Abstract
Chemokine CXCL12 and receptor CXCR4 control multiple steps in primary tumor growth and metastasis in breast cancer and more than 20 other human malignancies. Mechanisms that regulate availability of CXCL12 in tumor microenvironments will substantially impact cancer progression and ongoing efforts to target the CXCL12-CXCR4 pathway for cancer chemotherapy. We used dual luciferase imaging to investigate CXCR7 dependent scavenging of CXCL12 in breast tumors in vivo and quantify effects of CXCR7 on tumor growth and metastasis of a separate population of CXCR4+ breast cancer cells. In a mouse xenograft model of human breast cancer, in vivo imaging showed that malignant cells expressing CXCR7 reduced bioluminescent CXCL12 secreted in the primary tumor microenvironment. Capitalizing on sensitive detection of bioluminescent CXCL12, we also demonstrated that CXCR7+ cells reduced amounts of chemokine released from orthotopic tumors into the circulation. Immunofluorescence staining of human primary breast cancers showed expression of CXCR4 and CXCR7 on malignant cells in ≈ 30% of cases. In most cases, CXCR4 and CXCR7 predominantly were expressed on separate populations of malignant cells in a tumor. We modeled these cases of human breast cancer by co-implanting tumor xenografts with CXCR4+ breast cancer cells, human mammary fibroblasts secreting CXCL12, and CXCR7+ or control breast cancer cells. Bioluminescence imaging showed that CXCR7+ breast cancer cells enhanced proliferation of CXCR4+ breast cancer cells in orthotopic tumors and spontaneous metastases. Treatment with a small molecule inhibitor of CXCR7 chemokine scavenging limited growth of CXCR4+ breast cancer cells in tumors that also contained malignant CXCR7+ cells. These studies establish a new in vivo imaging method to quantify chemokine scavenging by CXCR7 in the tumor microenvironment and identify that CXCR7+ cells promote growth and metastasis of CXCR4+ breast cancer cells.
Keywords: chemokines, luciferase, bioluminescence, breast cancer, chemokine receptor
Introduction
CXCL12 signaling through CXCR4 promotes primary tumor growth and metastasis in breast cancer and more than 20 other malignancies. CXCL12 activates CXCR4 on cancer cells to directly increase proliferation and indirectly enhances tumor growth by increasing angiogenesis and establishing an immunosuppressive tumor microenvironment1-3. CXCL12-CXCR4 signaling promotes migration and invasion of malignant cells, leading to intravasation into the circulation4. Gradients of CXCL12 attract circulating tumor cells to secondary organs and support subsequent proliferation of malignant cells, contributing to characteristic patterns of metastasis in breast cancer and other malignancies5. Multiple functions of CXCL12-CXCR4 in tumor growth and metastasis account for studies showing CXCR4 as a marker of poor prognosis in cancers including breast, prostate, ovary, and leukemia6-9.
CXCR4 signaling depends on duration of exposure and gradients of CXCL12. When added as a one-time dose, CXCL12-CXCR4 activates signaling pathways, including AKT and MAPK, to promote multiple steps in primary and metastatic cancer. Ligand binding causes internalization of CXCR4, receptor degradation, and desensitization to further CXCL12-CXCR4 signaling10. CXCR4-dependent chemotaxis of breast cancer cells requires gradients of CXCL12 rather than constant levels of this chemokine as we demonstrated recently in a microchannel migration device11. Within the microenvironment of primary human breast cancers, carcinoma associated fibroblasts are a major source of CXCL12 (ref. 2, 12). Based on ex vivo analysis of isolated carcinoma associated fibroblasts, these cells appear to secrete CXCL12 constitutively, potentially leading to desensitization of CXCR4 signaling. Therefore, mechanisms that alter overall availability, distribution, and gradients of CXCL12 in tumor microenvironments will regulate functions of CXCR4 in tumor growth and metastasis.
CXCR7 is a second receptor for CXCL12 that binds this chemokine with greater affinity than CXCR4. Cell culture and ex vivo studies show that CXCR7 functions as a scavenger receptor for CXCL12, removing this chemokine from the extracellular space and degrading it in lysosomes13-15. By removing CXCL12 from the extracellular space, CXCR7 reduces amounts of chemokine available to activate CXCR4 signaling. This effect of CXCR7 could limit CXCL12-CXCR4-dependent effects on tumor growth. However, chemokine scavenging by CXCR7 may establish gradients of CXCL12 and maintain responsiveness of CXCR4 signaling and chemotaxis in response to these gradients. For example, expression of CXCR7 on somatic cells is necessary for proper directional migration of primordial germ cells during zebrafish development13. In the absence of CXCR7, CXCR4-expressing germ cells move randomly, likely because there is no effective gradient of CXCL12. These studies suggest that CXCR7+ cells may regulate growth and metastasis of a separate population of CXCR4+ tumor cells under conditions in which cells are exposed chronically to CXCL12, such as the microenvironment of primary breast cancers.
In this study, we developed an in vivo bioluminescence imaging assay to establish that CXCR7 scavenges chemokine CXCL12 in orthotopic human breast cancer xenografts and reduces systemic release of this chemokine from the tumor site. Immunofluorescence staining of primary human breast cancers showed that CXCR4 and CXCR7 frequently are expressed on separate populations of cells in the same tumor. When implanted as tumor xenografts with human mammary fibroblasts secreting CXCL12, proliferation and spontaneous metastasis of CXCR4+ breast cancer cells increased when these cells originated in tumors containing a separate population of malignant cells expressing CXCR7. Treatment with an inhibitor of CXCL12 scavenging by CXCR7 reversed effects of CXCR7+ cells on growth of CXCR4+ cells in orthotopic tumors. This study defines interdependent effects of cells expressing CXCR4 or CXCR7 in breast cancer and suggests new therapeutic opportunities to treat patients with this disease.
Results
Breast cancer cells expressing CXCR7 reduce extracellular CXCL12
To model primary human breast tumors and effects of CXCR7 on levels of CXCL12 in this microenvironment, we co-cultured breast cancer cells with fibroblasts. We used MDA-MB-231 breast cancer cells stably transduced with CXCR7 (231-CXCR7) or vector control (231-control). 231 cells do not express endogenous CXCR7 (ref.16). We stably transduced immortalized human mammary fibroblasts (HMF) or HT0180 cells with CXCL12 fused to Gaussia luciferase (CXCL12-GL). We previously established that bioluminescent CXCL12-GL is secreted from cells, retains signaling functions of unfused CXCL12, and provides sensitive detection of this chemokine in cell-based assays16. HT1080 cells are a fibrosarcoma cell line often used as a model of activated fibroblasts17. We selected these cells because they secrete higher levels of CXCL12-GL than HMF-CXCL12-GL cells, providing a more demanding test of CXCR7 scavenging in the tumor microenvironment. HT1080-CXCL12-GL and HMF-CXCL12-GL cells secrete ≈ 1.6 and 0.75 ng/ml/hour of CXCL12-GL. As controls, we generated HT1080 and HMF cells stably transduced with unfused Gaussia luciferase, which normally is secreted from mammalian cells 18. HMF and HT1080 cells also were transduced with firefly luciferase (FL) as a means to normalize for total numbers of cells in in live cell assays.
We plated equal numbers of various 231 and HT1080 cell types and quantified CXCL12-GL and unfused GL in supernatants at various times through 4 hours. Co-cultures of 231-CXCR7 and HT1080-CXCL12-GL/FL cells had lower levels of CXCL12-GL in supernatants relative to combinations of 231-control and HT1080-CXCL12 cells (Fig 1A). By comparison, levels of unfused GL in supernatants did not differ between co-cultures of 231-CXCR7 or 231-control cells with HT1080-GL/FL. 231-CXCR7 cells also depleted CXCL12-GL produced by HMF-CXCL12-GL cells without affecting amounts of GL in supernatants (Fig 1B).
Figure 1. CXCR7 removes CXCL12 from the extracellular space.
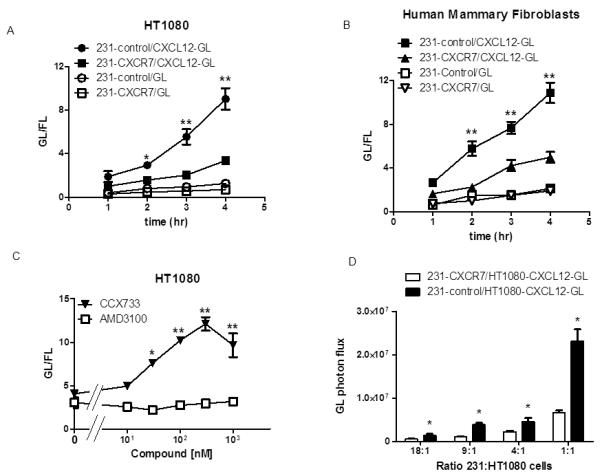
A) MD-MBA-231 breast cancer cells stably transduced with CXCR7 (231-CXCR7) or GFP (231-control) were co-cultured overnight with equal numbers of HT1080 cells secreting either CXCL12-GL or unfused GL, respectively. Amounts of bioluminescent CXCL12-GL and GL in the extracellular space were quantified at various times through 4 hours (n = 4 per condition). B) Co-cultures of 231-CXCR7 or 231-control cells with human mammary fibroblasts secreting either CXCL12-GL or unfused GL were analyzed as in A (n = 4 per condition). C) Co-cultures of 231-CXCR7 and HT1080-CXCL12-GL cells were treated with increasing concentrations of inhibitors of CXCR7 (CCX733) or CXCR4 (AMD3100) for 4 hours before quantifying amounts of CXCL12-GL in culture medium (n = 4 per condition). D) Various ratios of 231-CXCR7 or 231-control cells to HT1080-CXCL12-GL cells were incubated for 4 hours before quantifying CXCL12-GL in culture medium (n = 4 per condition). Data are presented as mean values + SEM. *, p < 0.05; **, p < 0.01.
To verify that CXCR7 depleted CXCL12 from supernatants, we co-cultured 231-CXCR7 and HT1080-CXCL12-GL cells with inhibitors of CXCR7 (CCX733) or CXCR4 (AMD3100). Treatment with CCX733 produced dose-dependent increases in CXCL12-GL present in culture medium (Fig 1C). Relative to vehicle control, 30 nM CCX733 significantly increased amounts of CXCL12-GL (p < 0.05), and the maximal effect occurred at 300 nM (p < 0.01). AMD3100 did not alter CXCL12-GL in supernatants, showing that scavenging was not mediated by CXCR4.
We further investigated CXCR7-dependent scavenging of CXCL12 using various ratios of 231 cell types to HT1080-CXCL12-GL/FL cells. We cultured 231-CXCR7 or 231-control cells with HT1080-CXCL12-GL/FL cells at ratios between 18:1 and 1:1. Under these conditions, amounts of CXCL12-GL produced by HT1080-CXCL12-GL/FL cells ranged from ≈ 0.5 ng/ml/hour – 16 ng/ml/hour. 231-CXCR7 cells reduced CXCL12-GL in supernatants by ≈ 30 – 40% relative to 231-control cells in all co-cultures, showing that CXCR7 sequesters CXCL12 across a wide range of concentrations (Fig 1D). These data establish that CXCR7 limits amounts of CXCL12 in co-cultures of breast cancer cells and stromal fibroblasts, an in vitro model of tumor-stromal interactions in primary breast cancer.
CXCR7 depletes CXCL12 in breast tumors
To investigate effects of CXCR7 on CXCL12 in tumors, we implanted orthotopic breast tumor xenografts of 231-CXCR7 or 231-control cells with HT1080-CXCL12-GL/FL cells. This model reproduces expression of CXCR7 by malignant cells in ≈ 30% of primary human breast cancers19. We selected HT1080-CXCL12-GL/FL cells for the initial experiment because these cells secrete higher levels of bioluminescent chemokine, improving detection of CXCL12-GL in small tumors. Using HT1080-CXCL12-GL/FL cells also tests chemokine scavenging by CXCR7 at levels greater than reported to be secreted ex vivo by carcinoma associated fibroblasts isolated from primary breast tumors2.
We used bioluminescence imaging for Gaussia luciferase as an in vivo assay for amounts of CXCL12-GL present in orthotopic breast tumor xenografts. To normalize for relative numbers of HT1080 cells in each tumor, we measured bioluminescence from firefly luciferase, which uses a different substrate than Gaussia luciferase. Imaging data showed significantly lower CXCL12-GL in tumors comprised of 231-CXCR7 and HT1080-CXCL12-GL/FL cells as compared with tumors containing 231-control cells (p < 0.05) (Fig 2A, B). We measured CXCL12-GL in serum to determine effects of CXCR7 on systemic release of this chemokine from primary tumors. Mice with 231-CXCR7/HT1080-CXCL12-GL/FL tumors had less CXCL12-GL in serum relative to animals bearing 231-control/HT1080-CXCL12-GL/FL tumors (Fig 2C). In mice with 231-CXCR7 tumors, bioluminescence from CXCL12-GL in serum also was lower when data were normalized to total weight of excised tumors (Figure 2D). We observed similar results when HMF-CXCL12-GL cells were implanted with either 231-CXCR7 or 231-control cells, respectively (Fig S1). Collectively, these data show that CXCR7 sequesters CXCL12 in the breast tumor microenvironment and establishes an imaging assay to quantify amounts of this chemokine in vivo.
Figure 2. CXCR7 reduces CXCL12 in primary breast tumors.
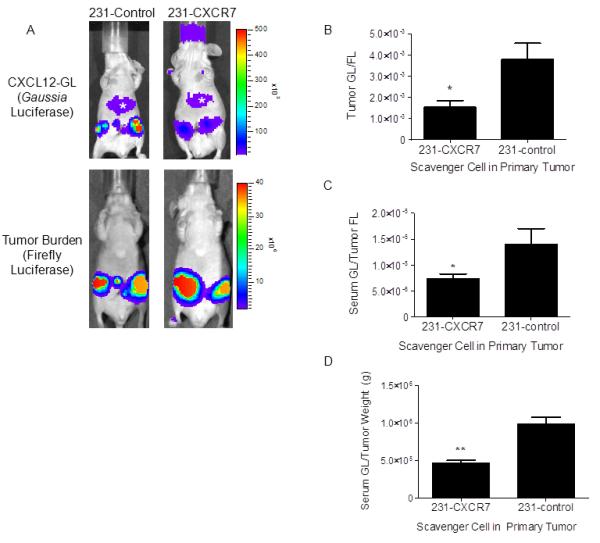
A) Mice were implanted with orthotopic breast tumor xenografts of 231-CXCR7 or 231-control cells with HT1080-CXCL12-GL/FL cells. Representative images are presented from Gaussia and firefly luciferase bioluminescence imaging for tumors with 231-CXCR7 or 231-control cells. Asterisk denotes bioluminescence from oxidation of coelenterazine in liver. Scale bar depicts range of photon flux values as pseudocolor display with red and blue representing high and low values, respectively. B) Photon flux from CXCL12-GL was normalized to firefly luciferase in each tumor and presented as mean values + SEM (n = 5 per group). C) CXCL12-GL in serum was quantified by bioluminescence. Data were normalized to photon flux from firefly luciferase in each mouse. D) Bioluminescence from CXCL12-GL in serum was normalized to weight of excised tumors. Data were graphed as mean values + SEM. *, p < 0.05; **, p < 0.01.
CXCR4 and CXCR7 expression in primary human breast cancers
We stained tissue microarrays of primary breast cancers for CXCR4 and CXCR7. Immunofluorescence showed that ≈ 30% of tumors had breast cancer cells with detectable CXCR4 and CXCR7 on either separate populations of cells or co-expressed on the same cell (Fig 3A, 3B). Receptors more commonly were present on separate populations of breast cancer cells within the same tumor (≈ 70% of positive tumors). We also detected CXCR7 on blood vessels in primary breast tumors, confirming previous studies showing this receptor on tumor vasculature in breast cancer and other malignancies (Fig 3C)19, 20. These results establish that both CXCR4 and CXCR7 are expressed by separate populations of malignant cells or co-expressed on the same tumor cells in primary breast cancers.
Figure 3. CXCR4 and CXCR7 expression in primary human breast cancers.
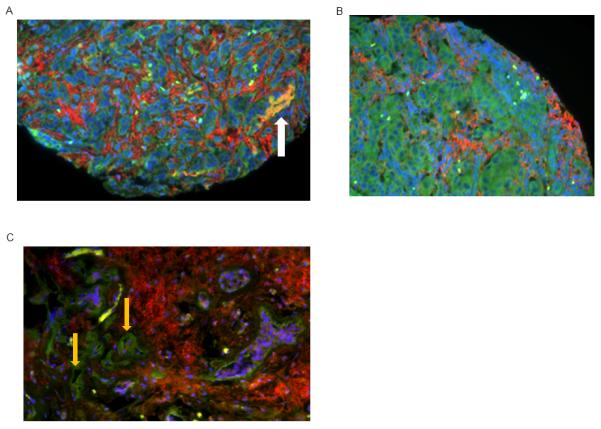
A-C) Tissue microarrays from primary human breast cancers were stained for CXCR4 (red) and CXCR7 (green). Nuclei were stained with DAPI (blue). Representative images from 3 different tumors are shown. A, B) CXCR4 and CXCR7 are present on separate populations of malignant cells, and white arrow in A shows malignant cells that co-express CXCR4 and CXCR7. C) CXCR4+ breast cancer cells are present with CXCR7 on endothelial cells in tumor blood vessels (orange arrows).
CXCR7 promotes primary tumor growth and metastasis of CXCR4-expressing breast cancer cells
CXCL12-CXCR4 signaling enhances growth of primary breast tumors by mechanisms including proliferation of cancer cells and angiogenesis1, 2, 21. Previous studies suggest potentially opposing effects of CXCR7 on CXCL12-CXCR4 signaling, making it difficult to predict how CXCR7 will affect functions of CXCL12-CXCR4 in tumor progression. CXCR7 reduces amounts of CXCL12 in the extracellular space, which limits acute signaling through CXCR4 (ref.15). This effect of CXCR7 would be expected to oppose functions of CXCL12-CXCR4 in tumor progression and metastasis. However, by reducing amounts of CXCL12 in the tumor microenvironment, CXCR7 may prevent desensitization of CXCR4 on breast cancer cells and maintain CXCR4 signaling over time. By analogy, genetic deficiency of CXCR7 in mice substantially elevates amounts of CXCL12 in the brain, resulting in internalization and degradation of CXCR4 and loss of CXCL12-CXCR4 signaling22, 23. Therefore, CXCR7 may be essential for maintaining functions of CXCR4 on breast cancer cells during chronic exposure to CXCL12 in the tumor microenvironment.
To test effects of CXCR7 on growth and metastasis of CXCR4+ breast cancer cells, we implanted orthotopic tumor xenografts with 231 cells stably transduced with CXCR4 and firefly luciferase (231-CXCR4-FL) and equal numbers of 231-CXCR7 or 231-control cells. We used 231 cells transduced with CXCR4 because we previously have shown that our parental 231 cells express very minimal CXCR4 and do not activate CXCR4-dependent signaling in cell culture24. We also co-implanted HMF-CXCL12-GL cells to reproduce secretion of CXCL12 by carcinoma associated fibroblasts in primary human breast tumors2. This strategy allowed selective detection of CXCR4+ breast cancer cells by firefly luciferase and levels of CXCL12-GL by Gaussia luciferase imaging. We implanted separate populations of breast cancer cells expressing either CXCR4 or CXCR7 because this was the condition we identified most commonly in human breast tumors. In addition, we previously demonstrated in a microchannel device that CXCR4+ cells migrated toward CXCL12 when CXCR7 was expressed on a separate population of cells but not when both receptors were co-expressed on the same cell11. For these experiments we used SCID mice lacking IL2Rγ (NOG) mice because 231 breast cancer cells metastasize more extensively in NOG mice relative to nude mice.
After 4 weeks of tumor growth, bioluminescence imaging showed significantly more 231-CXCR4-FL cells in orthotopic tumors with 231-CXCR7 cells as compared with tumors containing 231-control cells (p < 0.01) (Fig 4A). Only 231-CXCR4-FL cells produce firefly luciferase bioluminescence, so these results show enhanced proliferation of CXCR4+ breast cancer cells when co-implanted as tumors with 231-CXCR7 cells. Total weight of tumors with 231-CXCR4-FL, 231-CXCR7, and HMF-SDF-GL cells also was significantly greater than tumors with 231-control cells combined with 231-CXCR4-FL and HMF-CXCL12-GL cells, respectively (p < 0.005) (Fig 4B). These data are consistent with cell culture studies showing that CXCL12 increases growth of 231-CXCR4 cells relative to 231-control cells, although greater growth of 231-CXCR7 cells also contributes to increased overall tumor weight (Fig S2). To improve detection of metastases, we euthanized mice shortly after injecting luciferin and then imaged 231-CXCR4-FL cells in exposed internal organs. Bioluminescence imaging showed widespread metastasis of 231-CXCR4-FL cells to multiple organs and tissues, including lung, greater omentum, and lymph nodes (Fig 4C). Metastatic burden of 231-CXCR4-FL cells was significantly greater when these cells originated from primary tumors containing 231-CXCR7 cells (Fig 4C, D). However, the presence of 231-CXCR7 cells did not alter the anatomic distribution of metastatic 231-CXCR4-FL cells. These results establish that CXCR7+ cells in primary tumors promote overall growth and metastasis of a separate population of CXCR4+ breast cancer cells.
Figure 4. CXCR7 in the primary tumor microenvironment increases growth and metastasis of breast cancer cells expressing CXCR4.
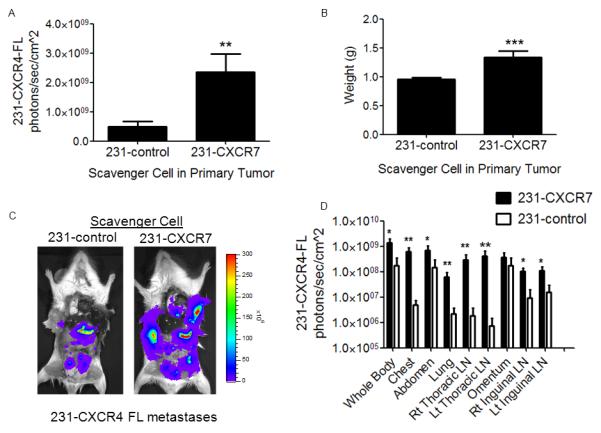
Mice were implanted with orthotopic breast tumor xenografts comprised of human mammary fibroblasts expressing CXCL12-GL (HMF-CXCL12-GL) and 231-CXCR4-FL cells combined with either 231-CXCR7 or 231-control cells (n = 10 per group). A) Mean values + SEM for firefly luciferase bioluminescence after 4 weeks of tumor growth from 231-CXCR4-FL cells in mice with either 231-control or 231-CXCR7 scavenger cells. B) Mean values for weights of excised tumors from each group of mice. C) Representative firefly luciferase bioluminescence images of metastases from 231-CXCR4-FL cells in a mouse from each group. Scale bar shows pseudocolor display for photon flux with red and blue representing highest and lowest values, respectively. D) Quantified data for 231-CXCR4-FL bioluminescence in metastatic sites for mice with either 231-control or 231-CXCR7 scavenger cells. Data were presented as mean values + SEM. *, p < 0.05; **, p < 0.01; ***, p < 0.005.
In addition to increasing proliferation of breast cancer cells expressing CXCR4, CXCL12 may promote tumor growth by recruiting circulating endothelial progenitor cells and enhancing angiogenesis2, 25. We quantified endothelial progenitor cells in tumors by flow cytometry for Sca-1+, CD-31+ cells2. We identified comparable numbers of endothelial progenitor cells in tumors with 231-CXCR7 or 231-control cells co-implanted with HMF-CXCL12-GL and 231-CXCR4-FL cells (Fig S3). Similarly, there were no differences in tumor blood vessels detected by immunohistochemistry for von Willebrand factor (data not shown). These data establish that effects of CXCR7 on growth and metastasis of CXCR4+ breast cancer cells were independent of tumor vasculogenesis and angiogenesis.
Inhibiting CXCR7 reduces primary tumor growth and metastasis of 231-CXCR4-FL breast cancer cells
To further investigate effects of CXCR7 on 231-CXCR4 breast cancer cells, we implanted HMF-CXCL12-GL, 231-CXCR4-FL, and 231-CXCR7 cells orthotopically into mammary fat pads of NOG mice. After palpable tumors formed one week after implantation, we treated mice daily with subcutaneous injections of a small molecule inhibitor of CXCL12 binding to CXCR7 (CCX771) or matched vehicle control. We used CCX771 for animal experiments because this compound can be formulated readily in a vehicle appropriate for in vivo studies, unlike CCX733. Treatment continued for approximately 3 weeks until mice were euthanized for tumor burden.
Treatment with CCX771 increased serum levels of CXCL12-GL by ≈ 70%, demonstrating inhibiting of chemokine scavenging by CXCR7 in orthotopic tumors (Fig 5A). The relative difference in serum levels of CXCL12-GL between CCX771 and vehicle control groups was less than differences between tumors with 231-CXCR7 and 231-control cells, indicating incomplete inhibition of chemokine scavenging. Bioluminescence imaging of mice treated with CCX771 showed decreased growth of 231-CXCR4-FL cells in orthotopic tumors, establishing that even partial inhibition of CXCL12 scavenging by CXCR7 was sufficient to limit growth of CXCR4+ breast cancer cells (Fig 5B). However, overall weight of excised primary tumors did not differ between treatment groups (Fig S4). We also analyzed effects on metastasis of 231-CXCR4-FL cells by imaging firefly luciferase in internal organs and tissues. Overall anatomic distribution of metastases was indistinguishable between mice treated with CCX771 and vehicle control. Quantitatively, both groups of mice had comparable firefly luciferase bioluminescence in all sites except the right inguinal LN, which showed significantly more CXCR4+ breast cancer cells in mice treated with CCX771 (Fig 5C, D). These data show that pharmacologic inhibition of chemokine scavenging by CXCR7 blocks local growth of CXCR4+ breast cancer cells combined with CXCR7+ cells in orthotopic primary tumors. However, inhibiting CXCR7 scavenging with CCX771 had minimal effects on metastasis of CXCR4+ tumor cells.
Figure 5. Inhibiting CXCR7 reduces growth of CXCR4+ breast cancer cells in orthotopic tumors.
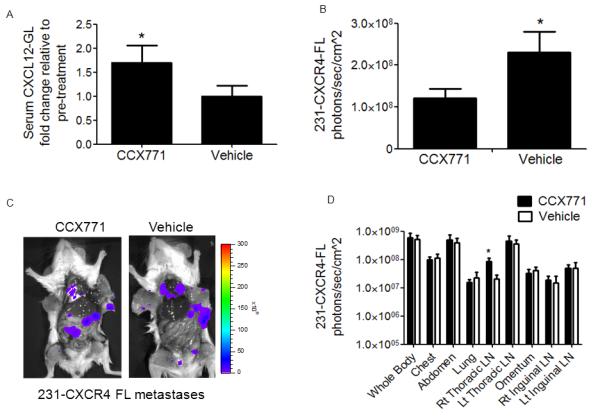
Mice were implanted with orthotopic breast tumor xenografts comprised of human mammary fibroblasts expressing CXCL12-GL (HMF-CXCL12-GL), 231-CXCR4-FL cells, and 231-CXCR7 cells (n = 10 per group). After palpable tumors developed one week after implantation, mice were treated daily with subcutaneous injections of CXCR7 inhibitor CCX771 (30 mg/kg) or vehicle control. A) CXCL12-GL in serum samples was quantified after one week of treatment. Data are presented as mean values + SEM. B) Quantified firefly luciferase bioluminescence from 231-CXCR4-FL cells in primary tumors after 4 weeks of tumor growth. Graph shows mean values + SEM. C) Representative firefly luciferase bioluminescence images of 231-CXCR4-FL metastases in mice treated with CCX771 or vehicle control. Scale bar depicts range of values for photon flux as a pseudocolor display. D) Quantified data for photon flux from 231-CXCR4-FL metastases in multiple anatomic sites. Graph displays mean values + SEM. *, p < 0.05.
CXCR7 Regulates Cell Surface Expression of CXCR4
Prior studies of neuronal development indicate that one critical function of CXCR7 is to prevent internalization and degradation of CXCR4, thereby maintaining CXCL12-CXCR4 signaling22, 23. To determine to what extent CXCR7 controls cell membrane expression of CXCR4 in cancer cells, we co-cultured 231-CXCR4-FL cells with either 231-CXCR7 or 231-control cells. As controls, we prepared parallel cultures of 231-CXCR4 cells alone. We simulated chronic exposure to CXCL12 in the tumor microenvironment by culturing cells for 2 days with 100 ng/ml CXCL12 for 2 days and then measured CXCR4 at the cell membrane by flow cytometry. Relative to incubation in medium alone, treatment with CXCL12 decreased cell surface CXCR4 by ≈ 60% in single cell type cultures of 231-CXCR4 cells (Fig 6A, B). Co-culturing 231-CXCR4 cells with 231-control cells did not prevent loss of cell surface CXCR4. However, 231-CXCR7 cells substantially reduced internalization of CXCR4 in response to CXCL12. In co-cultures of 231-CXCR4 and 231-CXCR7 cells, cell surface CXCR4 decreased by < 20% compared with 231-CXCR4 cells cultured without CXCL12. These data demonstrate that CXCR7 on a separate population of cells reduces internalization of CXCR4 in the presence of constant CXCL12, maintaining levels of CXCR4 at the cell surface to signal in response to extracellular CXCL12.
Figure 6. CXCR7 regulates cell surface expression of CXCR4 and CXCL12-dependent signaling.
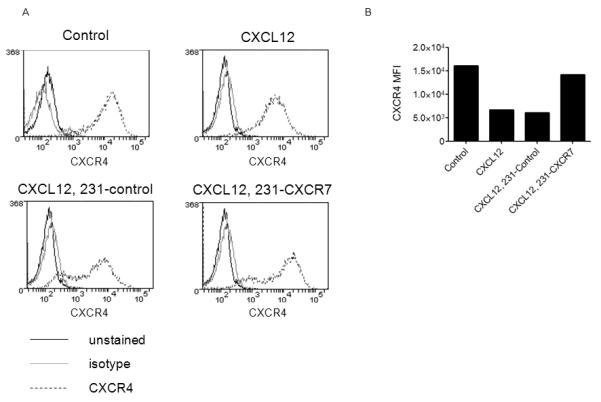
A) Flow cytometry plots for expression of cell surface CXCR4 in 231-CXCR4 cells cultured for 2 days in serum-free medium alone (control), 100 ng/ml CXCL12 (CXCL12), 100 ng/ml CXCL12 plus co-cultured 231-control cells (CXCL12, 231-control), or 100 ng/ml CXCL12 plus co-cultured 231-CXCR7 cells (CXCL12, 231-CXCR7). Black line, unstained cells; gray line, isotype antibody; dashed line, CXCR4 antibody. B) Graph displays mean fluorescence intensity for cell surface CXCR4 from flow cytometry.
Discussion
CXCL12 signaling is a promising therapeutic target in breast cancer and many other common human malignancies, based on numerous studies showing key functions of this chemokine in primary tumor growth and metastasis. While initial efforts focused on CXCR4 , identification of CXCR7 as a second receptor for CXCL12 increased the complexity of this chemokine in cancer biology and as a drug target26, 27. To understand functions of CXCL12 in cancer and optimize development and application of new chemotherapeutic agents, it is essential to determine interrelated effects of CXCR4 and CXCR7 in vivo.
Toward the objective of understanding functions of CXCR7 in living subjects, we developed a new bioluminescence imaging method based on Gaussia luciferase to quantify chemokine scavenging by CXCR7 in a mouse model of human breast cancer. Using this imaging technique, we established that CXCR7 reduced CXCL12 in primary breast tumors and significantly limited release of this chemokine into the circulation. These data build upon prior in vitro studies showing that CXCR7 scavenges and degrades chemokine ligands13-15. Furthermore, this imaging strategy allowed us to analyze effects of pharmacologic inhibition of CXCR7 chemokine scavenging in living mice and compare the extent of inhibition to complete absence of CXCR7+ breast cancer cells in orthotopic tumors. Using in vivo imaging and ex vivo quantification of CXCL12-GL levels in serum will inform and direct ongoing efforts to develop molecules and treatment protocols that effectively block CXCR7-dependent chemokine scavenging in vivo.
We used bioluminescence imaging to selectively monitor growth of CXCR4+ breast cancer cells in an orthotopic tumor xenograft model of human breast cancer. In this model, we used separate populations of breast cancer cells expressing either CXCR4 or CXCR7. This experimental strategy reproduces primary human breast cancers in which we found these receptors to be expressed on different populations of malignant cells in the same tumor. For studies of tumor growth and metastasis, we also implanted human mammary fibroblasts stably expressing CXCL12-GL, based on prior studies showing carcinoma associated fibroblasts as a key source of this chemokine in primary breast tumors2, 12. While 231 cells used in these studies do not endogenously express CXCL12 (data not shown), some breast cancer cells do secrete this chemokine. Other cell types in tumors, such as endothelium, also are potential sources of CXCL12 (ref.28). In these settings, CXCL12-GL secreted by stromal fibroblasts serves as a marker for localization and total amounts of this chemokine originating in tumor xenografts.
Using this mouse model, we discovered that a separate population of CXCR7+ breast cancer cells significantly increased overall growth of CXCR4+ cells, establishing important intercellular interactions between these receptors in cancer. CXCL12 signaling through CXCR4 promotes growth of primary tumors in breast cancer and other malignancies. These effects of CXCR4 occur through mechanisms including proliferation of malignant cells, vasculogenesis, and angiogenesis. Using bioluminescence imaging to selectively monitor CXCR4+ breast cancer cells, we established that CXCR7+ breast cancer cells in orthotopic tumors enhanced overall proliferation of CXCR4+ tumor cells. By comparison, we did not detect any effects of CXCR7-expressing breast cancer cells on recruitment of endothelial progenitor cells or tumor angiogenesis. Collectively, these data show that increased tumor growth was mediated by effects of CXCR7 on CXCR4+ breast cancer cells in the same local tumor microenvironment.
The presence of CXCR7+ breast cancer cells in a primary tumor also enhanced metastasis of CXCR4+ breast cancer cells. This result is consistent with prior studies showing that CXCR7 is required for normal chemotaxis of CXCR4+ cells in response to CXCL12. During migration of primordial germ cells in zebrafish, absence of CXCR7 disrupts normal trafficking of these CXCR4+ cells toward CXCL12 (ref.13). Similarly, genetic deficiency of CXCR7 prevents appropriate migration of CXCR4+ neurons toward CXCL12 during development22. Using a microchannel device to precisely pattern cell positions, we have shown that a separate population of CXCR7 cells is essential for migration of CXCR4+ breast cancer cells toward cells producing CXCL12 (ref.11). Furthermore, computational modeling revealed that formation of chemotactic gradients was critically dependent upon distances and relative positions of cells producing CXCL12, CXCR4+ cells, and CXCR7+ cells, respectively. These data establish that CXCL12 and CXCR7 cells function in a source-sink model of gradient formation for CXCR4-dependent chemotaxis. We propose that CXCR7 cells perform the same function in primary breast tumors, establishing gradients of CXCL12 that increase invasion and intravasation of CXCR4+ breast cancer cells into the circulation to form metastases.
In addition to generating chemotactic gradients, CXCR7 may promote tumor growth by maintaining cell surface levels of CXCR4 during chronic stimulation with CXCL12. When treated for extended time with CXCL12, 231-CXCR7 cells limited internalization of CXCR4 on co-cultured 231-CXCR4 breast cancer cells. These results parallel recent studies in mice showing that genetic deletion of CXCR7 elevates CXCL12 and causes loss of CXCR4 expression in developing neurons22, 23. By limiting desensitization of CXCR4 to chronic CXCL12, CXCR7 may maintain CXCL12-CXCR4 signaling and promote local growth and metastasis of a separate population of CXCR4+ breast cancer cells.
We established that treatment with a small molecule inhibitor of CXCL12-CXCR7 binding partially blocked chemokine scavenging in breast tumors and increased amounts of chemokine released systemically. Pharmacologic inhibition of CXCR7 chemokine scavenging limited proliferation of CXCR4+ breast cancer cells in orthotopic tumors that also contained CXCR7+ cancer cells. These results may be relevant to human breast cancers comprised of malignant cells expressing CXCR4 or CXCR7. Since CXCR7 is upregulated on blood vessels in many common cancers, compounds that block CXCL12 binding to this receptor also may benefit patients with tumors containing only CXCR4+ cancer cells. While pharmacologic inhibition of CXCR7 limited local growth of CXCR4+ cells in orthotopic primary tumors, treatment with CCX771 had minimal effects on metastasis of these cells. This result may be due to incomplete inhibition of chemokine scavenging. Relative differences in serum levels of CXCL12-GL between mice with or without CXCR7+ breast cancer cells in tumors were greater than differences between mice with CXCR7+ tumor cells treated with CCX771 or vehicle. Potentially, more complete inhibition of CXCR7 chemokine scavenging will be required to inhibit metastasis of CXCR4+ cells. Alternatively, since molecules and pathways other than CXCL12-CXCR4 are required for metastasis, the result also suggests that CXCR7+ cells may promote metastasis of CXCR4+ cells through mechanisms other than chemokine scavenging.
We used human breast cancer cells and mammary fibroblasts for these studies to directly link our mouse model to human breast cancer biology. This selection necessitates use of immunocompromised mice. Immunocompromised mouse models of breast cancer and other malignancies have substantially advanced knowledge of CXCL12, CXCR4, and other chemokine pathways in tumor growth and metastasis29, 30. An inherent limitation of immunocompromised mice is the inability to investigate effects of specific molecules and receptors on host immune responses to tumor progression at primary and metastatic sites. Further studies in immunocompetent mice or mice with partially humanized immune systems will be required to determine integrated effects of CXCL12, CXCR4, and CXCR7 on host immune responses to breast cancer.
In summary, we developed a new bioluminescence imaging method to analyze levels of CXCL12 in living subjects and used this technology to establish that CXCR7 limits amounts of CXCL12 in primary breast tumors in vivo. CXCR7+ breast cancer cells promote growth and metastasis of a separate population of CXCR4+ breast cancer cells in a mouse model of human breast cancer, indicating that CXCR7 enhances effects of CXCR4 to increase tumor progression. We note that functions of CXCR7 as a scavenger receptor for CXCL12 do not preclude other potential mechanisms of action reported for this receptor in cancer biology, including direct activation of signaling pathways, resistance to apoptosis, and/or enhanced cell migration31-33. These studies advance our understanding of intercellular regulation of CXCR4 by CXCR7 in breast cancer. This knowledge is essential for ongoing development and clinical translation of therapies targeting CXCL12 signaling in breast cancer and other malignancies.
Experimental Procedures
Cells
We stably transduced immortalized human mammary fibroblasts (provided by Dan Hayes, University of Michigan) and HT1080 fibroblasts (provided by Igor Matushansky, Columbia University) with recombinant lentiviruses for CXCL12 fused to Gaussia luciferase (CXCL12-GL) or unfused Gaussia luciferase (GL)1, 16, 34. We used lentiviruses to stably express firefly luciferase in HT1080 cells that already expressed CXCL12-GL (HT1080-CXCL12-GL), generating HT10801-CXCL12-GL/FL cells. We previously have described MDA-MB-231 human breast cancer cells (ATCC) expressing CXCR4 and firefly luciferase (231-CXCR4-FL), CXCR7 (231-CXCR7), or vector control (231-control)24. We cultured all cells in DMEM (Invitrogen), 10% fetal bovine serum, 1% glutamine, and 0.1% penicillin/streptomycin/gentamicin. We maintained cells in a 37° incubator with 5% CO2.
Co-culture experiments
We plated 231-CXCR7 or 231-control cells in black wall 96 well plates with HT1080 or HMF cells secreting either CXCL12-GL or unfused GL. We seeded a total of 2 × 104 cells per well with various ratios of cells as indicated in figures. We initially plated cells in standard culture medium and then switched to phenol red free DMEM (Invitrogen) without serum. In selected experiments, we incubated cells with small molecule inhibitors of CXCR7 (gift of ChemoCentryx, Mountain View, CA) or AMD3100 (Sigma). We quantified bioluminescence from cell-associated CXCL12-GL or GL as described previously and normalized these data to photon flux from firefly luciferase to account for numbers of CXCL12-GL or GL cells16.
Animal studies
The University of Michigan Committee for the Use and Care of Animals approved all animal procedures. We implanted breast tumor xenografts orthotopically into 4th inguinal mammary fat pads of 5-6 week-old NcrNu/Nu (Harlan) or NOD/SCID Il2rg-/- female mice (Taconic)35. For experiments with HT1080-CXCL12-GL/FL cells, we co-injected 1.5 × 105 fibroblast cells with 1.35 × 106 231-CXCR7 or 231-control cells, respectively. To investigate growth of 231-CXCR4-FL cells in breast tumors, we injected combinations of 3 cell types: 1) human mammary fibroblasts expressing CXCL12-GL (2.5 × 105 HMF-CXCL12-GL); 2) 231-CXCR4-GFP-FL cells (5 × 105); and either 3A) 231-CXCR7 cells (5 × 105) or 3B) 231-control cells (5 × 105). When palpable (≈ 2 mm) tumors formed after one week, we began treatment with CXCR7 inhibitor CCX771 (30 mg/kg CCX771 subcutaneously daily) or vehicle control.
We measured CXCL12-GL in serum by collecting 20 μL of blood by retro-orbital puncture. We mixed blood with 4 μL of 20 mM EDTA and then centrifuged 10 minutes at 10,000 × g to collect the serum fraction36. We quantified bioluminescence from Gaussia luciferase in serum as described above. We measured bioluminescence in serum collected from mice without cells producing CXCL12-GL and subtracted these values from measurements in tumor-bearing mice. To image CXCL12-GL in tumors, we injected mice intravenously with 4 mg/kg coelenterazine via tail vein injection18. We imaged firefly luciferase as described previously37. We imaged mice with an IVIS Spectrum instrument (Caliper) and quantified data as photon flux.
To quantify metastatic breast cancer cells in exposed organs and tissues, we injected luciferin i.p., euthanized mice 5 minutes after injection, and then performed a limited dissection to remove primary tumors and expose internal organs for imaging. We imaged firefly luciferase ≈ 10 minutes after injection of luciferin. We identified sites of metastatic 231-CXCR4-FL cells and quantified bioluminescence from defined anatomic sites and the entire mouse.
Flow cytometry
We digested tumors with type I collagenase (Sigma) for 1-2 hours at 37°C and then removed undigested tissues with 100 and 40 μm strainers (BD Biosciences). We stained recovered cells with antibodies to murine CD31 (clone 390) and Sca-1 (clone D7) (eBioscience) to identify endothelial progenitor cells2.
To quantify effects of CXCR7 on cell surface expression of CXCR4, we co-cultured 5 × 105 231-CXCR4 cells with 5 × 105 231-CXCR7 or 231-control cells in 60 mm dishes. We plated cells in complete medium with serum and then incubated cells overnight in serum-free medium. We cultured cells for an additional 2 days in serum-free medium without or with 100 ng/ml CXCL12 (R&D Systems). We stained cells with mAb 12G5 to CXCR4 (R&D Systems) and quantified levels of cell surface receptor by mean fluorescence intensity. We subtracted fluorescence from cells incubated with isotype control antibody. We recorded data from at least 1 × 105 live cell events for all conditions.
Immunohistochemistry
We detected and quantified tumor blood vessels in formalin-fixed, paraffin-embedded mouse tumor xenografts using an antibody to von Willebrand factor (Sigma)1. We used tissue microarrays generated at University of Michigan from 204 different patients with primary breast cancer and stained these tissues for expression of CXCR4 and CXCR7 using standard methods38. We used a 1:100 dilution of polyclonal rabbit antibody to CXCR4 (Imgenex) and 1:200 dilution of mouse monoclonal antibody to CXCR7 (clone 11G8, gift of ChemoCentryx). We used matched isotype antibodies as controls. To detect primary antibodies, we used PE-conjugated anti-rabbit and FITC-conjugated anti-mouse secondary antibodies. We counterstained nuclei with DAPI. We used an automated fluorescence microscope system (Aperio) to image tumor specimens.
Statistics
We plotted data as mean values with standard error of the mean (SEM). Data from cell-based assays are representative of 2-5 independent experiments, while animal studies show data representative of 1-2 independent experiments each. We analyzed pairs of data by Mann-Whitney U test to determine statistically significant differences (GraphPad Prism).
Supplementary Material
Acknowledgments
Research was supported by NIH grants R01CA136553, R01CA136829, P50CA093990, and R24CA083099. Support also was provided by Fashion Footwear Association of New York (FFANY)/QVC presents Shoes on Sale. We thank ChemoCentryx for small molecule inhibitors of CXCR7 and antibody 11G8.
Footnotes
Conflict of Interest The authors declare no conflicts of interest.
References
- 1.Smith M, Luker K, Garbow J, Prior J, Jackson E, Piwnica-Worms D, et al. CXCR4 regulates growth of both primary and metastatic breast cancer. Cancer Res. 2004;64:8604–8612. doi: 10.1158/0008-5472.CAN-04-1844. [DOI] [PubMed] [Google Scholar]
- 2.Orimo A, Gupta P, Sgroi D, Arenzana-Seisdedos F, Delaunay T, Naeem R, et al. Stromal fibroblasts present in invasive human breast carcinomas promote tumor growth and angiogenesis through elevated SDF-1/CXCL12 secretion. Cell. 2005;121:335–348. doi: 10.1016/j.cell.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 3.Zou W, Machelon V, Coulomb-L’Hermin A, Borvak J, Nome F, Isaeva T, et al. Stromal-derived factor-1 in human tumors recruits and alters the function of plasmacytoid precursor dendritic cells. Nat Med. 2001;7:1339–1346. doi: 10.1038/nm1201-1339. [DOI] [PubMed] [Google Scholar]
- 4.Lee B, Lee T, Avraham S, Avraham H. Involvement of the chemokine receptor CXCR4 and its ligand stromal cell-derived factor 1alpha in breast cancer cell migration through human brain microvascular endothelial cells. Mol Cancer Res. 2004;2:327–338. [PubMed] [Google Scholar]
- 5.Muller A, Homey B, Soto H, Ge N, Catron D, Buchanon M, et al. Involvement of chemokine receptors in breast cancer metastasis. Nature. 2001;410:50–56. doi: 10.1038/35065016. [DOI] [PubMed] [Google Scholar]
- 6.Jiang Y, Wu X, Shi B, Wu W, Yin G. Expression of chemokine CXCL12 and its receptor CXCR4 in human epithelial ovarian cancer: an independent prognostic factor for tumor progression. Gynecol Oncol. 2006;103:226–233. doi: 10.1016/j.ygyno.2006.02.036. [DOI] [PubMed] [Google Scholar]
- 7.Chu Q, Panu L, Holm N, Li B, Johnson L, Zhang S. High chemokine receptor CXCR4 level in triple negative breast cancer specimens predicts poor clinical outcome. J Surg Res. 2010;159:689–695. doi: 10.1016/j.jss.2008.09.020. [DOI] [PubMed] [Google Scholar]
- 8.Akashi T, Koizumi K, Tsuneyama K, Saiki I, Takano Y, Fuse H. Chemokine receptor CXCR4 expression and prognosis in patients with metastatic prostate cancer. Cancer Sci. 2008;99:539–542. doi: 10.1111/j.1349-7006.2007.00712.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Konoplev S, Jorgensen J, Thomas D, Lin E, Burger J, Kantarjian H, et al. Phosphorylated CXCR4 is associated with poor survival in adults with B-acute lymphoblastic leukemia. Cancer. 2011 doi: 10.1002/cncr.26113. doi: 10.1002/cncr.26113. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 10.Busillo J, Benovic J. Regulation of CXCR4 signaling. Biochim Biophys Acta. 2007;1768:952–963. doi: 10.1016/j.bbamem.2006.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Torisawa Y, Mosadegh B, Bersano-Begey T, Steele J, Luker K, Luker G, et al. Microfluidic platform for chemotaxis in gradients formed by CXCL12 source-sink cells. Integr Biol (Camb) 2010;2:680–686. doi: 10.1039/c0ib00041h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Allinen M, R B, Cai L, Brennan C, Lahti-Domerci J, Huang H, et al. Molecular characterization of the tumor microenvironment in breast cancer. Cancer Cell. 2004;6:17–32. doi: 10.1016/j.ccr.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 13.Boldajipour B, Mahabaleshwar S, Kardash E, Reichman-Fried M, Blaser H, Minina S, et al. Control of chemokine-guided cell migration by ligand sequestration. Cell. 2008;132:463–473. doi: 10.1016/j.cell.2007.12.034. [DOI] [PubMed] [Google Scholar]
- 14.Naumann U, Cameroni E, Pruenster M, Mahabaleshwar S, Raz E, Zerwes H, et al. CXCR7 functions as a scavenger for CXCL12 and CXCL11. PLoS One. 2010;5:e9175. doi: 10.1371/journal.pone.0009175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luker K, Steele J, Mihalko L, Luker G. Constitutive and chemokine-dependent internalization and recycling of CXCR7 in breast cancer cells to degrade chemokine ligands. Oncogene. 2010;29:4599–4610. doi: 10.1038/onc.2010.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luker K, Gupta M, Luker G. Bioluminescent CXCL12 fusion protein for cellular studies of CXCR4 and CXCR7. Biotechniques. 2009;47:625–632. doi: 10.2144/000113126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Albright C, Graciani N, Han W, Yue E, Stein R, Lai Z, et al. Matrix metalloproteinase–activated doxorubicin prodrugs inhibit HT1080 xenograft growth better than doxorubicin with less toxicity. Mol Cancer Ther. 2005;4:751–760. doi: 10.1158/1535-7163.MCT-05-0006. [DOI] [PubMed] [Google Scholar]
- 18.Tannous B, Kim D, Fernandez J, Weissleder R, Breakefield X. Codon-optimized Gaussia luciferase cDNA for mammalian gene expression in culture and in vivo. Mol Ther. 2005;11:435–443. doi: 10.1016/j.ymthe.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 19.Miao Z, Luker K, Summers B, Berahovich R, Bhojani M, Rehemtulla A, et al. CXCR7 (RDC1) promotes breast and lung tumor growth in vivo and is expressed on tumor-associated vasculature. Proc Natl Acad Sci U S A. 2007;104:15735–15740. doi: 10.1073/pnas.0610444104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takano S, Yamashita T, Ohneda O. Molecular therapeutic targets for glioma angiogenesis. J Oncol. 2010:351908. doi: 10.1155/2010/351908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Darash-Yahana M, Pikarsky E, Abramovitch R, Zeira E, Pal B, Karplus R, et al. Role of high expression levels of CXCR4 in tumor growth, vascularization, and metastasis. FASEB J. 2004;18:1240–1242. doi: 10.1096/fj.03-0935fje. [DOI] [PubMed] [Google Scholar]
- 22.Sanchez-Alcaniz J, Haege S, Mueller W, Pla R, Mackay F, Schulz S, et al. Cxcr7 controls neuronal migration by regulating chemokine responsiveness. Neuron. 2011;69:77–90. doi: 10.1016/j.neuron.2010.12.006. [DOI] [PubMed] [Google Scholar]
- 23.Wang Y, Li G, Stanco A, Long J, Crawford D, Potter G, et al. CXCR4 and CXCR7 have distinct functions in regulating interneuron migration. Neuron. 2011;69:61–76. doi: 10.1016/j.neuron.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song J, Cavnar S, Walker A, Luker K, Gupta M, Tung Y, et al. Microfluidic endothelium for studying the intravascular adhesion of metastatic breast cancer cells. PLoS One. 2009;4:e5756. doi: 10.1371/journal.pone.0005756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salcedo R, Wasserman K, Young H, Grimm M, Howard O, Avnver M, et al. Vascular endothelial growth factor and basic fibroblast growth factor induce expression of CXCR4 on human endothelial cells: In vivo neovascularization induced by stromal-derived factor-1alpha. Am J Pathol. 1999;154:1125–1134. doi: 10.1016/s0002-9440(10)65365-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Teicher B, Fricker S. CXCL12 (SDF-1)/CXCR4 pathway in cancer. Clin Cancer Res. 2010;16:2927–2931. doi: 10.1158/1078-0432.CCR-09-2329. [DOI] [PubMed] [Google Scholar]
- 27.Duda D, Kozin S, Kirkpatrick N, Xu L, Fukumura D, Jain R. CXCL12 (SDF1α) - CXCR4/CXCR7 Pathway Inhibition: An Emerging Sensitizer for Anti-Cancer Therapies? Clin Cancer Res. 2011;17:2074–2080. doi: 10.1158/1078-0432.CCR-10-2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pablos J, Amara A, Bouloc A, Santiago B, Caruz A, Galindo M, et al. Stromal-cell derived factor is expressed by dendritic cells and endothelium in human skin. Am J Pathol. 1999;155:1577–1586. doi: 10.1016/S0002-9440(10)65474-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karnoub A, Dash A, Vo A, Sullivan A, Brooks M, Bell G, et al. Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature. 2007;449:557–563. doi: 10.1038/nature06188. [DOI] [PubMed] [Google Scholar]
- 30.Zhang X, Wang Q, Gerald W, Hudis C, Norton L, Smid M, et al. Latent bone metastasis in breast cancer tied to Src-dependent survival signals. Cancer Cell. 2009;16:67–78. doi: 10.1016/j.ccr.2009.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang J, Shiozawa Y, Wang J, Wang Y, Jung Y, Pienta K, et al. The role of CXCR7/RDC1 as a chemokine receptor for CXCL12/SDF-1 in prostate cancer. J Biol Chem. 2008;283:4283–4294. doi: 10.1074/jbc.M707465200. [DOI] [PubMed] [Google Scholar]
- 32.Zabel B, Wang Y, Lewen S, Berahovich R, Penfold M, Zhang P, et al. Elucidation of CXCR7-mediated signaling events and inhibition of CXCR4-mediated tumor cell transendothelial migration by CXCR7 ligands. J Immunol. 2009;183:3204–3211. doi: 10.4049/jimmunol.0900269. [DOI] [PubMed] [Google Scholar]
- 33.Hattermann K, Held-Feindt J, Lucius R, Muerkoster S, Penfold M, Schall T, et al. The chemokine receptor CXCR7 is highly expressed in human glioma cells and mediates antiapoptotic effects. Cancer Res. 2010;70:3299–3308. doi: 10.1158/0008-5472.CAN-09-3642. [DOI] [PubMed] [Google Scholar]
- 34.Lois C, Hong E, Pease S, Brown E, Baltimore D. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science. 2002;295:868–872. doi: 10.1126/science.1067081. [DOI] [PubMed] [Google Scholar]
- 35.Luker K, Gupta M, Luker G. Imaging chemokine receptor dimerization with firefly luciferase complementation. FASEB J. 2009;23:823–834. doi: 10.1096/fj.08-116749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wurdinger T, Badr C, Pike L, de Kleine R, Weissleder R, Breakefield X, et al. A secreted luciferase for ex vivo monitoring of in vivo processes. Nat Methods. 2008;5:171–173. doi: 10.1038/nmeth.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luker G, Pica C, Song J, Luker K, Piwnica-Worms D. Imaging 26S proteasome activity and inhibition in living mice. Nat Med. 2003;9:969–973. doi: 10.1038/nm894. [DOI] [PubMed] [Google Scholar]
- 38.Meng H, Chen G, Zhang X, Wang Z, Thomas D, Giordano T, et al. Stromal LRP1 in Lung Adenocarcinoma Predicts Clinical Outcome. Clin Cancer Res. 2011;17:2426–2433. doi: 10.1158/1078-0432.CCR-10-2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


