansen AM, Anninga JK, Briaire-de Bruijn IH, Romeo S, Oosting J, Egeler RM, Gelderblom H, Taminiau AH, Hogendoorn PC 2009. Profiling of high-grade central osteosarcoma and its putative progenitor cells identifies tumourigenic pathways. Br J Cancer 101: 1909–1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui K, Zang C, Roh TY, Schones DE, Childs RW, Peng W, Zhao K 2009. Chromatin signatures in multipotent human hematopoietic stem cells indicate the fate of bivalent genes during differentiation. Cell Stem Cell 4: 80–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easwaran HP, Van Neste L, Cope L, Sen S, Mohammad HP, Pageau GJ, Lawrence JB, Herman JG, Schuebel KE, Baylin SB 2010. Aberrant silencing of cancer-related genes by CpG hypermethylation occurs independently of their spatial organization in the nucleus. Cancer Res 70: 8015–8024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fackler MJ, Umbricht C, Williams D, Argani P, Cruz LA, Merino VF, Teo WW, Zhang Z, Huang P, Visvanathan K, et al. 2011. Genome-wide methylation analysis identifies genes specific to breast cancer hormone receptor status and risk of recurrence. Cancer Res 71: 6195–6207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcon S, Gentleman R 2007. Using GOstats to test gene lists for GO term association. Bioinformatics 23: 257–258 [DOI] [PubMed] [Google Scholar]
- Fang F, Turcan S, Rimner A, Kaufman A, Giri D, Morris LG, Shen R, Seshan V, Mo Q, Heguy A et al. 2011. Breast cancer methylomes establish an epigenomic foundation for metastasis. Sci Transl Med 3: 75ra25 doi: 10.1126/scitranslmed.3001875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal-Yam EN, Egger G, Iniguez L, Holster H, Einarsson S, Zhang X, Lin JC, Liang G, Jones PA, Tanay A 2008. Frequent switching of Polycomb repressive marks and DNA hypermethylation in the PC3 prostate cancer cell line. Proc Natl Acad Sci 105: 12979–12984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm K, Hegardt C, Staaf J, Vallon-Christersson J, Jonsson G, Olsson H, Borg A, Ringner M 2010. Molecular subtypes of breast cancer are associated with characteristic DNA methylation patterns. Breast Cancer Res 12: R36 doi: 10.1186/bcr2590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal N, Haynesworth SE, Caplan AI, Bruder SP 1997. Osteogenic differentiation of purified, culture-expanded human mesenchymal stem cells in vitro. J Cell Biochem 64: 295–312 [PubMed] [Google Scholar]
- Jones PA, Baylin SB 2007. The epigenomics of cancer. Cell 128: 683–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Woo AJ, Chu J, Snow JW, Fujiwara Y, Kim CG, Cantor AB, Orkin SH 2010. A Myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell 143: 313–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, Presser A, Nusbaum C, Xie X, Chi AS, et al. 2008. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet 4: e1000242 doi: 10.1371/journal.pgen.1000242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25 doi: 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Lee YM, Kim MJ, Choi JY, Park EK, Kim SY, Lee SP, Yang JS, Kim DS 2006a. Methylation of the mouse DIx5 and Osx gene promoters regulates cell type-specific gene expression. Mol Cells 22: 182–188 [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al. 2006b. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell 125: 301–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levings PP, McGarry SV, Currie TP, Nickerson DM, McClellan S, Ghivizzani SC, Steindler DA, Gibbs CP 2009. Expression of an exogenous human Oct-4 promoter identifies tumor-initiating cells in osteosarcoma. Cancer Res 69: 5648–5655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lujambio A, Esteller M 2007. CpG island hypermethylation of tumor suppressor microRNAs in human cancer. Cell Cycle 6: 1455–1459 [PubMed] [Google Scholar]
- McGarvey KM, Greene E, Fahrner JA, Jenuwein T, Baylin SB 2007. DNA methylation and complete transcriptional silencing of cancer genes persist after depletion of EZH2. Cancer Res 67: 5097–5102 [DOI] [PubMed] [Google Scholar]
- McGarvey KM, Van Neste L, Cope L, Ohm JE, Herman JG, Van Criekinge W, Schuebel KE, Baylin SB 2008. Defining a chromatin pattern that characterizes DNA-hypermethylated genes in colon cancer cells. Cancer Res 68: 5753–5759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlo A, Herman JG, Mao L, Lee DJ, Gabrielson E, Burger PC, Baylin SB, Sidransky D 1995. 5′ CpG island methylation is associated with transcriptional silencing of the tumour suppressor p16/CDKN2/MTS1 in human cancers. Nat Med 1: 686–692 [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. 2007. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448: 553–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohm JE, McGarvey KM, Yu X, Cheng L, Schuebel KE, Cope L, Mohammad HP, Chen W, Daniel VC, Yu W, et al. 2007. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat Genet 39: 237–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohm JE, Mali P, Van Neste L, Berman DM, Liang L, Pandiyan K, Briggs KJ, Zhang W, Argani P, Simons B, et al. 2010. Cancer-related epigenome changes associated with reprogramming to induced pluripotent stem cells. Cancer Res 70: 7662–7673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perou CM, Jeffrey SS, van de Rijn M, Rees CA, Eisen MB, Ross DT, Pergamenschikov A, Williams CF, Zhu SX, Lee JC, et al. 1999. Distinctive gene expression patterns in human mammary epithelial cells and breast cancers. Proc Natl Acad Sci 96: 9212–9217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee I, Bachman KE, Park BH, Jair KW, Yen RW, Schuebel KE, Cui H, Feinberg AP, Lengauer C, Kinzler KW, et al. 2002. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature 416: 552–556 [DOI] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP 2011. Integrative genomics viewer. Nat Biotechnol 29: 24–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlesinger Y, Straussman R, Keshet I, Farkash S, Hecht M, Zimmerman J, Eden E, Yakhini Z, Ben-Shushan E, Reubinoff BE, et al. 2007. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet 39: 232–236 [DOI] [PubMed] [Google Scholar]
- Schuebel KE, Chen W, Cope L, Glockner SC, Suzuki H, Yi JM, Chan TA, Van Neste L, Van Criekinge W, van den Bosch S, et al. 2007. Comparing the DNA hypermethylome with gene mutations in human colorectal cancer. PLoS Genet 3: 1709–1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma SV, Lee DY, Li B, Quinlan MP, Takahashi F, Maheswaran S, McDermott U, Azizian N, Zou L, Fischbach MA, et al. 2010. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell 141: 69–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siclari VA, Qin L 2010. Targeting the osteosarcoma cancer stem cell. J Orthop Surg 5: 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sproul D, Nestor C, Culley J, Dickson JH, Dixon JM, Harrison DJ, Meehan RR, Sims AH, Ramsahoye BH 2011. Transcriptionally repressed genes become aberrantly methylated and distinguish tumors of different lineages in breast cancer. Proc Natl Acad Sci 108: 4364–4369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squazzo SL, O'Geen H, Komashko VM, Krig SR, Jin VX, Jang SW, Margueron R, Reinberg D, Green R, Farnham PJ 2006. Suz12 binds to silenced regions of the genome in a cell-type-specific manner. Genome Res 16: 890–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Gabrielson E, Chen W, Anbazhagan R, van Engeland M, Weijenberg MP, Herman JG, Baylin SB 2002. A genomic screen for genes upregulated by demethylation and histone deacetylase inhibition in human colorectal cancer. Nat Genet 31: 141–149 [DOI] [PubMed] [Google Scholar]
- Tchieu J, Kuoy E, Chin MH, Trinh H, Patterson M, Sherman SP, Aimiuwu O, Lindgren A, Hakimian S, Zack JA, et al. 2010. Female human iPSCs retain an inactive X chromosome. Cell Stem Cell 7: 329–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teschendorff AE, Menon U, Gentry-Maharaj A, Ramus SJ, Weisenberger DJ, Shen H, Campan M, Noushmehr H, Bell CG, Maxwell AP, et al. 2010. Age-dependent DNA methylation of genes that are suppressed in stem cells is a hallmark of cancer. Genome Res 20: 440–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari VK, McGarvey KM, Licchesi JD, Ohm JE, Herman JG, Schubeler D, Baylin SB 2008. PcG proteins, DNA methylation, and gene repression by chromatin looping. PLoS Biol 6: 2911–2927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP 1999. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci 96: 8681–8686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trumpp A, Wiestler OD 2008. Mechanisms of disease: Cancer stem cells—targeting the evil twin. Nat Clin Pract Oncol 5: 337–347 [DOI] [PubMed] [Google Scholar]
- Vire E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, Morey L, Van Eynde A, Bernard D, Vanderwinden JM, et al. 2006. The Polycomb group protein EZH2 directly controls DNA methylation. Nature 439: 871–874 [DOI] [PubMed] [Google Scholar]
- Wagner W, Ho AD 2007. Mesenchymal stem cell preparations–comparing apples and oranges. Stem Cell Rev 3: 239–248 [DOI] [PubMed] [Google Scholar]
- Wales MM, Biel MA, el Deiry W, Nelkin BD, Issa JP, Cavenee WK, Kuerbitz SJ, Baylin SB 1995. p53 activates expression of HIC-1, a new candidate tumour suppressor gene on 17p13.3. Nat Med 1: 570–577 [DOI] [PubMed] [Google Scholar]
- Weisenberger DJ, Siegmund KD, Campan M, Young J, Long TI, Faasse MA, Kang GH, Widschwendter M, Weener D, Buchanan D, et al. 2006. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet 38: 787–793 [DOI] [PubMed] [Google Scholar]
- Widschwendter M, Fiegl H, Egle D, Mueller-Holzner E, Spizzo G, Marth C, Weisenberger DJ, Campan M, Young J, Jacobs I, et al. 2007. Epigenetic stem cell signature in cancer. Nat Genet 39: 157–158 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol 9: R137 doi: 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]