Abstract
Neuronal injury elicits potent cellular responses from glia, but molecular pathways modulating glial activation, phagocytic function, and termination of reactive responses remain poorly defined. Here we show that positive or negative regulation of glial reponses to axon injury are molecularly encoded by unique isoforms of the Drosophila engulfment receptor Draper. Draper-I promotes engulfment of axonal debris through an immunoreceptor tyrosine-based activation motif (ITAM). In contrast, Draper-II, an alternative splice variant, potently inhibits glial engulfment function. Draper-II suppresses Draper-I signaling through a novel immunoreceptor tyrosine-based inhibitory motif (ITIM)-like domain and the tyrosine phosphatase Corkscrew (Csw). Intriguingly, loss of Draper-II/Csw signaling prolongs expression of glial engulfment genes after axotomy and reduces the ability of glia to respond to secondary axotomy. Our work highlights a novel role for Draper-II in inhibiting glial responses to neurodegeneration, and indicates a balance of opposing Draper-I/-II signaling events is essential to maintain glial sensitivity to brain injury.
Introduction
Glia are highly sensitive to nervous system insults, including acute injury, infection, and neurodegenerative disease. Neuronal injury triggers reactive gliosis, a robust glial response that includes dramatic changes in glial gene expression and cell morphology1,2. Reactive glia rapidly extend membrane processes toward damage sites and clear degenerating neurons through phagocytic engulfment and then return to a resting state in which pre-injury morphology and molecular profile is restored 3,4.
Damaged neurons release a battery of toxic factors that threaten the health of neighboring cells. Reactive glia counteract these effects by clearing excess glutamate to prevent excitotoxicity 5 and secreting protective molecules 6. Glial clearance of degenerating neurons prevents large-scale release of proteases and cytosolic antigens, which can activate inflammatory immune responses 7,8. However, some reactive glial responses are highly detrimental and can actively promote the destruction of healthy neurons. For example, chronic exposure to amyloid plaques, a characteristic component of Alzheimer’s disease, causes microglia to continuously release pro-inflammatory agents 9. In addition, activation of glial scavenger receptors and phagocytic activity stimulates secretion of harmful reactive oxygen species10–12. Given the powerful effects of reactive glia on neuronal survival, tight regulation of their interactions with neurons after trauma is essential to minimize damage.
In professional immune cells, such as macrophages, many immune responses are regulated by receptors that contain intracellular immunoreceptor tyrosine-based activation motifs (ITAMs), defined by a consensus sequence (YXXI/L-X6–12-YXXI/L), that signal through non-receptor tyrosine kinase cascades13. ITAM signaling can be negatively regulated by receptors that bear a related immunoreceptor tyrosine-based inhibitory motif (ITIM). ITIMs recruit the Src-homology 2 (SH2)-domain-containing tyrosine phosphatases (SHP) SHP-1 and/or SHP-2, which dephosphorylate ITAM domains, key downstream signaling kinases, or adaptor molecules14,15. Interestingly, ITAM and ITIM signaling molecules are expressed in mammalian microglia 16,17, but the role of ITAM/ITIM pathways during glial responses to trauma remains unclear.
The Drosophila engulfment receptor Draper, which regulates glial clearance of axon debris in the injured adult brain, has recently been shown to signal through an ITAM/Src/Syk-like signaling pathway. Extension of glial membranes to injury sites and glial clearance of axonal debris, requires the ITAM-bearing receptor Draper, the Src kinase Src42A, the non-receptor tyrosine kinase Shark8,18. Here we explored how glial responses to axon injury are encoded by Draper receptor isoforms and found that unique isoforms play strikingly different roles in glial responses to axonal injury: the Draper-I isoform activated engulfment activity, while the alternatively spliced Draper-II isoform potently blocked all glial responses to axonal injury. Draper-II associated with the protein tyrosine phosphatase Corkscrew (Csw) through an intracellular ITIM-like domain to negatively regulate glial responses to injury. Intriguingly, upon loss of Draper-II/Csw signaling, glia failed to terminate responses to axonal injury and exhibited a reduced ability to respond to secondary nerve lesions. These data argue that a balance of the opposing actions of Draper-I/ITAM and Draper-II/ITIM-like signaling fine-tune glial responses to neurodegeneration.
Results
Drosophila adult glia express three Draper isoforms
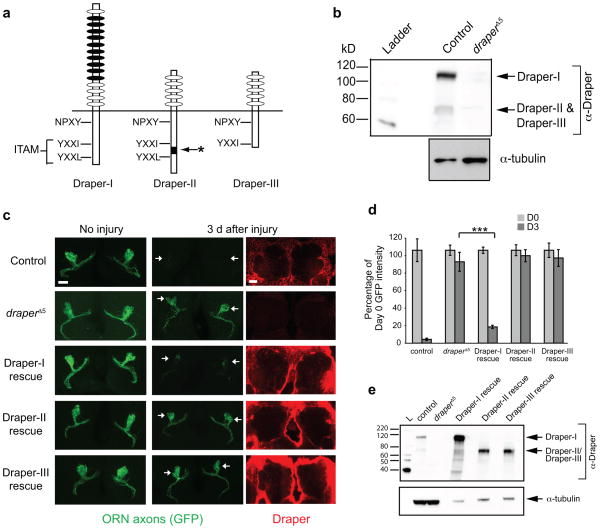
Three Draper receptor isoforms are generated through alternative splicing of the draper gene (Draper-I, Draper-II, and Draper-III) and each contains a unique combination of extracellular and intracellular sequences (Fig. 1a) 19. To determine which isoforms are present in the adult brain, we prepared head protein lysates from control yw and draperΔ5 null mutant flies and performed anti-Draper Western immunoblots using an antibody that recognizes all isoforms 19. We detected bands corresponding to at least two Draper isoforms (Fig. 1b): a larger predominant band representing Draper-I (113kD) and a smaller band at the predicted sizes of Draper-II and/or Draper-III (65kD and 59kD, respectively). Since it was difficult to resolve Draper-II and Draper-III proteins, we performed RT-PCR and detected transcripts for all three isoforms in whole adult heads and dissected brains. draper-I and draper-III are also expressed in embryos, larval brains, and larval body wall tissue. Notably, draper-II is selectively expressed in adults (Supplemental Fig. 2).
Figure 1. Draper-I is sufficient for glial clearance of degenerating axons in the adult Drosophila CNS.
(a) Draper isoform schematic. Ovals represent extracellular EGF-like motifs. NPXY is the predicted Ced-6 binding site. Draper-I contains an ITAM (YXXI-X11-YXXL), which binds Shark. Draper-II contains a unique insertion (asterisk) within the ITAM. Draper-III lacks an ITAM due to a frame shift and premature stop codon.
(b) Draper Western blot (WB) on control yw adult heads. At least two bands were detected: Draper-I (113 kD) and at least one smaller band corresponding to Draper-II (65 kD) and Draper-III (59 kD). Full-length blot is presented in Supplementary Figure 1.
(c) repo-Gal4 was used to express UAS-driven transgenes of each Draper isoform in draperΔ5 mutants that also carried OR85e-mCD8::GFP to label maxillary palp 85e ORNs. Projected confocal Z-stacks show GFP+ axons (green) in the antennal lobe before and after axotomy. Confocal slices of Draper immunostaining (red) show robust glial expression of Draper transgenes.
(d) Quantification of GFP+ axon material in Draper rescue experiments shown in (c). Bars depict mean ± S.E.M. ***p<0.001.
(e) Draper WB on head lysates of control yw, draperΔ5, and rescue flies to confirm expression of UAS-Draper transgenes. Blot was reprobed for tubulin. Less protein lysate was loaded in rescue lanes because GAL4/UAS drives robust expression. D0, Day 0; D3, Day 3. Full-length blots shown in Supplementary Figure 1.
Genotypes and N values in (c,d): control=w;OR85e-mCD8::GFP/+;repo-Gal4/+. (D0 N=12, D3 N=12).
draperΔ5=w; draperΔ5. (D0 N=20; D3 N=14).
Draper-I rescue=w;OR85e-mCD8::GFP/UAS-Draper-I;repo-Gal4,drprΔ5/drprΔ5. (D0 N=31; D3=39).
Draper-II rescue=w;OR85e-mCD8::GFP/UAS-Draper-II;repo-Gal4,drprΔ5/drprΔ5. (D0 N=16; D3 N=16).
Draper-III rescue=w;OR85e-mCD8::GFP/UAS-Draper-III;repo-Gal4,drprΔ5/drprΔ5. (D0 N=23; D3 N=26).
Draper-I is required for glial engulfment of severed axons
Surgical ablation of the antennae or maxillary palps severs olfactory receptor neuron (ORN) axons that project into the brain, triggering their destruction through Wallerian degeneration. Glia infiltrate the injury site and engulf ORN axonal debris, but in the absence of Draper, glia fail to react and axonal debris lingers in the brain 8,18. To determine how each Draper isoform modulates glial responses to injury in vivo, we used the pan-glial driver repo-Gal4 to drive expression of UAS-Draper-I, UAS-Draper-II, or UAS-Draper-III in draperΔ5 null mutants, severed ORN axons, and assayed glial clearance of axon debris (Fig. 1c,d). We confirmed expression of each UAS-Draper transgene by anti-Draper immunostaining (Fig. 1c) and immunoblots (Fig. 1e). Next, we quantified clearance of degenerating GFP+ axons (labeled with OR85e-mCD8::GFP) three days after axotomy. In control animals, GFP+ axonal debris was cleared from the brain, while virtually all GFP+ degenerating ORN axons persisted in draperΔ5 mutants (Fig. 1c,d) 8. Strikingly, glial expression of Draper-I rescued the engulfment defect of draper mutants to near control levels. In contrast, expression of Draper-II or Draper-III failed to rescue draper engulfment defects (Fig. 1c,d). Glia cleared severed axons normally in heterozygous UAS-Draper flies (Supplemental Fig. 3), indicating these phenotypes were not due to insertional transgene effects. Thus, Draper-I, but not Draper-II or Draper-III, is sufficient for glial phagocytic engulfment of degenerating axons in the adult brain.
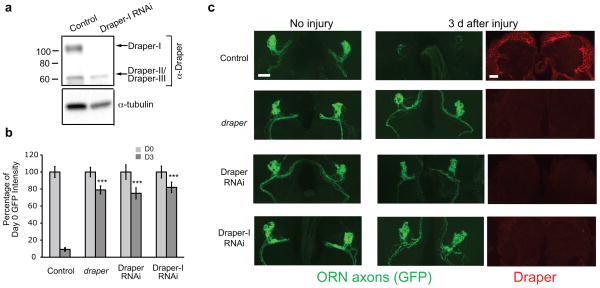
To determine if loss of Draper-I was sufficient to suppress engulfment activity, we generated an RNA interference (RNAi) construct (UAS-Draper-IRNAi) that targets the unique extracellular domain region in Draper-I (depicted by black ovals in Fig. 1a) 20. We first confirmed specificity by ubiquitously driving UAS-Draper-IRNAi and performing an anti-Draper immunoblot (Fig. 2a). Next, we used repo-Gal4 to drive UAS-DraperIRNAi and assessed glial clearance of severed axons. Clearance of degenerating axons was significantly inhibited to levels indistinguishable from those observed by knockdown of all Draper isoforms or in draper null mutants (Fig. 2b,c). We also noted that glial expression of UAS-Draper-IRNAi resulted in a loss of the vast majority of Draper staining (Fig. 2c), suggesting that Draper-I is the predominant isoform in adult glia. These clearance phenotypes were not an artifact of activating the RNAi machinery as repo-Gal4 driven RNAi targeting other genes did not affect clearance of severed axons (see Supplemental Fig. 4). Since glial expression of Draper-II and –III is retained in DraperIRNAi animals, we conclude these isoforms are not sufficient to drive glial engulfment of degenerating axons. Together, our findings suggest that Draper-I is necessary and sufficient for glial engulfment of axonal debris after axotomy.
Figure 2. Draper-I is required for glial clearance of severed axons.
(a) UAS-Draper-IRNAi, which targets the unique region in the extracellular region of Draper-I (see Fig. 1a) was driven ubiquitously with tubulin-Gal4. Draper WB was performed on adult head protein lysates to confirm specific knockdown of Draper-I (96.4 ± .3% reduction in Draper-I, p<0.01; 2.1 ± 25% reduction in Draper-II/-III, p=0.76). Full-length blots shown in Supplementary Figure 1.
(b) Quantification of OR85e-mCD8::GFP shown in (c). Bars represent mean ± S.E.M. ***p<0.001. D0, Day 0; D3, Day 3.
(c) repo-Gal4 was used to knock down Draper-I by driving UAS-Draper-IRNAi or to knock down all Draper isoforms with UAS-DraperRNAi. Projected confocal Z-stacks show that glial clearance of axonal debris (green) was inhibited by knockdown of all Draper isoforms and in draper mutants. Notably, glial clearance of severed axons was equally suppressed by selective knockdown of Draper-I. Draper (red) was barely detectable in the brain following expression of UAS-DraperRNAi or UAS-Draper-IRNAi in glia. The mean intensity of cortical Draper immunostaining in control, UAS-DraperRNAi, and UAS-Draper-IRNAi animals was 82.5 ± 4.7, 39 ± 1.2, and 36 ± 1.3, respectively.
Genotypes in (a): control=tubulin-Gal4/+, Draper-I RNAi=tubulin-Gal4/+;UAS-DraperIRNAi/+.
Genotypes and N values for (b) and (c):
control=w;OR85e-mCD8::GFP/+;repo-Gal4/+. (D0 N=22; D3 N=23).
draper =w; OR85e-mCD8::GFP/+;draperΔ5/draperΔ5
Draper RNAi=w;OR85e-mCD8::GFP/UAS-DraperRNAi;repo-Gal4/+. (D0 N=20; D3 N=24).
Draper-I RNAi=w;OR85e-mCD8::GFP/+;repo-Gal4/UAS-Draper-IRNAi. (D0 N=22; D3 N=19).
The Draper-I intracellular domain activates engulfment
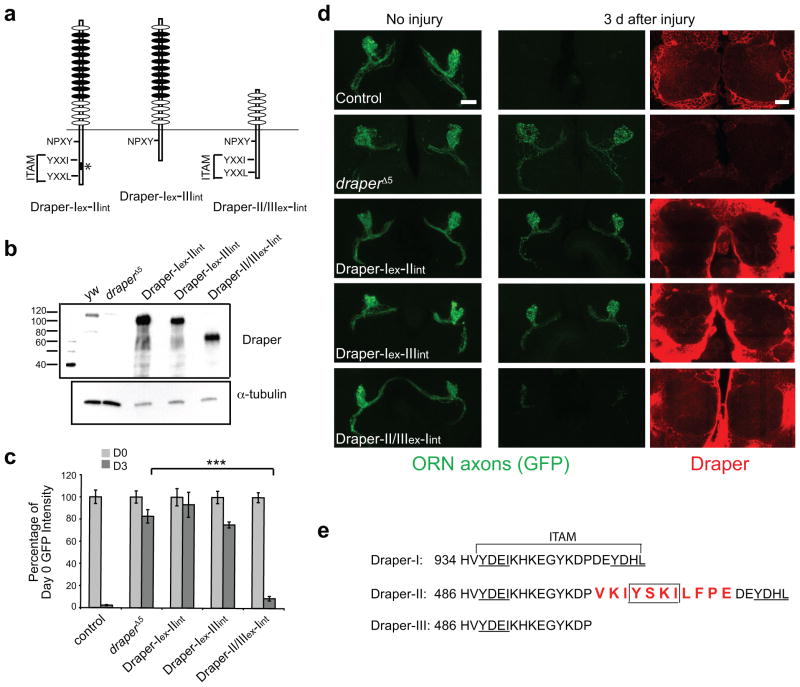
Since Draper-I is structurally unique in both its extracellular and intracellular domain compared to Draper-II and –III, engulfment-promoting activity could map to either region. To examine this, we generated chimeric receptors that contained every combination of extracellular (ex) and intracellular (int) domains from each Draper isoform (Fig. 3a) and tested the ability of each to promote glial engulfment of severed axons. We first confirmed that each construct was stably expressed in adult brains (Fig. 3b). Next, we examined the ability of each chimeric receptor to drive glial clearance of axonal debris in draper a mutant background. Surprisingly, glial expression of Draper-II/III(ex)-I(int) resulted in normal clearance of degenerating axons after axotomy (Fig. 3c,d). Thus, the shorter extracellular domain of Draper-II and -III can functionally replace that of Draper-I during glial clearance of severed axons.
Figure 3. Either Draper ECD with the ICD of Draper-I can trigger glial phagocytic activity.
(a) Schematic of chimeric Draper receptors, which consist of each possible combination of Draper extracellular (ex) and intracellular (in) domains. The name of each construct indicates the extracellular-intracellular domain within.
(b) repo-Gal4 was used to express each chimeric Draper receptor in glial cells of draper mutant flies carrying an OR85e-mCD8::GFP transgene. Anti-Draper WB was performed on adult head lysates to confirm expression of each domain swap construct. Full-length blot is presented in Supplementary Figure 1.
(c) Quantification of axon clearance experiment shown in (d) Bars represent mean ± S.E.M. ***p<0.0001.
(d) Projected confocal Z-stack images of GFP-labeled OR85e ORNs before and 3 days after injury. Single confocal images of Draper immunostaining (red) show robust glial expression of each chimeric Draper transgene.
Genotypes in (b)–(d) and N values for (d):
control=w;OR85e-mCD8::GFP/+;repo-Gal4/+. (D0 N=10; D3 N=12).
draperΔ5=w; draperΔ5. (D0 N=18; D3 N=24).
Draper-I-II rescue=w;OR85e-mCD8::GFP/UAS-Draper-I-II;repo-Gal4,drprΔ5/drprΔ5. (D0 N=16; D3 N=20).
Draper-I-III rescue=w;OR85e-mCD8::GFP/UAS-Draper-I-III;repo-Gal4,drprΔ5/drprΔ5. (D0 N=20; D3 N=32).
Draper-II/III-I rescue=w;OR85e-mCD8::GFP/UAS-Draper-II/III-I;repo-Gal4,drprΔ5/drprΔ5. (D0 N=18; D3 N=26).
(e) The intracellular domain of Draper-I contains a classic ITAM sequence (YXXI-(X)11-YXXL) with two immunoreceptor tyrosine phosphorylation sites (underlined) that are required for recruitment of Shark and subsequent Draper signaling. Draper-II contains a unique 11 amino acid insertion (red text) that interrupts the ITAM and introduces an additional immunoreceptor tyrosine phosphorylation site (YXXI) (box). A frame shift in the Draper-III transcript introduces a truncation in the intracellular domain, which deletes half of the ITAM sequence, leaving only one immunoreceptor tyrosine phosphorylation site (underlined).
We next asked whether the intracellular domain of Draper-I contains unique engulfment-promoting activity. All three isoforms contain an NPXY motif proposed to bind Ced-621–25, but because only Draper-I rescued the draper engulfment defect, this domain is not sufficient for clearance of degenerating axons. Draper-I also contains an essential intracellular ITAM motif (Fig. 3e) 18, while a C-terminal truncation in Draper-III deletes half of this ITAM sequence, including a key tyrosine residue. We overexpressed Draper-I(ex)-III(int) in draper mutant glia and found that this provided no rescue of the engulfment defect (Fig. 3c,d), supporting the notion that a complete ITAM domain is required for Draper-I-mediated engulfment of severed axons.
The Draper-II intracellular domain contains an 11 amino acid insertion in the middle of the ITAM present in Draper-I (Fig. 3e). When we expressed Draper-I(ex)-II(int) in draper mutant glia, we found that this chimeric isoform failed to rescue clearance of axonal debris (Fig. 3c,d). Thus, although the extracellular domains of Draper are fully interchangeable, only the Draper-I intracellular domain is sufficient to drive glial engulfment activity. Remarkably, the 11 amino acid insertion in the intracellular domain of Draper-II renders this isoform incapable of promoting glial engulfment activity.
Draper-II inhibits engulfment through an ITIM-like domain
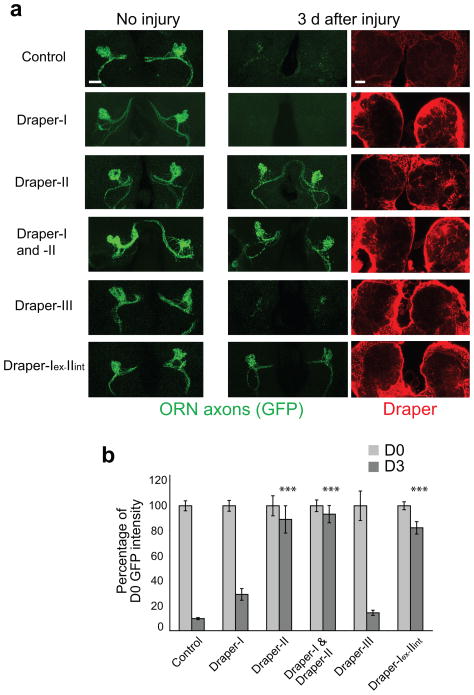
Upon closer inspection of Draper-II, we identified a motif that is reminiscent of an immunoreceptor tyrosine-based inhibitory motif (ITIM; box in Fig. 3e (VKIYXXI)). Mammalian ITIM-bearing receptors inhibit ITAM signaling cascades 14,15,26, which raised the intriguing possibility that Draper-II negatively regulates Draper-I. To test this idea, we first overexpressed Draper-II in glia of wild type flies and found that glial clearance of axonal debris was completely blocked (Fig. 4a,b). Likewise, glial overexpression of Draper-I(ex)-II(int) blocked glial engulfment of GFP+ debris (Fig. 4a,b). In contrast, we overexpressed Draper-I or Draper-III in wild type glia and discovered that engulfment activity was comparable to that of control animals (Fig. 4a,b). Thus, the inhibitory activity of Draper-II maps to the intracellular domain, and since the unique feature of this domain is the ITIM-like motif, we conclude this motif confers inhibitory signaling activity.
Figure 4. Draper-II contains an inhibitory motif in the ICD that inhibits glial engulfment activity.
(a) repo-Gal4 was used to overexpress UAS-draper transgenes in wild type flies that carried OR85e::mCD8-GFP. Projected confocal Z-stacks of OR85e axons (green) in the antennal lobe region are shown before injury and 3 days after injury. Overexpression of Draper-II or the chimeric receptor Draper-I-II completely blocked glial clearance of GFP-labeled degenerating axons. Co-overexpression of Draper-I and Draper-II in glia also robustly blocked clearance. Overexpression of Draper-I or Draper-III had little to no effect on glial engulfment of severed axons. Draper immunostaining (red) shown in single confocal slice images confirmed glial overexpression of each Draper construct.
(b) Graph of GFP quantification of glial clearance assay shown in (a). Bars represent mean ± S.E.M. **p<0.01, ***p<0.0001 as compared to 3 days after ablation in control animals. D0, Day 0; D3, Day 3.
Genotypes and N values:
control=w;OR85e-mCD8::GFP/+;repo-Gal4/+. (D0 N=40; D3 N=40).
Draper-I overexpression=w;OR85e-mCD8::GFP/UAS-Draper-I;repo-Gal4/+. (D0 N=41; D3 N=47).
Draper-II overexpression=w;OR85e-mCD8::GFP/UAS-Draper-II;repo-Gal4/+. (D0 N=8; D3 N=24).
Draper-I and –II overexpression=w;OR85e-mCD8::GFP,UAS-Draper-I/UAS-Draper-II;repo-Gal4/+. (D0 N=8; D3MP N=8).
Draper-III overexpression=w;OR85e-mCD8::GFP/UAS-Draper-III;repo-Gal4/+. (D0 N=16; D3 N=20).
Draper-I-II overexpression=w;OR85e-mCD8::GFP/UAS-Draper-I-II;repo-Gal4/+. (D0 N=18; D3 N=26).
Finally, since Gal4/UAS-driven Draper-II levels were much higher than endogenous Draper levels, we reasoned that providing a high level of Draper-I might overcome the inhibitory activity of Draper-II. We expressed Draper-I and Draper-II together in glia using repo-Gal4 but found that glial engulfment of axonal debris was still potently suppressed (Fig. 4a,b). Thus, providing roughly equivalent levels of Draper-I and Draper-II is insufficient to overcome the inhibitory activity of Draper-II, which may explain the predominance of Draper-I in wild type brains.
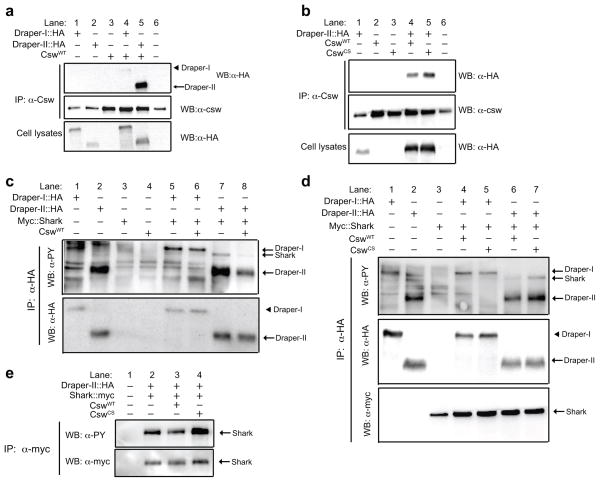
Corkscrew preferentially binds Draper-II
In vertebrates, the tyrosine phosphatases SHP-1 and SHP-2 typically bind ITIMs 14,27. We explored the possibility that Draper-II might associate with Csw, the Drosophila homolog of SHP-1/SHP-2. We transfected Drosophila S2 cells with plasmids for wild type Csw (CswWT), HA-tagged versions of Draper-I or Draper-II, immunoprecipitated with anti-Csw antibody, and performed anti-HA and anti-Csw Western blots on these samples. We consistently detected robust association of Csw with Draper-II, and, notably, significantly more HA-tagged Draper-II co-immunoprecipitated with Csw compared to Draper-I (Fig. 5a, lane 4 versus lane 5, 24 ± 1.7-fold increase, p<0.0001), indicating that Csw preferentially associates with Draper-II.
Figure 5. Corkscrew associates preferentially with and dephosphorylates Draper-II.
(a) HA-tagged Draper constructs and wild type Corkscrew (CswWT) were transfected into insect S2 cells. Anti-Csw immunoprecipitation (IP) samples were analyzed by anti-HA WB to compare Csw association with Draper-I and Draper-II. Anti-HA WB was performed on cell lysates to confirm Draper::HA transfection.
(b) Draper-II::HA was co-transfected with CswWT or CswCS. Anti-HA WB was performed on anti-Csw IP samples to compare Draper-II::HA association with CswWT versus CswCS. Blots were reprobed for Csw. Anti-HA WBs on lysates confirmed Draper-II::HA expression.
(c) Cells were transfected with indicated constructs and anti-HA IP was performed on cell lysates. Anti-phosphotyrosine and anti-HA WBs were performed on IP samples. Co-transfection of CswWT with Draper-II significantly reduced the tyrosine phosphorylation level of bands corresponding to the sizes of Shark and Draper-II (lane 7 versus 8). CswWT did not affect phosphorylation of Draper-I::HA or Shark in the absence of Draper-II::HA (lane 5 versus 6).
(d) Anti-phosphotyrosine, anti-HA, and anti-Myc WBs blots were performed on anti-HA IP samples from cells transfected with indicated constructs. CswCS increased the phosphorylation status of Shark and Draper-II when transfected together (lane 6 versus 7) but did not significantly alter Draper-I or Shark when expressed together in S2 cells (lanes 4 versus 5).
(e) Anti-Myc IP was performed on cells transfected with Draper-II::HA, Myc::Shark, and noted versions of Csw. Anti-phosphotyrosine, anti-HA, and anti-Myc WBs on IP samples are shown. Tyrosine phosphorylation of Shark was reduced by CswWT (lane 2 versus 3) but increased by CswCS (lane 2 versus 4).
Catalytically inactive phosphatases can bind target substrates but fail to release because they cannot dephosphorylate the phosphotyrosine residue required for dissociation, thereby “trapping” the substrate 28–30. As an alternative strategy to show Draper-II/Csw association, we used a substrate-trapping Csw, CswC583S (CswCS)30. We transfected plasmid for CswCS, CswWT, and/or Draper-II::HA, immunoprecipitated with anti-Csw, and performed a Western immunoblot with anti-HA to visualize Draper-II::HA. We found that more Draper-II::HA co-immunoprecipitated with the catalytically inactive CswCS as compared to CswWT (Fig. 5b, lane 4 versus lane 5, 1.7 ± 0.15-fold increase, p<0.05), suggesting that Draper-II is a Csw substrate requiring Csw catalytic activity for dissociation. These results, together with the presence of an ITIM-like sequence in Draper-II, and the ability of Draper-II to inhibit ITAM-mediated signaling during glial engulfment in vivo, strongly implicate Draper-II as a functional ITIM-like receptor that negatively regulates glial engulfment activity.
Draper-II is dephosphorylated by Corkscrew
To determine if Csw alters the tyrosine phosphorylation status of Draper, we transfected S2 cells with CswWT, CswCS and HA-tagged Draper-I or Draper-II and performed anti-HA immunoprecipitations followed by anti-phosphotyrosine immunoblots. Co-transfection of CswWT caused a striking reduction in the intensity of the anti-phosphotyrosine band for Draper-II::HA (Fig. 5c, lane 7 versus lane 8, 28 ± 4% decrease, p<0.01). In contrast, Draper-II phosphorylation was significantly higher when co-expressed with CswCS (Fig. 5d, lane 6 versus lane 7, 330 ± 90% increase, p<0.01), arguing that Draper-II is a phosphatase substrate for Csw. Tyrosine phosphorylation of Draper-I was unaffected by Csw, as we found no significant changes in the phosphorylation status of Draper-I::HA when co-transfected with CswWT (Fig. 5c, lane 5 versus lane 6, 6 ± 3% decrease, p=0.13) or CswCS (Fig. 5d, lane 4 versus lane 5, 40 ± 60% increase with CswCS, p= 0.85). We note that it remains possible Draper-I is a Csw substrate in vivo since expression of constructs in cultured cells does not fully mimic an engulfment event. Nevertheless, these results suggest that Draper-II is a physiological substrate for Csw, whereby Csw dephosphorylates the intracellular domain of Draper-II, perhaps to facilitate the dissociation of Csw or other signaling molecules.
We wondered whether DraperII::HA/Csw signaling might affect the tyrosine phosphorylation status of Shark, which is required for Draper-mediated engulfment in vivo 18. We therefore transfected Draper-I::HA or Draper-II::HA plus N-terminal myc-tagged Shark (Myc::Shark) with or without CswWT into S2 cells and performed anti-HA immunoprecipitation. Interestingly, we found that CswWT significantly reduced the intensity of the Shark anti-phosphotyrosine band in the presence of Draper-II::HA (Fig. 5c, lane 7 versus lane 8, 32 ± 2.6% decrease, p<0.01), but not in the presence of Draper-I::HA (Fig. 5c, lane 5 versus lane 6, 7 ± 12% decrease with CswWT, p=0.59). Anti-phosphotyrosine Shark levels were higher in the presence of CswCS as compared to CswWT (Fig. 5d, lane 6 versus lane 7, 520 ± 140%-fold greater for Shark, p<0.05) when co-transfected with Draper-II::HA. Importantly, tyrosine phosphorylation of Shark was not significantly altered by CswCS in the presence of Draper-I::HA (Fig. 5d, lane 5 versus lane 4, 9± 40% increase, p=0.82,). We also performed reciprocal experiments in which we transfected DraperII::HA and Myc::Shark plus CswWT or CswCS (or no Csw), immunoprecipitated Shark using anti-myc, and then analyzed the tyrosine phosphorylation status of Shark. Again, we found that phosphorylation of Myc::Shark was consistently reduced when co-transfected with CswWT (Fig. 5e, lane 2 versus lane 3, 10% ± 3 % decrease, p<0.05), but increased in the presence of CswCS (Fig. 5e, lane 2 versus 4, 21% ± 8% increase, p<0.05). Finally, we observed that Myc::Shark co-immunoprecipitated equally with Draper-I::HA and Draper-II::HA, regardless of the phosphorylation status of Myc::Shark (Fig. 5d, lanes 4–7). Together, these data support a model whereby Draper-II/Csw inhibits glial engulfment activity by reducing the activity of Draper-I signaling effectors, including Shark, through targeted dephosphorylation.
Corkscrew is required for Draper-II inhibitory activity
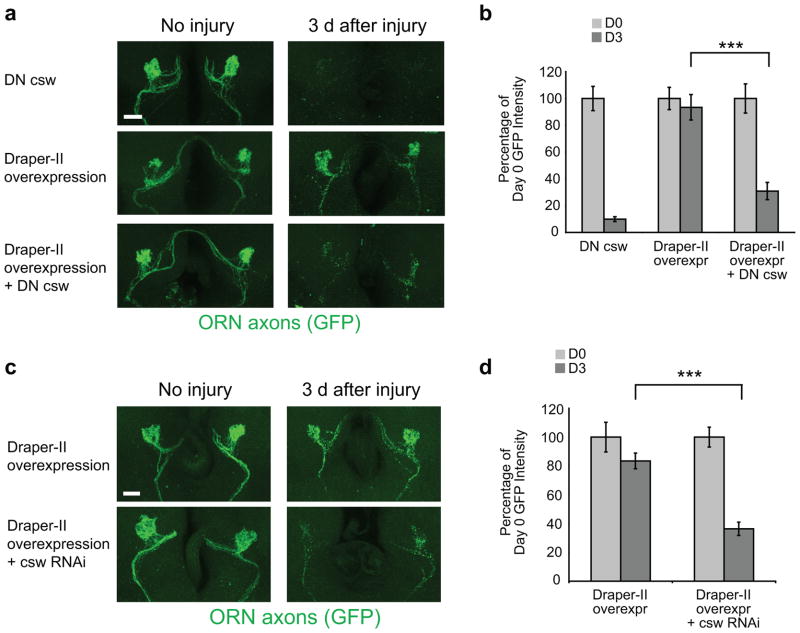
To determine whether Csw was required for Draper-II signaling in vivo, we assayed the effects of csw mutants and RNAi on Draper-II-dependent inhibition of glial engulfment. Glial overexpression of Draper-II completely blocked engulfment of severed axons, but this phenotype was significantly suppressed in heterozygous mutant cswva199 animals31,32 (Fig. 6a,b). Glial engulfment activity was normal in cswva199/+ animals, indicating that Csw is not a positive regulator of glial engulfment. To determine the cell autonomy of Csw function, we used repo-Gal4 to drive UAS-cswRNAi in glia (Supplemental Fig. 5) and found that this significantly suppressed the inhibitory activity of Draper-II (Fig. 6c,d). Thus, Csw, like Draper-II, functions in glia to inhibit engulfment activity.
Figure 6. Draper-II inhibition of glial engulfment of severed axons is mediated through Corkscrew.
(a) The pan-glial driver repo-Gal4 was used to overexpress UAS-Draper-II in a wild type background or in animals that carried one copy of cswva199, a dominant negative allele of corkscrew (DN csw). Reduced Csw activity in cswva199 heterozygotes partially rescued the engulfment phenotype resulting from glial overexpression of Draper-II. Images of GFP-labeled OR85e axons before and 3 days after maxillary palp ablation are all shown as confocal Z-stack projections.
(b) Quantification of experiment in (a). Bars represent mean S.E.M. ***p< 0.0001. D0, Day 0; D3, Day 3.
Genotypes and N values in (a,b):
DN csw = cswva199/+. (D0 N=10; D3 N=10).
Draper-II overexpression = w;OR85e-mCD8::GFP/UAS-Draper-II;repo-Gal4/+. (D0 N=14; D3 N=10).
Draper-II overexpression + DN csw= cswva199/w;OR85e-mCD8::GFP/UAS-Draper-II;repo-Gal4/+. (D0 N=10; D3 N=16).
(c) repo-Gal4 was used to drive UAS-Draper-II in animals that also carried UAS-cswRNAi to knock down Csw expression. Clearance of GFP-labeled OR85e axons was examined 3 days after maxillary palp ablation. Confocal Z-stack projections show GFP+ axonal material in the antennal lobes in each genotype before and after injury. Overexpression of Draper-II in wild type animals blocks glial engulfment of degenerating axons, but this engulfment phenotype is significantly reduced when Csw expression is knocked down by RNAi.
(d) Quantification of the experiment shown in (a). Error bars represent S.E.M. ***p < 0.001. D0, Day 0; D3, Day 3.
Genotypes and N values in (c,d):
Draper-II overexpression = w;OR85e-mCD8::GFP/UAS-Draper-II;repo-Gal4/+. (D0 N=10; D3 N=14).
Draper-II overexpression + cswRNAi = UAS-cswRNAi/w;OR85e-mCD8::GFP/UAS-Draper-II;repo-Gal4/+. (D0 N=12; D3 N=16).
Draper-II/Csw terminates glial responses to axon injury
Our findings suggest that Draper-I and Draper-II each play important, but opposing, roles in glial responses to axotomy. Previous work has shown that Draper immunoreactivity is robustly upregulated in antennal lobe glia within 24 hours after ORN axon injury 8. We performed real time quantitative PCR on central brains at 0, 1.5, 3, 4.5, 6, 12, and 24 hours, and 7 days after axon injury. We found that draper-I mRNA abundance was increased within 1.5 hours after axotomy (Fig. 7a). Interestingly, draper-II transcripts also increased, although these levels did not peak until 4.5 hours after ablation (Fig. 7a). csw levels also appear to be modulated by injury and were significantly decreased at 4.5 hours, coordinate with Draper-II increases (Supplemental Fig. 6). Thus, draper-I and draper-II transcript levels are regulated in response to injury, with up-regulation of the pro-engulfment isoform preceding that of the inhibitory Draper-II isoform.
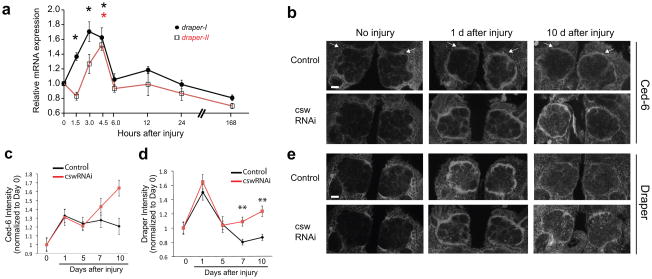
Figure 7. Csw signaling is required for normal termination of glial responses to axon degeneration.
(a) Real-time PCR analysis of normalized expression levels of draper-I and draper-II mRNA in adult central brains before and after antennal ablation (AA). Draper threshold cycle (Ct) values were normalized to ribosomal protein L32 and results are presented as fold-induction relative to uninjured levels. Values represent mean ± S.E.M. from at least 3 independent RNA isolations. draper-I transcript levels were significantly upregulated 1.5, 3.0, and 4.5 hours after injury. draper-II mRNA levels were significantly increased at 4.5 hours after injury (*p<0.05 ANOVA analysis, Dunnett’s post hoc test).
(b) Csw was knocked down in glia by driving UAS-cswRNAi with repo-Gal4. ORN axons were severed by ablating the third antennal segments. Brains were immunostained for Ced-6 0, 1, 5, 7, or 10 days after injury to compare activation of glia following axon injury in control and CswRNAi animals. Representative single confocal slices shown for days 0, 1, and 10.
(c) Quantification of experiment in (b). Bars represent mean ± S.E.M. **p<0.01.
(d) Quantification of experiment in (e). Bars represent mean ± S.E.M. **p<0.01.
(e) Brains were immunostained with anti-Draper to assess the time course of glial responses to AA in the presence and absence of Csw signaling. Representative single confocal slices shown for days 0, 1, and 10.
Genotypes and N values:
Control = w;repo-Gal4/+. (D0 N=20; D1AA N=20; D5AA N=24; D7AA N=20; D10AA N=20).
CswRNAi = UAS-cswRNAi/w; repo-Gal4/+. (D0 N=20; D1AA N=20; D5AA N=24; D7AA N=20; D10AA N=20).
Since Draper-II/Csw negatively regulates glial engulfment activity, we suspected Draper-II/Csw signaling might function to downregulate glial responses to injury after axon clearance. We therefore assayed the efficiency of initiation and termination of glial responses to antennal nerve axotomy in wild type and CswRNAi animals. We first asked whether the expression of key glial engulfment molecules (i.e. Draper and Ced-6) were regulated normally before and after injury. Glial knockdown of Csw had no effect on the basal expression of Ced-6 or Draper protein in uninjured animals (Fig. 7b–e). Both control and CswRNAi animals exhibited a robust increase in Ced-6 and Draper protein in antennal lobe glia 1 and 5 days after ORN axotomy (Fig. 7b–e), indicating glial engulfment responses were activated normally. Furthermore, we observed no difference in clearance of degenerating axons five days maxillary nerve injury in control versus CswRNAi animals (“single injury” Fig. 8a,b), indicating that glial engulfment of severed axons occurred normally in CswRNAi backgrounds. In controls, at 7 and 10 days after injury, while Ced-6 remained elevated, Draper levels returned to baseline, indicating that glia were terminating responses. However, in CswRNAi animals, Ced-6 levels were further elevated at 10 days, and Draper levels remained high at 7 and 10 days (Fig. 7b–e). These data argue that loss of Draper-II/Csw signaling results in a prolonged glial response and in turn a failure to properly terminate glial responses to axotomy.
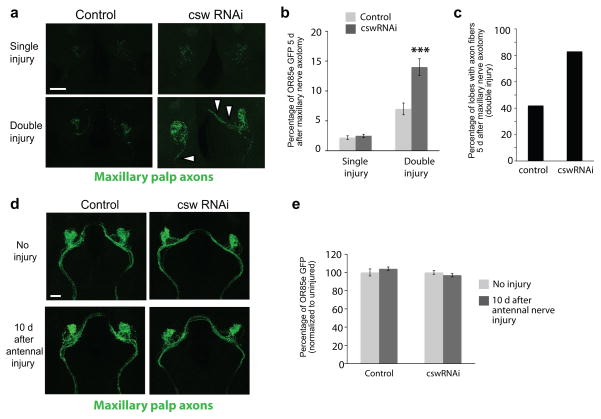
Figure 8. Csw signaling is required for proper glial clearance of severed axons.
(a) Csw was knocked down in glia by driving UAS-cswRNAi with repo-Gal4. A “single injury” entailed ablating the maxillary palps to sever ORN axons and then assessing clearance of GFP+ maxillary palp ORNs 5 days later. “Double injury” animals were pre-injured by ablating the third antennal segments 5 days prior to ablating the maxillary palps and then analyzing clearance of degenerating GFP+ maxillary palp ORNS after 5 days. Representative confocal Z-stack projections shown.
(b) Quantification of GFP intensity in OR85e-innervated maxillary palp glomeruli from experiment in (a). Bars represent mean ± S.E.M. ***p<0.001.
(c) The number of antennal lobes that contained visible tracts of GFP+ maxillary palp axonal debris were scored in control and CswRNAi animals at the end of the double injury experiment. GFP+ axon tracts were faintly visible in 42% of the control brains. In animals expressing glial CswRNAi, the majority of brains (83%) contained visible axonal tracts with persistent GFP+ debris.
Genotypes and N values:
Control = w;repo-Gal4/+ (single injury (D5MP) N=10; double injury (D10AA + D5MP) N=12).
CswRNAi = UAS-cswRNAi/w; repo-Gal4/+ (single injury (D5MP) N=20; double injury (D10AA + D5MP) N=18).
(d) Projected confocal Z-stacks of GFP-labeled OR85e ORNs in control flies and in flies in which Csw is knocked down in glia by RNAi. Images were collected from uninjured animals and 10 days after antennal ablation to compare the morphology of OR85e axons.
(e) Quantification of GFP levels in OR85e-innervated glomeruli in experiment depicted in (d). Bars represent mean ± S.E.M
Sustained expression of engulfment molecules after a primary injury might enhance glial responses to subsequent injuries. Alternatively, a failure to terminate glial responses to axotomy might negatively affect the responsiveness of glial cells to further injury events. To discriminate between these possibilities we developed a serial axotomy assay (Supplemental Fig. 7). Briefly, in animals expressing GFP in maxillary palp neurons (OR85e-mCD8::GFP) we generated a primary antennal nerve lesion to activate glial injury responses. Five days later, we performed a second (maxillary nerve) axotomy, and then assayed clearance of OR85e+ axonal debris. Interestingly, while control animals cleared most axonal debris within 5 days after the secondary injury, CswRNAi animals failed to efficiently clear GFP+ axonal debris at this time point (Fig. 8a–c) and we observed nearly twice as many brains that contained visible axonal tracts of fragmented GFP+ material (Fig. 8c; 42% in controls versus 83% in CswRNAi animals). Uninjured maxillary palp ORNs appeared morphologically normally after the initial injury in control and CswRNAi animals (Fig. 8d,e), arguing that activated wild type and Csw− glia do not promote the indiscriminant overt destruction of uninjured axons in the area of injury. Together these data argue strongly that Draper-II/Csw form a functional ITIM-like signaling cascade that suppresses Draper-I/ITAM signaling, which is essential for the proper termination of glial responses to axon injury in vivo. Furthermore, this work reveals a novel role for Draper and ITIM-like signaling in maintaining glial responsiveness to neural trauma, since efficient termination of an initial glial response appears critical for glia to respond subsequent neural injury.
DISCUSSION
Here we show how a single receptor, Draper, can positively or negatively influence glial responses to axon injury. Our work reveals a critical role for ITAM and ITIM-like signaling events in regulating the activation, termination, and maintenance of engulfment signal transduction cascades during glial responses to axonal injury. In addition, we provide direct evidence that negative regulation of glial responses to neurodegeneration is essential for glia to reset their responses after an initial injury, and thereby remain competent to respond efficiently to subsequent brain trauma.
The Draper intracellular domain directs engulfment
The unique intracellular domains of Draper-I and Draper-II determine their effects on glial responses to axonal injury. While Draper-I promoted engulfment of axonal debris, Draper-II completely inhibited glial clearance of degenerating axons and the inhibitory activity mapped to a Draper-II-specific intracellular motif that contains an ITIM-like domain. We note that this insertion also produces two novel ITAMs in Draper-II, raising the possibility that one or both of these may function as an inhibitory ITAM (ITAMi). Recent work has shown that some ITAMs function in a dual manner, recruiting activating or inhibitory effectors in response to changes in receptor configuration33. However we favor a model in which Draper-II acts exclusively as an inhibitory ITIM-like receptor, since the novel ITIM-like Draper-II domain is not a functional activator in any context we have examined in vivo.
There are two unique Draper extracellular domains that are likely involved in recognition of engulfment targets (Fig. 1a). These are fully interchangeable in our engulfment assay, indicating that neither extracellular domain contains inherent inhibitory activity. It is possible that both domains recognize the same molecule, perhaps a ligand presented by degenerating axons. Alternatively, each extracellular domain may recognize a unique ligand. Identifying specific factors that associate with the extracellular region of each Draper isoform after axotomy will provide key insight into these post-injury neuron-glia communication events.
Draper-II/Csw signaling reprimes glia after injury
Following axotomy, engulfment molecules (Draper and Ced-6) are robustly upregulated in responding glia 8,23 and return to baseline levels once axonal debris has been cleared (Fig. 7). Interestingly, we found that Csw signaling is essential to restore Draper and Ced-6 to basal levels as glia terminate responses to axotomy and that glia lacking Draper-II/Csw fail to respond to secondary injuries in the brain. These data highlight novel and important in vivo requirements for Draper-I and Draper-II/Csw signaling in coordinating the activation, termination, and maintenance of glial cellular responses to axonal injury.
We propose that Draper-I, acting via Src42A and Shark, promotes the expression of engulfment genes after axotomy and phagocytosis of degenerating axons; such upregulation of engulfment genes is likely essential for rapid clearance of degenerating axons. We also propose that Draper-II/Csw then negatively regulates Draper-I signaling to terminate reactive glial responses and allow glia to return to a resting state. Our data showing differential regulation of Draper-I and Draper-II transcripts, with Draper-I preceding Draper-II by several hours, supports the idea that Draper-II/Csw signaling may be delayed relative to Draper-I/ITAM signaling, thereby allowing for potent activation and temporal regulation of responses. Curiously, Csw transcript levels appear to decrease ~4 hrs after injury, suggesting that although Draper-II and Csw are both influenced by post-injury signaling, they are differentially regulated.
It will be important in the future to identify additional factors functioning within the Draper-II/Csw inhibitory signaling pathway. Following engagement of mammalian ITIM receptors, activated SHP-1/2 can target the Src family kinases that phosphorylate the critical ITAM tyrosine residues required for ITAM receptor signaling 34. Thus, Src42A, the kinase that phosphorylates the Draper-I ITAM, may also be an in vivo Csw target. SHP phosphatases can also influence gene transcription within immune cells, although it is largely unclear how this occurs at the molecular level 35.
Our discovery that unique Draper isoforms execute opposing functions during the engulfment of degenerating axons raises interesting questions regarding the putative function of Draper homologs in other species. The related mammalian receptors, Jedi-1 and MEGF10, are expressed in glia and play a conserved role in glial phagocytic activity 36–38. Jedi/MEGF10 drive engulfment in peripheral glia that clear large numbers of apoptotic neurons in the developing dorsal root ganglia 38. It is unknown whether splice variants of Jedi/MEGF10 exist. Notably, the role of Draper-II may be adult-specific; we did not detect Draper-II transcript at embyronic or larval stages, which may indicate that the exuberant phagocytic activity of glia that occurs during nervous system development does not require negative regulation.
We envision that inhibitory constraints on reactive gliosis will be shared by many organisms, and this will be particularly important in the context of neuropathological conditions. It is well known that reactive microglia can actively destroy neurons through phagocytic activity 10–12,39 and reactive glia release a number of factors to promote neuronal death or inhibit neuronal function 11,40,41. Maintaining glial cells in an activated state might exacerbate these effects. The role of MEGF10 and/or Jedi in glia responding to adult neural injury has not yet been examined, but there is evidence that ITIM-mediated pathways may provide neuroprotection. For example, in the SHP-1 knockout mouse motheaten there is striking evidence of enhanced microglial activation and neurodegeneration following acute neuronal damage 42.
Promoting glial termination of reactive responses may be particularly important in neurodegenerative disease, which amounts to a continuous series of neural injuries. Reactive gliosis is certainly a hallmark of nearly all neurodegenerative diseases 9,16 and there is growing evidence that glia help promote disease pathology in mouse models 43–46. The Draper-II/Csw signaling pathway is remarkably specific in its negative regulation of glial responses to axon injury, and provides and exciting molecular entry point to understanding how glial cells terminate cellular and molecular responses to neural trauma.
Methods and Materials
Fly lines and molecular biology
The following Drosophila strains were used: yw, draperΔ5 8, repo-Gal4 47, repo-Gal4, draperΔ5 21, OR85e-mCD8::GFP (gift from B. Dickson), UAS-Draper-IRNAi20, UAS-DraperRNAi 8, UAS-Draper-IRNAi 20, UAS-cswRNAi48, cswva199 32, UAS-neuroliginRNAi (VDRC transformant 44236), UAS-LDLRRNAi (VDRC transformant 27242). We confirmed efficacy of UAS-DraperRNAi by Western blot analysis (Supplemental Fig. 8).
To generate pUAST-Draper-I, pUAST-Draper-II, and pUAST-Draper-III fly lines, we obtained BDGP full length cDNAs (GH24127, GH03529, and RH13935, respectively) and cloned them into the XhoI site of pUAST. To generate the Draper extracellular/intracellular domain swap constructs pUAST-Draper-I-II, pUAST-Draper-I-III, and pUAST-Draper-II/III-I, each pUAST-Draper construct was cut with KpnI and SphI, which excised a fragment containing the 5′UTR, the extracellular domain and a portion of the intracellular domain common to all three isoforms just following the NPXY motif. Each Draper fragment was cloned into KpnI/SphI digested pUAST-Draper vectors to generate domain swap constructs. We sequence verified all constructs, and transgenic flies were generated by BestGene Inc.
We excised the full-length cDNA sequences for Draper-I, Draper-II, and Draper-III from pUAST with NotI/XbaI and cloned into pAc/V5-HisA to generate pAc-Draper-I, pAc-Draper-II, and pAc-Draper-III.
RT-PCR
We isolated total RNA from Drosophila embryos (stage 13–16), L3 larval brains, L3 larval body wall, whole adult heads, and adult dissected brains using Trizol reagent and generated first strand cDNA (Superscript). We performed PCR with Draper isoform specific primers as previously described 24.
Real time PCR
We manually dissected central brains in Jans saline (1.8 mM Ca2+) and immediately froze them on dry ice. We extracted total RNA in Trizol and aqueous phase was passed over an Omega Bio-Tek E.Z.N.A MicroElute RNA Clean Up Column. RNA concentration was determined on a Nanodrop 2000c spectrometer (Thermo Scientific). RNA was diluted to equal concentration, DNAse treated (Ambion DNA-free™ kit), and 125 ng of total RNA was reverse transcribed with the SuperScript® VILO™ cDNA synthesis Kit for 2 hours at 42 degrees °C.
Relative quantitation of gene expression was carried out on an ABI 7500 Fast Real-Time PCR machine. The following Taqman assays (Applied Biosystems) were used: 1) Ribosomal protein L32 (ABI pre-made assay Dm02151827_g1), 2) Draper-I custom assay, F primer-TGTGATCATGGTTACGGAGGAC; R primer-CAGCCGGGTGGGCAA; probe – CGCCTGCGATATAA, 3) Draper-II custom assay, F primer- CAAGCACAAGGAGGGCTACAA; R primer-CATCCTCGGGAAACAAAATCTT; probe – TCCCGTGAAAATATATAGC, and 4) Corkscrew (ABI pre-made assay Dm018211096_g1). Assay efficiencies were experimentally determined (RpL32- 102%, Draper-I – 103%, Draper-II – 102%, and Corkscrew- 98%) using a 5-point dilution series of cDNA spanning a 20-fold range in concentration. The raw threshold cycle (Ct) of the normalization control (RpL32) did not vary more than 0.5 cycles across all time points analyzed. Statistical analysis was performed on 2^-(ΔCt) values.
Olfactory receptor injury protocol, immunohistochemistry, and confocal microscopy
We performed maxillary palp and third antennal segment ablations, adult brain dissections, and antibody stainings using previously described methods 8. Confocal imaging and quantitation of GFP intensity within maxillary palp glomeruli using Image J were performed as described 8,23. We quantified cortical Draper staining for Figure 3B was performed with Image J; single confocal slices at the depth of OR85e-innervated glomeruli were identified, a fixed size rectangular region adjacent to each antennal lobe was selected, and total intensity measurements were calculated. The following antibodies were used: 1:200 mouse anti-GFP (Invitrogen), 1:500 rabbit anti-Draper 19, 1:500 rat anti-Ced-6 21, 1:200 FITC anti-mouse IgG, 1:200 Cy3 anti-rabbit IgG, 1:200 Cy5 anti-rabbit IgG, 1:200 Cy5 anti-rat IgG (Jackson ImmunoResearch).
Equipment and Settings
All immunostained brains were imaged on a Zeiss LSM 5 Pascal with a 63X 1.4NA apochromatic lens. Brains within a single experiment (being compared for quantification) were mounted in Vectashield and imaged on the same day with the same confocal settings (laser power, PMT gain, offset, filter configuration). Analysis and quantification of confocal and Western blot images was performed in ImageJ. See relevant Methods sections for more details on quantification procedures. No post acquisition filtering was performed on displayed images.
Immunoprecipitation and immunoblotting
Insect Schneider S2 cells were seeded into 6-well plates (2×106 cells/well) and transfected (Effectene; QIAGEN) twenty-hour hours later with the appropriate expression vectors: pAc-Draper-I, pAc-Draper-II, pAc-Draper-III, pUAS-myc::shark 50, pAc-Gal4, pAT-Hygro-cswWT, and pAT-Hygro-cswCS 30. Three wells were used for each experimental condition. Forty-eight hours after transfection, we lysed cells in buffer containing 1% Nonidet P-40, 10 mM Tris-HCl (pH 7.5), 50 mM NaCl, 30 mM Na4P2O7, 50 mM NaF, 100 uM Na3VO4, 5 uM ZnCL2, and protease inhibitor cocktail (Complete Tablets; Roche). For immunoprecipitation experiments, we precleared cell lysates with Protein G beads (Sigma) and then incubated with antibody-conjugated beads at 4° overnight. We washed beads the following day 4 times with lysis buffer and then resuspended in 45 ul of lysis buffer plus 10 ul of SDS loading buffer (60mM Tris pH 6.8, 10% glycerol, 2% SDS, 1% b-mercaptoethanol, 0.01% bromophenol blue) and then eluted protein complexes from beads by boiling for 5 minutes. For Western analysis, 15 ul of each sample were loaded onto 10% SDS-PAGE gels (BioRad), transferred to nitrocellulose membranes (BioRad), and probed with the appropriate antibodies: 1:1,000 rabbit a-Draper, 1:500 rat anti-HA (Roche), 1:1,000 mouse anti-phosphotyrosine (Millipore), 1:500 rabbit anti-csw 30, 1:1,000 mouse anti-myc (Covance), 1:10,000 mouse anti-tubulin (Sigma). HRP conjugated secondary antibodies (Abcam) were used at 1:6,000. Antibodies were diluted in PBS/0.01% Tween-20/5% BSA, with the exception of anti-phosphotyrosine, which we diluted in PBS/0.01% Tween-20/5% dry milk. We incubated blots overnight rocking at 4°C, washed 6 times for 20 minutes, probed with appropriate HRP conjugated secondary antibody for 2 hours at room temperature, washed 6 times for 20 minutes, developed using chemiluminescence (Amersham ECL Plus), and detected with a Fujifilm Luminescent Imager. Protein blots were stripped by rocking in mild stripping buffer (0.2M glycine, 0.1% sodium dodecyl sulfate, 1% Tween, pH 2.2) at room temperature for 10 minutes and washing in 1XPBS followed by 1XPBS + 0.01% Tween-20. All biochemical experiments were performed independently between three and six times. ImageJ was used to quantify protein and anti-phosphotyrosine band intensities. For Draper/Csw association experiments, the mean intensity of each Draper band was normalized to the mean intensity of the Csw band in each immunoprecipitation sample. The mean intensity of anti-phosphotyrosine bands for Draper and Shark were normalized to the mean intensity of anti-Draper or anti-myc bands, respectively. Draper Westerns were performed on whole head lysates as previously described20. For Figure 2A, Image J was used to normalize the intensity of Draper-I and Draper-II/-III bands to anti-tubulin band intensities for 3 independent experiments.
Statistical analysis
GraphPad Prism was used to perform ANOVA and two-tailed Student’s t-tests.
Supplementary Material
Supplemental Figure 1.
Full length blots of cropped images shown in manuscript.
Supplemental Figure 2.
Developmental time course of Draper isoform expression in Drosophila. RT-PCR using isoform specific primers was performed on cDNA generated from control yw embryo, larvae, and adults. Plasmids containing each Draper isoform were used as positive controls. Note that transcripts for Draper-I, II, and III were all detected in the adult head and brain. PCR amplification of Gapdh was performed as a positive control for cDNA synthesis.
Supplemental Figure 3.
UAS-Draper transgenes do not affect adult glial engulfment activity. Maxillary palp ablations were performed on flies that carried one copy of each UAS-Draper transgene and OR85e-mCD8::GFP. GFP+ axonal debris remaining in the antennal lobes was quantified 3 days later. In all animals carrying one copy of the Draper UAS rescue transgenes, glial engulfment of GFP+ degenerating axon fragments occurred normally and was indistinguishable from clearance in control animals. (a) Representative confocal images of Z-stack projections shown. (b) Quantification of the experiment shown in (a). Bars depict mean ± S.E.M. D0, Day 0; D3, Day 3.
N values for (D0, D3): control OR85e-mCD8::GFP/+ (10,8); UAS-Draper-I/OR85e-mCD8::GFP (6,8); UAS-Draper-II/OR85e-mCD8::GFP (10,8); UAS-Draper-III/OR85e-mCD8::GFP (8,8); UAS-Draper-I-II/OR85e-mCD8::GFP (6,10); UAS-Draper-I-III/OR85e-mCD8::GFP (8,10); UAS-Draper-II/III-I/OR85e-mCD8::GFP (10,10). Scale bar represents 20 μm.
Supplemental Figure 4.
Activation of RNAi machinery in glia does not inhibit phagocytic clearance of axon debris. Two representative examples of RNAi constructs (neuroligin and LDL-receptor related protein) driven in glia with repo-Gal4 that had no affect on glial clearance of severed axonal debris are shown here. (a) Quantification of GFP+ ORN debris clearance 5 days after maxillary palp ablation shows that clearance is unaffected by non-specific expression of dsRNA. Values represent mean ± S.E.M. (b) Representative confocal Z-stack images depict normal clearance in both RNAi genotypes. Scale bar represents 20 μm.
Supplemental Figure 5.
CswRNAi inhibits Csw-C expression in adult glia. To confirm effectiveness of CswRNAi, UAS-cswRNAi was driven with repo-gal4 and an anti-Csw Western blot was performed on adult brain lysates from cswRNAi (UAS-cswRNAi/+;;repo-Gal4/+) and control (repo-Gal4/+) animals. Expression of CswRNAi in glia results in robust knockdown of at least one isoform of Csw (Csw-C). Preferential knockdown of the smaller isoform suggests that there maybe interesting cell-type specific differences in Csw isoform expression.
Supplemental Figure 6.
csw is not upregulated in the adult brain after axotomy. Real-time PCR analysis of normalized expression levels of csw mRNA in adult central brains before and after antennal ORN axotomy. Threshold cycle (Ct) values were normalized to ribosomal L32 and results are presented as fold-induction relative to levels in uninjured brains. Values represent mean ± S.E.M. of at least 3 independent RNA isolations. Corkscrew transcript levels were significantly downregulated at 4.5 hours after injury (*p<0.05 ANOVA analysis, Dunnett’s post hoc test).
Supplemental Figure 7.
Single and double injury paradigm for experiments described in Figure 8.
Supplemental Figure 8.
UAS-DraperRNAi inhibits expression of all Draper isoforms in the adult brain. A UAS-DraperRNAi transgene that targets a sequence common to all three Draper isoforms was driven in all glia with repo-Gal4. Draper Western blot was performed on dissected adult brain protein lysates to confirm knockdown of Draper-I, Draper-II, and Draper-III.
Acknowledgments
We thank T. Awasaki, M. Simon, E. Perkins, B. Dickson, Vienna Drosophila RNAi Center for sharing fly lines and antibodies. We thank K. Kerr for technical advice and expertise. This work was supported by NIH Grant 1RO1NS053538 (M.R.F), NIH New Faculty Recruitment Grant P30NS069346 P30 (MAL), an American Cancer Society Postdoctoral Fellowship (PF-07-258-01-CSM) (M.A.L.) and the Medical Research Foundation of Oregon (M.A.L.). M.R.F. is a Howard Hughes Medical Institute Early Career Scientist.
Footnotes
Author Contributions:
M.A.L and M.R.F. developed the overall concept and design of the project. M.A.L. performed, analyzed, and interpreted the majority of the experiments and wrote the initial version of the manuscript. R.H. performed the immunoprecipitation and Western blot experiments with S2 cells. J.D. performed the experiments with cswva199 and provided intellectual input for the study. S.D.S performed and analyzed the real-time quantitative PCR time course of Draper-I, Draper-I, and Csw expression after injury and assisted with double injury experiments. A.S. generated the extracellular-intracellular Draper domain swap constructs.
References
- 1.Gebicke-Haerter PJ. Microarrays and expression profiling in microglia research and in inflammatory brain disorders. J Neurosci Res. 2005;81:327–341. doi: 10.1002/jnr.20479. [DOI] [PubMed] [Google Scholar]
- 2.Sofroniew MV. Molecular dissection of reactive astrogliosis and glial scar formation. Trends Neurosci. 2009;32:638–647. doi: 10.1016/j.tins.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Logan MA, Freeman MR. The scoop on the fly brain: glial engulfment functions in Drosophila. Neuron Glia Biol. 2007;3:63–74. doi: 10.1017/S1740925X07000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Neumann H, Kotter MR, Franklin RJ. Debris clearance by microglia: an essential link between degeneration and regeneration. Brain. 2009;132:288–295. doi: 10.1093/brain/awn109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rothstein JD, et al. Knockout of glutamate transporters reveals a major role for astroglial transport in excitotoxicity and clearance of glutamate. Neuron. 1996;16:675–686. doi: 10.1016/s0896-6273(00)80086-0. [DOI] [PubMed] [Google Scholar]
- 6.Lalancette-Hebert M, Gowing G, Simard A, Weng YC, Kriz J. Selective ablation of proliferating microglial cells exacerbates ischemic injury in the brain. J Neurosci. 2007;27:2596–2605. doi: 10.1523/JNEUROSCI.5360-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Block ML, Zecca L, Hong JS. Microglia-mediated neurotoxicity: uncovering the molecular mechanisms. Nat Rev Neurosci. 2007;8:57–69. doi: 10.1038/nrn2038. [DOI] [PubMed] [Google Scholar]
- 8.MacDonald JM, et al. The Drosophila cell corpse engulfment receptor Draper mediates glial clearance of severed axons. Neuron. 2006;50:869–881. doi: 10.1016/j.neuron.2006.04.028. [DOI] [PubMed] [Google Scholar]
- 9.Perry VH, Nicoll JA, Holmes C. Microglia in neurodegenerative disease. Nat Rev Neurol. 2010;6:193–201. doi: 10.1038/nrneurol.2010.17. [DOI] [PubMed] [Google Scholar]
- 10.Bamberger ME, Harris ME, McDonald DR, Husemann J, Landreth GE. A cell surface receptor complex for fibrillar beta-amyloid mediates microglial activation. J Neurosci. 2003;23:2665–2674. doi: 10.1523/JNEUROSCI.23-07-02665.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cho S, et al. The class B scavenger receptor CD36 mediates free radical production and tissue injury in cerebral ischemia. J Neurosci. 2005;25:2504–2512. doi: 10.1523/JNEUROSCI.0035-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Husemann J, Loike JD, Anankov R, Febbraio M, Silverstein SC. Scavenger receptors in neurobiology and neuropathology: their role on microglia and other cells of the nervous system. Glia. 2002;40:195–205. doi: 10.1002/glia.10148. [DOI] [PubMed] [Google Scholar]
- 13.Underhill DM, Goodridge HS. The many faces of ITAMs. Trends Immunol. 2007;28:66–73. doi: 10.1016/j.it.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 14.Daeron M, Jaeger S, Du Pasquier L, Vivier E. Immunoreceptor tyrosine-based inhibition motifs: a quest in the past and future. Immunol Rev. 2008;224:11–43. doi: 10.1111/j.1600-065X.2008.00666.x. [DOI] [PubMed] [Google Scholar]
- 15.Pinheiro da Silva F, Aloulou M, Benhamou M, Monteiro RC. Inhibitory ITAMs: a matter of life and death. Trends Immunol. 2008;29:366–373. doi: 10.1016/j.it.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 16.Napoli I, Neumann H. Microglial clearance function in health and disease. Neuroscience. 2009;158:1030–1038. doi: 10.1016/j.neuroscience.2008.06.046. [DOI] [PubMed] [Google Scholar]
- 17.Takahashi K, Prinz M, Stagi M, Chechneva O, Neumann H. TREM2-transduced myeloid precursors mediate nervous tissue debris clearance and facilitate recovery in an animal model of multiple sclerosis. PLoS Med. 2007;4:e124. doi: 10.1371/journal.pmed.0040124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ziegenfuss JS, et al. Draper-dependent glial phagocytic activity is mediated by Src and Syk family kinase signalling. Nature. 2008;453:935–939. doi: 10.1038/nature06901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Freeman MR, Delrow J, Kim J, Johnson E, Doe CQ. Unwrapping glial biology: Gcm target genes regulating glial development, diversification, and function. Neuron. 2003;38:567–580. doi: 10.1016/s0896-6273(03)00289-7. [DOI] [PubMed] [Google Scholar]
- 20.McPhee CK, Logan MA, Freeman MR, Baehrecke EH. Activation of autophagy during cell death requires the engulfment receptor Draper. Nature. 2010;465:1093–1096. doi: 10.1038/nature09127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Awasaki T, et al. Essential role of the apoptotic cell engulfment genes draper and ced-6 in programmed axon pruning during Drosophila metamorphosis. Neuron. 2006;50:855–867. doi: 10.1016/j.neuron.2006.04.027. [DOI] [PubMed] [Google Scholar]
- 22.Cuttell L, et al. Undertaker, a Drosophila Junctophilin, links Draper-mediated phagocytosis and calcium homeostasis. Cell. 2008;135:524–534. doi: 10.1016/j.cell.2008.08.033. [DOI] [PubMed] [Google Scholar]
- 23.Doherty J, Logan MA, Tasdemir OE, Freeman MR. Ensheathing glia function as phagocytes in the adult Drosophila brain. J Neurosci. 2009;29:4768–4781. doi: 10.1523/JNEUROSCI.5951-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fuentes-Medel Y, et al. Glia and muscle sculpt neuromuscular arbors by engulfing destabilized synaptic boutons and shed presynaptic debris. PLoS Biol. 2009;7:e1000184. doi: 10.1371/journal.pbio.1000184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Su HP, et al. Interaction of CED-6/GULP, an adapter protein involved in engulfment of apoptotic cells with CED-1 and CD91/low density lipoprotein receptor-related protein (LRP) J Biol Chem. 2002;277:11772–11779. doi: 10.1074/jbc.M109336200. [DOI] [PubMed] [Google Scholar]
- 26.Barrow AD, Trowsdale J. You say ITAM and I say ITIM, let’s call the whole thing off: the ambiguity of immunoreceptor signalling. Eur J Immunol. 2006;36:1646–1653. doi: 10.1002/eji.200636195. [DOI] [PubMed] [Google Scholar]
- 27.Olcese L, et al. Human and mouse killer-cell inhibitory receptors recruit PTP1C and PTP1D protein tyrosine phosphatases. J Immunol. 1996;156:4531–4534. [PubMed] [Google Scholar]
- 28.Blasioli J, Paust S, Thomas ML. Definition of the sites of interaction between the protein tyrosine phosphatase SHP-1 and CD22. J Biol Chem. 1999;274:2303–2307. doi: 10.1074/jbc.274.4.2303. [DOI] [PubMed] [Google Scholar]
- 29.Flint AJ, Tiganis T, Barford D, Tonks NK. Development of “substrate-trapping” mutants to identify physiological substrates of protein tyrosine phosphatases. Proc Natl Acad Sci U S A. 1997;94:1680–1685. doi: 10.1073/pnas.94.5.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Herbst R, et al. Daughter of sevenless is a substrate of the phosphotyrosine phosphatase Corkscrew and functions during sevenless signaling. Cell. 1996;85:899–909. doi: 10.1016/s0092-8674(00)81273-8. [DOI] [PubMed] [Google Scholar]
- 31.Perkins LA, Johnson MR, Melnick MB, Perrimon N. The nonreceptor protein tyrosine phosphatase corkscrew functions in multiple receptor tyrosine kinase pathways in Drosophila. Dev Biol. 1996;180:63–81. doi: 10.1006/dbio.1996.0285. [DOI] [PubMed] [Google Scholar]
- 32.Perrimon N, Engstrom L, Mahowald AP. Developmental genetics of the 2C-D region of the Drosophila X chromosome. Genetics. 1985;111:23–41. doi: 10.1093/genetics/111.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blank U, Launay P, Benhamou M, Monteiro RC. Inhibitory ITAMs as novel regulators of immunity. Immunol Rev. 2009;232:59–71. doi: 10.1111/j.1600-065X.2009.00832.x. [DOI] [PubMed] [Google Scholar]
- 34.Xie ZH, Zhang J, Siraganian RP. Positive regulation of c-Jun N-terminal kinase and TNF-alpha production but not histamine release by SHP-1 in RBL-2H3 mast cells. J Immunol. 2000;164:1521–1528. doi: 10.4049/jimmunol.164.3.1521. [DOI] [PubMed] [Google Scholar]
- 35.Lee SM, Kim EJ, Suk K, Lee WH. Synthetic peptides containing ITIM-like sequences of IREM-1 inhibit BAFF-mediated regulation of interleukin-8 expression and phagocytosis through SHP-1 and/or PI3K. Immunology. 2011;134:224–233. doi: 10.1111/j.1365-2567.2011.03481.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cahoy JD, et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci. 2008;28:264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singh TD, et al. MEGF10 functions as a receptor for the uptake of amyloid-beta. FEBS Lett. 2010;584:3936–3942. doi: 10.1016/j.febslet.2010.08.050. [DOI] [PubMed] [Google Scholar]
- 38.Wu HH, et al. Glial precursors clear sensory neuron corpses during development via Jedi-1, an engulfment receptor. Nat Neurosci. 2009;12:1534–1541. doi: 10.1038/nn.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qin L, et al. Microglial NADPH oxidase is a novel target for femtomolar neuroprotection against oxidative stress. FASEB J. 2005;19:550–557. doi: 10.1096/fj.04-2857com. [DOI] [PubMed] [Google Scholar]
- 40.Brambilla R, et al. Inhibition of astroglial nuclear factor kappaB reduces inflammation and improves functional recovery after spinal cord injury. J Exp Med. 2005;202:145–156. doi: 10.1084/jem.20041918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hamby ME, Hewett JA, Hewett SJ. TGF-beta1 potentiates astrocytic nitric oxide production by expanding the population of astrocytes that express NOS-2. Glia. 2006;54:566–577. doi: 10.1002/glia.20411. [DOI] [PubMed] [Google Scholar]
- 42.Zhao J, Brooks DM, Lurie DI. Lipopolysaccharide-activated SHP-1-deficient motheaten microglia release increased nitric oxide, TNF-alpha, and IL-1beta. Glia. 2006;53:304–312. doi: 10.1002/glia.20283. [DOI] [PubMed] [Google Scholar]
- 43.Chitnis T, et al. Elevated neuronal expression of CD200 protects Wlds mice from inflammation-mediated neurodegeneration. Am J Pathol. 2007;170:1695–1712. doi: 10.2353/ajpath.2007.060677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hoek RM, et al. Down-regulation of the macrophage lineage through interaction with OX2 (CD200) Science. 2000;290:1768–1771. doi: 10.1126/science.290.5497.1768. [DOI] [PubMed] [Google Scholar]
- 45.Jenmalm MC, Cherwinski H, Bowman EP, Phillips JH, Sedgwick JD. Regulation of myeloid cell function through the CD200 receptor. J Immunol. 2006;176:191–199. doi: 10.4049/jimmunol.176.1.191. [DOI] [PubMed] [Google Scholar]
- 46.Koning N, Swaab DF, Hoek RM, Huitinga I. Distribution of the immune inhibitory molecules CD200 and CD200R in the normal central nervous system and multiple sclerosis lesions suggests neuron-glia and glia-glia interactions. J Neuropathol Exp Neurol. 2009;68:159–167. doi: 10.1097/NEN.0b013e3181964113. [DOI] [PubMed] [Google Scholar]
- 47.Leiserson WM, Harkins EW, Keshishian H. Fray, a Drosophila serine/threonine kinase homologous to mammalian PASK, is required for axonal ensheathment. Neuron. 2000;28:793–806. doi: 10.1016/s0896-6273(00)00154-9. [DOI] [PubMed] [Google Scholar]
- 48.Dietzl G, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- 49.Lee YS, Carthew RW. Making a better RNAi vector for Drosophila: use of intron spacers. Methods. 2003;30:322–329. doi: 10.1016/s1046-2023(03)00051-3. [DOI] [PubMed] [Google Scholar]
- 50.Biswas R, Stein D, Stanley ER. Drosophila Dok is required for embryonic dorsal closure. Development. 2006;133:217–227. doi: 10.1242/dev.02198. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1.
Full length blots of cropped images shown in manuscript.
Supplemental Figure 2.
Developmental time course of Draper isoform expression in Drosophila. RT-PCR using isoform specific primers was performed on cDNA generated from control yw embryo, larvae, and adults. Plasmids containing each Draper isoform were used as positive controls. Note that transcripts for Draper-I, II, and III were all detected in the adult head and brain. PCR amplification of Gapdh was performed as a positive control for cDNA synthesis.
Supplemental Figure 3.
UAS-Draper transgenes do not affect adult glial engulfment activity. Maxillary palp ablations were performed on flies that carried one copy of each UAS-Draper transgene and OR85e-mCD8::GFP. GFP+ axonal debris remaining in the antennal lobes was quantified 3 days later. In all animals carrying one copy of the Draper UAS rescue transgenes, glial engulfment of GFP+ degenerating axon fragments occurred normally and was indistinguishable from clearance in control animals. (a) Representative confocal images of Z-stack projections shown. (b) Quantification of the experiment shown in (a). Bars depict mean ± S.E.M. D0, Day 0; D3, Day 3.
N values for (D0, D3): control OR85e-mCD8::GFP/+ (10,8); UAS-Draper-I/OR85e-mCD8::GFP (6,8); UAS-Draper-II/OR85e-mCD8::GFP (10,8); UAS-Draper-III/OR85e-mCD8::GFP (8,8); UAS-Draper-I-II/OR85e-mCD8::GFP (6,10); UAS-Draper-I-III/OR85e-mCD8::GFP (8,10); UAS-Draper-II/III-I/OR85e-mCD8::GFP (10,10). Scale bar represents 20 μm.
Supplemental Figure 4.
Activation of RNAi machinery in glia does not inhibit phagocytic clearance of axon debris. Two representative examples of RNAi constructs (neuroligin and LDL-receptor related protein) driven in glia with repo-Gal4 that had no affect on glial clearance of severed axonal debris are shown here. (a) Quantification of GFP+ ORN debris clearance 5 days after maxillary palp ablation shows that clearance is unaffected by non-specific expression of dsRNA. Values represent mean ± S.E.M. (b) Representative confocal Z-stack images depict normal clearance in both RNAi genotypes. Scale bar represents 20 μm.
Supplemental Figure 5.
CswRNAi inhibits Csw-C expression in adult glia. To confirm effectiveness of CswRNAi, UAS-cswRNAi was driven with repo-gal4 and an anti-Csw Western blot was performed on adult brain lysates from cswRNAi (UAS-cswRNAi/+;;repo-Gal4/+) and control (repo-Gal4/+) animals. Expression of CswRNAi in glia results in robust knockdown of at least one isoform of Csw (Csw-C). Preferential knockdown of the smaller isoform suggests that there maybe interesting cell-type specific differences in Csw isoform expression.
Supplemental Figure 6.
csw is not upregulated in the adult brain after axotomy. Real-time PCR analysis of normalized expression levels of csw mRNA in adult central brains before and after antennal ORN axotomy. Threshold cycle (Ct) values were normalized to ribosomal L32 and results are presented as fold-induction relative to levels in uninjured brains. Values represent mean ± S.E.M. of at least 3 independent RNA isolations. Corkscrew transcript levels were significantly downregulated at 4.5 hours after injury (*p<0.05 ANOVA analysis, Dunnett’s post hoc test).
Supplemental Figure 7.
Single and double injury paradigm for experiments described in Figure 8.
Supplemental Figure 8.
UAS-DraperRNAi inhibits expression of all Draper isoforms in the adult brain. A UAS-DraperRNAi transgene that targets a sequence common to all three Draper isoforms was driven in all glia with repo-Gal4. Draper Western blot was performed on dissected adult brain protein lysates to confirm knockdown of Draper-I, Draper-II, and Draper-III.