Abstract
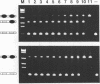
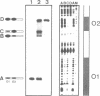
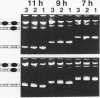
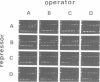
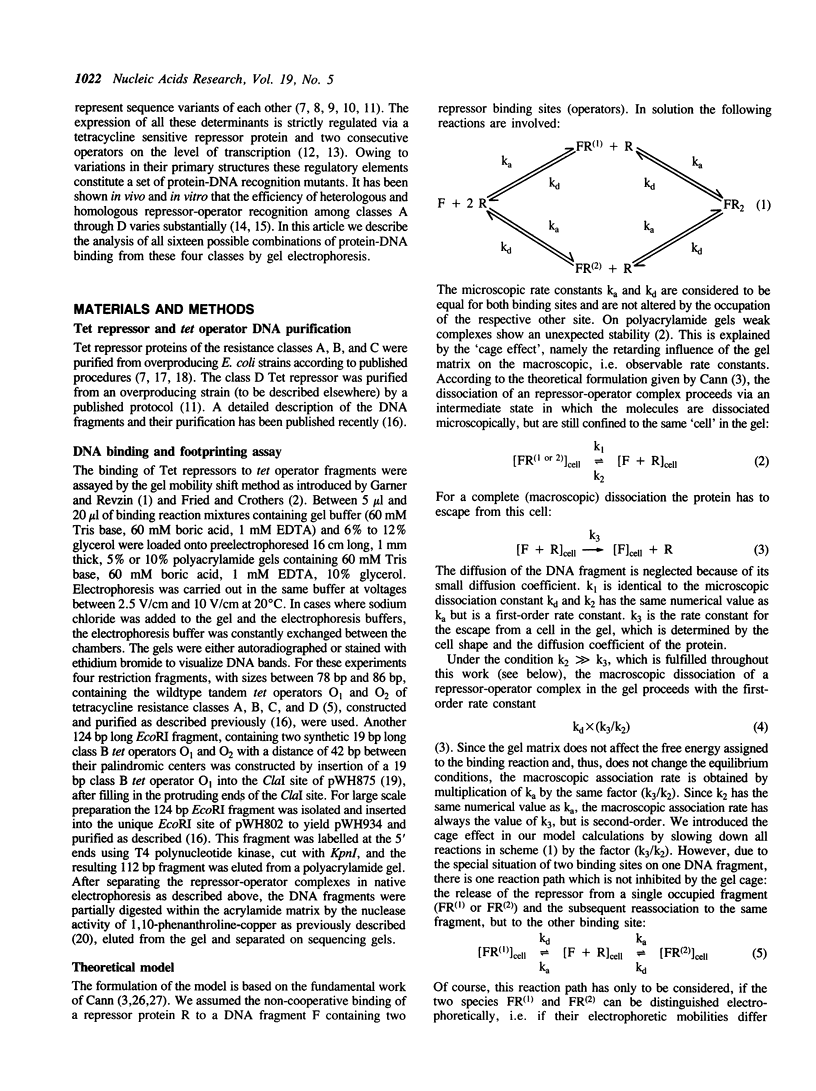
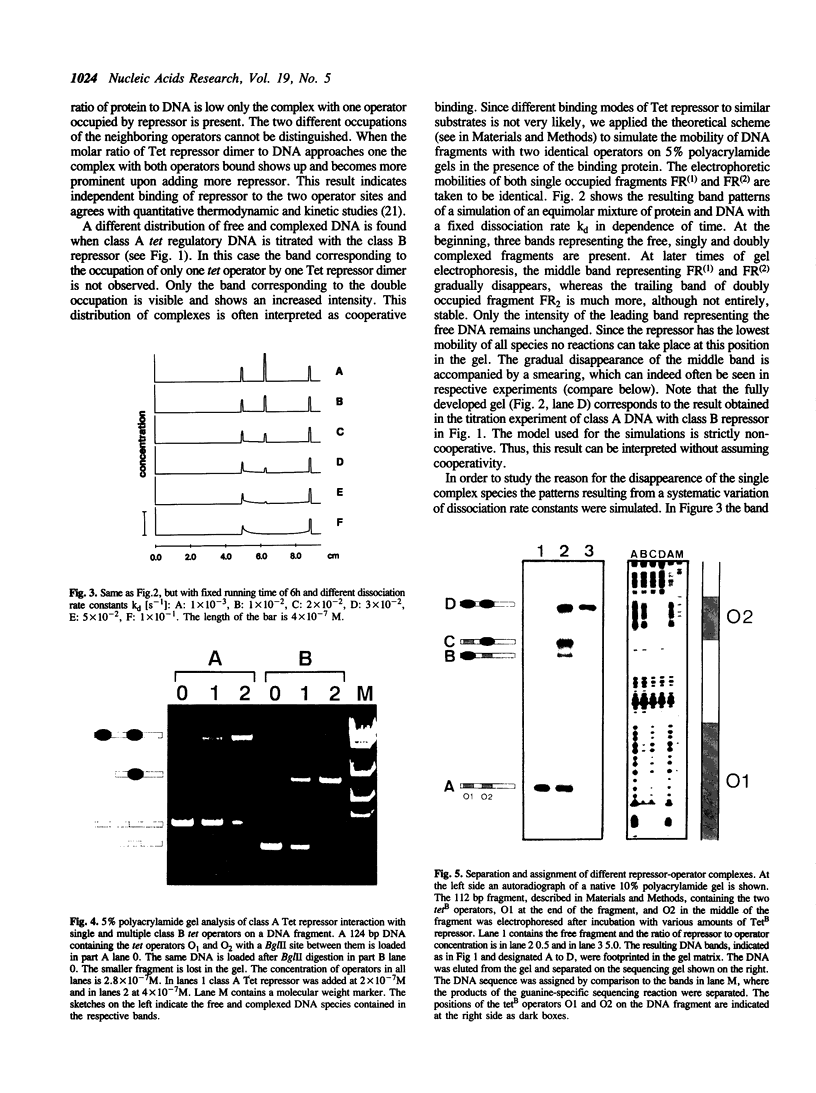
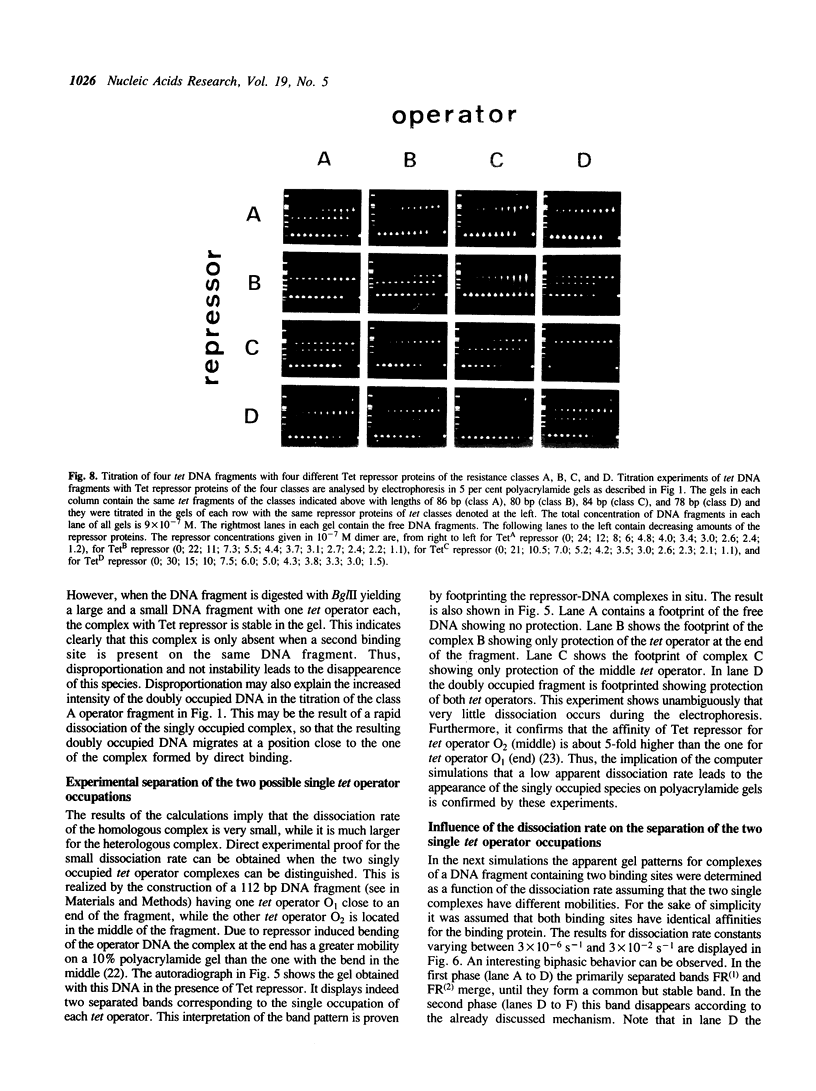
A series of computer simulations of gel patterns assuming non-cooperative binding of a protein to two targets on the same DNA fragment was performed and applied to interprete gel mobility shift experiments of Tet repressor-tet operator binding. While a high binding affinity leads to the expected distribution of free DNA, DNA bound by one repressor dimer and DNA bound by two repressor dimers, a lower affinity or an increased electrophoresis time results in the loss of the band corresponding to the singly occupied complex. The doubly occupied complex remains stable under these conditions. This phenomenon is typical for protein binding to DNA fragments with two identical sites. It results from statistical disproportionation of the singly occupied complex in the gel. The lack of the singly occupied complex is commonly taken to indicate cooperative binding, however, our analysis shows clearly, that cooperativity is not needed to interprete these results. Tet repressor proteins and small DNA fragments with two tet operator sites have been prepared from four classes of tetracycline resistance determinants. The results of gel mobility shift analyses of various complexes of these compounds confirm the predictions. Furthermore, calculated gel patterns assuming different gel mobilities of the two singly occupied complexes show discrete bands only if the electrophoresis time is shorter than the inverse of the microscopic dissociation rate constant. Simulations assuming increasing dissociation rates predict that the two bands first merge into one, which then disappears. This behavior was verified by gel mobility analyses of Tet repressor-tet operator titrations at increased salt concentrations as well as by direct footprinting of the complexes in the gel. It is concluded that comparison of the intensities of the single and the double occupation bands allow a rough estimation of the dissociation rate constant. On this basis the sixteen possible Tet repressor-tet operator combinations can be ordered with decreasing binding affinities by a simple gel shift experiment. The implications of these results for gel mobility analyses of other protein-DNA complexes are discussed.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altschmied L., Baumeister R., Pfleiderer K., Hillen W. A threonine to alanine exchange at position 40 of Tet repressor alters the recognition of the sixth base pair of tet operator from GC to AT. EMBO J. 1988 Dec 1;7(12):4011–4017. doi: 10.1002/j.1460-2075.1988.tb03290.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand K. P., Postle K., Wray L. V., Jr, Reznikoff W. S. Overlapping divergent promoters control expression of Tn10 tetracycline resistance. Gene. 1983 Aug;23(2):149–156. doi: 10.1016/0378-1119(83)90046-x. [DOI] [PubMed] [Google Scholar]
- CANN J. R., GOAD W. B. THEORY OF MOVING BOUNDARY ELECTROPHORESIS OF REVERSIBLY INTERACTING SYSTEMS. REACTION OF PROTEINS WITH SMALL UNCHARGED MOLECULES SUCH AS UNDISSOCIATED BUFFER ACID. J Biol Chem. 1965 Jan;240:148–155. [PubMed] [Google Scholar]
- Cann J. R. Phenomenological theory of gel electrophoresis of protein-nucleic acid complexes. J Biol Chem. 1989 Oct 15;264(29):17032–17040. [PubMed] [Google Scholar]
- Cann J. R. Theory of electrophoresis of hybridizing enzymes with kinetic control: implications for population genetics of electrophoretic markers. J Theor Biol. 1987 Aug 21;127(4):461–477. doi: 10.1016/s0022-5193(87)80143-1. [DOI] [PubMed] [Google Scholar]
- Fried M., Crothers D. M. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981 Dec 11;9(23):6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner M. M., Revzin A. A gel electrophoresis method for quantifying the binding of proteins to specific DNA regions: application to components of the Escherichia coli lactose operon regulatory system. Nucleic Acids Res. 1981 Jul 10;9(13):3047–3060. doi: 10.1093/nar/9.13.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillen W., Gatz C., Altschmied L., Schollmeier K., Meier I. Control of expression of the Tn10-encoded tetracycline resistance genes. Equilibrium and kinetic investigation of the regulatory reactions. J Mol Biol. 1983 Sep 25;169(3):707–721. doi: 10.1016/s0022-2836(83)80166-1. [DOI] [PubMed] [Google Scholar]
- Hillen W., Klock G., Kaffenberger I., Wray L. V., Reznikoff W. S. Purification of the TET repressor and TET operator from the transposon Tn10 and characterization of their interaction. J Biol Chem. 1982 Jun 10;257(11):6605–6613. [PubMed] [Google Scholar]
- Kleinschmidt C., Tovar K., Hillen W., Porschke D. Dynamics of repressor-operator recognition: the Tn10-encoded tetracycline resistance control. Biochemistry. 1988 Feb 23;27(4):1094–1104. doi: 10.1021/bi00404a003. [DOI] [PubMed] [Google Scholar]
- Klock G., Hillen W. Expression, purification and operator binding of the transposon Tn1721-encoded Tet repressor. J Mol Biol. 1986 Jun 20;189(4):633–641. doi: 10.1016/0022-2836(86)90493-6. [DOI] [PubMed] [Google Scholar]
- Klock G., Unger B., Gatz C., Hillen W., Altenbuchner J., Schmid K., Schmitt R. Heterologous repressor-operator recognition among four classes of tetracycline resistance determinants. J Bacteriol. 1985 Jan;161(1):326–332. doi: 10.1128/jb.161.1.326-332.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwabara M. D., Sigman D. S. Footprinting DNA-protein complexes in situ following gel retardation assays using 1,10-phenanthroline-copper ion: Escherichia coli RNA polymerase-lac promoter complexes. Biochemistry. 1987 Nov 17;26(23):7234–7238. doi: 10.1021/bi00397a006. [DOI] [PubMed] [Google Scholar]
- Marshall B., Morrissey S., Flynn P., Levy S. B. A new tetracycline-resistance determinant, class E, isolated from Enterobacteriaceae. Gene. 1986;50(1-3):111–117. doi: 10.1016/0378-1119(86)90315-x. [DOI] [PubMed] [Google Scholar]
- McMurry L., Petrucci R. E., Jr, Levy S. B. Active efflux of tetracycline encoded by four genetically different tetracycline resistance determinants in Escherichia coli. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3974–3977. doi: 10.1073/pnas.77.7.3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez B., Tachibana C., Levy S. B. Heterogeneity of tetracycline resistance determinants. Plasmid. 1980 Mar;3(2):99–108. doi: 10.1016/0147-619x(80)90101-8. [DOI] [PubMed] [Google Scholar]
- Oehmichen R., Klock G., Altschmied L., Hillen W. Construction of an E. coli strain overproducing the Tn10-encoded TET repressor and its use for large scale purification. EMBO J. 1984 Mar;3(3):539–543. doi: 10.1002/j.1460-2075.1984.tb01843.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogden S., Haggerty D., Stoner C. M., Kolodrubetz D., Schleif R. The Escherichia coli L-arabinose operon: binding sites of the regulatory proteins and a mechanism of positive and negative regulation. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3346–3350. doi: 10.1073/pnas.77.6.3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postle K., Nguyen T. T., Bertrand K. P. Nucleotide sequence of the repressor gene of the TN10 tetracycline resistance determinant. Nucleic Acids Res. 1984 Jun 25;12(12):4849–4863. doi: 10.1093/nar/12.12.4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson C. A., Nash H. A. Bending of the bacteriophage lambda attachment site by Escherichia coli integration host factor. J Biol Chem. 1988 Mar 15;263(8):3554–3557. [PubMed] [Google Scholar]
- Tovar K., Ernst A., Hillen W. Identification and nucleotide sequence of the class E tet regulatory elements and operator and inducer binding of the encoded purified Tet repressor. Mol Gen Genet. 1988 Dec;215(1):76–80. doi: 10.1007/BF00331306. [DOI] [PubMed] [Google Scholar]
- Tovar K., Hillen W. Tet repressor binding induced curvature of tet operator DNA. Nucleic Acids Res. 1989 Aug 25;17(16):6515–6522. doi: 10.1093/nar/17.16.6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unger B., Becker J., Hillen W. Nucleotide sequence of the gene, protein purification and characterization of the pSC101-encoded tetracycline resistance-gene-repressor. Gene. 1984 Nov;31(1-3):103–108. doi: 10.1016/0378-1119(84)90199-9. [DOI] [PubMed] [Google Scholar]
- Unger B., Klock G., Hillen W. Nucleotide sequence of the repressor gene of the RA1 tetracycline resistance determinant: structural and functional comparison with three related Tet repressor genes. Nucleic Acids Res. 1984 Oct 25;12(20):7693–7703. doi: 10.1093/nar/12.20.7693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters S. H., Rogowsky P., Grinsted J., Altenbuchner J., Schmitt R. The tetracycline resistance determinants of RP1 and Tn1721: nucleotide sequence analysis. Nucleic Acids Res. 1983 Sep 10;11(17):6089–6105. doi: 10.1093/nar/11.17.6089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wissmann A., Meier I., Hillen W. Saturation mutagenesis of the Tn10-encoded tet operator O1. Identification of base-pairs involved in Tet repressor recognition. J Mol Biol. 1988 Aug 5;202(3):397–406. doi: 10.1016/0022-2836(88)90273-2. [DOI] [PubMed] [Google Scholar]