Abstract
In Saccharomyces cerevisiae, 59 of the 78 ribosomal proteins are encoded by duplicated genes that, in most cases, encode identical or very similar protein products. However, different sets of ribosomal protein genes have been identified in screens for various phenotypes, including life span, budding pattern, and drug sensitivities. Due to potential suppressors of growth rate defects among this set of strains in the ORF deletion collection, we regenerated the entire set of haploid ribosomal protein gene deletion strains in a clean genetic background. The new strains were used to create double deletions lacking both paralogs, allowing us to define a set of 14 nonessential ribosomal proteins. Replicative life-span analysis of new strains corresponding to ORF deletion collection strains that likely carried suppressors of growth defects identified 11 new yeast replicative aging genes. Treatment of the collection of ribosomal protein gene deletion strains with tunicamycin revealed a significant correlation between slow growth and resistance to ER stress that was recapitulated by reducing translation of wild-type yeast with cycloheximide. Interestingly, enhanced tunicamycin resistance in ribosomal protein gene deletion mutants was independent of the unfolded protein response transcription factor Hac1. These data support a model in which reduced translation is protective against ER stress by a mechanism distinct from the canonical ER stress response pathway and further add to the diverse yet specific phenotypes associated with ribosomal protein gene deletions.
THE yeast ribosome consists of two subunits, the 40S (small) and 60S (large), which together contain four discrete rRNA species and 78 ribosomal proteins (RPs). In Saccharomyces cerevisiae, 59 of the 78 ribosomal proteins are encoded by a pair of paralogous genes, most of which arose through a genome-wide duplication event roughly 100 million years ago (Wolfe and Shields 1997). Only ∼12% of the duplicated genome remains, and of the paralogous gene pairs present, a majority of ribosomal proteins genes (RPGs) are in a class that exhibits little or even decelerated evolution (Kellis et al. 2004). Remarkably, 21 of the 59 RPG pairs encode identical proteins, and the others are highly similar (Supporting Information, Table S1). The prevalence of synthetic lethality among RPG paralogs indicates that the two protein products are generally redundant for at least one essential function (Dean et al. 2008).
Despite the significant similarity among RPG paralogs, many reports have described differential effects of deleting only one, and such instances have been observed even in cases where the encoded protein product is identical (Briones et al. 1998). One explanation for this is that the two genes contribute different amounts of protein, and neither is alone sufficient to support wild-type growth. In the case of Rpl16, for example, expression of either RPL16A or RPL16B can rescue the growth defect of cells lacking RPL16B (Rotenberg et al. 1988). Consistently, the RPL16B transcript accumulates to twice the level of the RPL16A transcript, suggesting that under normal conditions, cells lacking RPL16B have a greater deficit in Rpl16 than cells lacking RPL16A. Paralog-specific defects are not uncommon among RPG paralogs and have often been attributed to differences in expression (Abovich and Rosbash 1984; Leer et al. 1984; Herruer et al. 1987; Lucioli et al. 1988; Rotenberg et al. 1988; Briones et al. 1998; Simoff et al. 2009).
More complex relationships between paralogous RPs have also been reported. A study by Komili et al. (2007) showed that transcriptomes from cells in which RPG paralogs had been deleted were considerably different, and mining published data sets for phenotypic effects among cells lacking RPG paralogs also revealed significant differences. Screens for such varied phenotypes as bud site selection (Ni and Snyder 2001), growth of diploid cells haploinsufficient for actin (Haarer et al. 2007), or replicative life span (Kaeberlein et al. 2005; Chiocchetti et al. 2007; Managbanag et al. 2008; Smith et al. 2008; Steffen et al. 2008) are among the many that have identified deletions of one RPG paralog and not the other. In addition, some paralogous RPs have different genetic requirements for their assembly and exhibit paralog-specific aberrant localizations when GFP tagged in certain genetic backgrounds (Komili et al. 2007; Kim et al. 2009). These data support a role for functional specificity among RP paralogs that is difficult to explain by a simple gene dosage model. Instead, Komili et al. (2007) proposed the existence of a ribosomal code in which ribosomes of particular composition preferentially translate subsets of mRNAs. Interestingly, ribosome-mediated translational control of specific mRNAs has recently been reported in mammals; mice heterozygous for a deletion in RPL38 exhibit extensive patterning defects arising from perturbed translation of several homeobox mRNAs, although global protein synthesis remains unchanged in these animals (Kondrashov et al. 2011).
A majority of screens for phenotypes associated with deletion of single genes have employed the yeast ORF deletion collection (Winzeler et al. 1999), in which 107 of the 137 total RPGs are represented. We previously screened the set of haploid rpgΔ strains for replicative life span and observed several instances in which faster-growing colonies would appear when slow-growing strains were streaked for single colonies (Steffen et al. 2008). Consistently, tetrad analysis of spores from diploids generated by mating these strains could yield both slow- and faster-growing colonies, suggesting the presence of genetic suppressors of growth defects. Given the fact that many of these strains are significantly slow-growing and encode a gene paralogous to that deleted, it is possible that selective pressure enhances the frequency of suppression of growth defects among this set of strains. Suppression of the growth defect could presumably arise by increased expression of the present paralog; indeed, rpgΔ mutants have previously been shown to be aneuploid for segments of chromosomes on which the paralagous RPG resides, resulting in enhanced expression of the given protein (Hughes et al. 2000).
To avoid potential confounding effects caused by suppressors of growth rate defects among the RPG deletions in the existing haploid yeast deletion collection, we created in the deletion set background a new collection of haploid RPG deletions, as well as all viable double deletions, i.e., lacking both paralogs. Here we describe the initial characterization of this collection for growth, identifying 14 RPs that are nonessential, a conclusion based on the viability of haploid cells lacking both paralagous RPGs. We previously identified a correlation between slow growth and replicative life-span extension among 60S rpΔ strains; therefore, in cases where the newly generated haploid deletions differed in growth rate from analogous strains in the ORF deletion collection, we repeated replicative life-span analysis. From these studies we identified 11 new long-lived rpΔs, bringing the total to 23.
We also report that a subset of RPG deletions is resistant to the ER stress-inducing agent tunicamycin, and that lowering overall translation in wild-type yeast via cycloheximide treatment can recapitulate this resistance. This ER stress resistance occurs through an uncommon mechanism that is distinct from the canonical ER stress response pathway.
Materials and Methods
Strains and media
All yeast strains were derived from the parent strains of the haploid yeast ORF deletion collections (Winzeler et al. 1999), BY4742 (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0) and BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0).
Cells were grown in standard YPD containing 1% yeast extract, 2% peptone, and 2% glucose. For tetrad dissection, standard YPD plates with agar were used. For tunicamycin growth assays, tunicamycin (Sigma, T7765) was added to liquid YPD to a final concentration of 2 μg/ml. Tunicamycin stock concentration was stored at 10 mg/ml in DMSO. DMSO vehicle controls were performed during all experiments.
Creating a new set of haploid RPG deletion strains
When possible, new haploid strains were created by sporulating the heterozygous diploid strain from the yeast deletion collection. For sporulation, 300 μl of saturated yeast culture was added to 3 ml sporulation medium (0.3% potassium acetate, 0.02% raffinose), which was incubated at 30° with shaking for at least 5 days. The resulting spores were dissected with a micromanipulator after zymolyase digestion. In other cases, heterozygous diploid strains were generated by standard PCR-mediated gene disruption, then sporulated and dissected as above. This set included seven strains for which heterozygous diploid deletions were not present in our laboratory collection (RPL4B, RPL14B, RPL11A, RPL36B, RPL42B, RPS8B, and RPS26A). Additionally, this method was also used to generate five strains for which tetrad dissection was inexplicable (RPL17B, RPL20B, RPL27A, RPL31A, and RPS20), and five strains for which PCR verification (see below) indicated that the gene of interest was still present in the genome (RPL16B, RPL24A, RPL37B, RPS23A, and RPL33B). We note that these cases could potentially result from aneuploidy, the presence of other mutations in the genetic background, or could simply be due to error in replication or use of the collection.
Creating strains lacking single RPs
In cases where an RP was encoded by duplicate RPGs, two haploid strains lacking the respective genes were mated, and the resulting diploids were sporulated and dissected as above. All strains that were called inviable were given at least 7 days at 30° to form colonies. Due to the propensity of RPG mutants to accumulate likely growth rate suppressors, frozen stocks of each strain were created immediately after verification. For every case where it appeared that loss of both RPG paralogs simultaneously resulted in viable cells, both gene deletions were verified by PCR. At this step, we identified five cases where the relevant markers segregated properly but the gene of interest was not actually deleted (RPL16B, RPL24A, RPL33B, RPL37B, and RPS23A). In these cases, we remade the heterozygous diploid deletion strain using standard PCR-mediated gene disruption methods and continued as described above.
Growth rate analysis
Growth curves for the ribosomal protein deletion strains were generated using a Bioscreen C machine (Growth Curves USA). Overnight cultures of the strains were grown in 250 μl YPD in 96-well plates (inoculated from single colonies). The next day, 5 μl of overnight culture was added to 145 μl fresh YPD medium in 100-well Bioscreen C Honeycomb microplates and cultures were grown in the Bioscreen C at 30° for 24 to 72 hr. Optical density measurements were taken every 30 min and the plates were shaken constantly. YODA (Olsen et al. 2010) was used to analyze the growth data; the generation time was defined as the average of the three adjacent lowest doubling times (steepest part of the growth curve). The average generation time ± SD for at least three independent assays is given in Table S2.
Determining the growth rate of the strains required culturing them in liquid media, where it is not generally possible to visually assess the presence of growth rate suppressors. For the purposes of growth rate determination, instances where strains grew significantly faster than their colony size by original tetrad dissection (Figure S1) would suggest they should, the data point was removed from the set used for average growth rate determination displayed in Table S2. Results for which the standard deviation was >15% of the average generation time are noted in red on Table S2 and have been omitted from the analysis in Figure 5.
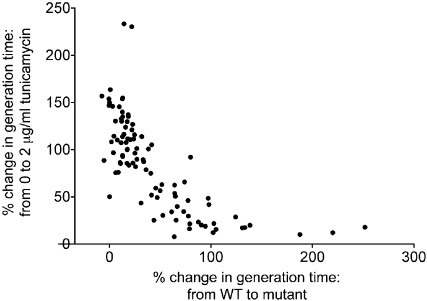
Figure 5 .
Percentage of change in generation time from wild-type to rpΔ vs. percentage of change in growth from 0 to 2 μg/ml tunicamycin.
For growth in tunicamycin, the data were more variable. For some strains, we were unable to obtain reliable growth rate data; data points where the standard deviation was >15% of the average generation time are noted in red on Table S2 and have been omitted from the analysis in Figure 5. We performed DMSO-only controls for all tunicamycin growth assays. Our analysis indicated that the DMSO control had no effect on growth rate in any of our strains, therefore data from YPD and YPD + DMSO were pooled for the average growth rates shown in Table S2.
Polysome analysis
Polysome analysis was carried out as described previously (MacKay et al. 2004). Briefly, log-phase yeast cultures were quick chilled with crushed frozen YPD containing 100 μg/ml cycloheximide. Cells were harvested by centrifugation, washed with 10 ml lysis buffer (25 mM Tris-HCl, pH 7.5, 40 mM KCl, 7.5 mM MgCl2, 1 mM DTT, 0.5 mg/ml heparin, 100 μg/ml cycloheximide) and resuspended in 1 ml lysis buffer. Cells were lysed by vortexing with glass beads. Triton X-100 and sodium deoxycholate were added (1% final concentration each) with vortexing and the samples stood on ice for 5 min before the supernatant was clarified by centrifugation. All reagents were ice-cold and all steps were done in a 4° cold room. For separation on gradients, 1 ml containing 20 A260 units of lysate was loaded onto an 11-ml linear 7–47% sucrose gradient in 50 mM Tris-HCl, pH 7.5, 0.8 M KCl, 15 mM MgCl2, 0.5 mg/ml heparin, 100 μg/ml cycloheximide, and sedimented at 39,000 rpm at 4° in an SW40 Ti swinging bucket rotor (Beckman) for 1.5 hr. Gradients were collected from the top and profiles were monitored at 254 nm.
Results
Generation of haploid strains lacking single RPGs
A majority of the 137 RPGs are present as deletions in the heterozygous diploid yeast ORF collection. For these cases, we constructed the new haploid strains by sporulating the heterozygous diploid RPG deletion strains from the yeast ORF collection and dissecting the tetrads. For 18 of 134 cases, this resulted in 2:2 segregation of viable to inviable spores (Figure S1). Genetic analysis of the viable spores confirmed that the inviable spores lacked the particular RPG and these were categorized as essential RPGs (Table 1). The majority of remaining cases yielded haploid spores in which the relevant markers segregated 2:2 as expected; the haploid strains were recovered from the tetrad plates. In five cases, differential growth phenotypes of the resulting spores indicated the likely presence of a growth rate suppressor in the heterozygous diploid strain. For these instances, we used standard PCR-mediated gene disruption to construct a new heterozygous diploid deletion strain, and then sporulated the heterozygote and dissected the tetrads. The same method was used to construct heterozygous diploids for the strains that were not present in the collection or for cases where PCR verification showed that a particular RPG was not actually deleted (see Materials and Methods).
Table 1 . Essential riobosomal protein genes.
| Gene | ORF | Paralog |
|---|---|---|
| RPL3 | YOR063W | None |
| RPL5 | YPL131W | None |
| RPL10 | YLR075W | None |
| RPL15A | YLR029C | RPL15B |
| RPL18A | YOL120C | RPL18B |
| RPL25 | YOL127W | None |
| RPL28 | YGL103W | None |
| RPL30 | YGL030W | None |
| RPL32 | YBL092W | None |
| RPL42A | YNL162W | RPL42B |
| RPP0 | YLR340W | None |
| RPS2 | YGL123W | None |
| RPS3 | YNL178W | None |
| RPS5 | YJR123W | None |
| RPS13 | YDR064W | None |
| RPS15 | YOL040C | None |
| RPS20 | YHL015W | None |
| RPS28A | YOR167C | RPS28B |
| RPS30B | YOR182C | RPS30A |
| RPS31 | YLR167W | None |
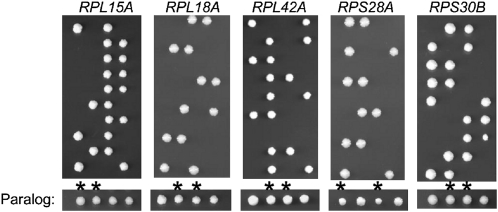
In total, we found that 20 of the 137 RPGs were essential (Table 1, Figure S1). Of the 20 essential genes, 15 do not have paralogs and 5 do (Figure 1). Three of the essential genes with paralogs, RPL15A, RPL18A, and RPL42A, were previously reported to be inviable (Giaever et al. 2002), and in the case of RPL15A, it has been shown that its paralog is not actively transcribed (Simoff et al. 2009); thus RPL15A acts similarly to a single nonduplicated gene. Consistently, rpl18bΔ and rpl42bΔ appear to grow similarly to wild type (Figure 1), suggesting that their essential paralogs may be more important for contributing sufficient amounts of protein. RPS28A and RPS30B, on the other hand, have previously been reported to be nonessential (Giaever et al. 2002); it is unclear why our results differ from those previously reported, although we note that these are among a set of 96 gene deletions that contain a second mutation (Lehner et al. 2007).
Figure 1 .
Essential ribosomal protein genes with paralogs. Sporulation of heterozygous diploids lacking the specific gene (text above panel), followed by ascus digestion, dissection, and analysis, yielded 2:2 segregation of viable to inviable spores, thus indentifying essential RPGs. A representative tetrad from the corresponding paralog heterozygous diploid is shown below with asterisks designating colonies lacking the corresponding paralog.
Identification of nonessential ribosomal proteins
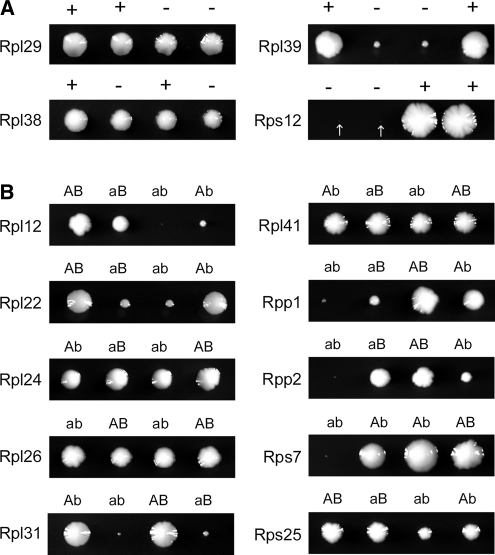
In cases where a ribosomal protein is encoded by a single RPG, sporulation of the heterozygous diploid followed by tetrad analysis allowed us to determine whether the particular RP is required for viability. In the majority of these cases (15 of 19), the RP encoded by a single gene was essential; however, we found that 4 ribosomal proteins, RPL29 (DeLabre et al. 2002), RPL38 (Giaever et al. 2002), RPL39 (Sachs and Davis 1990), and RPS12 (Giaever et al. 2002), are dispensable for viability. Loss of RPL39 or RPS12 severely limits the growth of these colonies, but cells lacking RPL29 or RPL38 grow similarly to wild type (Figure 2A).
Figure 2 .
Nonessential ribosomal proteins. (A) Representative tetrads dissected from sporulated heterozygous diploids lacking the specified gene are shown, with “+” indicating presence and “−” indicating absence of the specific gene. Arrows denote especially small (rps12Δ) colonies. (B) Representative tetrads dissected from sporulated doubly heterozygous diploids lacking one copy of each of the paralogous RPGs are shown, with uppercase “A” or “B” indicating that paralog A or B is present; lowercase “a” or “b” indicate that paralog A or B is absent. Genotypes were confirmed by PCR.
To determine whether the RPs encoded by duplicate genes are essential, we mated haploid strains lacking single RPGs with haploid strains lacking their paralogs (e.g., rpl1aΔ MATa × rpl1bΔ MATα). The resulting diploids were sporulated and tetrads were dissected and genotyped to identify the double mutants. The majority of these cases (48 of 59) resulted in the predicted pattern, indicating that loss of both copies of duplicated RPGs simultaneously is lethal. Unexpectedly, we observed 10 cases where loss of both paralogs simultaneously resulted in viable spores (Figure 2B). In total, our analysis identified 14 RPs that are not essential for viability (Table 2), 4 of which are encoded by single genes (Figure 2A) and 10 encoded by duplicate genes (Figure 2B).
Table 2 . Nonessential riobosomal proteins.
| Nonessential RP | Number of genes | Growth defect | Conservationa | Original reference |
|---|---|---|---|---|
| Rpl12 | 2 | Severe | BAE | Briones et al. (1998) |
| Rpl22 | 2 | Moderate | E | Costanzo et al. (2010) |
| Rpl24 | 2 | Slight | AE | Baronas-Lowell and Warner (1990) |
| Rpl26 | 2 | Slight | BAE | Costanzo et al. (2010) |
| Rpl29 | 1 | Slight | E | DeLabre et al. (2002) |
| Rpl31 | 2 | Severe | AE | Peisker et al. (2008) |
| Rpl38 | 1 | None | AE | Giaever et al. (2002) |
| Rpl39 | 1 | Moderate | AE | Sachs and Davis (1990) |
| Rpl41 | 2 | None | AE | Yu and Warner (2001) |
| Rpp1 | 2 | Severeb | BAE | Remacha et al. (1995) |
| Rpp2 | 2 | Severeb | BAE | Remacha et al. (1995) |
| Rps7 | 2 | Severec | E | This studyd |
| Rps12 | 1 | Severe | E | Giaever et al. (2002) |
| Rps25 | 2 | Moderate | AE | Costanzo et al. (2010) |
The three domains of life are denoted as B (bacteria), A (archae) and E (eukarya) and defined according to Lecompte et al. (2002).
Restreaking of colonies from cells lacking Rpp1 or Rpp2 invariably resulted in faster growing colonies.
Cells lacking Rps7 are extremely slow growing and we were unable to generate reliable growth rate data in liquid culture.
Rps7 was previously reported to be essential in the W303 background (Synetos et al. 1992).
Analyzing growth rates of RPG or RP deletion strains
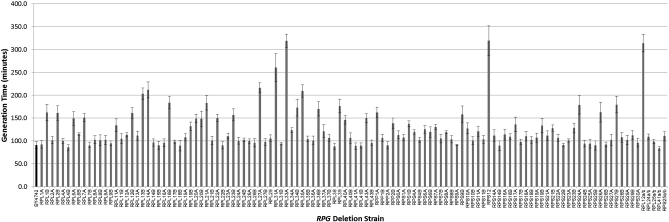
Growth rates for the 107 RPG deletion strains present in the MATα yeast deletion collection have been previously reported (Steffen et al. 2008), and we now report the growth rates for the set of rpgΔ strains generated and described here (Figure 3, Table S2) (Olsen et al. 2010). As expected, a majority of the mutants were significantly slower growing than wild type. There appears to be no relationship between the growth rates of strains lacking duplicate RPGs; both cases where growth rate of strains lacking RPG paralogs were very different (i.e., rpl27aΔ and rpl27bΔ), and cases where the growth rates were nearly identical (i.e., rpl26aΔ and rpl26bΔ) were observed. Comparison with growth rates determined for the RPG deletion strains from the MATα deletion collection (Steffen et al. 2008) reveals that at least 30% of the strains from the MATα deletion collection showed a ≥15% increase in generation time when remade, suggesting that the corresponding rpgΔ strains from the deletion collection may have carried suppressors of growth defects (Figure S2).
Figure 3 .
Average generation time of all remade RPG deletion strains in YPD, ±SD. Solid bar is BY4742 wild-type control. See also Table S2.
Cells lacking nonessential proteins generally have generation times that reflect their growth on solid YPD plates (Figure 2B, Table S2). In yeast, generation time is closely coupled to translation; therefore, we examined polysome profiles and observed that overall translation is also affected in a manner that corresponds to growth rate. For example, polysome profiles for rpl12aΔ rpl12bΔ cells appear most severely affected, followed by rpl12bΔ and then rpl12aΔ (Figure S3). A similar relationship between the extent to which polysome profiles were affected and growth on solid and in liquid medium was observed for cells lacking nonessential proteins Rpl26 (Figure S4) and Rpl22 (data not shown) as well. We expect that in most cases, polysome profiles for cells lacking other RPGs or RPs will be reflective of their growth rates, consistent with reduced translation being the primary cause of a growth rate defect. However, it is also possible that decreased growth rate is due to accumulation of damaged translation products produced from defective ribosomes assembled with a missing RP. In such a case, the polysome profile may appear less affected than expected, as is the case for rpl1bΔ (McIntosh et al. 2011).
Replicative life span
We have reported previously that many rplΔ strains are long lived in the replicative life-span assay, which determines the number of daughter cells that one mother can produce (Kaeberlein et al. 2005; Steffen et al. 2008). Of the 107 rpΔ strains examined from the ORF deletion collection, 14 were identified as long lived, a substantial enrichment when compared to nonribosomal deletions. Because many of these deletion strains could be harboring growth rate suppressors that may also affect life span either independently or by interfering with the effect of the rpΔ in question, we analyzed the life span of any newly created rpΔ that differed in growth rate from the corresponding strain in the ORF collection by at least 15%. Of the 31 strains tested, 11 newly made strains were identified as long lived (Figure S5. Table 3). In contrast, only 5 deletions originally identified as long lived no longer displayed the phenotype in the newly made strain. These findings indicate that growth rate suppressors may mask long life-span phenotypes in some rpΔ strains but rarely were required for enhanced longevity.
Table 3 . Replicative life-span data for ribosomal protein gene deletion strains from the ORF deletion collection and remade strains.
| ORF collection | Remade strain | |||||
|---|---|---|---|---|---|---|
| RPG deletion strain | % change in RLS | P-value | % change in RLS | P-value | ll+ | ll− |
| rpl1b | 20.9 | 0.0245 | 33.1 | <0.0001 | ||
| rpl2b | 10.1 | 0.1265 | 35.4 | <0.0001 | 1 | |
| rpl6a | −8.6 | 0.9161 | 39.8 | <0.0001 | 1 | |
| rpl7a | 30.9 | 0.0011 | 7.0 | 0.394 | 1 | |
| rpl12b | 13.3 | 0.192 | 21.6 | 0.0054 | 1 | |
| rpl13b | −29.9 | <0.0001 | 32.4 | <0.0001 | 1 | |
| rpl14a | −9.9 | 0.0656 | 4.8 | 0.1541 | ||
| rpl16b | 10.8 | 0.1957 | 20.6 | 0.024 | 1 | |
| rpl19b | 10.2 | 0.0036 | 45.7 | <0.0001 | ||
| rpl20a | −58.1 | <0.0001 | 40.9 | <0.0001 | 1 | |
| rpl20b | 3.0 | 0.6789 | 36.4 | <0.0001 | 1 | |
| rpl21a | −9.3 | 0.0144 | 11.9 | 0.1165 | 1 | |
| rpl22a | 30.2 | <0.0001 | 38.3 | <0.0001 | ||
| rpl23b | 16.8 | 0.0034 | 7.7 | 0.3753 | 1 | |
| rpl31a | 35.3 | <0.0001 | 28.8 | <0.0001 | ||
| rpl34a | −25.0 | 0.0037 | 27.4 | 0.0003 | 1 | |
| rpl34b | 13.7 | 0.1466 | 45.4 | <0.0001 | 1 | |
| rpl35a | −10.9 | 0.4406 | 37.8 | <0.0001 | 1 | |
| rpl39 | −18.5 | 0.0028 | −23.3 | 0.0129 | ||
| rpl40a | 19.8 | 0.0008 | 36.6 | <0.0001 | ||
| rpl43a | −63.7 | <0.0001 | 16.4 | 0.0844 | ||
| rpp1a | −13.3 | 0.0852 | 30.8 | <0.0001 | 1 | |
| rps4a | −17.4 | 0.0067 | 7.8 | 0.4443 | 1 | |
| rps8a | 7.6 | 0.0843 | −0.4 | 0.9883 | ||
| rps9b | −1.8 | 0.8504 | −22.2 | 0.0008 | ||
| rps11a | 28.0 | <0.0001 | −7.4 | 0.1253 | 1 | |
| rps18a | 1.9 | 0.8402 | −13.8 | 0.0921 | ||
| rps21b | −21.7 | 0.0097 | −24.9 | 0.0046 | ||
| rps23b | 7.5 | 0.5273 | −8.6 | 0.578 | ||
| rps24a | −30.2 | <0.0001 | −43.1 | <0.0001 | ||
| rps27b | −26.9 | <0.0001 | −14.2 | 0.0156 | ||
| Totals: | 11 | 5 | ||||
ll+, remade strain is significantly longlived while deletion set strain was not; ll−, remade strain is not longlived while deletion set strain was.
Response to tunicamycin
Tunicamycin inhibits N-linked glycosylation in the ER and is often used experimentally to elicit the unfolded protein response (UPR). The accumulation of unfolded proteins in the ER activates Ire1. Once active, Ire1 promotes noncanonical splicing of the HAC1 transcript to yield the active transcription factor. Genes that are transcriptionally activated by Hac1 include ER-resident chaperones, phospholipid biosynthetic genes, and those involved in ER-associated degradation (Travers et al. 2000).
Generation times for the set of RP and RPG deletion strains generated in this study were determined in the presence of 2 μg/ml tunicamycin, which increases the generation time of wild-type cells by ∼2.5-fold (Figure 4A, Table S2) and, on average, this dosage of tunicamycin decreased growth by ∼2-fold. However, the RPG deletion strains varied dramatically in their response to tunicamycin.
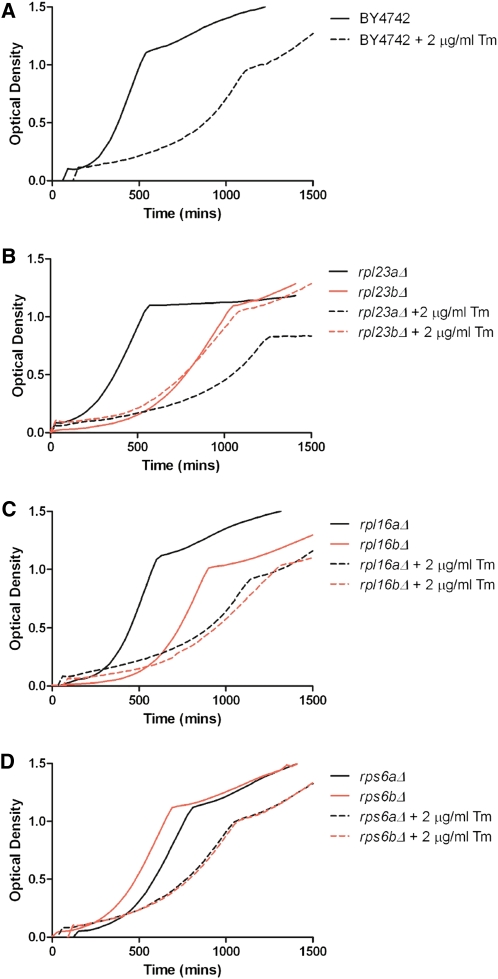
Figure 4 .
Growth in YPD vs. YPD + 2 μg/ml tunicamycin. (A) Generation time of BY4742 in tunicamycin is ∼2.5-fold greater than in YPD. (B) Cells lacking RPL23A are more severely affected by tunicamycin treatment than cells lacking RPL23B. (C) Cells lacking RPL16A are more severely affected by tunicamycin treatment than cells lacking RPL16B. (D) Cells lacking RPS6A or RPS6B respond similarly to tunicamycin treatment. See also Table S2.
Similar to other phenotypic screens, we often found that strains lacking RPG paralogs responded differently to tunicamycin. For example, the addition of tunicamycin increased the generation time of cells lacking RPL23A by 3.3-fold, but cells lacking RPL23B only exhibited a 1.3-fold increase in generation time (Figure 4B). In this case, it would be difficult to attribute the differential phenotypes to functional specificity, as the protein products encoded by these genes are identical (although differential regulation of the two paralogs cannot be ruled out). We also observed cases where paralogs encoding nonidentical proteins exhibited significantly different responses to tunicamycin; for example, rpl16aΔ and rpl16bΔ exhibited a 130 and a 22% increase in generation time, respectively (Figure 4C). There are also cases where two paralogs respond similarly to tunicamycin treatment: the generation time of both rps6aΔ and rps6bΔ approximately double in response to tunicamycin (Figure 4D).
The most obvious correlation regarding the response to tunicamycin is that strains that grew slowly in the absence of tunicamycin exhibited less growth inhibition in the presence of tunicamycin; likewise, strains with growth rates that were similar to wild type in the absence of tunicamycin tended to exhibit tunicamycin-induced growth inhibition that was similar to wild type (Figure 5, Table S2). Indeed, the percentage of change in generation time when an RPG is deleted in wild-type cells compared to the percentage of change in generation time from 0 to 2 μg/ml tunicamycin reveals a strong correlation (linear regression, R2 = 0.52) among this set of RPGs (Figure 5). To determine whether reduced translation generally results in tunicamycin resistance, we treated wild-type yeast with cycloheximide, and found that this association held true (Figure S6).
In some cases, the growth rates of particular strains were inconsistent among multiple biological replicates, suggesting that the cells may be adapting to growth in tunicamycin. In general, these seem to occur in cases where the growth rates are most significantly affected, although the variability prevents accurate analysis. Cases in which the replicates were highly inconsistent (standard deviation >15% of the average generation time) were eliminated from the analysis in Figure 5 (see Materials and Methods).
HAC1-independent resistance to tunicamycin
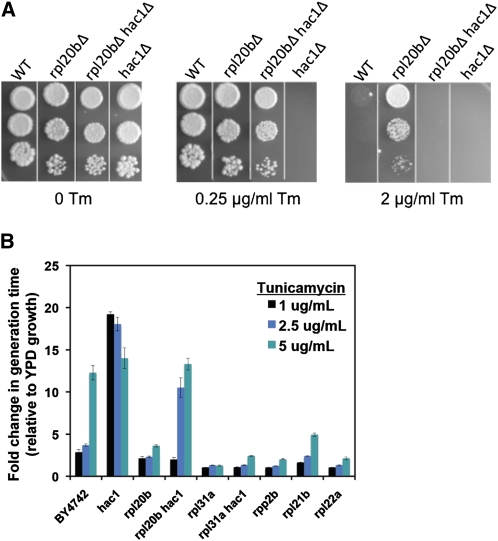
Cells lacking the UPR transcription factor Hac1 or its splicing factor Ire1 are unaffected by these deficiencies under normal conditions, but are not viable in conditions that cause ER stress, including growth in media containing tunicamycin. On solid media, we performed spotting assays with concentrations of tunicamycin as low as 0.25 μg/ml and still observed complete absence of growth for cells lacking HAC1. Remarkably, however, cells lacking both HAC1 and RPL20B (which is alone resistant to tunicamycin) are able to grow on media containing 0.25 μg/ml tunicamycin (Figure 6A). Thus, rpl20bΔ is resistant to tunicamycin-induced ER stress in a manner at least partially independent of HAC1, and a similar phenotype was observed for other RPG deletion strains (Figure 6B). One possible explanation for this finding is that ER stress is significantly decreased in rpl20bΔ cells, allowing them to withstand an increased dosage of tunicamycin before necessitating activation of the UPR. Consistently, on higher concentrations of tunicamycin, the rpl20bΔ hac1Δ cells are unable to grow, while rpl20bΔ cells are still resistant (Figure 6).
Figure 6 .
Cells lacking RPL20B are resistant to tunicamycin (Tm) in a manner at least partially independent of HAC1. (A) Tenfold serial dilutions of saturated yeast cultures were spotted on YPD plates containing 0, 0.25, or 2 μg/ml tunicamycin. (B) Fold change in generation time in liquid medium containing 1, 2.5, or 5 μg/ml Tm relative to YPD alone.
Discussion
Slow growth among ribosomal protein gene deletion strains
The generation of a new set of RPG deletion strains allowed us to confidently attribute specific growth rate defects to loss of particular RPGs, the majority of which have growth defects. We presume that in most cases, slow growth is a result of limited production of an RP required for the assembly of functioning ribosomes. It is also possible, however, (and perhaps likely in instances where both genes encoding a RP are deleted), that production of defective ribosomes lacking a particular RP is the underlying cause for the growth defect. In cells that lack Rpl1, ribosomes lacking the protein are nonetheless assembled, exported to the cytoplasm and incorporated into polysomes (McIntosh et al. 2011). In this instance, the cells are hypersensitive to defects in the ubiquitin–proteasome system (McIntosh et al. 2011), consistent with speculation that incompetent ribosomes produce defective translation products, causing stress to the cell’s degradation machinery. Interestingly, elevated capacity of the ubiquitin–proteasome system positively affects yeast replicative life span (Kruegel et al. 2011).
While a majority of ribosomal protein gene deletion strains exhibit growth rate defects, cells lacking Rpl26, Rpl24, or Rpl29 are relatively unaffected and cells lacking Rpl41 and Rpl38 have growth rates that are not significantly different than wild type. Mass spectrometry analysis confirms that these proteins are indeed incorporated into ribosomes (Lee et al. 2002), indicating that perhaps they are important for regulated translation of specific mRNAs under conditions other than those used in this study. This possibility is supported by recent findings that Rpl38, the mouse ortholog of yeast RPL38, acts as a regulatory component of the ribosome to facilitate selective translation of homeobox genes during developmental regulation in mice (Kondrashov et al. 2011).
Suppressors of growth defects
Comparison with growth rate data generated from strains present in the MATα haploid ORF collection (Steffen et al. 2008) indicates that rpgΔ strains in the deletion collection may frequently carry suppressors of slow growth (Figure S2). Presumably, spontaneous suppressors of growth rate defects among RPG paralog deletion strains could be due to increased expression of the remaining paralog, possibly by duplication of a chromosomal fragment encoding the paralogous gene, as previously described (Hughes et al. 2000). Consistently, we often observed fast-growing colonies on plates where slow-growing rpl22aΔ cells were streaked, but never observed them when rpl22aΔ rpl22bΔ double mutants were streaked (data not shown), suggesting that the faster-growing cells resulted from enhanced expression of RPL22B.
Essential RPGs with paralogs
Our study identified 20 essential RPGs, 5 of which have paralogs: RPL15A, RPL18A, RPL42A, RPS28A, and RPS30B. That these genes are essential suggests that their paralogs do not alone contribute enough protein to support viability of the cell; this has been confirmed in the case of RPL15B, which is not transcribed and thus contributes nothing to the essential pool of Rpl15 (Simoff et al. 2009). It is possible that RPL15B is expressed, but only under conditions different from those used for laboratory growth; indeed, it is important to note that this, as well as many prior studies, examined growth in rich medium with excess glucose, a condition that is likely rare for wild yeast. It is also important to note that a RP that is essential does not necessarily mean that cells lacking it are inviable due to the inability to translate; these cells may be translation competent but inviable for another reason. The collection of strains described here should prove useful for addressing these questions.
Nonessential RPs
Our data indicate that the functions of the RPs listed in Table 2 are not required to support cell growth. Assigning exclusive functions to particular RPs, however, is complicated due to the highly cooperative nature of the interactions between RPs and the rRNA in the ribosome. For example, Rpl26, Rpl31, and Rpl39 all localize to the polypeptide tunnel exit of the ribosome (Ban et al. 2000; Peisker et al. 2008), and are each individually dispensable for viability. However, strains lacking both Rpl31 and Rpl39 are inviable (Peisker et al. 2008), suggesting that these proteins function somewhat redundantly. Rpl39 also has a role in subunit assembly (Sachs and Davis 1990) and is important for translational fidelity (Dresios et al. 2000), despite this function normally being attributed to the 40S subunit. Similarly, Rpl24 and Rpl41 affect peptidyltransferase activity even though they are localized away from the 25S rRNA catalytic center (Dresios et al. 2003). The highly cooperative nature of RPs is also highlighted by the large number of negative synthetic genetic interactions among RPGs (Costanzo et al. 2010).
Together with Rpp0, the nonessential acidic proteins Rpp1 and Rpp2 form the ribosomal stalk and are the only RPs generally present in multiple copies on the ribosome. These proteins, together with Rpl12, have a key role in stimulating elongation factor binding and GTP hydrolysis (Gonzalo and Reboud 2003). Loss of either Rpp1 or Rpp2 significantly affects growth, and in fact, we were unable to recover and restreak cells lacking Rpp1 or Rpp2 from tetrad plates that continued to grow as slowly as the colony formed on the tetrad plate. That P0 is essential suggests that while cells can survive with severely impaired stalk function, it must be maintained to at least some extent for viability.
RPs, tunicamycin resistance, and life span
Upon measuring resistance to tunicamycin, we observed that slow growth among RPG deletion strains correlates with enhanced resistance to tunicamycin-induced growth inhibition, and that reducing translation with cycloheximide was able to recapitulate this effect. Reduced growth rate is also protective against heat stress (Lu et al. 2009) and some slow-growing rpgΔ strains may be broadly resistant to chemical treatments (Hillenmeyer et al. 2008).
That the tunicamycin resistance we observed is at least partially Hac1 independent is noteworthy because Hac1-independent resistance to ER stress in yeast has been described only in the case of SIN4 alleles, which are thought to activate a transcriptional response to ER stress in a manner dependent on an interaction between RNA Pol II and the core promoter of ER chaperone genes (Schroder et al. 2003). Interestingly, deletion of some RPGs has been shown to alter the transcriptional response to tunicamycin (Zhao et al. 2003). Hac1-independent resistance in rpΔs could also be a result of enhanced translation of chaperones or other factors that aid in folding in the ER, as deletion of particular RPGs is known to result in enhanced translation of at least one specific message, GCN4 (Foiani et al. 1991; Martin-Marcos et al. 2007; Steffen et al. 2008), and the generality of this phenomenon has not been globally assessed. Our data are also consistent with a model whereby reduced translation, caused by deletion of an RPG and indicated by slow growth, is protective against ER stress due to decreased protein load in the ER. In support of this model, it has been proposed that a nitrogen-stimulated increase in translation results in ER stress and activation of the UPR (Schroder et al. 2000). Likewise, a decrease in translation could relieve ER stress.
Interestingly, both stress resistance (see Kourtis and Tavernarakis 2011 for review) and reduced translation are correlated with increased longevity in model organisms, including yeast (Kaeberlein et al. 2005; Chiocchetti et al. 2007; Steffen et al. 2008), worms (Hamilton et al. 2005; Chen et al. 2007; Curran and Ruvkun 2007; Hansen et al. 2007; Pan et al. 2007; Syntichaki et al. 2007), and flies. Interestingly, the conserved ER stress regulator Ire1 is required for dietary restriction-mediated longevity in Caenorhabditis elegans (Chen et al. 2009). In yeast cells, life-span extension is mainly limited to rplΔ rather than rpsΔ strains and is largely dependent on GCN4, a translationally regulated transcription activator that is induced by reduction of 60S subunits (Foiani et al. 1991; Martin-Marcos et al. 2007; Steffen et al. 2008). The data here suggest that while reduced ER stress may be an important feature of life-span extension by inhibition of translation, ER stress resistance is not sufficient to confer enhanced longevity in yeast cells.
Functional specificity of RP paralogs and extraribosomal functions
The possibility for functional specificity among RP paralogs is intriguing and could arise through ribosomal specificity whereby ribosomes of different composition have preference for specific mRNAs (Komili et al. 2007) or through RPs having extraribosomal functions. In S. cerevisiae, extraribosomal functions for Rpl2, Rps14, Rpl30, and Rps28 in autoregulation of their own synthesis have been demonstrated (Eng and Warner 1991; Presutti et al. 1991; Fewell and Woolford 1999; Badis et al. 2004). Two other known cases of extraribosomal functions are for Rps20 and Rpl6, proteins that are capable of influencing Pol III transcription (Hermann-Le Denmat et al. 1994; Dieci et al. 2009). Given their abundance (Warner 1999) and the fact that most RPGs in yeast are present in duplicate copies, it seems feasible that RPs in S. cerevisiae would have evolved extraribosomal functions more frequently than in other eukaryotes. However, the lack of verified cases of RPs being recruited for functions unrelated to the ribosome or its synthesis is surprising (Warner and McIntosh 2009).
Several cases of extraribosomal functions for RPs have been reported in multicellular organisms (see Warner and McIntosh 2009 for review), including inhibition of mRNA translation (human L13a) (Mazumder et al. 2003), DNA endonuclease activity (human and fruit fly S3) (Wilson et al. 1994), NFkB binding (human S3) (Wan et al. 2007), and c-jun binding (human L10) (Imafuku et al. 1999). It is becoming increasingly clear that RPs can dramatically affect human pathology regardless of whether their phenotypes are due to extraribosomal functions. Of note are instances of RPs interacting with p53 (reviewed in Deisenroth and Zhang 2010), a process primarily thought to be a result of the cell’s complex ribosome surveillance mechanisms, which can result in cell cycle arrest via p53, and may be the reason for RPs being associated with cancer. In addition to a variety of cancers, RPs have been implicated in a number of diverse pathologies (Narla and Ebert 2010) including Diamond-Blackfan anemia (Boria et al. 2010) and Turner syndrome (Fisher et al. 1990). Furthermore, the complex tissue-specific expression patterns of individual RPs in the developing mouse embryo and the finding that Rpl38 specifically regulates translation of particular Hox mRNAs (Kondrashov et al. 2011) hints that ribosomal proteins may commonly influence selection of mRNAs undergoing translation.
Our data do not support or directly refute the hypothesis of a ribosomal code, proposed by Komili et al. (2007). The strong correlation between reduced growth rate and tunicamycin resistance suggests that, in this case, drug resistance is largely a property of a general decline in protein synthesis. Nevertheless, with respect to replicative life span, we have identified several cases in which reduced growth rate among paralog deletions is discordant with enhanced longevity, raising the possibility that a more complex explanation is required in this setting. Screens like this, in which phenotypes associated with RPG deletions are not correlated with their effect on translation, may serve as a good starting point for uncovering ribosomal specificity and/or extraribosomal functions of RPs.
Conclusions
We have generated a new set of RPG deletion strains and defined the set of essential RPGs and essential RPs. Growth rate analysis for this set of strains can serve as a reference for researchers working with RPG deletion strains, and may help identify cases where suppressors of growth rate defects could be clouding the data.
This study highlights the protective nature of reduced translation against ER stress, but it may extend to other forms of cellular stress as well. RPs have been identified in a wide variety of phenotypic screens, implicating their function in both resistance and predisposition to a wide variety of cellular stresses. The set of strains described here will be useful in determining whether reduced translation is an underlying cause for such associations or whether particular RPs have properties that affect certain cellular processes. Importantly, understanding the underlying causes for RP-associated phenotypes in yeast will lead to a better understanding of the complex relationships between RPs and human disease.
Supplementary Material
Acknowledgments
We thank Brady Olsen for assistance with using YODA for growth rate analysis; Elroy An, Marissa Fletcher, Monica Jelic, Soumya Kotireddy, Dan Lockshon, Rick Moller, Brian Muller, Joe Peng, Brett Robison, Michael Sage, Katie Snead, and Scott Tsuchiyama for technical assistance with yeast life-span analysis; and members of the Kennedy and Kaeberlein laboratories for technical assistance and helpful discussion. This study was supported by National Institutes of Health (NIH) grants R01AG025549 and R01AG033373 to B. K. Kennedy. K. K. Steffen has been supported by NIH training grant T32AG00057. J. R. Delaney is supported by NIH training grant T32AG000057. M. Kaeberlein is an Ellison Medical Foundation New Scholar in Aging.
Footnotes
Communicating editor: C. Boone
Literature Cited
- Abovich N., Rosbash M., 1984. Two genes for ribosomal protein 51 of Saccharomyces cerevisiae complement and contribute to the ribosomes. Mol. Cell. Biol. 4: 1871–1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badis G., Saveanu C., Fromont-Racine M., Jacquier A., 2004. Targeted mRNA degradation by deadenylation-independent decapping. Mol. Cell 15: 5–15. [DOI] [PubMed] [Google Scholar]
- Ban N., Nissen P., Hansen J., Moore P. B., Steitz T. A., 2000. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science 289: 905–920. [DOI] [PubMed] [Google Scholar]
- Baronas-Lowell D. M., Warner J. R., 1990. Ribosomal protein L30 is dispensable in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 10: 5235–5243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boria I., Garelli E., Gazda H. T., Aspesi A., Quarello P., et al. , 2010. The ribosomal basis of Diamond-Blackfan Anemia: mutation and database update. Hum. Mutat. 31: 1269–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briones E., Briones C., Remacha M., Ballesta J. P., 1998. The GTPase center protein L12 is required for correct ribosomal stalk assembly but not for Saccharomyces cerevisiae viability. J. Biol. Chem. 273: 31956–31961. [DOI] [PubMed] [Google Scholar]
- Chen D., Pan K. Z., Palter J. E., Kapahi P., 2007. Longevity determined by developmental arrest genes in Caenorhabditis elegans. Aging Cell 6: 525–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., Thomas E. L., Kapahi P., 2009. HIF-1 modulates dietary restriction-mediated lifespan extension via IRE-1 in Caenorhabditis elegans. PLoS Genet. 5: e1000486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiocchetti A., Zhou J., Zhu H., Karl T., Haubenreisser O., et al. , 2007. Ribosomal proteins Rpl10 and Rps6 are potent regulators of yeast replicative life span. Exp. Gerontol. 42: 275–286. [DOI] [PubMed] [Google Scholar]
- Costanzo M., Baryshnikova A., Bellay J., Kim Y., Spear E. D., et al. , 2010. The genetic landscape of a cell. Science 327: 425–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran S. P., Ruvkun G., 2007. Lifespan regulation by evolutionarily conserved genes essential for viability. PLoS Genet. 3: e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean E. J., Davis J. C., Davis R. W., Petrov D. A., 2008. Pervasive and persistent redundancy among duplicated genes in yeast. PLoS Genet. 4: e1000113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deisenroth C., Zhang Y., 2010. Ribosome biogenesis surveillance: probing the ribosomal protein-Mdm2-p53 pathway. Oncogene 29: 4253–4260. [DOI] [PubMed] [Google Scholar]
- DeLabre M. L., Kessl J., Karamanou S., Trumpower B. L., 2002. RPL29 codes for a non-essential protein of the 60S ribosomal subunit in Saccharomyces cerevisiae and exhibits synthetic lethality with mutations in genes for proteins required for subunit coupling. Biochim. Biophys. Acta 1574: 255–261. [DOI] [PubMed] [Google Scholar]
- Dieci G., Ruotolo R., Braglia P., Carles C., Carpentieri A., et al. , 2009. Positive modulation of RNA polymerase III transcription by ribosomal proteins. Biochem. Biophys. Res. Commun. 379: 489–493. [DOI] [PubMed] [Google Scholar]
- Dresios J., Derkatch I. L., Liebman S. W., Synetos D., 2000. Yeast ribosomal protein L24 affects the kinetics of protein synthesis and ribosomal protein L39 improves translational accuracy, while mutants lacking both remain viable. Biochemistry 39: 7236–7244. [DOI] [PubMed] [Google Scholar]
- Dresios J., Panopoulos P., Suzuki K., Synetos D., 2003. A dispensable yeast ribosomal protein optimizes peptidyltransferase activity and affects translocation. J. Biol. Chem. 278: 3314–3322. [DOI] [PubMed] [Google Scholar]
- Eng F. J., Warner J. R., 1991. Structural basis for the regulation of splicing of a yeast messenger RNA. Cell 65: 797–804. [DOI] [PubMed] [Google Scholar]
- Fewell S. W., Woolford J. L., Jr, 1999. Ribosomal protein S14 of Saccharomyces cerevisiae regulates its expression by binding to RPS14B pre-mRNA and to 18S rRNA. Mol. Cell. Biol. 19: 826–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher E. M., Beer-Romero P., Brown L. G., Ridley A., McNeil J. A., et al. , 1990. Homologous ribosomal protein genes on the human X and Y chromosomes: escape from X inactivation and possible implications for Turner syndrome. Cell 63: 1205–1218. [DOI] [PubMed] [Google Scholar]
- Foiani M., Cigan A. M., Paddon C. J., Harashima S., Hinnebusch A. G., 1991. GCD2, a translational repressor of the GCN4 gene, has a general function in the initiation of protein synthesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 11: 3203–3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever G., Chu A. M., Ni L., Connelly C., Riles L., et al. , 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418: 387–391. [DOI] [PubMed] [Google Scholar]
- Gonzalo P., Reboud J. P., 2003. The puzzling lateral flexible stalk of the ribosome. Biol. Cell 95: 179–193. [DOI] [PubMed] [Google Scholar]
- Haarer B., Viggiano S., Hibbs M. A., Troyanskaya O. G., Amberg D. C., 2007. Modeling complex genetic interactions in a simple eukaryotic genome: actin displays a rich spectrum of complex haploinsufficiencies. Genes Dev. 21: 148–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton B., Dong Y., Shindo M., Liu W., Odell I., et al. , 2005. A systematic RNAi screen for longevity genes in C. elegans. Genes Dev. 19: 1544–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M., Taubert S., Crawford D., Libina N., Lee S. J., et al. , 2007. Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans. Aging Cell 6: 95–110. [DOI] [PubMed] [Google Scholar]
- Hermann-Le Denmat S., Sipiczki M., Thuriaux P., 1994. Suppression of yeast RNA polymerase III mutations by the URP2 gene encoding a protein homologous to the mammalian ribosomal protein S20. J. Mol. Biol. 240: 1–7. [DOI] [PubMed] [Google Scholar]
- Herruer M. H., Mager W. H., Woudt L. P., Nieuwint R. T., Wassenaar G. M., et al. , 1987. Transcriptional control of yeast ribosomal protein synthesis during carbon-source upshift. Nucleic Acids Res. 15: 10133–10144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillenmeyer M. E., Fung E., Wildenhain J., Pierce S. E., Hoon S., et al. , 2008. The chemical genomic portrait of yeast: uncovering a phenotype for all genes. Science 320: 362–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes T. R., Roberts C. J., Dai H., Jones A. R., Meyer M. R., et al. , 2000. Widespread aneuploidy revealed by DNA microarray expression profiling. Nat. Genet. 25: 333–337. [DOI] [PubMed] [Google Scholar]
- Imafuku I., Masaki T., Waragai M., Takeuchi S., Kawabata M., et al. , 1999. Presenilin 1 suppresses the function of c-Jun homodimers via interaction with QM/Jif-1. J. Cell Biol. 147: 121–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M., Powers R. W., 3rd, Steffen K. K., Westman E. A., Hu D., et al. , 2005. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science 310: 1193–1196. [DOI] [PubMed] [Google Scholar]
- Kellis M., Birren B. W., Lander E. S., 2004. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature 428: 617–624. [DOI] [PubMed] [Google Scholar]
- Kim T. Y., Ha C. W., Huh W. K., 2009. Differential subcellular localization of ribosomal protein L7 paralogs in Saccharomyces cerevisiae. Mol. Cells 27: 539–546. [DOI] [PubMed] [Google Scholar]
- Komili S., Farny N. G., Roth F. P., Silver P. A., 2007. Functional specificity among ribosomal proteins regulates gene expression. Cell 131: 557–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov N., Pusic A., Stumpf C. R., Shimizu K., Hsieh A. C., et al. , 2011. Ribosome-mediated specificity in Hox mRNA translation and vertebrate tissue patterning. Cell 145: 383–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourtis N., Tavernarakis N., 2011. Cellular stress response pathways and ageing: intricate molecular relationships. EMBO J. 30: 2520–2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruegel U., Robison B., Dange T., Kahlert G., Delaney J. R., et al. , 2011. Elevated proteasome capacity extends replicative lifespan in Saccharomyces cerevisiae. PLoS Genet. 7: e1002253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecompte O., Ripp R., Thierry J. C., Moras D., Poch O., 2002. Comparative analysis of ribosomal proteins in complete genomes: an example of reductive evolution at the domain scale. Nucleic Acids Res. 30: 5382–5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. W., Berger S. J., Martinovic S., Pasa-Tolic L., Anderson G. A., et al. , 2002. Direct mass spectrometric analysis of intact proteins of the yeast large ribosomal subunit using capillary LC/FTICR. Proc. Natl. Acad. Sci. USA 99: 5942–5947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leer R. J., van Raamsdonk-Duin M. M., Mager W. H., Planta R. J., 1984. The primary structure of the gene encoding yeast ribosomal protein L16. FEBS Lett. 175: 371–376. [DOI] [PubMed] [Google Scholar]
- Lehner K. R., Stone M. M., Farber R. A., Petes T. D., 2007. Ninety-six haploid yeast strains with individual disruptions of open reading frames between YOR097C and YOR192C, constructed for the Saccharomyces genome deletion project, have an additional mutation in the mismatch repair gene MSH3. Genetics 177: 1951–1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C., Brauer M. J., Botstein D., 2009. Slow growth induces heat-shock resistance in normal and respiratory-deficient yeast. Mol. Biol. Cell 20: 891–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucioli A., Presutti C., Ciafre S., Caffarelli E., Fragapane P., et al. , 1988. Gene dosage alteration of L2 ribosomal protein genes in Saccharomyces cerevisiae: effects on ribosome synthesis. Mol. Cell. Biol. 8: 4792–4798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKay V. L., Li X., Flory M. R., Turcott E., Law G. L., et al. , 2004. Gene expression analyzed by high-resolution state array analysis and quantitative proteomics: response of yeast to mating pheromone. Mol. Cell. Proteomics 3: 478–489. [DOI] [PubMed] [Google Scholar]
- Managbanag J. R., Witten T. M., Bonchev D., Fox L. A., Tsuchiya M., et al. , 2008. Shortest-path network analysis is a useful approach toward identifying genetic determinants of longevity. PLoS ONE 3: e3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Marcos P., Hinnebusch A. G., Tamame M., 2007. Ribosomal protein L33 is required for ribosome biogenesis, subunit joining, and repression of GCN4 translation. Mol. Cell. Biol. 27: 5968–5985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumder B., Sampath P., Seshadri V., Maitra R. K., DiCorleto P. E., et al. , 2003. Regulated release of L13a from the 60S ribosomal subunit as a mechanism of transcript-specific translational control. Cell 115: 187–198. [DOI] [PubMed] [Google Scholar]
- McIntosh K. B., Bhattacharya A., Willis I. M., Warner J. R., 2011. Eukaryotic cells producing ribosomes deficient in Rpl1 are hypersensitive to defects in the ubiquitin-proteasome system. PLoS ONE 6: e23579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narla A., Ebert B. L., 2010. Ribosomopathies: human disorders of ribosome dysfunction. Blood 115: 3196–3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni L., Snyder M., 2001. A genomic study of the bipolar bud site selection pattern in Saccharomyces cerevisiae. Mol. Biol. Cell 12: 2147–2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen B., Murakami C. J., Kaeberlein M., 2010. YODA: software to facilitate high-throughput analysis of chronological life span, growth rate, and survival in budding yeast. BMC Bioinformatics 11: 141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan K. Z., Palter J. E., Rogers A. N., Olsen A., Chen D., et al. , 2007. Inhibition of mRNA translation extends lifespan in Caenorhabditis elegans. Aging Cell 6: 111–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peisker K., Braun D., Wolfle T., Hentschel J., Funfschilling U., et al. , 2008. Ribosome-associated complex binds to ribosomes in close proximity of Rpl31 at the exit of the polypeptide tunnel in yeast. Mol. Biol. Cell 19: 5279–5288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presutti C., Ciafre S. A., Bozzoni I., 1991. The ribosomal protein L2 in S. cerevisiae controls the level of accumulation of its own mRNA. EMBO J. 10: 2215–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remacha M., Jimenez-Diaz A., Bermejo B., Rodriguez-Gabriel M. A., Guarinos E., et al. , 1995. Ribosomal acidic phosphoproteins P1 and P2 are not required for cell viability but regulate the pattern of protein expression in Saccharomyces cerevisiae. Mol. Cell. Biol. 15: 4754–4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotenberg M. O., Moritz M., Woolford J. L., Jr, 1988. Depletion of Saccharomyces cerevisiae ribosomal protein L16 causes a decrease in 60S ribosomal subunits and formation of half-mer polyribosomes. Genes Dev. 2: 160–172. [DOI] [PubMed] [Google Scholar]
- Sachs A. B., Davis R. W., 1990. Translation initiation and ribosomal biogenesis: involvement of a putative rRNA helicase and RPL46. Science 247: 1077–1079. [DOI] [PubMed] [Google Scholar]
- Schroder M., Chang J. S., Kaufman R. J., 2000. The unfolded protein response represses nitrogen-starvation induced developmental differentiation in yeast. Genes Dev. 14: 2962–2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder M., Clark R., Kaufman R. J., 2003. IRE1- and HAC1-independent transcriptional regulation in the unfolded protein response of yeast. Mol. Microbiol. 49: 591–606. [DOI] [PubMed] [Google Scholar]
- Simoff I., Moradi H., Nygard O., 2009. Functional characterization of ribosomal protein L15 from Saccharomyces cerevisiae. Curr. Genet. 55: 111–125. [DOI] [PubMed] [Google Scholar]
- Smith E. D., Tsuchiya M., Fox L. A., Dang N., Hu D., et al. , 2008. Quantitative evidence for conserved longevity pathways between divergent eukaryotic species. Genome Res. 18: 564–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen K. K., MacKay V. L., Kerr E. O., Tsuchiya M., Hu D., et al. , 2008. Yeast life span extension by depletion of 60s ribosomal subunits is mediated by Gcn4. Cell 133: 292–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Synetos D., Dabeva M. D., Warner J. R., 1992. The yeast ribosomal protein S7 and its genes. J. Biol. Chem. 267: 3008–3013. [PubMed] [Google Scholar]
- Syntichaki P., Troulinaki K., Tavernarakis N., 2007. eIF4E function in somatic cells modulates ageing in Caenorhabditis elegans. Nature 445: 922–926. [DOI] [PubMed] [Google Scholar]
- Travers K. J., Patil C. K., Wodicka L., Lockhart D. J., Weissman J. S., et al. , 2000. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell 101: 249–258. [DOI] [PubMed] [Google Scholar]
- Wan F., Anderson D. E., Barnitz R. A., Snow A., Bidere N., et al. , 2007. Ribosomal protein S3: a KH domain subunit in NF-kappaB complexes that mediates selective gene regulation. Cell 131: 927–939. [DOI] [PubMed] [Google Scholar]
- Warner J. R., 1999. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 24: 437–440. [DOI] [PubMed] [Google Scholar]
- Warner J. R., McIntosh K. B., 2009. How on are extraribosomal functions of ribosomal proteins? Mol. Cell 34: 3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson D. M., 3rd., Deutsch W. A., Kelley M. R., 1994. Drosophila ribosomal protein S3 contains an activity that cleaves DNA at apurinic/apyrimidinic sites. J. Biol. Chem. 269: 25359–25364. [PubMed] [Google Scholar]
- Winzeler E. A., Shoemaker D. D., Astromoff A., Liang H., Anderson K., et al. , 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285: 901–906. [DOI] [PubMed] [Google Scholar]
- Wolfe K. H., Shields D. C., 1997. Molecular evidence for an ancient duplication of the entire yeast genome. Nature 387: 708–713. [DOI] [PubMed] [Google Scholar]
- Yu X., Warner J. R., 2001. Expression of a micro-protein. J. Biol. Chem. 276: 33821–33825. [DOI] [PubMed] [Google Scholar]
- Zhao Y., Sohn J. H., Warner J. R., 2003. Autoregulation in the biosynthesis of ribosomes. Mol. Cell. Biol. 23: 699–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.