Abstract
Planar cell polarity (PCP) is a common feature of many epithelia and epithelial organs. Although progress has been made in the dissection of molecular mechanisms regulating PCP, many questions remain. Here we describe a screen to identify novel PCP regulators in Drosophila. We employed mild gain-of-function (GOF) phenotypes of two cytoplasmic Frizzled (Fz)/PCP core components, Diego (Dgo) and Prickle (Pk), and screened these against the DrosDel genome-wide deficiency collection for dominant modifiers. Positive genomic regions were rescreened and narrowed down with smaller overlapping deficiencies from the Exelixis collection and RNAi-mediated knockdown applied to individual genes. This approach isolated new regulators of PCP, which were confirmed with loss-of-function analyses displaying PCP defects in the eye and/or wing. Furthermore, knockdown of a subset was also sensitive to dgo dosage or dominantly modified a dishevelled (dsh) GOF phenotype, supporting a role in Fz/PCP-mediated polarity establishment. Among the new “PCP” genes we identified several kinases, enzymes required for lipid modification, scaffolding proteins, and genes involved in substrate modification and/or degradation. Interestingly, one of them is a member of the Meckel-Gruber syndrome factors, associated with human ciliopathies, suggesting an important role for cell polarity in nonciliated cells.
PLANAR cell polarity (PCP) controls the orientation of single cells or groups of cells within a plane of tissue and is conserved throughout the animal kingdom (Seifert and Mlodzik 2007; Wang and Nathans 2007; Bayly and Axelrod 2011). In Drosophila, for example, PCP manifests in each wing cell as a single distally pointing hair, or in the compound eye in the arrangement of photoreceptor cells (Adler 2002; Strutt 2003; Klein and Mlodzik 2005; Seifert and Mlodzik 2007). When PCP establishment is perturbed in the wing, hairs can point in random directions and/or several wing hairs form in single cells. In the eye, PCP controls two aspects of ommatidial orientation: photoreceptor R3 and R4 cell fate determination and a subsequent 90° rotation of an entire ommatidium, which together establish a mirror image symmetry along the dorsoventral boundary, the equator (Mlodzik 1999; Strutt and Strutt 1999). In this context, PCP defects can produce random chiral arrangements of photoreceptors and symmetrical and misrotated ommatidia (Adler 2002; Strutt 2003; Klein and Mlodzik 2005; Seifert and Mlodzik 2007).
A conserved core set of proteins is critical for PCP establishment. These include the multipass trans-membrane proteins Frizzled (Fz), Strabismus/Van Gogh (Stbm/Vang), and Flamingo/Starry night (Fmi/Stan, an atypical cadherin), and the cytoplasmic factors Dishevelled (Dsh), Prickle (Pk), and Diego (Dgo) (Adler 2002; Strutt 2003; Klein and Mlodzik 2005; Seifert and Mlodzik 2007; Wang and Nathans 2007; Bayly and Axelrod 2011). The Fz receptor recruits and signals through Dsh, a component shared with the canonical wingless (wg)/Wnt signaling pathway (Boutros and Mlodzik 1999; Wallingford and Habas 2005). The other core PCP factors are thought to regulate Fz/Dsh activity and/or localization: Dgo promotes Fz/Dsh complex formation, whereas Stbm/Vang and Pk antagonize it (Tree et al. 2002; Jenny et al. 2005); Fmi/Stan is thought to promote the function of both complexes by stabilizing their membrane association (Sahai et al. 1998; Usui et al. 1999; Das et al. 2002; Lawrence et al. 2004; Klein and Mlodzik 2005; Casal et al. 2006; Chen et al. 2008; Strutt and Strutt 2008, 2009). As a result of their interactions, the core components localize asymmetrically in Drosophila tissues, forming two complexes on opposite sides of any given cell. In the wing, Stbm/Vang and Pk accumulate in complexes on the proximal side of each cell, whereas Fz, Dsh, and Dgo form a complex that localizes distally. Fmi/Stan is part of both complexes (Lawrence et al. 2004; Klein and Mlodzik 2005; Casal et al. 2006; Chen et al. 2008; Strutt and Strutt 2009). Whereas the interactions among the core factors are beginning to be understood, less is known about potential upstream long-range signaling input (Wu and Mlodzik 2009) or downstream cellular interactions/effectors of the complexes.
Besides the Fz/PCP core group, a parallel pathway anchored around the protocadherins Fat (Ft) and Dachsous (Ds) also acts in PCP establishment (Casal et al. 2006; Lawrence et al. 2007). In certain contexts Fat/Ds and Fz/PCP signaling act redundantly, though the exact relationship between these pathways remains unclear (Casal et al. 2006; Donoughe and Dinardo 2011). Similarly, although apical–basal (A/B)-polarity determinants can interact with Fz/PCP factors (Djiane et al. 2005; Courbard et al. 2009) and A/B polarity in epithelia is generally a prerequisite for PCP-type polarity, interactions among A/B-polarity factors and PCP core components are not well defined.
To gain insight into the regulatory interactions among the core Fz/PCP genes, their relationship with other polarity determinants, and to identify novel effectors of the core PCP complexes, we designed a genetic screen employing mild core PCP factor overexpression. We selected Pk and Dgo, because they act at the level of Dsh, compete for Dsh binding in vitro, and antagonize each other in the context of PCP establishment (Jenny et al. 2005). Gain-of-function (GOF) backgrounds of Dgo and Pk were used in a genome-wide modifier screen to select for genetic interactions with both complexes. Furthermore, we took advantage of several recently available genetic tools in Drosophila: collections of overlapping deficiencies generated by recombinant techniques in genetically identical animals, including the DrosDel (Ryder et al. 2007) and Exelixis (Parks et al. 2004) deficiency collections, as well as transgenic RNAi tools available for most genes [Vienna Drosophila RNAi Collection (VDRC), Dietzl et al. 2007, and Nippon Institute of Genetics (NIG)]. This was combined with in vivo imaging of GFP-labeled rhabdomeres (animals carried a rhodopsin1-GFP transgene, referred to as Rh1-GFP; Pichaud and Desplan 2001) allowing for rapid verification of modifiers of the core PCP phenotypes.
In this study, we isolated several new regulators of PCP that either enhanced or suppressed the pk or dgo GOF phenotype in the adult eye and/or wing. Of the 195 deficiencies initially screened, 11 were confirmed by smaller deficiencies, RNAi knockdowns, and/or mutant alleles. Two deficiencies harbored dachsous (ds) and Delta (Dl), which are known PCP factors acting in the parallel Fat/Ds pathway or as an eye-specific effector of Fz/PCP signaling, respectively. The new PCP regulators include kinases, scaffolding proteins, lipid modification enzymes, and factors involved in covalent protein modification and degradation. Loss-of-function analyses of many of these genes indicated that they indeed function in PCP establishment in the eye and/or wing. Furthermore, knockdown of a subset of these genes was shown to be sensitive to dgo dosage or could dominantly modify dsh GOF phenotypes, supporting a role in PCP establishment.
Materials and Methods
Fly stocks and genetic screen
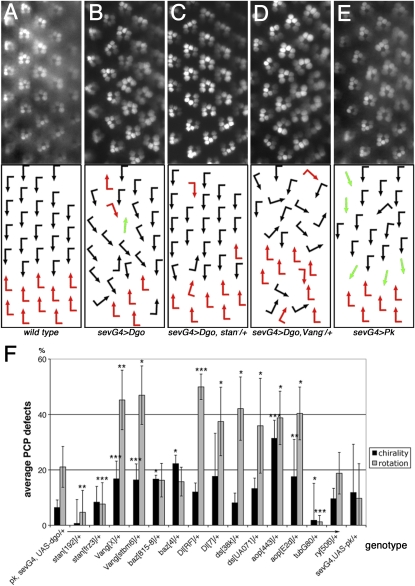
Recombinants of Rh1-GFP, sev-GAL4, and UAS–dgo or UAS-pksple (the Sple isoform of Pk (Gubb et al. 1999; Jenny et al. 2005) (referred to as sev-GAL4, UAS-dgo, or sev-GAL4, UAS-pk, respectively) were established and tested for reproducible chirality and ommatidial rotation defects. Testcrosses to pk13 and pksple1 alleles revealed that the w; UAS-dgo, Rh1-GFP; sev-GAL4 recombinant flies carry a strong pk (pk-sple) mutation, originating from the Rh1-GFP chromosome, affecting all adult tissues. The sev-GAL4 driver (expressed during PCP establishment in a subset of R cells including the R3/R4 precursors) has basal expression in other tissues due to the presence of a heat-shock promoter (from hsp70). sev-GAL4 expression of PCP proteins has been observed to induce wing hair orientation defects (U. Weber and M. Mlodzik, unpublished results). In the pilot screen, we noted dosage-sensitive modifications of the phenotype with Vangstbm-6 and Vangstbm-X, fmi/stanfrz3, fmi/stan192, and fmi/stanE59, while most other core PCP genes did not modify the phenotype significantly as heterozygotes. Alleles tested that did not modify were fz: R52 or 23, P21 or 21, R54 or 25, and F31 or 13; dsh: V26 or 3, A3, 477, and A21; dgo: 380, 308, and 269; nemo (nmo; Nlk in vertebrates): E33, DB, and P1; fat: l(2)fd or 8, G-rv, and k07918; par-6Δ226; aPKC: k06403; scribbled (scrib): 673 and 1; lethal(2)giant larvae (l(2)gl): 4w3 and 4; Epidermal growth factor receptor (Egfr): top-18A and CO or f24; kayak (kay)/Dfos: 1 and Df(3R)ED6315; pnt: Δ88, 2, and 19099; Jra/Djun: RC46 and 2 or IA109; and Notch (N): 55e11 and Df(1)N-5419. All crosses were grown at 25° unless otherwise noted. bazooka (baz)/D-Par3, Delta (Dl), ds, and anterior open (aop)/yan showed significant modification of the eye phenotype, but only with some alleles (see also below). The photoreceptor arrangement/rhabdomere pattern was visualized by the Rh1-GFP transgene (Pichaud and Desplan 2001) (see Figure 1, A–E for examples).
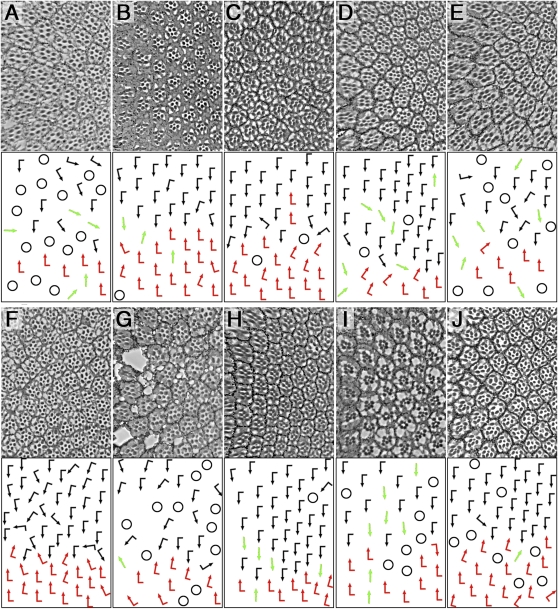
Figure 1 .
Ommatidial PCP orientation defects induced by Dgo or Pk overexpression and dominant modifications in the pilot screen. (A–E) Rhodopsin1-GFP (Rh1-GFP) pictures of rhabdomere patterns are shown on top and schematic arrows representing ommatidial orientation at bottom; dorsal is up and anterior to the left in these and all subsequent figures. The following genotypes are shown: wild type (A), pk/+, UAS-dgo/+, Rh1-GFP/+; sev-GAL4/+ (referred to as sev-GAL4, UAS-dgo) (B), stanfrz3/+, pk/+, UAS-dgo/+, Rh1-GFP/+; sev-GAL4/+ (C), Vangstbm-X/+, pk/+, UAS-dgo/+, Rh1-GFP/+; sev-GAL4/+ (D), and UAS-Pk/+, sev-GAL4/+, Rh1-GFP/+ (referred to as sev-GAL4, UAS-pk) (E). Quantifications of interactions are shown in F. Black and red arrows indicate the two chiral ommatidial forms, and green arrows represent nonchiral, symmetrical ommatidia. Note that stanfrz3/+ suppressed sev-GAL4, UAS-dgo rotation defects (C and F) and Vangstbm-X/+ enhanced sev-GAL4, UAS-dgo (D and F) chirality and rotation defects. Quantifications of additional candidate genes tested in pilot screen, showing significant modification: bazooka (baz)/D-Par3, Delta (Dl), dachsous (ds), and aop/yan (F) (*P < 0.06, **P < 0.01, and ***P < 0.005; three to four eyes and 63–173 ommatidia were scored for each genotype). Chirality and rotation defects were counted independently; negative controls did not modify sev-GAL4, UAS-dgo and GAL80ts abolished the phenotype (F).
In the wing, stbm/Vang and fmi/stan showed modification of pk−/+, sev-GAL4, UAS-dgo/+ wing PCP defects with two alleles each. No robust modification was found with fz, dgo, dsh, fat, ds, scrb, par-6, l(2)gl, aPKC, nmo/Nlk, baz/D-Par3 and Notch (for alleles see above).
In the deficiency screen, 32% of the DrosDel deficiencies (Ryder et al. 2007) showed an external modification. A total of 5.6% of these were excluded due to non-PCP effects in the Rh1-GFP assay. Regions of the genome that showed robust modification in two independent experiments were further screened with smaller, subdividing, and/or partially overlapping deficiencies (including the Exelixis collection, Parks et al. 2004, or cytologically mapped deficiencies). Complementation crosses between deficiencies and mutant alleles were performed as controls. For some DrosDel deficiencies the interacting genes were identified by exclusion, as not all genomic regions within the original deficiency were uncovered by the Exelixis collection or other deficiencies. UAS-RNAi stocks from VDRC (Dietzl et al. 2007) and NIG were subsequently used to test individual genes within the respective genomic regions via loss-of-function (LOF) studies. The candidate genes were knocked down by UAS-RNAi trangenes using decapentaplegic (dpp)-GAL4, engrailed (en)-GAL4, and apterous (ap)-GAL4 drivers for the wing and sev-GAl4 for the eye. If tissue-specific RNAi knockdown produced PCP defects, we increased the phenotype by coexpressing UAS-dicer2 and/or, increasing transgene copy numbers or by elevating the temperature to enhance the activity of the GAL4/UAS system.
Specific fly strains used:
sev-GAL4 (Basler et al. 1989), gift from K. Basler;
Rh1-GFP, gift from F. Pichaud (Pichaud and Desplan 2001);
UAS-dgo; sev-GAL4 (K. Gaengel and M. Mlodzik, unpublished results);
UAS-pksple, gift from D. Gubb, (Gubb et al. 1999);
VangstbmX gift from Nuria Paricio;
DrosDel deficiencies were received from Szeged (now distributed by the Bloomington Stock Center, Ryder et al. 2007);
Exelixis deficiencies (distributed by FlyBase, were from Harvard University/Exelixis) (Parks et al. 2004);
sev-Dsh (2x) and sev-Fz (Boutros et al. 1998).
UAS-RNAi lines were obtained from VDRC (Dietzl et al. 2007) and the NIG Fly Stock Centers. All other stocks were received from the Bloomington Stock Center.
Imaging and histology
Adult flies were initially inspected and evaluated on a Zeiss stereo microscope at ×66 magnification for external eye and wing phenotype modification. The internal rhabdomere pattern was documented on a Zeiss Axioscope2 plus with a ×40 water immersion lens at ×400 magnification with UV light illumination. Concentric pictures of four individual eyes were evaluated and a total of 80–160 ommatidia were scored independently for rotation and chirality defects and statistically analyzed by t test. Wings were mounted in 80% glycerol in 1× PBS (Wu et al. 2004) and wing hair orientation defects analyzed for both surfaces of 4–10 wings. Tangential eye sections were prepared as described (Tomlinson and Ready 1987).
Results
Establishment of genotypes and pilot screen
Several genetic modifier screens addressing PCP have been performed in the past with a focus on the trans-membrane proteins fz and stbm/Vang (for example Rawls and Wolff 2003; Strutt and Strutt 2003), while the cytoplasmic components have been less explored. In particular, dgo and pk have not yet been used as screening tools, but are of specific interest due to their antagonistic relationship and opposing effects on Fz–Dsh/PCP signaling (Jenny et al. 2005).
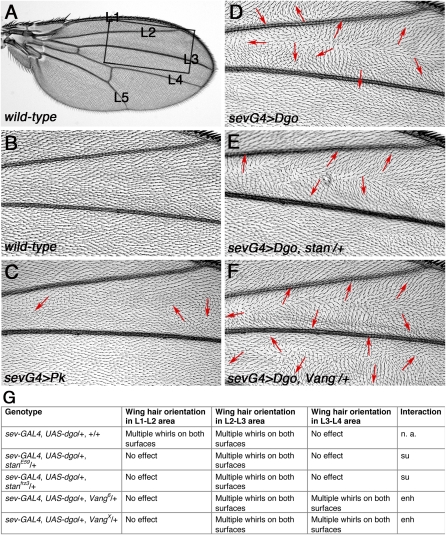
To identify new regulatory factors related to PCP establishment that could either act on Dgo or Pk, affect Fz–Dsh/PCP signaling in general, or also function as effectors of the PCP pathway, we employed the GAL4-UAS system (Brand and Perrimon 1993) to overexpress Dgo and Pk. To screen both eye and wing tissues, we used a sevenless (sev-enhancer) heat-shock (hs)-promoter–GAL4 (see Materials and Methods) (Basler et al. 1989) that drives expression in the eye transiently at high levels in the R3/R4 photoreceptor precursor pair at a time when PCP is being established (Strutt et al. 1997; Boutros et al. 1998) and at low levels in other tissues including the wing (due to the presence of basal level of expression from the hs-promoter, Figure 2, C and D). To test whether these genotypes were sensitive and specific enough for such an assay, we tested mutant alleles (see Materials and Methods) of the known Fz/PCP core components (reviewed in Adler 2002; Strutt 2003; Klein and Mlodzik 2005; Seifert and Mlodzik 2007), the parallel acting Fat/Ds PCP pathway (Matakatsu and Blair 2004; Casal et al. 2006; Lawrence et al. 2007; Simon et al. 2010), or the apicobasal (A/B)-polarity factors that are known to interact with PCP proteins (Djiane et al. 2005; Courbard et al. 2009). Furthermore, we tested the consistency of our imaging and scoring methods by examining the rhodopsin1-GFP rhabdomere marker (Pichaud and Desplan 2001) (Materials and Methods) for PCP-associated rhabdomere orientation defects (Figure 1, A–F) and hair orientation defects by microscopic inspection (Figure 2, C–F). In sev-GAL4, UAS-dgo, and sev-GAL4, UAS-pk eyes (referred to as the screen genotypes), ommatidia showed chirality and rotation defects (Figure 1, B, E, and F and Materials and Methods). In sev-GAL4, UAS-dgo wings, cellular hairs were misoriented in the region between the anterior wing margin and longitudinal vein L3 (Figure 2D), while in sev-GAL4, UAS-pk wing hairs were partially misoriented between L2 and L3 on one surface (Figure 2C).
Figure 2 .
Wing hair orientation defects induced by Dgo or Pk overexpression and dominant modifications by Vang/stbm and stan/fmi in the pilot screen. (A and B) Wild-type wing overview (A) and detail (B, boxed area from A) as represented in all following panels. (C–F) Wing hair orientation defects between veins L1 and L4, highlighted by arrows in sev-GAL4, UAS-dgo (D), which are suppressed by stanE59/+ (E) and enhanced by Vangstbm6 /+ (F). Wing hair defects in sev-GAL4, UAS-pk are milder (C). (G) Table summarizing dominant modification of wing hair orientations in sev-GAL4, UAS-dgo by candidate genes. Two alleles each of stan/fmi (E and G) and Vang/stbm showed the effect (F and G).
In the pilot screen, among the core PCP factors, removing a copy of stbm/Vang enhanced (Jenny et al. 2005) and, of fmi/stan suppressed, both eye and wing defects significantly (Figures 1, C, D, and F and 2, D–G). The fact that stbm/Vang modified the dgo GOF background is consistent with earlier data (Jenny et al. 2005). The observation that fmi/stan acted as a dominant suppressor in both tissues and the genetic effect of fmi/stan acting in opposition to stbm/Vang has not been observed but is consistent with current models (see Discussion). Of the other PCP-related genes tested, we observed that some but not all alleles of ds and Delta (Dl), acting in R3/R4 specification (Cooper and Bray 1999; Fanto and Mlodzik 1999; Tomlinson and Struhl 1999) modified the sev-GAL4, UAS-dgo eye phenotype (Figure 1F; also below). Furthermore, the apicobasal determinant bazooka (baz)/D-Par3 and the epidermal growth factor receptor (Egfr) effector aop/yan were found to enhance eye PCP defects significantly (Figure 1F). It has been shown independently that baz/D-Par3 acts specifically on Fz: it positively regulates Fz levels/signaling in the eye and is upregulated during PCP signaling (Djiane et al. 2005) and that dgo overexpression in photoreceptor R4 promotes there the incorrect R3 fate (Jenny et al. 2005). In our experiment, reduction of baz/D-Par3 function in sev-GAL4, UAS-dgo, presumably further abolished the difference between R3 and R4 cell fate and therefore enhances PCP chirality defects (Figure 1F). The Aop/Yan transcription factor is required in photoreceptor R3 to inhibit R4 fate (Weber et al. 2008). Here, reduction of aop/yan function in the context of dgo overexpression further reduced the cell fate difference between R3 and R4, causing more severe PCP defects (Figure 1F). Negative controls showed no effect in this assay (Figure 1F and data not shown), nor did other mutants tested (see Materials and Methods). As further control, GAL80 abolished all effects, as it suppresses GAL4-mediated expression (Duffy 2002). No modifications were observed with ds, Dl, baz/D-Par3, and aop/yan in the wing, which is consistent with eye-specific PCP functions of Dl, baz/D-Par3, and aop/yan (Figure 1, legend and Materials and Methods).
Taken together, we have established effective tools to screen for novel PCP factors, which are both sensitive enough to identify suppressor- and enhancer-type interactions, but also stringent enough to only detect specific modifiers in the eye and the wing. We therefore utilized this set of tools to screen the genome of Drosophila melanogaster for new PCP regulators by lowering the copy number of gene intervals with deficiencies generated by DrosDel (Ryder et al. 2007).
Deficiency screen
To identify new PCP regulatory factors, we screened both genotypes, sev-GAL4, UAS-Dgo and sev-GAL4, UAS-Pk (see Materials and Methods for details), testing these against the DrosDel deficiency collection for dominant modifications. In the primary screening, we examined the external appearance of adult compound eyes and wings for phenotypic suppression or enhancement (Table 1, Figure 3). For deficiencies displaying robust external modification(s) in either tissue, we examined in detail PCP orientation in the eye, using the internal Rh1-GFP rhabdomere pattern and wing hair orientation patterns in mounted wings (Table 1). A total of 195 deficiencies were screened in this manner, covering ∼80% of the genome. Of these, 21% modified the internal PCP eye phenotype and 21% modified the wing phenotype. A total of 11% showed effects in both tissues and 8.5% affected both genotypes (Table 1). Those deficiencies affecting both genotypes were considered high-priority candidates and were analyzed further. A total of 68% of the deficiencies did not show an effect (listed in Supporting Information, Table S1).
Table 1 . List of DrosDel deficiencies, which showed external modification of the dgo or pk induced PCP GOF phenotypes in the eye and/or wing and their analysis by Rh1-GFP and wing hair orientation assessment.
| sev-GAL4, UAS-dgo/+ | sev-GAL4, UAS-pk/+ | |||||
|---|---|---|---|---|---|---|
| DrosDel Df | External eye | Rh1-GFP eye | Wing | External eye | Rh1-GFP eye | Wing |
| Df(1)ED6720 | Enh 2× | Enh rota 2× | M enh 2× | ND | Enh rota, chir | M su |
| Df(1)ED6727 | Enh | Enh rota 2× | No | ND | Enh rota | ND |
| Df(1)ED6957 | Enh 2× | Enh rota 2× | Enh 2× | ND | No | Su |
| Df(1)ED7161 | Enh 2× | Enh rota, chir 2× | Enh 2× | ND | ND | ND |
| Df(1)ED447 | Enh 2× | Enh rota 2× | No 2× | ND | No | M su |
| Df(1)ED7441 | No | Su chir | ND | Enh | Su chir | ND |
| Df(1)ED7374 | Enh | Enh rota | ND | ND | ND | ND |
| Df(1)ED7147 | Enh | Enh rota | ND | No | Su chir | ND |
| Df(1)ED7344 | No | Enh rota | ND | Enh | No | ND |
| Df(1)ED6521 | Enh | Enh rota | ND | Enh | No | ND |
| Df(1)ED6443 | No | No | ND | Enh | No | ND |
| Df(2L)ED62 | Enh 2× | Enh rota, chir 2× | M enh | ND | No | Su |
| Df(2L)ED623 | Enh 2× | Enh rota, chir 2× | Enh 2× | ND | No | M su |
| Df(2L)ED775 | Enh | Variable | No | ND | ND | ND |
| Df(2L)ED784 | M enh | Variable | M enh | ND | Enh rota, chir | M su |
| Df(2L)ED793 | M enh 2× | Enh rota | No 2× | ND | Enh rota, chir | M su |
| Df(2L)ED1203 | Enh 2× | No | M enh | ND | Not scorable | No |
| Df(2L)ED1315 | M enh | Enh rota 2× | Enh 2× | ND | Enh rota, chir | M su |
| Df(2R)ED1552 | Enh | No | No | ND | ND | ND |
| Df(2R)ED1618 | Enh 2× | Enh rota | S enh 2× | ND | Enh rota, chir | Enh |
| Df(2R)ED1673 | S enh 2× | Enh rota, chir 2× | Enh 2× | ND | Enh rota, chir | M enh |
| Df(2R)ED1715 | S enh 2× | ND | No | ND | ND | ND |
| Df(2R)ED1742 | No 2× | No | Enh 2× | ND | No | Su |
| Df(2R)ED2308 | M enh | Enh rota, chir | Enh | ND | No | M enh |
| Df(2R)ED3791 | Enh 2× | Variable | Enh 2× | ND | No | No |
| Df(2R)Exel6076 | Nd | Enh rota | ND | ND | Su chir | ND |
| Df(2R)ED2247 | Nd | Enh rota | ND | ND | No | ND |
| Df(2R)ED1725 | Nd | Enh rota | ND | ND | Su chir | ND |
| Df(2R)ED1791 | Nd | Enh rota | ND | ND | Enh chir | ND |
| Df(3L)ED201 | Enh 2× | Enh rota 2× | No 2× | ND | No | Su |
| Df(3L)ED207 | Enh 3× | Enh rota 2× | Enh | ND | No | Su |
| Df(3L)ED4284 | M enh 2× | Variable | Enh 2× | ND | No | M su |
| Df(3L)ED4293 | Enh 2× | No | M enh | ND | ND | M su |
| Df(3L)ED4483 | No 2× | No | Enh 2× | ND | No | No |
| Df(3L)ED4502 | Enh | ND | Enh | ND | ND | ND |
| Df(3L)ED4543 | M enh | Variable | Enh | ND | ND | ND |
| Df(3L)ED220 | Enh 2× | Enh rota | M enh | ND | No | M su |
| Df(3L)ED230 | M enh 2× | Enh rota 2× | Enh 2× | ND | Lethal | Lethal |
| Df(3L)ED5017 | M enh 2× | Enh rota 2× | Enh 2× | ND | No | Su |
| Df(3L)ED6279 | No | Enh rota | ND | Enh | Enh rota | ND |
| Df(3L)ED4177 | No | No | ND | Enh | Enh rota, chir | No |
| Df(3R)ED7665 | Enh 2× | Enh chir 2× | Enh 2× | ND | No | No |
| Df(3R)ED5454 | Su | No | Su | ND | ND | ND |
| Df(3R)ED5516 | Su 2× | No | No | ND | ND | ND |
| Df(3R)ED5559 | M enh | Enh chir | Enh | ND | ND | ND |
| Df(3R)ED5577 | Enh 2× | Enh rota, chir 2× | Enh 2× | ND | No | M su |
| Df(3R)ED5623 | Enh 2× | Enh rota var | M enh 2× | ND | No | Enh |
| Df(3R)ED5644 | Enh 2× | Enh rota, chir | M enh 2× | ND | Su chir | Su |
| Df(3R)ED5705 | Enh 2× | Variable | Enh 2× | ND | Enh rota, chir | ND |
| Df(3R)ED10639 | Enh 3× | Enh rota | M enh 3× | ND | Su chir + rota | Su |
| Df(3R)ED5938 | No | Variable | Enh | ND | Enh rota | ND |
| Df(3R)ED5942 | Enh 2× | Enh chir 2× | Enh 2× | ND | Enh rota, chir | M su |
| Df(3R)ED10845 | Enh 2× | No | M enh | ND | No | Su |
| Df(3R)ED6076 | Enh 2× | Enh rota, chir | No | ND | No | No |
| Df(3R)ED6096 | Enh | No | S enh | ND | Enh rota, chir | ND |
| Df(3R)ED6361 | M enh 2× | Variable | M enh | ND | No | No |
| Df(3R)ED6085 | No | No | Enh | Enh | No | ND |
| Df(3R)ED5296 | Enh | Enh rota, chir | Enh | ND | No | ND |
| Df(3R)ED6052 | ND | Su rota, chir | Enh | ND | No | ND |
| Df(3R)ED7665 | ND | Enh rota | ND | ND | ND | ND |
| Df(3R)ED5612 | ND | Enh rota | ND | ND | ND | ND |
| Df(3R)ED6220 | ND | Enh rota | ND | ND | Su chir | ND |
Each DrosDel deficiency that showed an interaction was listed and strong interactors were analyzed further in detail for interaction with sev-GAL4, UAS-dgo and sev-GAL4, UAS-pk in the eye and wing. Some interactions were tested repeatedly, indicated by 2×. Interactions were categorized as Enh, enhanced; M, mildly; S, strongly; Su, suppressed; No, no interaction; ND, not determined. Rh1-GFP interactions were listed as enhanced or suppressed for rotation (rota) and/or chirality (chir) defects, and some effects were variable.
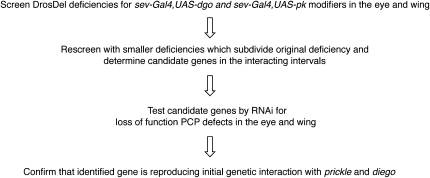
Figure 3 .
Overview of the PCP screen. Flow chart of the screening procedure, starting from the large DrosDel deficiencies.
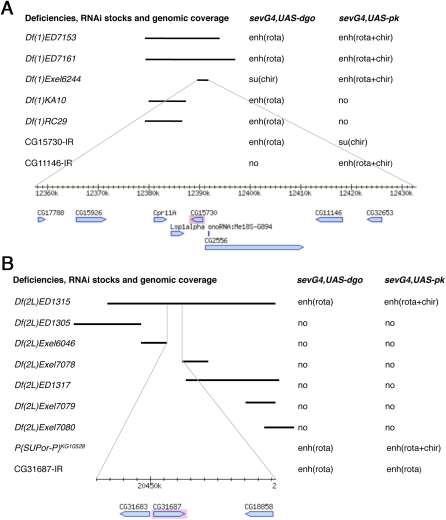
The initial genomic region responsible for an interaction was narrowed down by using smaller, subdividing deficiencies from the Exelixis collection (Parks et al. 2004) or partially overlapping cytologically mapped deficiencies (Figures 3 and 4). The same Rh1-GFP eye patterning analysis was used to confirm and refine the genomic region of interaction (see Figure 1). Many initially defined interactions were confirmed by such rescreening with smaller deficiencies (Table 2 and example shown in Figure 4A). In a small number of cases, however, we could not isolate the interaction in that manner, e.g., the large deficiency was fully covered by subdividing deficiencies, but none of these reproduced the original interaction (Table 2, Df(1) ED6957, Df(3L)ED207, and Df(3R)ED5623). In such cases, it is likely that a combination of two or more genes caused the modification, which could not be mapped down to a single locus or the deficiencies harbored mutations outside their assigned coverage that were responsible for the initial effect.
Figure 4 .
Single gene identification strategies for two DrosDel deficiencies, Df(1)ED7161 and Df(2L)ED1315. Deficiencies and their coverage indicated by black bars are shown on the left and their genetic interactions are listed on the right. Fine lines at the bottom of each panel connect to a genomic map for the area that was tested by RNAi’s (based on “MapBrowse, FlyBase”). (A) Mapping several interaction areas in Df(1)ED7161, which enhanced ommatidial rotation defects of sev-GAL4, UAS-dgo and enhanced rotation and chirality defects of sev-GAL4, UAS-pk. Two different genomic areas accounted for these effects, as identified by Df(1)KA10 and Df(1)RC29 for one of the interaction and Df(1)Exel6244 for the other. Df(1)Exel6244 was further studied since it modified both screen genotypes. CG15730 and CG11446 (now fused with CG32653 and called CG42251) were found to reproduce, each a subset of the original genetic interaction. (B) For Df(2L)ED1315, mapping to a single gene was done by exclusion. DrosDel deficiency Df(2L)ED1315 enhanced ommatidal rotation defects in sev-GAL4, UAS-dgo and enhanced both rotation and chirality defects in sev-GAL4, UAS-pk. Several overlapping or subdividing deficiencies were assayed for genetic interaction, but showed no modification of either genotype. This left three genes near the center of Df(2L)ED1315 that were not covered by any of the smaller deficiencies. We therefore tested these three genes and found that RNAi for CG31687 (an APC8 paralog) interacted similarly to Df(2L)ED1315 with the two screening genotypes, as did a P-element insertion KG10528 for CG2508 (APC8/cdc23).
Table 2 . Smaller deficiencies and individual genes, which confirmed original interactions.
| DrosDel deficiency | Subdividing deficiencies | Genes identified | sev-GAL4, UAS-dgo interaction | sev-GAL4, UAS-pk interaction |
|---|---|---|---|---|
| Df(1)ED6957 | Nonea | ND | ||
| Df(1)ED7161 | Df(1)ED7153 | CG17788 (no RNAi), CG15926 (no RNAi), CG2556, CG15730, CG11146, CG32653 | CG15730: enh(rota) | Su (chir)b |
| Df(1)Exel6244 | CG11146: nob | Enh (rota + chir) | ||
| Df(1)ED447 | Nonec | CG6461, CG6470, CG6335, CG10548, CG6481, CG15042, CG15047 | ||
| Df(2L)ED62 | Df(2L)Exel8003 | ds, CG2863 | ||
| Df(2L)ED94 | ||||
| Df(2L)ED49 | ||||
| dsUA071 | ||||
| Df(2L)ED623 | Df(2L)ED611 | CG13388, CG13399, CG13400 | Su (chir)b | Su (rota + chir)b |
| Df(2L)ED647 | Enh (rota) | |||
| Df(2L)Exel8021 | ||||
| Df(2L)ED793 | Df(2L)b87a25 | CG15283, CG4491, noc, CG15284, CG3474 | Enh (rota) | Nob |
| Df(2L)Exel6036 | ||||
| Df(2L)Exel6035 | ||||
| Df(2L)Exel8033 | ||||
| Df(2L)ED1315 | Nonec | CG31683, CG31687, CG18858 | Enh (rota) | Enh (rota + chir) |
| Df(2R)ED1618 | Df(2R)ED1673 | Pk | ||
| Df(2R)ED1673 | Df(2R)ED1618 | Pk | ||
| Df(3L)ED201 | Df(3L)Exel6084 | CG7004 | Enh (rota) | Enh (rota + chir)b |
| fwd Df(3L)Exel9057 | ||||
| fwdneo1 | ||||
| Df(3L)ED207 | Nonea | ND | ||
| Df(3L)ED220 | Df(3L)st[b11]d | ND | ||
| Df(3L)ED230 | Df(3L)Al29 | CG11438, CG11426 | ND | ND |
| Df(3L)AK1 | ||||
| Df(3L)ED5017 | Nonee | ND | ||
| Df(3R)ED7665 | Nonec | CG14612 (no RNAi), CG1070 (no RNAi), CG1019, CG10098 (ND), CG10068 | Enh (rota)f | No |
| Df(3R)ED5577 | Df(3R)Exel7316 | ND | ||
| Df(3R)ED5623 | Nonea | ND | ||
| Df(3R)ED5644 | Df(3R)Exel6267 | ND | ||
| Df(3R)ED5942 | Df(3R)Cha9 | Dl | ||
| Df(3R)Dl[RF] | ||||
| Df(3R)Dl[7] | ||||
| Df(3R)ED10639 | Df(3R)Exel7329 | CG6889, CG6815, CG6814, CG6864, CG12785, CG6963, CG31283 | Enh (rota) | ND |
The DrosDel and smaller deficiencies are listed in the first and second columns, respectively. The third column lists all genes that were tested by RNAi knockdown for PCP phenotypes. Genes that affected PCP when knocked down (compare Table 3) and reproduced the effects of the original deficiency are indicated in boldface type. Df(2L)ED62 and Df(3R)ED5642 covered ds and Dl, respectively (compare Figure S1). Columns 4 and 5 list genes and the quality of their genetic interactions that reproduced the original genetic interactions with screen genotypes.
Full coverage by other dfs.
Genetic interaction is different from what was observed with initial deficiency.
Genes tested based on exclusion from smaller Dfs.
Approximately 60 gene overlap.
No smaller Dfs, close to centromere.
Genetic interaction is a subset of what was observed originally.
Additionally, we observed that some larger deficiencies were not fully covered by smaller ones, in which case we postulated that the remaining uncovered genes were responsible for the original interaction. This was indeed the case and was confirmed for Df(1)ED447, Df(2L)ED1315, and Df(3R)ED7665 (Table 2 and example shown in Figure 4B; see also below). Among the deficiencies identified as modifiers, two uncovered genes with a previously known PCP function: ds and Dl (an eye-specific PCP factor) within Df(2L)ED62 and Df(3R)ED5942, respectively. Their LOF alleles, dsUA071 and DlRF, confirmed the genetic interaction originally found with the large deficiencies (Table 2, Figure S1), which was consistent with data observed in the pilot screen (Figure 1F, see above). We did not recover other known PCP genes in the deficiency screen (see Discussion).
Since pk and dgo act at the level of dsh, we further tested whether any of the identified deficiencies (Table 1) interacted with sev-dsh (Boutros et al. 1998) in the eye. Unfortunately, the Rh1-GFP rhabdomere pattern did not resolve into sharp pictures in this genetic background and therefore only external modifications could be used as selection criteria. Two deficiencies, Df(2L)ED793 and ED(3R)5644, displayed an interaction and were confirmed by eye sections to be suppressors of sev-dsh (Figure S2). In analogy, we also tested for modifications of sev-fz, Rh1-GFP, but none of the isolated deficiencies displayed a strong modification of this background.
For further analysis, we focused on the interacting genomic regions that showed an effect with both screen genotypes and/or in both tissues.
Identification and loss-of-function analyses of new PCP candidate regulators
The narrowing down of the initial genomic regions via subsequent screening with smaller deficiencies allowed us to define a small set of potential candidate genes for most interacting regions (Figure 3 and Table 2). These were then tested individually, each with UAS-RNAi transgenic flies in direct LOF function studies for PCP defects. Each candidate gene was first analyzed microscopically for eye PCP defects, with UAS-RNAi, sev-GAL4, Rh1-GFP. To increase the phenotypes observed, further crosses were set up in combination with UAS-dcr2, or the copy numbers of the GAL4/UAS constructs and/or the temperature were increased. Such flies were then analyzed in more detail in adult eye sections (Figure 5). For phenotypic analyses in the wing, specific GAL4 drivers (see Materials and Methods) were used in combinations as described above for the eye (Table 3). The individual genes that showed PCP related LOF defects were then tested via UAS-RNAi in the original genetic backgrounds employed in the screen to confirm that the interaction within a given genomic region was caused by gene dosage reduction of the respective factor (Table 2). In this manner, within the 13 DrosDel deficiencies identified originally, we confirmed and isolated the interaction to 11 individual genes (Table 2).
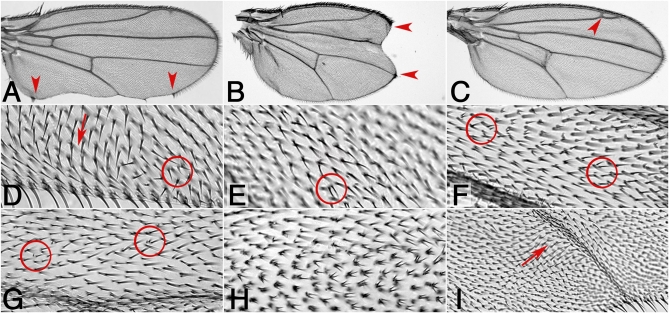
Figure 5 .
Eye PCP phenotypes of the novel candidate genes isolated in the screen. Top panels show tangential eye sections and bottom panels show a schematic of ommatidial orientation (arrows) (compare Figure 1A for wild type). Ommatidia with loss of photoreceptors or unscorable ommatidia are indicated by black circles in schematic. The following genotypes are shown: (A) CG1019IR; sev-GAL4 29°, (B) UAS-dcr-2/+; CG10068IR/+; sev-GAL4/+ 25°, (C) CG15730IR; sev-GAL4, escaper at 19°, (D) CG11146IR; sev-GAL4 29°, (E) CG15283IR/Y; sev-GAL4/+ 29°, (F) UAS-dcr-2/Y; CG31687IR/+; sev-GAL4/+ 29°, (G) CG7004IR; sev-GAL4/+ escapers 18°, (H) UAS-dcr-2/Y; CG11438IR/+; sev-GAL4/+ 29°, (I) UAS-dcr-2/+; CG13388IR/+; sev-GAL4/+ 25°, and (J) UAS-dcr-2/Y; CG6963IR/+; sev-GAL4/+ 29°.
Table 3 . Summary of PCP defects in the eye and wing of candidate genes studied.
| DrosDel deficiency | Gene/RNAi transf. ID | Vertebrate homolog, molecular signature | sev-GAL4 | dpp-GAL4 | en-GAL4 | ap-GAL4 |
|---|---|---|---|---|---|---|
| Df(1)ED7161 | CG15730/19583 | Mks1, Basal body component | Rota, chir, notches, hair oria | NE | NE | Mch |
| Df(1)ED7161 | CG42251/29634 | SH3/SH2 adaptor activity | Rota, chirb | NE | Vein defectsc | NE |
| Df(2L)ED62 | CG2863 (Nle)/33574 | Regulator of Notch pathway, WD40 repeats | NE | Lethal | Lethal | Mch |
| Df(2L)ED793 | CG15283/ 28550 | — | Rota, chirb | Hair oric | Hair oric | NE |
| Df(2L)ED793 | CG15284 (pburs)/27141/2 | insect spec. partner of bursicone, ligand for G prot. coupled rec. | Chird | Mch | Mch | Mchc |
| Df(2L)ED1315 | CG31687/21393 | APC8 paralog, component of the APC/C | Rota, chir | Mch | Hair ori | Mchc |
| Df(3L)ED201 | CG7004 (fwd)/27785/6 | PI4 kinase β | Rota, chird | Lethal | Lethal/notchese | Mchf |
| Df(3L)ED230 | CG11438 (PAP2)/11438 | PAP2 type phosphatase | Rota, chirc | ND | ND | Veinc |
| Df(3L)ED230 | CG11426 (PAP2)/11426 | PAP2 type phosphatase | NE | ND | ND | Mchc |
| Df(3R)ED7665 | CG1019 (Mlp84B)/18594 | CSRP1, LIM domains | Loss, rota, chirbd | NE | NE | NE |
| Df(3R)ED7665 | CG10068/15948 | — | Chird | NE | Mch | NE |
| Df(3R)ED10639 | CG6963 (gish)/26003 | Casein kinase1 gamma | Loss, rotaf | Mchc | Mchf | ND |
| Df(2L)ED623 | CG13388 (Akap200)/5647 | AKAP200/MESR2, protein kinase A binding | Loss, chir | NE | Vein defectsd | NE |
The first two columns list the deficiency and gene/RNAi. Third column indicates potential vertebrate homologs and/or molecular function. Further columns indicate which GAL4 driver caused phenotypes. Rota, ommatidial rotation defects; chir, ommatidial chirality defects; hair ori, wing hair orientation defects; mch, multiple cellular hairs; NE, no effect; ND, not done.
Increased copy number 19° escapers.
Increased copy number 29°.
29°.
With UAS-dcr2.
Escapers with a 25°–18° temperature shift.
UAS-dcr2 at 29°.
Phenotypes observed in the eye included classical PCP defects represented by either rotation (Figure 5F) or chirality (Figure 5H) defects, or both (Figure 5, A, B, and D); they often occurred in combination with loss of photoreceptors, which is also observed in dsh LOF eyes, for example (Boutros et al. 1998). In the wing, phenotypic analyses again revealed classical PCP defects, like misoriented cellular hairs (Figure 6, D and I) and/or clustered wing hairs (Figure 6, E–H). In addition, we observed wing margin defects/notches (Figure 6, A and B) and vein defects (Figure 6C). Both of the latter phenotypes might be associated with Notch-signaling (see Discussion). Most genes identified in this screen affected PCP in both tissues, the eye and wing, and few affected only one of the tissues (Table 3).
Figure 6 .
Wing PCP phenotypes of novel candidate genes. Wing phenotypes such as wing hair orientation defects, multiple cellular hairs, and notches were observed. For wild-type wing, compare Figure 2, A and B. Red arrowheads point to margin or vein defects. Wing hair orientation defects and multiple cellular hairs are indicated by arrows and circles, respectively. The following genotypes are shown: (A) CG15730IR; sev-GAL4 escapers 19°, (B) en-GAL4/+; CG7004IR /+ escapers 18–25°, (C) UAS-dcr-2/Y; ap-GAL4/+; CG1143IR/+ escapers 29°, (D) CG15730IR; sev-GAL4 19°, (E) UAS-dcr-2/+; ap-GAL4/+; CG7004IR/+ 29°, (F) UAS-dcr-2/Y; en-GAL4/+; CG11146IR/+ 29°, (G) CG31687IR/+; dpp-GAL4/+ 29°, (H) en-GAL4/+; CG6963IR/+ 29°C, and (I) CG15283IR /Y; en-GAL4/+ 29°. Wing defects with sev-GAL4 driver are due to low level activity/overexpression in all tissues due to the hs-promoter in sev-GAL4 (also Materials and Methods). Compare Table 3 for a summary of phenotypes for several GAL4 drivers for each gene/RNAi of an original genetic interaction.
The molecular nature and features of these new PCP regulatory genes ranged from kinases, phosphatases, enzymes required for lipid biosynthesis or modification, to proteins involved in substrate degradation or modification. Among the kinases, CG7004 (four wheel drive/fwd) encodes a PI4kinaseß, which has been shown to be required for male germ-line development (Polevoy et al. 2009), and CG6963 (gilgamesh/gish), a casein kinase1γ, known to be involved in many processes including Wnt signaling (Davidson et al. 2005), glial cell migration (Hummel et al. 2002), and spermatogenesis (Nerusheva et al. 2009). Phosphatases of the PAP2 family, CG11426 and CG11438, were also identified. These belong to the same subfamily as wunen and wunen2, which control female germ-line development (Starz-Gaiano et al. 2001). Fwd and the PAP2 phosphatases are likely to act as lipid modification enzymes.
Interestingly, CG31687, an APC8 paralog, and CG15283 encode proteins potentially involved in protein modification and/or degradation, possibly affecting substrate trafficking or localization, two features of likely importance in PCP establishment (Narimatsu et al. 2009). Scaffolding proteins encoded by CG11146, with SH3/SH2 domains, CG1019 (Muscle LIM protein, Mlp), a LIM domain-containing protein (Clark et al. 2007) and CG13388 (a kinase anchoring protein, Akap200) (Jackson and Berg 2002) were also isolated. Finally, proteins of unknown molecular nature encoded by CG15730, which is the fly ortholog of Mks1 (Meckel-Gruber syndrome 1), a gene mutated in human cilliary disease, and CG10068, which has been associated with cytokinesis in a cell-based genome-wide screen (Echard et al. 2004) round out the molecular features of the new PCP regulators (see Discussion for more details).
Genetic interactions with core PCP factors and Notch
The screen identified new PCP regulators as modifiers of a dgo or pk GOF phenotype. We also isolated factors that potentially modulate Notch signaling, as a few candidates showed notches or wing vein defects when knocked down with UAS-RNAi (Figure 6, A–C; Table 3). As Notch signaling interacts with Fz/PCP core factors in the eye via Dl upregulation (and Dl was indeed one of the genes identified in the screen) this was consistent. We thus tested these candidate genes in a Notch−/+ background. However, we did not observe an enhancement of the UAS-RNAi induced eye or wing margin/vein phenotype in a Notch heterozygous background.
To get further insight into the potential role(s) of the newly identified PCP regulatory factors and/or to corroborate that some of them might act at the level of dgo, pk, and dsh within the PCP hierarchy, we next tested whether their function was sensitive to endogenous levels of dgo, pk, or dsh. To this end UAS-RNAi; GAL4 stocks of the respective genes (with an eye- or wing-specific GAL4 driver, see above) were analyzed for genetic requirements by crossing these to dgo, pk, dsh, fat, and ds mutants. Two genes were sensitive to halving the genomic dose of dgo and showed an enhancement of PCP defects in a heterozygous dgo background: CG15730 and CG15283 in the wing (Figure 7). The others did not display an equivalent interaction with either dgo or the other genes tested. Intercrossing UAS-RNAi transgenes of the new candidate PCP regulators/modifiers that caused similar wing defects or fell into similar molecular function classes did not reveal specific enhancements, suggesting that they are not clustered functionally within a molecular complex.
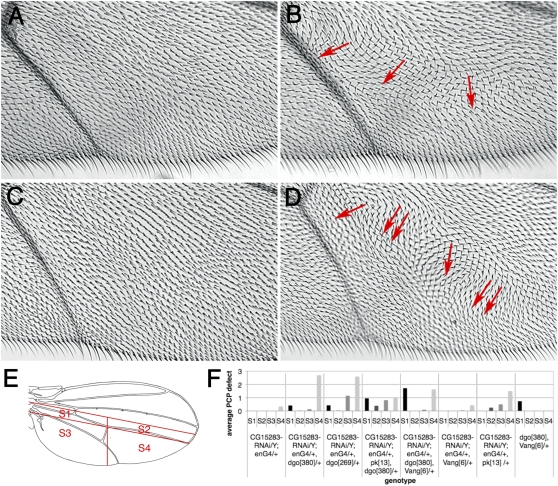
Figure 7 .
CG15283 and CG15730 dominantly interact with dgo loss of function. (A–D) Wing hair orientation in an area between longitudinal vein L4 and L5, controls (left, A and C) and dominant enhancement (dgo−/+) (right, B and D) are shown. Red arrows highlight wing hair orientation defects. Double arrow indicates defects on both wing surfaces. (A) CG15283IR/Y; en-GAL4/+, pk13/+ (n = 9) displayed wild-type orientation of hairs, whereas removal of one copy of dgo caused wing hair whirls in 89% of CG15283IR/Y; en-GAL4/+, dgo380/+ (n = 9) (B). (C) en-GAL4/+, pk13/+; CG15730IR/+ (n = 4) showed wild-type wing hair polarity, whereas reduction of dgo copy number enhanced the PCP defects (D) in 44% of en-GAL4/+, dgo380/+; CG15730IR/+ wings (n = 9). (E–F) Quantification of the dominant interactions of CG15283IR. Phenotypes were assessed in four sectors of the wing as indicated in drawing (E). Graph summarizing dominant interactions between CG15283, two dgo LOF alleles, and further modification by pk and Vang/stbm (n = 2–33 wings) at 30°. PCP defect of “1” corresponds to 10–20 misoriented wing hairs at a 45°–180° angle compared to surrounding wild-type hairs.
The enhancement of CG15283 by dgo heterozygosity was particularly robust (Figure 7, B and F) and thus we tested whether dsh, the genetic and molecular binding partner of dgo, also displayed interactions with CG15283. To this end we used dsh GOF and LOF eye phenotypes (as these are dosage sensitive and quantifiable) and asked whether levels of CG15283 affect these. Strikingly, the sev-Dsh GOF phenotype is markedly suppressed by lowering the CG15283 function via RNAi (Figure 8, A, B, and E) and comparable to the effect seen with Df(2L)ED793, which led us to identify CG15283 (Figure S2). Accordingly, the PCP-specific hypomorphic dsh1 loss-of-function allele phenotype is strongly enhanced by sevGAL4, CG15283IR gene knock down (Figure 8, C, D, and F). Thus, in both genetic scenarios, CG15283 promotes Dsh function, being positively required for Dsh (this conclusion is also consistent with its genetic interactions in the original screen genotypes; see above).
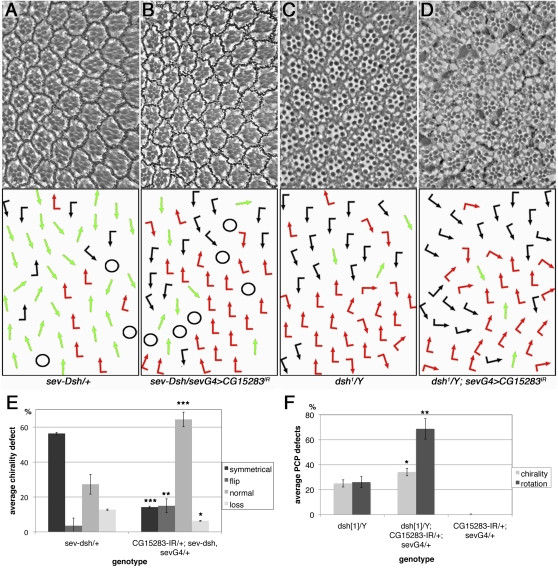
Figure 8 .
CG15283 acts at the level of dsh promoting its activity. (A–D) Top panels show tangential eye sections and bottom panels, the respective schematic of ommatidial chirality/orientation (arrows) (compare Figure 1A for wild type and Figure 5). The following genotypes are shown: (A) sev-dsh/+, (B) CG15283IR/+; sev-GAL4/+, sev-dsh/+, (C) dsh1/Y, (D) dsh1/Y; CG15283IR/+; sev-GAL4/+. (E and F) Quantification of the above interactions, note suppression of dsh GOF PCP defects in sev-dsh (A, B, and E) and enhancement of the PCP-specific hypomorphic dsh1 LOF phenotype (C, D, and F) by CG15283IR knockdown. (E) *P < 0.007, **P < 0.003, and ***P < 0.0001, with n = 568–893 ommatidia in four to five eyes. (F) *P < 0.005 and **P < 0.001, with n = 536–806 ommatidia in three to four eyes.
gish/CK1γ regulates ommatidial rotation
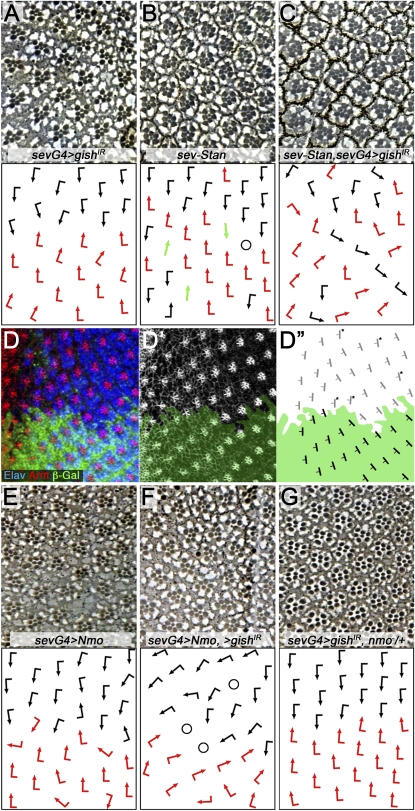
Phenotypic analyses of CG6963, the gene encoding CK1γ (gilgamesh/gish in Drosophila) revealed specific features in eye PCP establishment. Moderate gene knockdown of CG6963/gish appeared to primarily affect ommatidial rotation (Figure 9A). Stronger knockdown of gish caused additional defects in photoreceptor specification including rare symmetrical ommatidia and R-cell loss (Figure 5J). These data are consistent with a role for gish/CK1γ in canonical Wnt signaling, as established in vertebrates for CK1γ (Davidson et al. 2005), and a potential (partially) redundant role (with CK1α and CK1ε) in PCP signaling as suggested earlier (Davidson et al. 2005; Zeng et al. 2005; Klein et al. 2006; Strutt et al. 2006).
Figure 9 .
CG6963/CK1γ affects rotation in the developing eye and interacts with stan/fmi and nemo/Nlk. (A–C and E–F) Tangential eye sections in top panels and the respective schematic representation of ommatidial orientation is shown at bottom (arrows are as in Figures 1 and 5). All crosses were performed at 25°. (D–D′′) Eye imaginal disc containing a gishe01759 clone (marked by absence of GFP) stained for Elav (blue; marking all photoreceptor precursors) and Arm (red in D monochrome in D′, highlighting cellular architecture by labeling the adherens junctions). D′′ shows a schematic of rotation angles with the wild-type area indicated by green. Note several severely underrotated clusters marked by asterisks. Eye sections of the following genotypes are shown: (A) sev-GAL4/+, CG6963IR/+, (B) sev-Fmi/+, (B) sev-Fmi/+; sev-GAL4/+, CG6963IR/+. Note enhanced rotation defects in C; for quantification see Figure S3B. (E) sev-GAL4/+, UAS-Nmo/+, (F) sev-GAL4/+, UAS-Nmo/+, CG6963IR/+ (note enhancement of rotation defects as compared to E; for quantification see Figure S3B), (G) sev-GAL4/+, CG6963IR/+ nmoDB/+, which is suppressed (compare to A).
To get more insight into the potential role of gish in ommatidial rotation, we (1) wished to confirm and refine the rotation phenotype in third instar imaginal discs during the actual process by analyzing a gish LOF allele and (2) tested for genetic interactions with other genes involved in ommatidial rotation. First, analyses of LOF clones in eye imaginal discs revealed that many clusters are underrotated in gish mutant tissue (Figure 9, D–D′′).
Second, we asked whether a gish knockdown can affect other genes involved in ommatidial rotation. Among the core PCP factors, RNAi-mediated gish knockdown enhanced sev-Fmi (Figure 9, B and C; quantification in Figure S3B) and sevG4, UAS-Fz (not shown); in both cases the enhancement was specific to the rotation defects observed. This is consistent with the interaction detected in the original screen genotype (a GOF dgo background) and it suggested a specific role for gish in ommatidial rotation. Next we asked whether any of the “rotation-specific PCP genes,” including nemo (nmo, Nlk in mammals) (Choi and Benzer 1994; Fiehler and Wolff 2008; Mirkovic et al. 2011), Rho-kinase (dRok in Drosophila) (Winter et al. 2001), or zipper (zip, myosin II) (Fiehler and Wolff 2007), display a specific interaction with gish in this process. Of these, nmo displayed an antagonistic interaction with gish; whereas gishIR knockdown enhanced the sevGAL4, UAS-Nmo phenotype (Figure 9, E–F), the respective sevGAL4, UAS-gishIR knockdown phenotype was suppressed by dosage reduction of nmo (nmoDB/+; Figure 9G, see also Figure S3B for quantification). These data suggest that Gish/CK1γ acts in opposition to Nmo/Nlk and that a fine balance between the activities of these two kinases is required for normal ommatidial rotation to occur.
Discussion
Here we described a genetic screen to identify novel modifiers and regulatory factors linked to Fz/PCP signaling. We have used two mild GOF backgrounds of the cytosolic core components Dgo and Pk as screening tools, because they act antagonistically within the core Fz/PCP cassette. The screen relied on dosage sensitive interactions, a frequently used feature of mild PCP overexpression/GOF phenotypes (Strutt et al. 1997; Boutros et al. 1998; Strutt and Strutt 2003), and we used a combination of deficiency collections and transgenic RNAi strains to identify new PCP regulators.
We first tested the genotypes of interest in a pilot screen with known PCP factors, other associated signaling pathway components (e.g., Dl), and negative controls. The positive candidates from the pilot screen indicated that the phenotypes were well within the appropriate sensitivity range for a dosage-dependent screen. In particular, the fact that stbm/Vang enhanced the dgo GOF phenotype supports previous studies and the existing model that Dgo promotes Fz–Dsh signaling activity, whereas stbm/Vang antagonizes it (Jenny et al. 2005). The fact that stan/+ suppressed the dgo GOF phenotype is novel, but consistent with existing models, as Stan/Fmi is thought to contribute to anchoring Dgo at the membrane as part of the Fz–Dsh complex (Feiguin et al. 2001; Das et al. 2004; Strutt and Strutt 2007; Wu et al. 2008).
Our genome-wide screen has identified a set of genes with a broad range of functions, some of which were known to affect PCP (ds and Dl, see above for references) and a group of novel genes in the PCP context (see below). Generally, most of the genes identified were “hits” associated with dgo, and several of these not only modified the dgo GOF phenotype, but were also enhanced by dgo heterozygosity when knocked down via RNAi (Figure 7). We trust that all the genes identified via RNAi are specific (and not due to potential off-target effects), as they largely reproduced the genetic interaction(s) seen with the deficiencies (Table 2). As we narrowed down the genomic areas responsible for the genetic interactions, we sometimes observed that the quality of a genetic effect changed. This could be due to separating several independent interacting genes that were initially causing an “additive effect” within the larger deficiencies or by separating genomic regulatory sequences that modify effects (Figure 4A, Table 2, and Figure S1).
Through the design of the screen we not only isolated dosage-sensitive interactors, but our approach directly led to the identification of genes that display PCP phenotypes in a loss-of-function (knockdown) scenario. As such, every new gene isolated is indeed required for PCP establishment and most of them act in eye and wing tissue, suggesting a general requirement. There might be tissue-specific hits in our screen (Table 1), which have not yet been characterized.
Besides pk, we were not able to identify other core Fz/PCP genes in the deficiency screen that showed a dose-dependent interaction in the pilot screen (Vang/stbm and stan/fmi). A possible explanation is that a single deficiency can harbor genes that act antagonistically and thus neutralize each other in a modification assay, e.g., fmi/stan resides near other genes that can affect PCP (e.g., dgo and pipsqueak/psq; Weber et al. 1995; Feiguin et al. 2001; Weber et al. 2008) and no interaction was seen with deficiency Df(2R)ED2098, which removes fmi/stan. Several of the newly identified genes fall into interesting functional categories that are either novel within the PCP regulatory machinery or may provide insight into new regulatory mechanisms. These include for example lipid modifications, factors associated with mammalian ciliopathies, and, presumably, protein stability/trafficking via ubiquitination (see below).
Functional features of new PCP regulators
The types of new genes recovered in the screen fall into several categories expanding the biochemical functions associated with PCP establishment in Drosophila. Three of the new genes, CG11426, CG11438, and fwd, encode lipid modification enzymes, with fwd encoding a PI4Kinaseß (Polevoy et al. 2009) and the other two being PAP2-type phosphatases (Starz-Gaiano et al. 2001). These are intriguing new factors as lipid kinases and phosphatases could modulate the composition of the lipid microenvironment of the core PCP complexes and thus regulate their trafficking and/or stability at the plasma membrane among others (Weber et al. 2003; Simons et al. 2009). An aspect of lipid modification affecting core Fz/PCP signaling has recently been shown to be important for the stabilization of Dsh membrane association and thus stabilization of the Fz–Dsh complex (Simons et al. 2009). As the phosphorylation state of the lipid heads often contributes to signaling by serving as protein binding sites, the identification of two lipid phosphatases and a lipid kinase suggests that some core PCP factors may prefer specific phosphorylation states of lipids, for either direct lipid binding or stabilization of the associated complexes.
A second functionally linked group consists of CG31687 and CG15283. CG31687 encodes a Drosophila APC8 paralog (cdc23), a component of the anaphase promoting complex/cyclosome (APC/C) (reviewed in Peters 2006). Strikingly, CG15283 genetically also interacted not only with core PCP factors but also with CG31687, thus forming a potential pair. While it has been suggested that ubiquitin-mediated modifications can regulate PCP in mice, with Smurf1/2 leading to Pk ubiquitination (Narimatsu et al. 2009), a ubiquitin link has not yet been made in Drosophila. The potential involvement of the APC/C in noncell-cycle–associated cellular aspects like cell polarity (Peters 2002) is intriguing. In addition to its interaction with CG31687, CG15283 appears to be an interesting factor in its own right. Its phenotypic and genetic interactions with dgo and dsh (Figures 7 and 8) suggest that it functions in either modifying Dsh activity or affecting the balance between Dgo and Pk, which promote and antagonize Dsh function, respectively. Structure–function studies will be needed to reveal the molecular features of CG15283.
Three isolated genes fall into the category of scaffold proteins, CG1019 (Mlp1, a LIM domain-containing protein), CG11146 (an SH2–SH3 domain-containing factor), and CG13388 (Drosophila Akap200). Scaffold proteins are common factors in protein complexes and thus the formation or stabilization of a PCP-specific complex or an associated complex is likely to require other such proteins. Akap200, although by name a PKA-associated factor in some contexts, also has non–PKA-mediated functions (Jackson and Berg 2002) and thus could well be involved in Fz/PCP signaling.
CG6963 is Drosophila CK1γ, called gilgamesh (gish) in flies. Members of the casein kinase 1 family have been implicated in several aspects of canonical Wnt signaling (Davidson et al. 2005; Zeng et al. 2005; Klein et al. 2006; Strutt et al. 2006) as well as in Fz/PCP regulation, in particular CK1ε (Klein et al. 2006; Strutt et al. 2006) and CK1α (Strutt et al. 2006). CK1 family members have been shown to phosphorylate Dsh (Klein et al. 2006; Strutt et al. 2006), and gish might act (at least) partially in a redundant manner in Dsh (core Fz/PCP) regulation and hence was identified in the screen. In addition, we observed two specific PCP-associated phenotypes with CG6963/gish knockdown, which were also confirmed with LOF clones of existing mutant alleles. First, we detect a function in ommatidial rotation and ommatidial clusters appear to rotate less in gish mutant tissue. This defect is accompanied by genetic interactions with Fz and Stan/Fmi among the core PCP genes. Furthermore, gish interacts with nemo (Mirkovic et al. 2011) in this context in a specific manner: Gish/CK1γ and Nemo act in opposition to each other (Figure 9), suggesting that a fine balance between the activities of these two kinases is required for normal ommatidial rotation to occur. Second, gish/CG6963 knockdown displays a high frequency of multiple cellular hairs, which is independent of a direct interaction with the core PCP factors (Gault et al. 2012).
The identification of CG15730 with a connection to PCP signaling is intriguing, as it encodes the Drosophila homolog of the Mks1 gene, a member of the Meckel-Gruber syndrome factors that are generally linked to ciliopathies in humans (Simons and Mlodzik 2008). PCP establishment in vertebrates has been linked to cilia positioning and function in several contexts (e.g., Wallingford 2006; Park et al. 2008; Borovina et al. 2010). As epithelial cells in Drosophila are not ciliated, our observation that dMks1/CG15730 interacts with core Fz/PCP factors and displays PCP defects by itself when knocked down suggests that cilia-associated factors are likely to be generally required for aspects of PCP establishment unrelated to ciliogenesis. As most of the genes linked to ciliopathies (e.g., those associated with the Meckel-Gruber and Bardett-Biedl syndromes) are conserved in Drosophila, this observation suggests that analyses of other such genes in PCP establishment in Drosophila are warranted. CG10068, a novel protein of unknown function, has been linked to cytokinesis (Echard et al. 2004) and could thus also be interacting with cytoskeletal elements or associated proteins.
Supplementary Material
Acknowledgments
We thank K. Basler, C. Desplan, K. Gaengel, D. Gubb, N. Paricio, F. Pichaud, and the Nippon Institute of Genetics and Bloomington Stock Centers for fly strains, and Sophy Okello for technical help. We are grateful to Mei-Ling Chin and Lindsay Kelly for critical reading of the manuscript and all Mlodzik lab members for helpful comments throughout the course of this work. W. J. Gault was in part supported by T32 grants GM08553 and CA78207; P. Olguin was supported by a Pew Latin American Fellows postdoctoral fellowship. This work was supported by National Institutes off Health RO1 grant GM62917 to M. Mlodzik.
Footnotes
Communicating editor: T. Schupbach
Literature Cited
- Adler P. N., 2002. Planar signaling and morphogenesis in Drosophila. Dev. Cell 2: 525–535. [DOI] [PubMed] [Google Scholar]
- Basler K., Siegrist P., Hafen E., 1989. The spatial and temporal expression pattern of sevenless is exclusively controlled by gene-internal elements. EMBO J. 8: 2381–2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayly R., Axelrod J. D., 2011. Pointing in the right direction: new developments in the field of planar cell polarity. Nat. Rev. Genet. 12: 385–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borovina A., Superina S., Voskas D., Ciruna B., 2010. Vangl2 directs the posterior tilting and asymmetric localization of motile primary cilia. Nat. Cell Biol. 12: 407–412. [DOI] [PubMed] [Google Scholar]
- Boutros M., Mlodzik M., 1999. Dishevelled: at the crossroads of divergent intracellular signaling pathways. Mech. Dev. 83: 27–37. [DOI] [PubMed] [Google Scholar]
- Boutros M., Paricio N., Strutt D. I., Mlodzik M., 1998. Dishevelled activates JNK and discriminates between JNK pathways in planar polarity and wingless signaling. Cell 94: 109–118. [DOI] [PubMed] [Google Scholar]
- Brand A. H., Perrimon N., 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401–415. [DOI] [PubMed] [Google Scholar]
- Casal J., Lawrence P. A., Struhl G., 2006. Two separate molecular systems, Dachsous/Fat and Starry night/Frizzled, act independently to confer planar cell polarity. Development 133: 4561–4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W. S., Antic D., Matis M., Logan C. Y., Povelones M., et al. , 2008. Asymmetric homotypic interactions of the atypical cadherin flamingo mediate intercellular polarity signaling. Cell 133: 1093–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K.-W., Benzer S., 1994. Rotation of photoreceptor clusters in the developing Drosophila eye requires the nemo gene. Cell 78: 125–136. [DOI] [PubMed] [Google Scholar]
- Clark K. A., Bland J. M., Beckerle M. C., 2007. The Drosophila muscle LIM protein, Mlp84B, cooperates with D-titin to maintain muscle structural integrity. J. Cell Sci. 120: 2066–2077. [DOI] [PubMed] [Google Scholar]
- Cooper M. T. D., Bray S. J., 1999. Frizzled regulation of Notch signalling polarizes cell fate in the Drosophila eye. Nature 397: 526–529. [DOI] [PubMed] [Google Scholar]
- Courbard J. R., Djiane A., Wu J., Mlodzik M., 2009. The apical/basal-polarity determinant Scribble cooperates with the PCP core factor Stbm/Vang and functions as one of its effectors. Dev. Biol. 333: 67–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das G., Reynolds-Kenneally J., Mlodzik M., 2002. The Atypical Cadherin Flamingo Links Frizzled and Notch Signaling in Planar Polarity Establishment in the Drosophila Eye. Dev. Cell 2: 656–666. [DOI] [PubMed] [Google Scholar]
- Das G., Jenny A., Klein T. J., Eaton S., Mlodzik M., 2004. Diego interacts with Prickle and Strabismus/Van Gogh to localize planar cell polarity complexes. Development 131: 4467–4476. [DOI] [PubMed] [Google Scholar]
- Davidson G., Wu W., Shen J., Bilic J., Fenger U., et al. , 2005. Casein kinase 1 gamma couples Wnt receptor activation to cytoplasmic signal transduction. Nature 438: 867–872. [DOI] [PubMed] [Google Scholar]
- Dietzl G., Chen D., Schnorrer F., Su K. C., Barinova Y., et al. , 2007. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448: 151–156. [DOI] [PubMed] [Google Scholar]
- Djiane A., Yogev S., Mlodzik M., 2005. The apical determinants aPKC and dPatj regulate Frizzled-dependent planar cell polarity in the Drosophila eye. Cell 121: 621–631. [DOI] [PubMed] [Google Scholar]
- Donoughe S., DiNardo S., 2011. dachsous and frizzled contribute separately to planar polarity in the Drosophila ventral epidermis. Development 138: 2751–2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy J. B., 2002. GAL4 system in Drosophila: a fly geneticist’s Swiss army knife. Genesis 34: 1–15. [DOI] [PubMed] [Google Scholar]
- Echard A., Hickson G. R., Foley E., O’Farrell P. H., 2004. Terminal cytokinesis events uncovered after an RNAi screen. Curr. Biol. 14: 1685–1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanto M., Mlodzik M., 1999. Asymmetric Notch activation specifies photoreceptors R3 and R4 and planar polarity in the Drosophila eye. Nature 397: 523–526. [DOI] [PubMed] [Google Scholar]
- Feiguin F., Hannus M., Mlodzik M., Eaton S., 2001. The Ankyrin repeat protein Diego mediates Frizzled-dependent planar polarization. Dev. Cell 1: 93–101. [DOI] [PubMed] [Google Scholar]
- Fiehler R. W., Wolff T., 2007. Drosophila Myosin II, Zipper, is essential for ommatidial rotation. Dev. Biol. 310: 348–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiehler R. W., Wolff T., 2008. Nemo is required in a subset of photoreceptors to regulate the speed of ommatidial rotation. Dev. Biol. 313: 533–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gault W. J., Olguin P., Weber U., Mlodzik M., 2012. Drosophila CK1γ, gilgamesh, Controls PCP-mediated Morphogenesis Through Regulation of Vesicle Trafficking. J. Cell Biol. 196: 605–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubb D., Green C., Huen D., Coulson D., Johnson G., et al. , 1999. The balance between isoforms of the prickle LIM domain protein is critical for planar polarity in Drosophila imaginal discs. Genes Dev. 13: 2315–2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummel T., Attix S., Gunning D., Zipursky S. L., 2002. Temporal control of glial cell migration in the Drosophila eye requires gilgamesh, hedgehog, and eye specification genes. Neuron 33: 193–203. [DOI] [PubMed] [Google Scholar]
- Jackson S. M., Berg C. A., 2002. An A-kinase anchoring protein is required for protein kinase A regulatory subunit localization and morphology of actin structures during oogenesis in Drosophila. Development 129: 4423–4433. [DOI] [PubMed] [Google Scholar]
- Jenny A., Reynolds-Kenneally J., Das G., Burnett M., Mlodzik M., 2005. Diego and Prickle regulate Frizzled planar cell polarity signalling by competing for Dishevelled binding. Nat. Cell Biol. 7: 691–697. [DOI] [PubMed] [Google Scholar]
- Klein T. J., Mlodzik M., 2005. Planar cell polarization: an emerging model points in the right direction. Annu. Rev. Cell Dev. Biol. 21: 155–176. [DOI] [PubMed] [Google Scholar]
- Klein T. J., Jenny A., Djiane A., Mlodzik M., 2006. CKIepsilon/discs overgrown promotes both Wnt-Fz/beta-catenin and Fz/PCP signaling in Drosophila. Curr. Biol. 16: 1337–1343. [DOI] [PubMed] [Google Scholar]
- Lawrence P. A., Casal J., Struhl G., 2004. Cell interactions and planar polarity in the abdominal epidermis of Drosophila. Development 131: 4651–4664. [DOI] [PubMed] [Google Scholar]
- Lawrence P. A., Struhl G., Casal J., 2007. Planar cell polarity: One or two pathways? Nat. Rev. Genet. 8: 555–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matakatsu H., Blair S. S., 2004. Interactions between Fat and Dachsous and the regulation of planar cell polarity in the Drosophila wing. Development 131: 3785–3794. [DOI] [PubMed] [Google Scholar]
- Mirkovic I., Gault W. J., Rahnama M., Jenny A., Gaengel K., et al. , 2011. Nemo kinase phosphorylates beta-catenin to promote ommatidial rotation and connects core PCP factors to E-cadherin-beta-catenin. Nat. Struct. Mol. Biol. 18: 132–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlodzik M., 1999. Planar polarity in the Drosophila eye: a multifaceted view of signaling specificity and cross-talk. EMBO J. 18: 6873–6879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narimatsu M., Bose R., Pye M., Zhang L., Miller B., et al. , 2009. Regulation of planar cell polarity by Smurf ubiquitin ligases. Cell 137: 295–307. [DOI] [PubMed] [Google Scholar]
- Nerusheva O. O., Dorogova N. V., Gubanova N. V., Yudina O. S., Omelyanchuk L. V., 2009. A GFP trap study uncovers the functions of Gilgamesh protein kinase in Drosophila melanogaster spermatogenesis. Cell Biol. Int. 33: 586–593. [DOI] [PubMed] [Google Scholar]
- Park T. J., Mitchell B. J., Abitua P. B., Kintner C., Wallingford J. B., 2008. Dishevelled controls apical docking and planar polarization of basal bodies in ciliated epithelial cells. Nat. Genet. 40: 871–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks A. L., Cook K. R., Belvin M., Dompe N. A., Fawcett R., et al. , 2004. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat. Genet. 36: 288–292. [DOI] [PubMed] [Google Scholar]
- Peters J. M., 2002. The anaphase-promoting complex: proteolysis in mitosis and beyond. Mol. Cell 9: 931–943. [DOI] [PubMed] [Google Scholar]
- Peters J. M., 2006. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat. Rev. Mol. Cell Biol. 7: 644–656. [DOI] [PubMed] [Google Scholar]
- Pichaud F., Desplan C., 2001. A new visualization approach for identifying mutations that affect differentiation and organization of the Drosophila ommatidia. Development 128: 815–826. [DOI] [PubMed] [Google Scholar]
- Polevoy G., Wei H. C., Wong R., Szentpetery Z., Kim Y. J., et al. , 2009. Dual roles for the Drosophila PI 4-kinase four wheel drive in localizing Rab11 during cytokinesis. J. Cell Biol. 187: 847–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawls A. S., Wolff T., 2003. Strabismus requires Flamingo and Prickle function to regulate tissue polarity in the Drosophila eye. Development 130: 1877–1887. [DOI] [PubMed] [Google Scholar]
- Ryder E., Ashburner M., Bautista-Llacer R., Drummond J., Webster J., et al. , 2007. The DrosDel deletion collection: a Drosophila genomewide chromosomal deficiency resource. Genetics 177: 615–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahai E., Alberts A. S., Treisman R., 1998. RhoA effector mutants reveal distinct effector pathways for cytoskeletal reorganization, SRF activation and transformation. EMBO J. 17: 1350–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert J. R., Mlodzik M., 2007. Frizzled/PCP signalling: a conserved mechanism regulating cell polarity and directed motility. Nat. Rev. Genet. 8: 126–138. [DOI] [PubMed] [Google Scholar]
- Simon M. A., Xu A., Ishikawa H. O., Irvine K. D., 2010. Modulation of fat:dachsous binding by the cadherin domain kinase four-jointed. Curr. Biol. 20: 811–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons M., Mlodzik M., 2008. Planar cell polarity signaling: from fly development to human disease. Annu. Rev. Genet. 42: 517–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons M., Gault W. J., Gotthardt D., Rohatgi R., Klein T. J., et al. , 2009. Electrochemical cues regulate assembly of the Frizzled/Dishevelled complex at the plasma membrane during planar epithelial polarization. Nat. Cell Biol. 11: 286–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starz-Gaiano M., Cho N. K., Forbes A., Lehmann R., 2001. Spatially restricted activity of a Drosophila lipid phosphatase guides migrating germ cells. Development 128: 983–991. [DOI] [PubMed] [Google Scholar]
- Strutt D., 2003. Frizzled signalling and cell polarisation in Drosophila and vertebrates. Development 130: 4501–4513. [DOI] [PubMed] [Google Scholar]
- Strutt D., Strutt H., 2007. Differential activities of the core planar polarity proteins during Drosophila wing patterning. Dev. Biol. 302: 181–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strutt D. I., Weber U., Mlodzik M., 1997. The role of RhoA in tissue polarity and Frizzled signalling. Nature 387: 292–295. [DOI] [PubMed] [Google Scholar]
- Strutt H., Strutt D., 1999. Polarity determination in the Drosophila eye. Curr. Opin. Genet. Dev. 9: 442–446. [DOI] [PubMed] [Google Scholar]
- Strutt H., Strutt D., 2003. EGF signaling and ommatidial rotation in the Drosophila eye. Curr. Biol. 13: 1451–1457. [DOI] [PubMed] [Google Scholar]
- Strutt H., Strutt D., 2008. Differential stability of flamingo protein complexes underlies the establishment of planar polarity. Curr. Biol. 18: 1555–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strutt H., Strutt D., 2009. Asymmetric localisation of planar polarity proteins: mechanisms and consequences. Semin. Cell Dev. Biol. 20: 957–963. [DOI] [PubMed] [Google Scholar]
- Strutt H., Price M. A., Strutt D., 2006. Planar polarity is positively regulated by casein kinase Iepsilon in Drosophila. Curr. Biol. 16: 1329–1336. [DOI] [PubMed] [Google Scholar]
- Tomlinson A., Ready D. F., 1987. Cell fate in the Drosophila Ommatidium. Dev. Biol. 123: 264–275. [DOI] [PubMed] [Google Scholar]
- Tomlinson A., Struhl G., 1999. Decoding vectorial information from a gradient: sequential roles of the receptors Frizzled and Notch in establishing planar polarity in the Drosophila eye. Development 126: 5725–5738. [DOI] [PubMed] [Google Scholar]
- Tree D. R., Shulman J. M., Rousset R., Scott M. P., Gubb D., et al. , 2002. Prickle mediates feedback amplification to generate asymmetric planar cell polarity signaling. Cell 109: 371–381. [DOI] [PubMed] [Google Scholar]
- Usui T., Shima Y., Shimada Y., Hirano S., Burgess R. W., et al. , 1999. Flamingo, a seven-pass transmembrane cadherin, regulates planar cell polarity under the control of Frizzled. Cell 98: 585–595. [DOI] [PubMed] [Google Scholar]
- Wallingford J. B., 2006. Planar cell polarity, ciliogenesis and neural tube defects. Hum. Mol. Genet. 15(Spec No 2): R227–R234. [DOI] [PubMed] [Google Scholar]
- Wallingford J. B., Habas R., 2005. The developmental biology of Dishevelled: an enigmatic protein governing cell fate and cell polarity. Development 132: 4421–4436. [DOI] [PubMed] [Google Scholar]
- Wang Y., Nathans J., 2007. Tissue/planar cell polarity in vertebrates: new insights and new questions. Development 134: 647–658. [DOI] [PubMed] [Google Scholar]
- Weber U., Siegel V., Mlodzik M., 1995. pipsqueak encodes a novel nuclear protein required downstream of seven-up for the development of photoreceptors R3 and R4. EMBO J. 14: 6247–6257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber U., Eroglu C., Mlodzik M., 2003. Phospholipid membrane composition affects EGF receptor and Notch signaling through effects on endocytosis during Drosophila development. Dev. Cell 5: 559–570. [DOI] [PubMed] [Google Scholar]
- Weber U., Pataki C., Mihaly J., Mlodzik M., 2008. Combinatorial signaling by the Frizzled/PCP and Egfr pathways during planar cell polarity establishment in the Drosophila eye. Dev. Biol. 316: 110–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter C. G., Wang B., Ballew A., Royou A., Karess R., et al. , 2001. Drosophila Rho-associated kinase (Drok) links Frizzled-mediated planar cell polarity signaling to the actin cytoskeleton. Cell 105: 81–91. [DOI] [PubMed] [Google Scholar]
- Wu J., Mlodzik M., 2009. A quest for the mechanism regulating global planar cell polarity of tissues. Trends Cell Biol. 19: 295–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Klein T. J., Mlodzik M., 2004. Subcellular localization of frizzled receptors, mediated by their cytoplasmic tails, regulates signaling pathway specificity. PLoS Biol. 2: 1004–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Jenny A., Mirkovic I., Mlodzik M., 2008. Frizzled-Dishevelled signaling specificity outcome can be modulated by Diego in Drosophila. Mech. Dev. 125: 30–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X., Tamai K., Doble B., Li S., Huang H., et al. , 2005. A dual-kinase mechanism for Wnt co-receptor phosphorylation and activation. Nature 438: 873–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.