RNA silencing pathways in worms are still poorly understood at a molecular level. Here, Yang et al. discover novel components of the C. elegans RNAi pathway. The RDE-10/RDE-11 complex associates with target mRNAs and is identified as acting parallel to the nuclear RNAi pathway. The complex affects secondary siRNA abundance and is implicated in amplifying the exogenous RNAi response.
Keywords: RDE-10, RDE-11, RNAi, C. elegans
Abstract
The molecular mechanisms for target mRNA degradation in Caenorhabditis elegans undergoing RNAi are not fully understood. Using a combination of genetic, proteomic, and biochemical approaches, we report a divergent RDE-10/RDE-11 complex that is required for RNAi in C. elegans. Genetic analysis indicates that the RDE-10/RDE-11 complex acts in parallel to nuclear RNAi. Association of the complex with target mRNA is dependent on RDE-1 but not RRF-1, suggesting that target mRNA recognition depends on primary but not secondary siRNA. Furthermore, RDE-11 is required for mRNA degradation subsequent to target engagement. Deep sequencing reveals a fivefold decrease in secondary siRNA abundance in rde-10 and rde-11 mutant animals, while primary siRNA and microRNA biogenesis is normal. Therefore, the RDE-10/RDE-11 complex is critical for amplifying the exogenous RNAi response. Our work uncovers an essential output of the RNAi pathway in C. elegans.
RNAi is a gene silencing mechanism found in most eukaryotes (Mello and Conte 2004). Central to the execution of the RNAi pathway is the processing of dsRNA triggers into siRNA by the RNase III-related enzyme Dicer and its associated proteins (Ketting et al. 2001; Knight and Bass 2001; Tabara et al. 2002; Duchaine et al. 2006; Carthew and Sontheimer 2009). In Caenorhabditis elegans, this initial processing step yields primary siRNA that are bound by the Argonaute protein RDE-1 (Tabara et al. 1999; Yigit et al. 2006), which is followed by pioneering rounds of target mRNA degradation. The RNAi response is then amplified through generation of secondary siRNA by RNA-dependent RNA polymerases (RdRPs) such as RRF-1 (Sijen et al. 2001, 2007; Pak and Fire 2007). The secondary siRNA are bound by secondary Argonaute proteins (WAGOs) in C. elegans, which are limiting for RNAi-mediated gene silencing (Yigit et al. 2006).
The RNA-induced silencing complex (RISC) was originally defined biochemically as essential for siRNA-directed target mRNA recognition and degradation (Hammond et al. 2000). Besides post-transcriptional gene silencing, RISC can also mediate chromatin remodeling and transcriptional gene silencing in yeast, C. elegans, Drosophila, and plants (Mette et al. 2000; Pal-Bhadra et al. 2002; Volpe et al. 2002; Guang et al. 2008). The slicer activity of RISC is responsible for target mRNA degradation (Zamore et al. 2000). Argonaute proteins, the key components of RISCs, contain an RNase H-related PIWI domain that has been shown to slice target mRNAs (Liu et al. 2004; Baumberger and Baulcombe 2005; Miyoshi et al. 2005). The slicer activity requires three conserved catalytic residues in the PIWI domain, the so-called DDH motif (Song et al. 2004). There are >20 C. elegans Argonaute proteins (Yigit et al. 2006). Although RDE-1 possesses the DDH motif, the RNase H activity of RDE-1 is not required for triggering the silencing response at the target mRNA level (Steiner et al. 2009). The secondary Argonautes (WAGOs) are required for silencing, but most of them lack the conserved DDH motif, suggesting that they may trigger target mRNA degradation by mechanisms other than RNase H-mediated cleavage (Yigit et al. 2006). Indeed, it was demonstrated that CSR-1 (but not RDE-1) is the only Argonaute that can cleave target mRNA in vitro (Aoki et al. 2007). Another secondary Argonaute, NRDE-3, inhibits transcription elongation by recruiting NRDE-2 (Guang et al. 2008, 2010). Therefore, open questions remain regarding the precise mechanisms of RNAi-triggered target mRNA degradation in C. elegans and the differential contribution of various Argonaute protein family members.
In C. elegans, high-copy transgenic arrays created by microinjection are prone to silencing, which is considered to be initiated by aberrant mRNAs and antisense RNAs from cryptic transcription (Kelly et al. 1997; Montgomery and Fire 1998). Many RNAi pathway genes have been shown to be required for transgene silencing (Grishok et al. 2005; Kim et al. 2005). Here, we used a unique transgene that exhibited reiterative silencing and desilencing to discover novel components of the RNAi pathway in C. elegans. We report a divergent RDE-10/RDE-11 complex, which associates with target mRNA and triggers their degradation in parallel to the nuclear RNAi pathway. Therefore, RNAi engages multiple nonredundant outputs for gene silencing.
Results
rde-10 and rde-11 are required for transgene silencing and RNAi in C. elegans
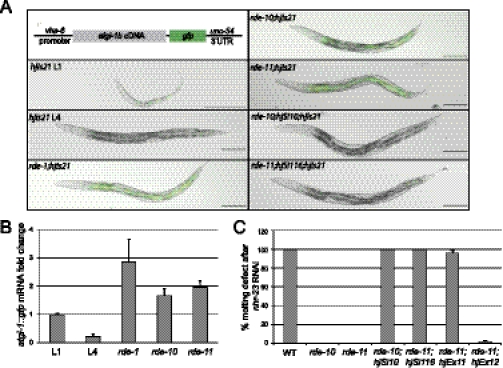
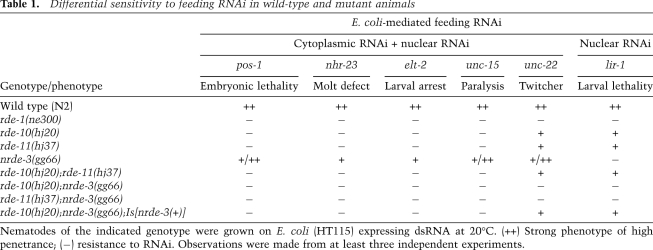
We established a transgene (hjIs21) in C. elegans that expressed an ATGL-1::GFP fusion protein under the control of the vha-6 promoter. The vha-6 promoter is active in intestinal cells from embryonic stage to adult (Oka et al. 2001). Interestingly, hjIs21 is gradually silenced during larval development such that the ATGL-1::GFP fusion protein is visible in L1 but not L4 animals (Fig. 1A). We measured the mRNA transcript level from the hjIs21 transgene by quantitative PCR (qPCR) and found that it was reduced by fivefold in L4 animals (Fig. 1B). The hjIs21 transgene undergoes a unique cycle of desilencing and silencing in every generation. We reasoned that identification of genes that are required for silencing hjIs21 may reveal novel regulators of transgene silencing. Therefore, we mutagenized animals carrying the hjIs21 transgene with ethyl methane-sulfonate (EMS) and screened for mutant alleles that conferred ATGL-1::GFP expression in adult animals. From this screen, we recovered two new alleles of rde-1, which suggests that the RNAi pathway is required for silencing the hjIs21 transgene. Accordingly, mutant animals that carried the rde-1(ne300) reference allele could also increase the mRNA transcript level of the hjIs21 transgene and restored ATGL-1::GFP expression in the larval L4 stage (Fig. 1A,B). Our results suggest that the canonical RNAi pathway is indeed involved in silencing hjIs21 and triggers degradation of atgl-1::gfp mRNA. We also identified two novel genes, rde-10 and rde-11, from our genetic screen. The hjIs21 mRNA levels in these two mutants were elevated in the L4 stage (Fig. 1A,B). In addition, rde-10 and rde-11 mutants failed to undergo RNAi by feeding Escherichia coli that expressed dsRNA targeting the endogenous pos-1 and nhr-23 genes (Fig. 1C; Table 1). Molecular cloning revealed lesions of three recessive alleles of rde-10 and one recessive allele of rde-11 (Supplemental Fig. 1A,B). The transgene silencing (Fig. 1A) and Rde phenotypes (Fig. 1C) of rde-10 and rde-11 mutant animals can be rescued by single-copy transgenes expressing wild-type RDE-10 and RDE-11, respectively. Taken together, our results strongly suggest that rde-10 and rde-11 are two novel genes that are required for transgene silencing and RNAi in C. elegans.
Figure 1.
rde-10 and rde-11 are required for RNAi and transgene silencing. (A) Schematic representation of the vha-6p::atgl-1b::gfp transgene (hjIs21). Images of larval stage L1 or L4 wild-type animals carrying hjIs21 are shown, together with rde-1(ne300), rde-10(hj20), rde-11(hj37), rde-10(hj20);hjSi10[rde-10(+)], and rde-11(hj37);hjSi116[rde-11(+)] L4 mutant animals carrying the same transgene. Bar, 100 μm. (B) Quantitation of atgl-1::gfp mRNA level by real-time PCR. The mRNA level in wild-type L1 animals was set as 1. Mean + standard deviation from triplicate reactions are shown. (C) Molting defect after nhr-23 RNAi in wild-type, rde-10(hj20), rde-11(hj37), rde-10(hj20);hjSi10[rde-10(+)], rde-11(hj37);hjSi116[rde-11(+)], rde-11(hj37);hjEx11[rde-11(+)], and rde-11(hj37);hjEx12[rde-11(C203S)]. Mean + standard deviation from three independent experiments are shown.
Table 1.
Differential sensitivity to feeding RNAi in wild-type and mutant animals
The rde-10 and rde-11 genes do not have orthologs outside the Caenorhabditis genus. rde-10 encodes a protein without any conserved domain, while rde-11 encodes a protein with a RING-type zinc finger domain, based on sequence homology search by SMART (Letunic et al. 2009). The rde-10 and rde-11 mutants have no defect in fertility, no chromosome nondisjunction (Him) phenotype, and no apparent defect in reproductive development, which suggests that it is not likely that rde-10 and rde-11 are required for microRNA (miRNA) and PIWI-interacting RNA (piRNA) pathways. In rde-10 mutant animals, we also detected NRDE-3::GFP in the nucleus (Supplemental Fig. 2), suggesting that the nuclear RNAi pathway is intact.
rde-10 and rde-11 act in parallel to the nuclear RNAi pathway
To address whether rde-10 and rde-11 are required for RNAi ubiquitously, we performed RNAi to silence genes that are expressed in different tissues. Surprisingly, rde-10 and rde-11 mutant animals show variable degrees of RNAi resistance (Table 1). They are resistant to RNAi against pos-1, nhr-23, and unc-15 but are only partially resistant to RNAi against unc-22 and lir-1, which caused a twitcher phenotype and larval lethality, respectively (Table 1). Incomplete RNAi resistance in rde-10 and rde-11 mutants was not dependent on target tissues, since both unc-15 and unc-22 were expressed in body wall muscles. We hypothesize that rde-10 and rde-11 act in parallel to additional RNAi effector pathways that contribute differentially toward gene silencing on a gene-specific or RNAi trigger-specific basis.
RNAi-mediated gene silencing in C. elegans can occur in both the cytoplasm and the nucleus (Guang et al. 2008). It has been reported that nrde-3 mutants are defective for nuclear RNAi but not cytoplasmic RNAi (Guang et al. 2008). Accordingly, nrde-3 mutants are sensitive to unc-22 RNAi (Table 1). However, rde-10;nrde-3 and rde-11;nrde-3 double mutants were resistant to unc-22 RNAi, while rde-10;rde-11 double mutants remained partially sensitive to unc-22 RNAi (Table 1). Our results suggest that rde-10 and rde-11 act in the same genetic pathway for cytoplasmic RNAi and in parallel to the nuclear RNAi pathway. Consistent with this notion, we found that lir-1 RNAi caused lethality in rde-10 and rde-11 mutants as a result of nuclear RNAi against the nuclear-localized lir-1::lin-26 polycistronic RNA and a loss of lin-26 function (Table 1).
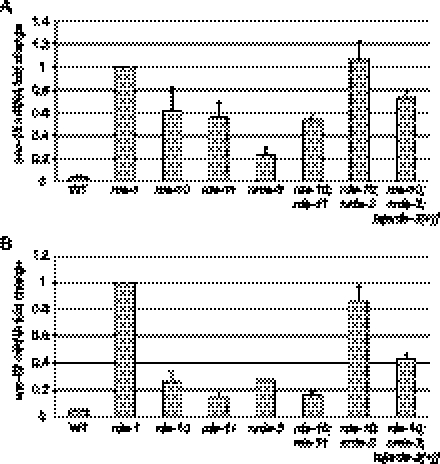
To further clarify the relative contribution of nrde genes, rde-10, and rde-11 for RNAi, we quantitated the endogenous mRNA levels of unc-15 in different genetic backgrounds after unc-15 RNAi. Using the unc-15 mRNA level in the rde-1(ne300) mutant as a reference, we observed >90% reduction of unc-15 mRNA in wild-type animals (Fig. 2A). However, in rde-10 and rde-11 mutants, the mRNA levels were ∼19-fold higher than that in wild-type animals. This indicates that rde-10 and rde-11 are required for efficient silencing of endogenous targets by RNAi. We noted that the nrde-3 mutant was still proficient in cytoplasmic RNAi, which caused an ∼80% reduction of unc-15 mRNA after RNAi (Fig. 2A). The rde-10;rde-11 double mutant had levels of residual unc-15 mRNA after RNAi similar to rde-10 and rde-11 single mutants (Fig. 2A). In contrast, we observed a significant additive effect of RNAi deficiency in the rde-10;nrde-3 double mutants. As a result, the level of unc-15 mRNA in the rde-10;nrde-3 mutant is comparable with that in the rde-1 mutant (Fig. 2A). This additive effect can be reversed by introducing a wild-type nrde-3 transgene into the rde-10;nrde-3 double mutant (Fig. 2A). Additional experiments using unc-22 RNAi support the same conclusion, although the unc-22 RNAi trigger seemed to induce nuclear RNAi more efficiently (Fig. 2B). Taken together, we conclude that rde-10 and rde-11 function in the same genetic pathway that acts in parallel to the nuclear RNAi pathway.
Figure 2.
rde-10 and rde-11 act in parallel to the nuclear RNAi pathway to silence target genes. (A) Quantitation of endogenous unc-15 mRNA levels after RNAi in wild-type (WT), rde-1(ne300), rde-10(hj20), rde-11(hj37), nrde-3(gg66), rde-10(hj20);rde-11(hj37), rde-10(hj20);nrde-3(gg66), and rde-10(hj20);nrde-3(gg66);Is[nrde-3(+)] animals. The mRNA level in rde-1 animals was set as 1. Mean + standard deviation from three independent samples assayed in triplicates are shown. (B) Quantitation of endogenous unc-22 mRNA levels after RNAi in wild-type and mutant animals as in A. Data are presented as in A.
RDE-10 and RDE-11 form a novel protein complex
We used a proteomics approach to address whether RDE-10 interacts with previously characterized RNAi pathway components. Using a single-copy transgenic strain that expressed a Flag epitope-tagged RDE-10, we affinity-purified RDE-10 from a worm population enriched with L4 stage animals, followed by multidimensional protein identification technology (MudPIT) analysis. Consistent with our genetic analysis, RDE-11 was identified as a major RDE-10-interacting protein (Fig. 3A). We also detected RSD-2, which is required for systemic RNAi (Tijsterman et al. 2004), and ERGO-1, which is required for biogenesis of some endogenous 26G siRNA (Gent et al. 2010; Vasale et al. 2010). Identification of these two proteins suggests that RDE-10 may be involved in both exogenous and endogenous RNAi pathways.
Figure 3.
RDE-10 directly interacts with RDE-11 to form a stable complex. (A) MudPIT analysis on proteins coimmunoprecipitated with Flag-tagged GFP::RDE-10 or GFP from worm extracts. dNASF (distributed normalized spectral abundance factors) indicates the relative abundance of proteins. Results are mean from two independent biological samples for GFP::RDE-10 and three independent biological samples for GFP. (B) Coimmunoprecipitation of HA-tagged RDE-11 with Flag-tagged GFP::RDE-10 but not Flag-tagged GFP from worm lysates. (C) Schematic representation of RDE-11. The RING-type zinc finger is shaded in light gray, and the zinc-coordinating Cys and His residues are enlarged. Cys203 is boxed. (D) Coimmunoprecipitation of in vitro translated HA-tagged RDE-11 with Flag-tagged RDE-10 in the presence or absence of Zn2+. Direct interaction between RDE-10 and RDE-11 was strongly enhanced by Zn2+. (E) Coimmunoprecipitation of in vitro translated wild-type and mutant HA-tagged RDE-11 with Flag-tagged RDE-10 in the presence of Zn2+. Interaction between RDE-10 and RDE-11 was impaired by the C203S mutation in the RING finger domain.
The physical association between RDE-10 and RDE-11 was further confirmed by co-immunoprecipitation assays. We found that Flag epitope-tagged RDE-10 could efficiently bind to HA epitope-tagged RDE-11 in worm lysates (Fig. 3B, lane 4). To address whether RDE-10 can directly interact with RDE-11, we performed the same assay using in vitro translated proteins. In the absence of Zn2+, we detected a very weak but reproducible interaction between RDE-10 and RDE-11 (Fig. 3D, lane 5). However, incubation of RDE-10 and RDE-11 in a buffer with 50 μM Zn2+ strongly enhanced their interaction in vitro (Fig. 3D, lane 7). Since RDE-11 possesses a RING-type zinc finger domain, we speculate that RDE-11 requires zinc for proper folding and subsequent interaction with RDE-10. Indeed, the RING finger domain is required for RDE-11 function, since mutation in a single conserved cysteine in the RING finger could reduce its interaction with RDE-10 in vitro (Fig. 3C,E, lane 7). The same mutation also abolished the rescuing activity of an rde-11 transgene in vivo (Fig. 1C, hjEx12) without affecting protein stability (Supplemental Fig. 1C). Our results indicate that RDE-10 directly interacts with RDE-11 in a zinc-dependent manner to form a stable complex.
The RDE-10/RDE-11 complex associates with target mRNAs that are undergoing degradation
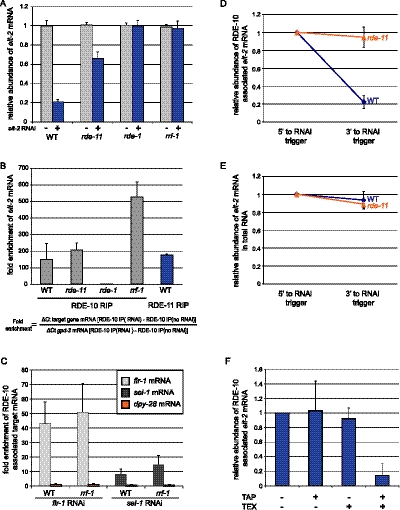
To further test the hypothesis that RDE-10 and RDE-11 are required for cytoplasmic mRNA degradation, we asked whether the RDE-10/RDE-11 complex could associate with mRNAs targeted by RNAi. To this end, we generated rescuing single-copy transgenes that expressed epitope-tagged RDE-10 or RDE-11 (Fig. 1C). We chose elt-2 as the target transcript for RNA coimmunoprecipitation assays (Supplemental Fig. 3A). Animals were fed E. coli that expressed dsRNA against elt-2 or an unrelated gene, prior to immunoprecipitation of RDE-10 or RDE-11. Wild-type animals treated with elt-2 RNAi showed an 80% reduction in elt-2 mRNA level (Fig. 4A). Despite such low abundance of elt-2 mRNA, we detected ∼170-fold enrichment of RDE-11 binding to elt-2 mRNA (Fig. 4B). We also found that RDE-10 preferentially associated with elt-2 mRNA in the presence or absence of RDE-11 (Fig. 4B). Similar observations were made when we used dpy-28 as the target gene (Supplemental Fig. 3B). These results indicate that RDE-10 can be recruited to target mRNAs independent of RDE-11 and suggest that RDE-11 is not required for target mRNA recognition. As rde-11 mutant animals are RNAi-defective (Fig. 4A), these results also indicate that the recruitment of RDE-10 alone to target mRNAs is not sufficient to trigger RNAi.
Figure 4.
The RDE-10/RDE-11 complex associates with mRNA targeted by RNAi. (A) Quantitation of elt-2 mRNA levels by real-time PCR in wild-type (WT), rde-11(hj37), rde-1(ne300), or rrf-1(pk1417) animals undergoing elt-2 RNAi. Each strain also carried rde-10(hj20) and the rescuing single-copy transgene hjSi10[rde-10(+)]. The mRNA levels of elt-2 in animals without RNAi treatment were set as 1 for each genotype. (B) Target mRNA preferentially associated with RDE-10 in wild-type (WT), rde-11(hj37), and rrf-1(pk1417) but not rde-1(ne300) animals as shown by real-time PCR. Preferential association of target mRNA to RDE-11 was also observed in wild-type animals. The levels of elt-2 mRNA 5′ to the RNAi trigger were measured. The formula for calculating fold enrichment is shown. (C) Wild-type (WT) and rrf-1(pk1417) animals carrying hjSi10 were subjected to flr-1 and sel-1 RNAi. Target mRNAs, but not the nonspecific dpy-28 mRNA, preferentially associated with RDE-10 in wild-type and rrf-1animals. (D) Depletion of the 3′ end of target mRNA associated with RDE-10 in wild-type but not rde-11(hj37) animals. The levels of elt-2 mRNA 5′ to the RNAi trigger were set as 1. (E) Comparison of relative abundance of elt-2 mRNA 5′ and 3′ to the RNAi trigger. Total RNA extracted from wild-type or rde-11(hj37) animals undergoing elt-2 RNAi was used. (F) RDE-10-associated elt-2 mRNA was resistant to terminator 5′–3′ exonuclease (TEX) unless pretreated with tobacco acid pyrophosphatase (TAP). RNA samples were subjected to TAP and/or TEX treatments and were converted to cDNA and quantified with qPCR. The abundance of elt-2 mRNA (RNAi target) in the sample without any treatment was set to 1. All results are shown as mean + standard deviation from at least three independent samples assayed in triplicates.
To test whether RDE-10 requires siRNA for target mRNA recognition, we also performed RDE-10 RNA coimmunoprecipitation in rde-1(−) and rrf-1(−) animals. While rde-1(−) animals are completely defective in primary and secondary siRNA biogenesis (Tabara et al. 2002), rrf-1(−) animals are unable to generate secondary siRNA in somatic tissues (Sijen et al. 2001, 2007). We found that RDE-10 failed to associate with target mRNAs in rde-1(−) animals (Fig. 4B; Supplemental Fig. 3B). In contrast, the association of RDE-10 with elt-2 target mRNAs appeared to increase in rrf-1(−) animals (Fig. 4B). Such enhancement was not universal, since the association of RDE-10 with two additional somatically expressed mRNA targets—flr-1 (Take-Uchi et al. 1998) and sel-1 (Pak and Fire 2007)—was comparable in wild-type and rrf-1(−) animals (Fig. 4C). Taken together, our results indicate that RDE-10 depends on primary but not secondary siRNA to engage target mRNAs and support a model that RDE-10 and, by extension, RDE-11 act downstream from RDE-1 and upstream of RRF-1 (Fig. 6, see below).
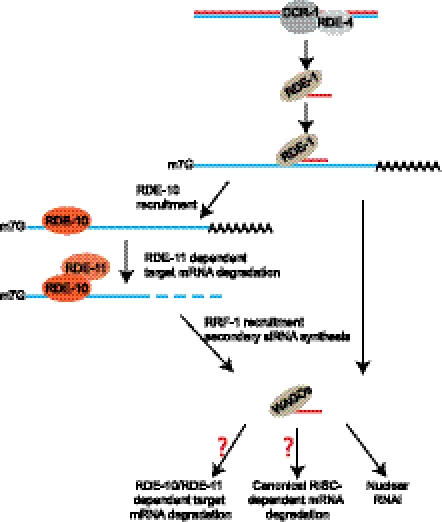
Figure 6.
A model: RDE-10/RDE-11 complex in the exogenous RNAi pathway in C. elegans.
Next, we examined the integrity of target mRNAs that were associated with the RDE-10/RDE-11 complex. We performed qPCR to measure the relative abundance of regions that are 5′ or 3′ to the RNAi trigger in RDE-10-associated elt-2 mRNAs. Interestingly, we observed a 70% reduction in their 3′ region in an RDE-11-dependent manner (Fig. 4D). Such preferential 3′ depletion was specific to elt-2 mRNA that associated with RDE-10 and was not apparent in total RNA samples (Fig. 4E). Similar results were also observed in animals undergoing RNAi against dpy-28 (Supplemental Fig. 3C,D). Since mature mRNAs possess a 5′ cap and a 3′ poly(A) tail, we next asked whether these structures were preserved in RDE-10-associated target mRNAs. Capped mRNA is resistant to the terminator 5′–3′ exonuclease (TEX) unless pretreated with the tobacco acid pyrophosphatase (TAP). We found that mRNA transcripts of a housekeeping gene, gpd-3, and RDE-10 associated target elt-2 mRNA could both be degraded by TEX only after TAP pretreatment (Fig. 4F; Supplemental Fig. 4). Therefore, our results suggest that RDE-10-associated target mRNAs are capped at the 5′ end.
The instability of the 3′ region of RDE-10-associated target mRNAs could be caused by deadenylation. To directly test this possibility, we repeated our RDE-10 RNA coimmunoprecipitation, followed by reverse transcription of bound mRNAs using random hexamer or oligo(dT) primers. The former allowed detection of RNAs with or without a poly(A) tail, while the latter allowed selective detection of poly(A) mRNAs. The relative abundance of RDE-10-associated elt-2 mRNA was decreased by 40% when the oligo(dT) primer was used (Supplemental Fig. 5). This decrease was observed in wild-type but not in rde-11 mutant animals. Our results imply that the 3′ end of RDE-10-associated target mRNAs undergoes deadenylation and degradation in an RDE-11-dependent manner. The retention of a 5′ cap of such mRNAs may confer transient stability of a region 5′ to the RNAi trigger, which can serve as a template for secondary siRNA synthesis.
RDE-10 and RDE-11 are required for secondary siRNA biogenesis
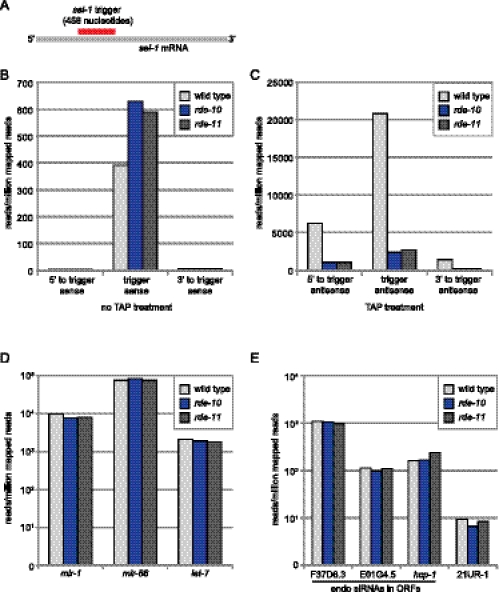
Our analysis so far placed the RDE-10/RDE-11 complex at a step between RDE-1 and RRF-1 in the exogenous RNAi pathway in C. elegans. It is conceivable that the RDE-10/RDE-11 complex is required for amplification of the RNAi response. To test this hypothesis, we subjected wild-type and rde-10 and rde-11 mutant animals to RNAi against a somatic gene, sel-1, using a well-characterized trigger (Fig. 5A; Pak and Fire 2007). We then purified small RNAs (21–24 nucleotides) from duplicate populations of animals and sequenced them using the Illumina platform. Each biological sample yielded, on average, 11 million reads. The abundance of primary siRNAs was comparable in wild-type and rde-10 and rde-11 mutant animals (Fig. 5B; Supplemental Table 1). In contrast, we found a fivefold decrease in secondary siRNAs in rde-10 and rde-11 mutant animals (Fig. 5C; Supplemental Table 1). This was primarily due to a reduction of 22G RNAs, which were the major secondary siRNAs (Supplemental Fig. 6). It is conceivable that the residual secondary siRNAs can be loaded to secondary Argonautes (WAGOs) to sustain a low level of cytoplasmic and nuclear RNAi. Unlike the clear deficit in secondary siRNAs, we detected comparable levels of miRNAs (let-7, mir-1, and mir-66) (Reinhart et al. 2000; Lau et al. 2001; Lee and Ambros 2001) and a piRNA (21UR-1) (Ruby et al. 2006; Batista et al. 2008) in wild-type and rde-10 and rde-11 mutant animals (Fig. 5D,E). In addition, the levels of endogenous siRNAs complementary to F37D6.3 (WAGO-1 target) (Gu et al. 2009), E01G4.5 (NRDE-3 target) (Guang et al. 2008), and hcp-1 (CSR-1 target) (Claycomb et al. 2009) were unchanged (Fig. 5E). Taken together, our results strongly suggest that the RDE-10/RDE-11 complex is required for secondary siRNA synthesis, which is critical for amplifying the exogenous RNAi response.
Figure 5.
RDE-10 and RDE-11 are required for secondary siRNA synthesis. (A) Schematic representation of sel-1 RNAi trigger and sel-1 mRNA. (B–E) The mean abundance of small RNAs from two independent populations of wild-type, rde-10(hj20), and rde-11(hj37) animals undergoing sel-1 RNAi is shown. (B) Sense sel-1 siRNAs without TAP treatment. (C) Antisense sel-1 siRNAs with TAP treatment. (D) Selected miRNAs (mir). (E) Selected endogenous siRNAs and one 21U-RNA. Reads per million (RPM) was calculated by (reads aligned to region/all aligned reads for sample) × 1,000,000. A 5′ ligation-dependent method was used in library preparations. Small RNAs with 5′ monophosphate were better represented in libraries without TAP treatment. Small RNAs with 5′ triphosphate were better represented in libraries with TAP treatment.
Discussion
In this study, we identified the divergent RDE-10/RDE-11 complex as an essential output for exogenous RNAi in C. elegans. Our genetic, proteomic, and biochemical data indicate that the RDE-10/RDE-11 complex is recruited to target mRNAs for their degradation downstream from the Argonaute RDE-1 and primary siRNA biogenesis. Recruitment of the RDE-10/RDE-11 complex also plays an important role in supporting secondary siRNA synthesis. Taken together, the RDE-10/RDE-11 complex links the amplification of the RNAi response to the initial processing and utilization of the exogenous dsRNA trigger (Fig. 6).
Multiple parallel pathways may operate downstream from the primary RNAi response that is dependent on DCR-1/dicer, RDE-1, and RDE-4, and their associated proteins (Tabara et al. 2002; Duchaine et al. 2006; Yigit et al. 2006). Although the RNAi response in rde-10 and rde-11 mutant animals is severely blunted, quantitative analysis of target mRNAs suggests that additional RNAi outputs remain operational. This can be attributed to residual secondary siRNA synthesis in rde-10 and rde-11 mutant animals. We postulate that recruitment of RRF-1 to target mRNAs, which is required for secondary siRNA synthesis, is strongly promoted by prior engagement of the RDE-10/RDE-11 complex to target mRNAs. However, RRF-1 may be recruited by additional mechanisms, perhaps through recognition of RDE-1 that is bound to target mRNAs. We note that high concentrations of exogenous dsRNA triggers introduced by injection (C Zhang and G Ruvkun, pers. comm.) and certain RNAi triggers introduced by feeding (e.g., unc-22) can partially bypass the requirement for RDE-10 and RDE-11. It is plausible that enhancement of RDE-1 association with target mRNAs, either through unusually strong primary siRNA–mRNA base-pairing or by high concentration of primary siRNAs, is sufficient for recruitment of RRF-1 to target mRNAs.
How does the RDE-10/RDE-11 complex promote secondary siRNA synthesis? RRF-1 may be recruited to the target mRNAs through protein–protein interactions. Such interactions may be transient, since we did not detect RRF-1 in our proteomic analysis of RDE-10-associated proteins. We also consider an alternative scenario. At the initial phase of exogenous RNAi, a fine balance must be maintained between degradation and retention of target mRNAs. This is because if target mRNAs are degraded too rapidly, no template will be available for RRF-1 to synthesize secondary siRNAs. The RDE-10/RDE-11 complex preferentially associates with the region 5′ to the RNAi trigger in target mRNAs that are 5′-capped (Fig. 4F) but lack a poly(A) tail (Fig. 4D; Supplemental Fig. 5). We therefore hypothesize that the RDE-10/RDE-11 complex generates and stabilizes partially degraded target mRNAs, which facilitates RRF-1 recruitment and/or its synthesis of secondary siRNAs.
Our deep-sequencing analysis of small RNAs indicates that RDE-10 and RDE-11 are required for secondary siRNA synthesis, which is crucial for exogenous RNAi. In contrast, RDE-10 and RDE-11 are dispensable for the synthesis of selected endogenous siRNAs and miRNAs (Fig. 5D,E). This is in agreement with a more comprehensive survey of endogenous small RNAs in rde-10 and rde-11 mutant animals, which reveals the depletion of a small subset of endogenous siRNAs (C Zhang and G Ruvkun, pers. comm.).
The specificity of the RNAi pathway is dictated by base-pairing between target mRNAs and siRNA bound by Argonaute proteins. Both RDE-10 and RDE-11 can be recruited to target mRNA specifically. However, we did not detect any Argonaute family members in the exogenous RNAi pathway as RDE-10-interacting proteins in our proteomic analysis. One possible reason is that the interaction between RDE-10 and Argonautes is too transient to be captured under our immunoprecipitation conditions, or this could suggest a “drop-off” model: Once recognized by an Argonaute-containing complex, target mRNAs are labeled and then dropped off at a subcellular compartment where RDE-10/RDE-11 dependent degradation takes place. The “label” could be a protein (e.g., RSD-2 or RDE-10) or modifications of target mRNAs at their 5′ or 3′ end.
Although multiple models exist in other organisms, it is currently unknown whether target mRNA degradation by RNAi in C. elegans is initiated by deadenylation, which is followed by 3′ → 5′ degradation by the exosome and/or 5′ → 3′ degradation initiated by decapping. Alternatively, endonucleolytic cleavage within the target mRNA could be the initial step of degradation. We found that RDE-10-associated target mRNAs retained a 5′ cap structure (Fig. 4F). Therefore, it was unlikely that the RDE-10/RDE-11 complex would promote decapping. It is plausible that RDE-11 can instigate target mRNA degradation through recruitment of additional factors. Although a handful of conserved RISC components have been identified in C. elegans, they have only been reported to act in miRNA-mediated gene silencing but not in RNAi (Caudy et al. 2003). Therefore, the RDE-10/RDE-11 complex may operate in conjunction with RISC in C. elegans. This is compatible with a previously proposed two-step model for RNAi in C. elegans (Yigit et al. 2006). The pioneering rounds of target mRNA degradation can be carried out by the primary siRNA-directed RDE-10/RDE-11 complex (Fig. 6), which is followed by full gene silencing by the secondary siRNA-directed RISC complex and nuclear RNAi apparatus. Nevertheless, we cannot rule out the possibility that the RDE-10/RDE-11 complex is required in additional gene silencing mechanisms subsequent to secondary siRNA synthesis (Fig. 6).
In C. elegans and plants, amplification of small RNAs is critical for potent endogenous and exogenous RNAi response and antiviral immunity (Chapman and Carrington 2007; Ding 2010). Although conserved proteins such as Argonautes and RdRPs are clearly involved, the precise mechanism of amplification differs. We propose that the divergence of the exogenous RNAi pathway in C. elegans can in part be attributed to its dependence on the RDE-10/RDE-11 complex. Further analysis of the C. elegans RNAi pathways should yield insights into how small RNA-based regulation of gene expression can drift through evolution and provide clues on how to improve the efficiency of RNAi in heterologous systems.
Materials and methods
Strains and transgenes
Details of strains and transgenes used in this study can be found in the Supplemental Material.
Genetic screen
We mutagenized hjIs21 animals with EMS using standard procedures for mutants that expressed ATGL-1::GFP in the adult stage. We screened ∼20,000 haploid genomes and isolated mutants that fell into at least three complementation groups. One of them was identified to be rde-1. Genetic mapping with the Hawaiian isolate CB4856 placed rde-10(hj18) on LGI between snp_Y47G6A and snp_T28F2. RNAi screens and candidate gene sequencing identified a C-to-T mutation that causes a single amino acid substitution (S228F) in Y47G6A.4. rde-10(hj19) and rde-10(hj20), both carrying a premature stop codon in Y47G6A.4, were later identified from this complementation group. We used hj20 as the reference allele. rde-11(hj37) was mapped to an interval in LGIV that included B0564.11, which encoded a strong interacting partner of GFP::RDE-10. Sequencing of B0564.11 in rde-11(hj37) identified a G-to-A mutation that yields a premature stop codon. rde-10(hj20) and rde-11(hj37) were rescued by single-copy transgenes hjSi10 and hjSi116, respectively.
RNAi experiments
RNAi was carried out as previously reported (Timmons et al. 2001). Bacterial clones expressing dsRNA were obtained from the Ahringer RNAi library except for dpy-28 (PCR product of 5′-CGAAACGTGCTTCAACTAG-3′ and 5′-ATGTCCATGTCGATTATTATCC-3′), sel-1 (PCR product of 5′-TGCATGGAGCCGGAATCGGA-3′ and 5′-TGCATCGGCACTTCCTGACT-3′), and flr-1 (PCR product of 5′-TTGGCAGGGAAAAGCTACAT-3′ and 5′-CTCCTCCTCAGCAACTGCAT-3′) RNAi clones. For unc-15 and unc-22 RNAi experiments, 10 OD600 bacteria were seeded on 100-mm RNAi plates. Plates were left overnight at room temperature for dsRNA expression. Six-thousand eggs from egg preps were seeded on plates in triplicate for each strain. After 48 h at 20°C, worms were washed extensively with M9, and RNA was extracted with TRI reagent (Molecular Research Center). For elt-2 RNAi experiments, 4000 synchronized L1 animals from egg preps were seeded on 100-mm RNAi plates. Worms were collected after 46 h at 20°C, and RNA was extracted with TRI reagent.
Real-time PCR
RNA was extracted using TRI reagent (Molecular Research Center) according to manufacturer's instructions. Genomic DNA contamination was removed using the TURBO DNA-free kit (Ambion, AM1907). cDNAs were synthesized using ImProm-II Reverse Transcription system (Promega). The cDNA was subjected to real-time PCR analysis using the IQ SYBR Green SuperMix (Bio-Rad) on an iCycler (Bio-Rad). Primer sequences are described in the Supplemental Material.
Worm lysate preparation and immunoprecipitation for MudPIT
Mixed-stage worms grown in liquid culture with E. coli HB101 were used for preparation of worm lysate and immunoprecipitation. Worms were collected and washed extensively with M9, ground in liquid nitrogen, and added to immunoprecipitation buffer [20 mM HEPES-KOH at pH 7.5, 10 mM KOAc, 2 mM Mg(OAc)2, 110 mM KCl, 0.1% NP-40, 1× protease inhibitor (Sigma, P8340)]. 3xFlag-tagged RDE-10 was purified by immunoprecipitation with agarose beads conjugated with anti-Flag antibodies. Immunoprecipitations were carried out for 4 h at 4°C. GFP::RDE-10 or GFP was eluted from beads by TEV protease treatment overnight at 4°C in immunoprecipitation buffer.
RNA immunoprecipitation (RIP)
RDE-10 and RDE-11 RIPs were performed as previously described (Guang et al. 2008) with modifications. To isolate RDE-10-associated total RNAs after RNAi, synchronized L1 animals (from egg preps) were seeded onto 100-mm RNAi plates and collected at L4 stage (∼44–46 h at 20°C). Animals were sonicated in lysis buffer (20 mM Tris-HCl at pH 7.5, 200 mM NaCl, 2.5 mM MgCl2, 0.5% NP-40, 10% glycerol, proteinase inhibitor tablet EDTA-free [Roche], RNaseOUT [Invitrogen]). 3xFlag::GFP::RDE-10 was immunoprecipitated with Dynabeads M280 precoated with anti-Flag antibody (Sigma, F1804). After extensive washes, 3xFlag::GFP::RDE-10 was eluted with 150 μg/mL 3xFlag peptide. Eluates were first incubated with Turbo DNase I (Ambion) for 20 min at 37°C. The RNAs were then purified by TRI reagent, followed by isopropanol precipitation. cDNAs were generated from RNAs with ImProm-II Reverse Transcription system using a mixture of oligo(dT) and random hexamer primers. For Supplemental Figure 5, RNA samples were split into two, and reverse transcription was performed with either oligo(dT) or random hexamer primers. qPCRs were performed as described above. Target mRNA enrichment was normalized by nonspecific binding of gpd-3 mRNA. RDE-11 RIP was performed in the same way as RDE-10 RIP.
Additional methods are described in Supplemental Material.
Acknowledgments
We thank Scott Kennedy and Shouhong Guang for reagents and advice; Kendra Walton and Kate Malanowski for processing of deep-sequencing samples; Marco Blanchette, Peter Baumann, and Lars Dreier for advice; and Gary Ruvkun and Chi Zhang for communicating unpublished results and critical reading of the manuscript. This work was performed to fulfill, in part, requirements for H.Y.'s PhD thesis research as a student registered with the University of Kansas Medical Center. This work was funded by the Stowers Institute for Medical Research. H.Y. was responsible for the design and execution of experiments in all of the figures, Table 1, and Supplemental Table 1. Y.Z. and L.F. performed MudPIT analysis for Figure 3A. J.V. and H.L. performed deep-sequencing data analysis for Figure 5, Supplemental Figure 6, and Supplemental Table 1. H.Y.M. conducted the genetic screen and contributed to experimental design. H.Y. and H.Y.M. wrote the manuscript.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.180679.111.
References
- Aoki K, Moriguchi H, Yoshioka T, Okawa K, Tabara H 2007. In vitro analyses of the production and activity of secondary small interfering RNAs in C. elegans. EMBO J 26: 5007–5019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista PJ, Ruby JG, Claycomb JM, Chiang R, Fahlgren N, Kasschau KD, Chaves DA, Gu W, Vasale JJ, Duan S, et al. 2008. PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans. Mol Cell 31: 67–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumberger N, Baulcombe DC 2005. Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc Natl Acad Sci 102: 11928–11933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carthew RW, Sontheimer EJ 2009. Origins and mechanisms of miRNAs and siRNAs. Cell 136: 642–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caudy AA, Ketting RF, Hammond SM, Denli AM, Bathoorn AM, Tops BB, Silva JM, Myers MM, Hannon GJ, Plasterk RH 2003. A micrococcal nuclease homologue in RNAi effector complexes. Nature 425: 411–414 [DOI] [PubMed] [Google Scholar]
- Chapman EJ, Carrington JC 2007. Specialization and evolution of endogenous small RNA pathways. Nat Rev Genet 8: 884–896 [DOI] [PubMed] [Google Scholar]
- Claycomb JM, Batista PJ, Pang KM, Gu W, Vasale JJ, van Wolfswinkel JC, Chaves DA, Shirayama M, Mitani S, Ketting RF, et al. 2009. The Argonaute CSR-1 and its 22G-RNA cofactors are required for holocentric chromosome segregation. Cell 139: 123–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding SW 2010. RNA-based antiviral immunity. Nat Rev Immunol 10: 632–644 [DOI] [PubMed] [Google Scholar]
- Duchaine TF, Wohlschlegel JA, Kennedy S, Bei Y, Conte D Jr, Pang K, Brownell DR, Harding S, Mitani S, Ruvkun G, et al. 2006. Functional proteomics reveals the biochemical niche of C. elegans DCR-1 in multiple small-RNA-mediated pathways. Cell 124: 343–354 [DOI] [PubMed] [Google Scholar]
- Gent JI, Lamm AT, Pavelec DM, Maniar JM, Parameswaran P, Tao L, Kennedy S, Fire AZ 2010. Distinct phases of siRNA synthesis in an endogenous RNAi pathway in C. elegans soma. Mol Cell 37: 679–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishok A, Sinskey JL, Sharp PA 2005. Transcriptional silencing of a transgene by RNAi in the soma of C. elegans. Genes Dev 19: 683–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Shirayama M, Conte D Jr, Vasale J, Batista PJ, Claycomb JM, Moresco JJ, Youngman EM, Keys J, Stoltz MJ, et al. 2009. Distinct argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline. Mol Cell 36: 231–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guang S, Bochner AF, Pavelec DM, Burkhart KB, Harding S, Lachowiec J, Kennedy S 2008. An Argonaute transports siRNAs from the cytoplasm to the nucleus. Science 321: 537–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guang S, Bochner AF, Burkhart KB, Burton N, Pavelec DM, Kennedy S 2010. Small regulatory RNAs inhibit RNA polymerase II during the elongation phase of transcription. Nature 465: 1097–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond SM, Bernstein E, Beach D, Hannon GJ 2000. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404: 293–296 [DOI] [PubMed] [Google Scholar]
- Kelly WG, Xu S, Montgomery MK, Fire A 1997. Distinct requirements for somatic and germline expression of a generally expressed Caernorhabditis elegans gene. Genetics 146: 227–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH 2001. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev 15: 2654–2659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JK, Gabel HW, Kamath RS, Tewari M, Pasquinelli A, Rual JF, Kennedy S, Dybbs M, Bertin N, Kaplan JM, et al. 2005. Functional genomic analysis of RNA interference in C. elegans. Science 308: 1164–1167 [DOI] [PubMed] [Google Scholar]
- Knight SW, Bass BL 2001. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science 293: 2269–2271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau NC, Lim LP, Weinstein EG, Bartel DP 2001. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294: 858–862 [DOI] [PubMed] [Google Scholar]
- Lee RC, Ambros V 2001. An extensive class of small RNAs in Caenorhabditis elegans. Science 294: 862–864 [DOI] [PubMed] [Google Scholar]
- Letunic I, Doerks T, Bork P 2009. SMART 6: Recent updates and new developments. Nucleic Acids Res 37: D229–D232 doi: 10.1093/nar/gkn808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ 2004. Argonaute2 is the catalytic engine of mammalian RNAi. Science 305: 1437–1441 [DOI] [PubMed] [Google Scholar]
- Mello CC, Conte D Jr 2004. Revealing the world of RNA interference. Nature 431: 338–342 [DOI] [PubMed] [Google Scholar]
- Mette MF, Aufsatz W, van der Winden J, Matzke MA, Matzke AJ 2000. Transcriptional silencing and promoter methylation triggered by double-stranded RNA. EMBO J 19: 5194–5201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi K, Tsukumo H, Nagami T, Siomi H, Siomi MC 2005. Slicer function of Drosophila Argonautes and its involvement in RISC formation. Genes Dev 19: 2837–2848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery MK, Fire A 1998. Double-stranded RNA as a mediator in sequence-specific genetic silencing and co-suppression. Trends Genet 14: 255–258 [DOI] [PubMed] [Google Scholar]
- Oka T, Toyomura T, Honjo K, Wada Y, Futai M 2001. Four subunit a isoforms of Caenorhabditis elegans vacuolar H+-ATPase. Cell-specific expression during development. J Biol Chem 276: 33079–33085 [DOI] [PubMed] [Google Scholar]
- Pak J, Fire A 2007. Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science 315: 241–244 [DOI] [PubMed] [Google Scholar]
- Pal-Bhadra M, Bhadra U, Birchler JA 2002. RNAi related mechanisms affect both transcriptional and posttranscriptional transgene silencing in Drosophila. Mol Cell 9: 315–327 [DOI] [PubMed] [Google Scholar]
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G 2000. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403: 901–906 [DOI] [PubMed] [Google Scholar]
- Ruby JG, Jan C, Player C, Axtell MJ, Lee W, Nusbaum C, Ge H, Bartel DP 2006. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell 127: 1193–1207 [DOI] [PubMed] [Google Scholar]
- Sijen T, Fleenor J, Simmer F, Thijssen KL, Parrish S, Timmons L, Plasterk RH, Fire A 2001. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell 107: 465–476 [DOI] [PubMed] [Google Scholar]
- Sijen T, Steiner FA, Thijssen KL, Plasterk RH 2007. Secondary siRNAs result from unprimed RNA synthesis and form a distinct class. Science 315: 244–247 [DOI] [PubMed] [Google Scholar]
- Song JJ, Smith SK, Hannon GJ, Joshua-Tor L 2004. Crystal structure of Argonaute and its implications for RISC slicer activity. Science 305: 1434–1437 [DOI] [PubMed] [Google Scholar]
- Steiner FA, Okihara KL, Hoogstrate SW, Sijen T, Ketting RF 2009. RDE-1 slicer activity is required only for passenger-strand cleavage during RNAi in Caenorhabditis elegans. Nat Struct Mol Biol 16: 207–211 [DOI] [PubMed] [Google Scholar]
- Tabara H, Sarkissian M, Kelly WG, Fleenor J, Grishok A, Timmons L, Fire A, Mello CC 1999. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell 99: 123–132 [DOI] [PubMed] [Google Scholar]
- Tabara H, Yigit E, Siomi H, Mello CC 2002. The dsRNA binding protein RDE-4 interacts with RDE-1, DCR-1, and a DExH-box helicase to direct RNAi in C. elegans. Cell 109: 861–871 [DOI] [PubMed] [Google Scholar]
- Take-Uchi M, Kawakami M, Ishihara T, Amano T, Kondo K, Katsura I 1998. An ion channel of the degenerin/epithelial sodium channel superfamily controls the defecation rhythm in Caenorhabditis elegans. Proc Natl Acad Sci 95: 11775–11780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tijsterman M, May RC, Simmer F, Okihara KL, Plasterk RH 2004. Genes required for systemic RNA interference in Caenorhabditis elegans. Curr Biol 14: 111–116 [DOI] [PubMed] [Google Scholar]
- Timmons L, Court DL, Fire A 2001. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene 263: 103–112 [DOI] [PubMed] [Google Scholar]
- Vasale JJ, Gu W, Thivierge C, Batista PJ, Claycomb JM, Youngman EM, Duchaine TF, Mello CC, Conte D Jr 2010. Sequential rounds of RNA-dependent RNA transcription drive endogenous small-RNA biogenesis in the ERGO-1/Argonaute pathway. Proc Natl Acad Sci 107: 3582–3587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA 2002. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297: 1833–1837 [DOI] [PubMed] [Google Scholar]
- Yigit E, Batista PJ, Bei Y, Pang KM, Chen CC, Tolia NH, Joshua-Tor L, Mitani S, Simard MJ, Mello CC 2006. Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell 127: 747–757 [DOI] [PubMed] [Google Scholar]
- Zamore PD, Tuschl T, Sharp PA, Bartel DP 2000. RNAi: Double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 101: 25–33 [DOI] [PubMed] [Google Scholar]