Abstract
Bone mineral density (BMD) is the most important predictor of fracture risk. We performed the largest meta-analysis to date on lumbar spine and femoral neck BMD, including 17 genome-wide association studies and 32,961 individuals of European and East Asian ancestry. We tested the top-associated BMD markers for replication in 50,933 independent subjects and for risk of low-trauma fracture in 31,016 cases and 102,444 controls. We identified 56 loci (32 novel)associated with BMD atgenome-wide significant level (P<5×10−8). Several of these factors cluster within the RANK-RANKL-OPG, mesenchymal-stem-cell differentiation, endochondral ossification and the Wnt signalling pathways. However, we also discovered loci containing genes not known to play a role in bone biology. Fourteen BMD loci were also associated with fracture risk (P<5×10−4, Bonferroni corrected), of which six reached P<5×10−8 including: 18p11.21 (C18orf19), 7q21.3 (SLC25A13), 11q13.2 (LRP5), 4q22.1 (MEPE), 2p16.2 (SPTBN1) and 10q21.1 (DKK1). These findings shed light on the genetic architecture and pathophysiological mechanisms underlying BMD variation and fracture susceptibility.
Osteoporosis is a disease characterized by low bone mass and microarchitectural deterioration of bone tissue leading to increased risk of fracture. The disease accounts for approximately 1.5 million new fracture cases each year representing a huge economic burden on health care systems, with annual costs estimated to be $17 billion in the USA alone and expected to rise 50% by the year 2025.1 Osteoporosis is defined clinically through the measurement of bone mineral density (BMD), which remains the single best predictor of fracture.2,3
Twin and family studies have shown that 50%–85% of the variance in BMD is genetically determined.4 Osteoporotic fractures are also heritable by mechanisms that are partly independent of BMD.5 Over the past 5 years, genome-wide association studies (GWAS) have revolutionized the understanding of the genetic architecture of common, complex diseases.6 This strategy is providing key insights into the mechanisms of disease with prospects of designing effective strategies for risk assessment and development of new interventions.7
Previous GWAS have identified to-date 24 loci which influence BMD variation.8–14 While several variants in these BMD loci have also been nominally-associated with fracture risk15,16, none have shown robust association at genome-wide significant levels (P<5×10−8). We report here the results of the largest effort to date searching for BMD loci in >80,000 subjects and testing them for association with fracture in >130,000 cases and controls. In addition, we employed bioinformatics tools and gene expression analyses to place the identified variants in the context of pathways relevant to bone biology.
Results
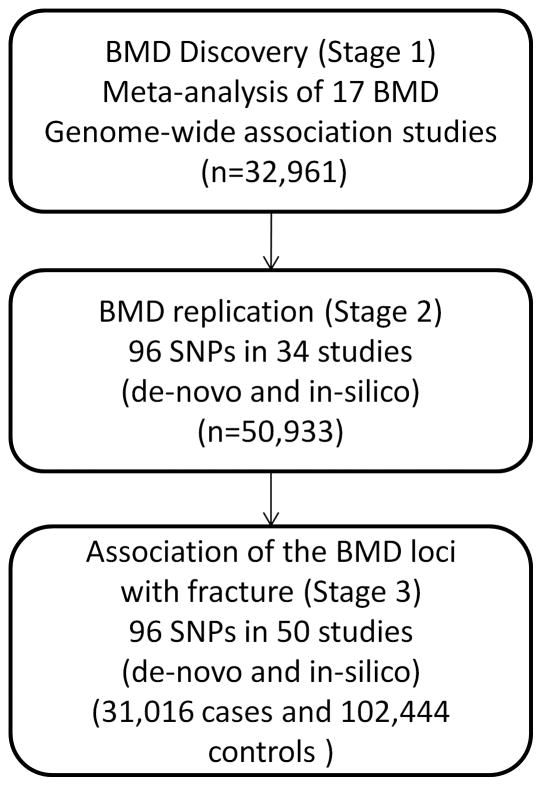
This study was performed across three main stages (Fig. 1): 1) discovery of BMD loci, 2) follow-up replication and 3) association of the BMD loci with fracture.
Figure 1. Description of Study design.
Stage 1: Meta-analysis of 17 Genome-wide association studies for BMD. Stage 2: 96 top independent SNPs (82 autosomal SNPs with P<5×10−6 + 5 X-chromosome SNPs + 9 SNPs from conditional analysis) were followed-up for de-novo and in-silico replication of the BMD association in 34 studies. Stage 3: the same 96 SNPs were tested for association with fracture in 50 studies with de-novo and in-silico data.
Discovery of BMD Loci (Stage 1)
We first performed a meta-analysis of GWAS for BMD of the femoral neck (FN-BMD; n=32,961) and lumbar spine (LS-BMD; n=31,800) including ~2.5 million autosomal genotyped or imputed SNPs from 17 studies from populations across North America, Europe, East Asia and Australia, with a variety of epidemiological designs and patient characteristics (see Online Methods). We also performed meta-analysis in men and women separately to identify sex-specific associations. The quantile-quantile (Q-Q) plots of the discovery meta-analysis displayed strong (and not early) deviation of the observed statistics from the null distribution for both BMD traits (Supplementary Fig. 1). After double Genomic Control (GC) correction of the overall (λFN_POOLED=1.112; λLS_POOLED=1.127) and sex-stratified analyses (λFN_WOMEN=1.091; λFN_MEN=1.059; λLS_WOMEN=1.086; λLS_MEN=1.061), SNPs in 34 loci surpassed GWS level while a total of 82 loci were associated at P<5×10−6 (Supplementary Fig. 2–3). Thirty eight loci were associated with FN-BMD, 25 with LS-BMD and 19 with both. The overlap reflects the correlation between the femoral neck and lumbar spine measurements (Pearson correlation = 0.53). Of these 82 loci, 59, 18 and 5 were prioritized from the analysis in the sex-combined, women and men sample sets, respectively (Supplementary Table 1). The meta-analysis was extended to include the evaluation of 76,253 X-chromosome imputed markers across 14 of the discovery GWAS including 31,801 participants (see Online Methods). Five X-chromosome loci were associated at P<5×10−5 four of which were derived from the sex-combined analysis and one from the analysis in men only (Supplementary Table 1). We further performed genome-wide conditional analyses in all sex-combined stage 1 studies. Each study repeated the GWAS analysis but additionally adjusted for 82 SNPs representing the autosomal loci associated at P<5×10−6 (see Online Methods). We then meta-analyzed these studies in the same way as for the primary GWA study meta-analysis. Nine loci showed at least two independent association signals arising from this conditional analysis (Supplementary Fig. 4 and Supplementary Table 2) suggesting that allelic heterogeneity is underlying BMD variation. We also assessed all possible pairwise interactions of the 82 SNPs, but none were significant after adjusting for the number of tests (Supplementary Fig. 5 and Supplementary Table 3). A total of 96 independent SNPs (82 autosomal SNPs with P<5×10−6 + 9 SNPs from conditional analysis and 5 X-chromosome SNPS) from 87 genomic loci were selected for further replication (Fig. 1).
Follow-up replication (Stage 2)
We de-novo genotyped these 96 SNPs and tested them for association with BMD in up to 50,933 additional participants from 34 studies (see Online Methods). The meta-analysis of the 96 SNPs in the discovery and replication studies (n=83,894) yielded 64 replicating SNPs from 56 associated loci. Of these loci, 32 were novel (Table 1 and Supplementary Table 4A) and 24 were reported previously8–14 (Supplementary Table 4B). Thirty two SNPs did not reach genome-wide significance after replication (Supplementary Table 4C) and included 10 markers remaining associated at a suggestive level. Of all analyzed SNPs only one (rs9533090 mapping to 13q14.11 near RANKL) displayed high degree of heterogeneity of effects (I2>50%) across studies, despite being the marker with highest significance (P=4.82×10−68) in the fixed-effect meta-analysis (Supplementary Table 4B). After applying random effects meta-analysis, this marker was still genome-wide significant (P=3.98×10−13).
Table 1.
Estimated effects of novel genome-wide significant SNPs on bone mineral density of femoral neck (FN-) BMD and lumbar spine (LS-) BMD across stages.
| SNP | Locus | Closest Gene/Candidate | Functional evidence | Aa | Freqb | FN-BMD
|
LSBMD
|
Phet Sited | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| eQTL | KO mouse | OMIM | Tags funct. | GRAIL | Pathway | STAGE 1 (32,961) | STAGE 2 (50,933) | STAGE 1 + STAGE 2 (83,894) | STAGE 1 (31,800) | STAGE 2 (45,708) | STAGE 1 + STAGE 2 (77,508) | ||||||||
|
|
|
||||||||||||||||||
| Betac | P | P | P | Betac | P | P | P | ||||||||||||
| rs479336 | 1q24.3 | DNM3 | T | 0.74 | −0.04 | 1.1×10−7 | 1.3×10−8 | 8.5×10−15 | −0.03 | 0.01 | 5.0×10−4 | 2.1×10−5 | 0.05 | ||||||
| rs7584262 | 2p21 | PKDCC | • | T | 0.23 | 0.03 | 1.4×10−7 | 3.4×10−4 | 1.3×10−9 | 0.01 | 0.13 | 0.28 | 0.07 | 0.01 | |||||
| rs17040773 | 2q13 | ANAPC1 | • | A | 0.76 | 0.03 | 4.3×10−6 | 6.1×10−5 | 1.5×10−9 | 0.01 | 0.61 | 0.21 | 0.19 | 5.2×10−3 | |||||
| rs1878526 | 2q14.2 | INSIG2 | A | 0.22 | 0.00 | 0.70 | 0.97 | 0.79 | 0.04 | 7.3×10−6 | 3.4×10−6 | 1.2×10−10 | 8.6×10−5 | ||||||
| rs1026364 | 3q13.2 | KIAA2018 | • | T | 0.37 | 0.03 | 2.0×10−6 | 2.5×10−5 | 4.1×10−10 | 0.02 | 0.04 | 7.3×10−3 | 7.6×10−4 | 0.11 | |||||
| rs344081 | 3q25.31 | LEKR1 | T | 0.87 | 0.03 | 1.1×10−4 | 2.5×10−3 | 2.2×10−6 | 0.06 | 2.8×10−5 | 3.5×10−8 | 4.5×10−12 | 0.12 | ||||||
| rs3755955 | 4p16.3 | IDUA | • | • | A | 0.16 | −0.05 | 3.9×10−7 | 6.1×10−9 | 1.5×10−14 | −0.05 | 1.4×10−7 | 5.5×10−9 | 5.2×10−15 | 0.80 | ||||
| rs11755164 | 6p21.1 | SUPT3H/RUNX2 | • | • | • | T | 0.40 | −0.01 | 0.23 | 0.12 | 0.05 | −0.03 | 3.5×10−7 | 9.2×10−6 | 5.6×10−11 | 2.1×10−3 | |||
| rs9466056 | 6p22.3 | CDKAL1/SOX4 | A | 0.38 | −0.03 | 1.8×10−8 | 1.6×10−6 | 2.7×10−13 | −0.03 | 6.5×10−5 | 1.1×10−4 | 3.6×10−8 | 0.34 | ||||||
| rs3801387 | 7q31.31 | WNT16 | • | • | • | A | 0.74 | −0.08 | 4.2×10−14 | 2.0×10−27 | 5.0×10−40 | −0.10 | 1.4×10−16 | 1.5×10−36 | 3.2×10−51 | 0.09 | |||
| rs13245690 g | 7q31.31 | C7orf58 | A | 0.62 | 0.00 | 8.6×10−5 | 0.69 | 8.2×10−4 | 0.03 | 1.1×10−9 | 1.3×10−3 | 6.0×10−11 | 0.05 | ||||||
| rs7812088 | 7q36.1 | ABCF2 | A | 0.13 | 0.04 | 1.2×10−6 | 4.4×10−4 | 7.3×10−9 | 0.04 | 2.9×10−5 | 1.1×10−3 | 2.2×10−7 | 0.86 | ||||||
| rs7017914e | 8q13.3 | XKR9/LACTB2 | • | • | A | 0.49 | 0.02 | 4.7×10−8 | 7.1×10−3 | 1.9×10−8 | −0.01 | 0.35 | 0.41 | 0.98 | 9.1×10−5 | ||||
| rs7851693 | 9q34.11 | FUBP3 | C | 0.64 | 0.05 | 3.1×10−8 | 1.4×10−15 | 3.4×10−22 | 0.04 | 0.06 | 6.7×10−8 | 6.1×10−8 | 0.02 | ||||||
| rs3905706 | 10p11.23 | MPP7 | T | 0.22 | −0.02 | 0.63 | 1.7×10−3 | 0.03 | 0.05 | 2.9×10−9 | 6.7×10−9 | 2.4×10−16 | 5.8×10−11 | ||||||
| rs1373004 | 10q21.1 | MBL2/DKK1 | • | • | T | 0.13 | −0.04 | 1.4×10−5 | 1.5×10−4 | 1.5×10−8 | −0.05 | 5.4×10−8 | 2.2×10−6 | 1.6×10−12 | 0.28 | ||||
| rs7071206 | 10q22.3 | KCNMA1 | T | 0.78 | 0.01 | 0.29 | 0.26 | 0.81 | −0.05 | 1.5×10−12 | 6.2×10−9 | 5.0×10−19 | 5.9×10−9 | ||||||
| rs7084921 | 10q24.2 | CPN1 | T | 0.39 | 0.03 | 1.4×10−4 | 1.6×10−6 | 9.0×10−10 | 0.03 | 0.01 | 1.9×10−5 | 9.2×10−7 | 0.58 | ||||||
| rs10835187 | 11p14.1 | LIN7C | • | T | 0.55 | −0.01 | 0.17 | 0.08 | 0.03 | −0.02 | 3.0×10−5 | 2.4×10−4 | 4.9×10−8 | 0.03 | |||||
| rs7953528 | 12p11.22 | KLHDC5/PTHLH | • | • | • | • | A | 0.18 | 0.04 | 5.8×10−8 | 2.4×10−6 | 1.9×10−12 | −0.02 | 0.94 | 0.05 | 0.13 | 2.3×10−7 | ||
| rs2887571 | 12p13.33 | ERC1/WNT5B | • | • | A | 0.76 | −0.03 | 1.1×10−4 | 1.6×10−5 | 6.5×10−9 | −0.04 | 2.2×10−7 | 2.9×10−6 | 5.6×10−12 | 0.37 | ||||
| rs12821008 | 12q13.12 | DHH | • | T | 0.39 | 0.03 | 1.9×10−4 | 5.2×10−4 | 3.3×10−7 | 0.05 | 1.5×10−7 | 1.9×10−9 | 1.2×10−15 | 0.06 | |||||
| rs1053051 | 12q23.3 | C12orf23 | • | T | 0.52 | −0.03 | 1.4×10−5 | 1.8×10−5 | 9.6×10−10 | −0.02 | 2.5×10−6 | 2.4×10−3 | 7.9×10−8 | 0.76 | |||||
| rs1286083 | 14q32.12 | RPS6KA5 | T | 0.81 | −0.05 | 2.9×10−8 | 9.3×10−9 | 2.0×10−15 | −0.04 | 1.7×10−11 | 7.1×10−6 | 1.8×10−14 | 0.92 | ||||||
| rs4985155 | 16p13.11 | NTAN1 | • | • | A | 0.67 | −0.03 | 3.5×10−4 | 1.4×10−7 | 1.7×10−10 | −0.03 | 8.7×10−7 | 1.8×10−4 | 2.2×10−9 | 0.98 | ||||
| rs9921222 | 16p13.3 | AXIN1 | • | • | • | T | 0.48 | −0.03 | 2.5×10−7 | 2.4×10−6 | 5.2×10−12 | −0.04 | 2.2×10−8 | 8.3×10−10 | 1.0×10−16 | 0.26 | |||
| rs13336428 | 16p13.3 | C16orf38/CLCN7 | • | • | A | 0.43 | −0.04 | 2.9×10−7 | 1.1×10−10 | 1.5×10−16 | −0.04 | 5.9×10−5 | 5.8×10−10 | 1.7×10−13 | 0.80 | ||||
| rs1566045 | 16q12.1 | SALL1/CYLD | • | T | 0.80 | −0.06 | 5.0×10−12 | 3.0×10−12 | 1.9×10−22 | −0.01 | 7.8×10−3 | 0.55 | 0.04 | 7.5×10−6 | |||||
| rs1564981 g | 16q12.1 | CYLD | • | A | 0.47 | −0.02 | 1.1×10−3 | 0.01 | 4.4×10−5 | −0.03 | 6.2×10−8 | 5.4×10−4 | 2.0×10−10 | 0.50 | |||||
| rs4790881 | 17p13.3 | SMG6 | A | 0.69 | 0.05 | 1.7×10−8 | 1.2×10−11 | 9.8×10−19 | 0.04 | 6.0×10−4 | 1.7×10−6 | 3.4×10−9 | 0.13 | ||||||
| rs7217932 | 17q24.3 | SOX9 | • | • | • | • | A | 0.46 | 0.03 | 3.7×10−8 | 2.7×10−5 | 1.9×10−11 | 0.01 | 0.31 | 0.15 | 0.08 | 3.8×10−3 | ||
| rs4796995 | 18p11.21 | C18orf19 | A | 0.63 | 0.02 | 3.2×10−6 | 1.1×10−3 | 4.9×10−8 | 0.01 | 5.2×10−4 | 0.11 | 6.7×10−4 | 0.29 | ||||||
| rs10416218 | 19q13.11 | GPATCH1 | • | T | 0.73 | −0.02 | 5.7×10−6 | 7.1×10−4 | 5.5×10−8 | −0.03 | 9.2×10−9 | 1.2×10−4 | 6.6×10−11 | 0.38 | |||||
| rs5934507f | Xp22.31 | FAM9B/KAL1 | A | 0.74 | −0.08 | 0.01 | 8.3×10−4 | 1.6×10−4 | −0.09 | 5.7×10−6 | 3.2×10−4 | 1.2×10−8 | 0.17 | ||||||
Boldface indicates P < 5 × 10−8 or Site-specificity P<5×10−4.
The effect estimates (Beta) are expressed as standardized values per copy of the SNP allele (A)from fixed effects meta-analysis.
Freq is the allele frequency of A.
Effect estimates were calculated in the Stage 2 samples.
Site-specificity null hypothesis, i.e., βLS = βFN.
rs7017914 was discovered in the meta-analysis of women-only, the effects and p-value for this marker are for the meta-analysis of women samples.
rs5934507 was discovered in the meta-analysis of men-only, the effects and p-value for this marker are for the meta-analysis of men samples.
The snps rs13245690 and rs1564981 were independently associated to their main signals in conditional analysis. Black dots in the six functional evidence columns indicate that: first: SNP is an expression quantitative trait loci (eQTL); second: there is a knockout mouse with skeletal phenotypes (MGI database 2011); third: candidate gene has been involved in a monogenic syndrome with skeletal phenotypes (OMIM database 2011); fourth: most significant SNP tags a SNP predicted to have impact on function of the candidate gene; fifth: gene is best candidate in GRAIL analysis; and sixth: candidate gene is part of a bone active pathway.
Two of the novel loci were discovered in the sex-stratified meta-analysis: 8q13.3 in women and Xp22.31 in men; however, only the association at Xp22.31 showed significant evidence for sex-specificity as reflected by significant heterogeneity of effects across sex strata (Phet=1.62×10−8). Yet, we acknowledge that the association at 8q13.3 in women may be driven by a lower number of men in the discovery and replication datasets (Table 1 and Supplementary Table 5). Furthermore, evidence for BMD site-specificity (Phet<5×10−4) was observed in a fraction of the loci including 6 of the 32 novel and 4 of the 24 known loci (Table 1 and Supplementary Fig. 6). Among the newly identified loci, 2q14 (INSIG2), 12p11.22 (PTHLH) and 16q12.1 (CYLD) displayed site-specificity with FN-BMD while 8q13.3 (LACTB2), 10p11.23 (MPP7) and 10q22.3 (KCNMA1) displayed site-specificity with LS-BMD.
After replication, the conditional analysis provided significant evidence of association (P<5×10−8) in 8 of the 9 loci containing secondary signals (Supplementary Fig. 4 and Supplementary Table 2). Three loci included variants localized less than 40 Kb from the initial main signal suggesting allelic heterogeneity and included the 1p31.3 (represented by rs17482952 near WLS), 6q25.1 (rs7751941 near ESR1) and the 16q12.1 (rs1564981 near CYLD) loci. The secondary signal in 16q12.1 (rs1564981) showed a strong association with LS-BMD, while the main signal in this locus (rs1566045) was only associated with FN-BMD. The other five secondary signals were represented by variants localized at more than 180kb away from the initial main signal and contained different candidate genes including the 1p36.12 (rs7521902 near WNT4), 7p14.1 (rs10226308 near SFRP4), 7q31.31 (rs13245690 near C7orf58), 12q13.13 (rs736825 near HOXC6) and the 17q21.31 (rs4792909 near SOST) loci. The secondary signal mapping to the 13q14.11 locus (rs7326472) did not achieve genome-wide significance after replication.
Association of the BMD loci with fracture (Stage 3)
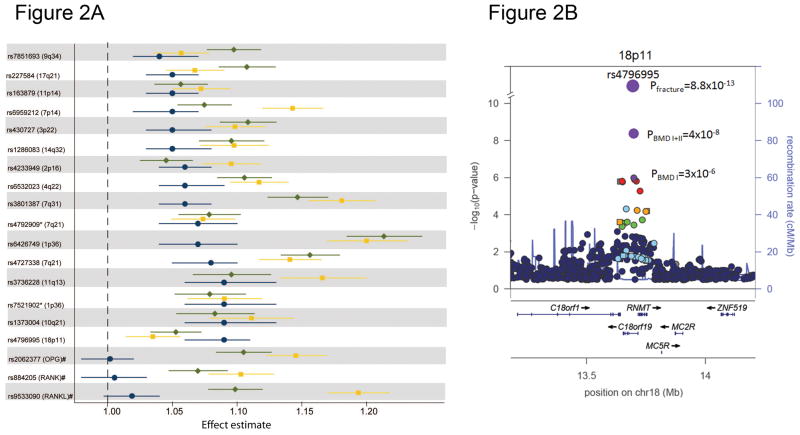
We tested the 96 markers for association with fracture in 31,016 cases and 102,444 controls from 50 studies with fracture information. This collection included: 5,411 cases and 21,909 controls tested in the BMD GWAS discovery samples, 9,187 cases and 45,057 controls tested by in-silico replication and 16,418 cases and 35,478 controls tested by de-novo genotyping (Figure 1 and Online Methods). In this fracture meta-analysis fourteen loci were significantly associated with any type of fracture at a Bonferroni level (P=5×10−4), of which five included novel BMD loci. None of the markers displayed large estimates of heterogeneity (Table 2, Supplementary Table 6 and Supplementary Fig. 7). Markers at six of these loci reached P<5×10−8 including 18p11.21 (C18orf19), 7q21.3 (SLC25A13), 11q13.2 (LRP5), 4q22.1 (MEPE), 2p16.2 (SPTBN1) and 10q21.1 (DKK1). The proportion of the overall fracture risk explained by BMD ranged between 0.09 and 0.40 across markers (Supplementary Table 7) and was estimated in a subset of Stage 2 samples (including n=8,594 cases and 23,218 controls) by modeling the BMD SNP effect on fracture risk with and without the inclusion of BMD as covariate. In general, the effect of these SNPs on BMD was larger than on fracture risk (Fig. 2A) except for the most significantly associated fracture locus 18p11.21 (Fig. 2B). SNPs in genes of the RANK-RANKL-OPG pathway (TNFRSF11A-TNFSF11-TNFRSF11B) despite being the strongest-associated BMD loci were not significantly associated with fracture. All 31 BMD loci with nominal association with fracture risk (P<0.05) showed consistent direction (decreasing BMD allele increased risk of fracture). When we performed subgroup analyses using “cleaner” phenotype definitions such as limiting to clinically-validated fractures and stratifying by anatomical site (i.e. “non-vertebral” fractures and “vertebral” fractures), we did not gain any additional signals (Supplementary Table 8). At a nominally significant level (P<0.05) only three loci were associated with vertebral fracture and all 14 BMD loci were associated with non-vertebral fracture, but these difference in effects between fracture sites were not significant. Therefore, the power of our study did not benefit from improving phenotype definition at expense of (a lower) sample size.
Table 2.
Association of identified BMD loci with risk for any type of low-trauma fracture
| SNP | Locus | Closest Gene/Candidate | Functional evidence
|
Meta-analysis without studies included in BMD discovery
|
Combined meta-analysis results
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| eQTL | KO mouse | OMIM | Tags funct. | GRAIL | Pathway | 25,605 cases, 80,535 controls
|
31,016 cases, 102,444 controls
|
|||||||||
| Risk Allele | Freqb | OR (95% CI) | P | OR (95%CI) | P | Qhet P | I2 | |||||||||
|
Loci significantly associated with fracture risk at P < 5 × 10−8
| ||||||||||||||||
| rs4233949 | 2p16.2 | SPTBN1 | • | G | 0.63 | 1.07 (1.04–1.09) | 1.4×10−7 | 1.06 (1.04–1.08) | 2.6×10−8 | 0.36 | 6 | |||||
| rs6532023 | 4q22.1 | MEPE/SPP1 | • | • | • | • | G | 0.67 | 1.06 (1.04–1.09) | 8.8×10−7 | 1.06 (1.04–1.09) | 1.7×10−8 | 1.00 | 0 | ||
| rs4727338 | 7q21.3 | SLC25A13 | G | 0.32 | 1.08 (1.05–1.10) | 1.0×10−8 | 1.08 (1.05–1.10) | 5.9×10−11 | 0.03 | 31 | ||||||
| rs1373004 | 10q21.1 | MBL2/DKK1 | • | • | T | 0.13 | 1.09 (1.06–1.13) | 7.2×10−7 | 1.10(1.06–1.13) | 9.0×10−9 | 0.64 | 0 | ||||
| rs3736228 | 11q13.2 | LRP5 | • | • | • | • | • | T | 0.15 | 1.09 (1.05–1.12) | 2.1×10−6 | 1.09 (1.06–1.13) | 1.4×10−8 | 0.78 | 0 | |
| rs4796995 | 18p11.21 | C18orf19 | G | 0.39 | 1.06 (1.04–1.09) | 6.4×10−7 | 1.08 (1.06–1.10) | 8.8×10−13 | 0.12 | 20 | ||||||
|
Other significant loci associated with fracture risk at P < 5 × 10−4 (Bonferroni)
| ||||||||||||||||
| rs6426749 | 1p36.12 | ZBTB40 | G | 0.83 | 1.06 (1.03–1.09) | 2.4×10−4 | 1.07 (1.04–1.10) | 3.6×10−6 | 0.07 | 24 | ||||||
| rs7521902 | 1p36.12a | WNT4 | • | • | • | A | 0.27 | 1.10(1.06–1.14) | 3.5×10−6 | 1.09 (1.06–1.13) | 1.4×10−7 | 0.87 | 0 | |||
| rs430727 | 3p22.1 | CTNNB1 | • | • | • | T | 0.47 | 1.05 (1.03–1.08) | 2.4×10−5 | 1.06 (1.03–1.08) | 2.9×10−7 | 0.93 | 0 | |||
| rs6959212 | 7p14.1 | STARD3NL | T | 0.33 | 1.04 (1.02–1.07) | 1.0×10−3 | 1.05 (1.02–1.07) | 7.2×10−5 | 0.43 | 2 | ||||||
| rs3801387 | 7q31.31 | WNT16 | • | • | • | A | 0.74 | 1.08 (1.05–1.11) | 4.9×10−9 | 1.06 (1.04–1.08) | 2.7×10−7 | 0.69 | 0 | |||
| rs7851693 | 9q34.11 | FUBP3 | G | 0.37 | 1.04 (1.01–1.06) | 1.9×10−3 | 1.05 (1.02–1.07) | 3.5×10−5 | 0.65 | 0 | ||||||
| rs163879 | 11p14.1 | DCDC5 | T | 0.66 | 1.06(1.03–1.09) | 6.4×10−6 | 1.05 (1.03–1.07) | 3.3×10−5 | 0.05 | 28 | ||||||
| rs1286083 | 14q32.12 | RPS6KA5 | T | 0.81 | 1.05 (1.02–1.08) | 9.8×10−4 | 1.05 (1.03–1.08) | 7.2×10−5 | 0.01 | 34 | ||||||
| rs4792909 | 17q21.31a | SOST | • | • | • | G | 0.62 | 1.07 (1.04–1.11) | 4.0×10−5 | 1.07 (1.04–1.10) | 6.9×10−6 | 0.31 | 10 | |||
| rs227584 | 17q21.31 | C17orf53 | • | A | 0.67 | 1.05 (1.02–1.08) | 2.2×10−4 | 1.05 (1.03–1.07) | 4.1×10−5 | 0.49 | 0 | |||||
Odds Ratios (OR)estimatedper risk allele copy for any low-trauma fracture among cases as compared with controls. Qhet is the Cochran’s Q statistic and I2 is the measure of heterogeneity.
rs7521902 and rs4792909 are secondary independent signals.
Freq is the frequency ofthe risk allele. Boldface indicates gene names from novel loci and/or P<5×10−8. Black dots in the six functional evidence columns indicate that: first: SNP is an expression quantitative trait loci (eQTL); second: there is a knockout mouse with skeletal phenotypes (MGI database 2011); third: candidate gene has been involved in a monogenic syndrome with skeletal phenotypes (OMIM database 2011); fourth: most significant SNP tags a SNP predicted to have impact on function of the candidate gene; fifth: gene is best candidate in GRAIL analysis; and sixth: candidate gene is part of a bone active pathway.
Figure 2.
A) Phenotype-wide effect for the BMD loci associated with fracture and those part of the OPG/RANK/RANKL pathway. Genetic effect estimates (± 95%CI) are shown for fracture (blue circles), lumbar-spine BMD (yellow rectangles) and femoral-neck BMD (green diamonds) for the 14 loci associated with fracture risk. Horizontal lines represent 95% confidence limits. Effect estimates are shown after transformation of the standardized mean difference(SMD) in the BMD effect to odds ratio equivalents34 (e.g. a 0.02 SMD in the BMD effect corresponds to an OR of 1.04). Secondary signals for markers rs227584 and rs6426749 are marked with an asterisk and the signals mapping to the OPG (rs2062377), RANK (rs884205), and RANKL (rs9533090) genes are marked with a hash. B) Regional association plot for the 18p11.21 locus displaying the P-values for the top SNP associated with fracture (rs4796995) together with P-values of the BMD discovery (Stage 1) and combined with the BMD replication (Stage 1 + 2). SNPs are plotted by position in a 500Kb window of chromosome 18 against association with FN-BMD (−log10 P). Estimated recombination rates (from HapMap) are plotted in cyan to reflect the local LD structure. SNPs surrounding most significant SNP are color-coded reflecting LD between markers (pairwise r2). Genes, exons and transcription direction are derived from the UCSC genome browser.
Allele Risk Modelling for Osteoporosis and Fracture
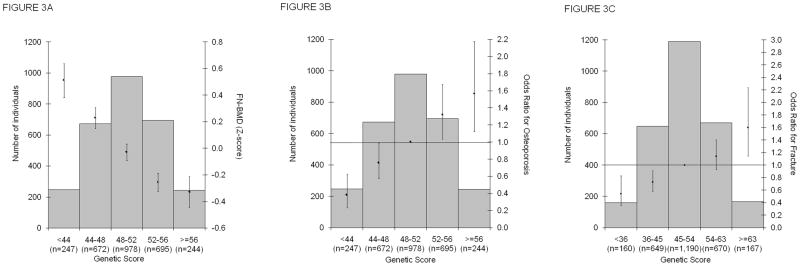
The combined effect of all significant autosomal SNPs on BMD, osteoporosis and any type of fracture was modelled in the PERF study (n=2,836), a prospective study in postmenopausal Danish women aged 55–86 years.17 This study comprises an independent validation setting since it was excluded from the overall meta-analysis for this purpose (see Supplementary Note for details). Risk alleles in the score (i.e., BMD-decreasing alleles) were weighted by their individual effect on BMD and grouped in 5 bins (Supplementary Table 9). The difference in mean FN-BMD between individuals in the highest bin of the risk score (9% of the population; n=244) and those in the middle bin (34% of the population; n=978) was −0.33 SDs (Fig. 3A). This analysis was based on 63 SNPs and explained 5.8% (95%CI [4.0–7.6]) of the total genetic variance in FN-BMD.
Figure 3.
Combined effect of BMD-decreasing alleles and fracture risk-increasing risk alleles modelled in the population-based Prospective Epidemiological Risk Factor (PERF) study (n=2,836 women) on A) Baseline FN-BMD standardized residuals (Z-scores), B) Risk for Osteoporosis and C) Risk for Any type of fracture. The genetic score of each individual for A) and B) was based on the 63 SNPs displaying genome-wide significant association with BMD (55 main and 8 secondary signals), and for C) was based on the 16 BMD SNPs associated with fracture. Both genetic scores are weighted for relative effect sizes estimated without the PERF study. Weighted allele counts summed for each individual were divided by the mean effect size making them equivalent to the percent of alleles carried by each individual and binned into 5 categories. Histograms describe counts of individuals in each genetic score category (left axis scale). Diamonds (right axis scale) represent A) mean FN-BMD standardized levels, risk estimates in the form of odds ratio for B) Osteoporosis (defined as NHANES T-score<−2.5) and for C) Any type of Fracture using the middle category as reference (OR=1). Vertical lines represent 95% confidence limits.
The ability of this genetic score to predict the risk for osteoporosis (defined as T-score<−2.5) and for fracture was modelled in the PERF study using the middle bin as reference (OR=1). Women in the highest bin had 1.56 (95%CI [1.12–2.18]) increased odds for osteoporosis (Fig. 3B), while women in the lowest bin were protected for both osteoporosis (OR=0.38 (95%CI [0.23–0.63])). A model based on the 16 BMD SNPs associated with fracture risk showed that women in the highest bin had 1.60 (95%CI [1.15–2.24]) increased odds for fracture, while women in the lowest bin had a decreased risk for fracture (OR=0.54 (95%CI [0.36–0.83])) (Fig. 3C). Despite serving as a robust proof of principle of the relation between the BMD-decreasing alleles and the risk of osteoporosis and fracture, prediction ability was modest. The ROC analysis showed a significant but relatively small discrimination ability of the genetic score alone with an area under the curve (AUC) of 0.59 (95%CI [0.56–0.62]) for osteoporosis (Supplementary Fig. 8). Adding this score to a model with age and weight alone (AUC 0.75 (95%CI [0.73–0.77])) did not substantially increase discrimination (AUC 0.76 (95%CI [0.74–0.78])). A similar pattern was observed for fracture discrimination with an AUC of 0.57 (95%CI [0.55–0.59]) in a model with the score alone and of 0.62 (95%CI [0.60–0.64]) in a model with age, weight and height. A model considering all 63 SNPs did not change the AUC for fracture risk prediction (0.57 (95%CI [0.54–0.59])).
Functional annotations and pathway analyses
1000 Genomes Project (1KGP)
For the purpose of fine-mapping and identifying additional SNPs with putative functional implication using linkage disequilibrium (LD), a subset of nine discovery studies (FN-BMD n=21,699; LS-BMD n=20,835) used 1KGP data (Release June/2010) to re-impute genotypes contained in the 55 autosomal BMD loci (see Supplementary note for details). In 13 of the 55 BMD loci (X-chromosome SNP not included) we identified markers in a surrounding 1 Mb region that were imputed from 1KGP, and that were more significant than the original HapMap signal (Supplementary Tables 10 and 11) highlighting the benefit of using a denser reference panel of markers. All HapMap markers in LD with variants with functional annotation and displaying higher significance in the 1KGP meta-analysis are shown on Supplementary Table 12. In 14 of the 56 discovered BMD loci a marker from the HapMap imputation was highly correlated (r2 >0.8) with at least one putative functional variant annotated in the 1KG reference. Three of the 14 BMD loci associated with fracture contained putative functional variants tagged by the top SNPs of the BMD meta-analysis. These included the known rs3736228 LRP5 (Ala→Val) functional marker,16,18 the intronic marker rs3779381 within a promoter/regulatory region of WNT16, and one intronic marker (rs4305309) within a promoter/regulatory region of SPTBN1.
eQTL analyses
Expression profiles at the GWS BMD loci were analyzed within four datasets (see Supplementary Note). In trans-iliac bone biopsies, expression of five genes correlated with LS-BMD and/or FN-BMD of the donors with P < 0.001 including PSME4 (2p16.2), DKK1 (10q21.1), C17orf91 (17p13.3), SOST (17q21.31_1) and DUSP3 (17q21.31_1) (Supplementary Table 13). Among them DKK1 (10q21.1) was the most significantly correlated with FN-BMD (P=1.3×10−5) and LS-BMD (P=3.2×10−4). Variants in all these BMD loci (with exception of 17p13.3) were also associated with fractures.
The SNP-eQTL analyses were performed across diverse tissues examining the correlation between marker alleles and transcript levels at the associated BMD loci. Fourteen of the BMD-associated SNPs correlated with the expression of one or more of the nearby genes with P < 5×10−5 and were either the strongest cis-variants, or good surrogates thereof, for those genes (Supplementary Tables 14 and 15). The most significant BMD-SNP eQTL was observed for rs10835187[T] with reduced expression of the LIN7C gene at the 11p14.1 locus (P = 2.8×10−39 in adipose tissue). Of particular interest were BMD-SNP cis-variants at three loci that were also associated with fracture including: 1p36.12, 4q22.1 and 17q21.31. At 1p36.12, rs6426749[G] correlated with reducedWNT4 expression in fibroblast, osteoblast and adipose tissue; at 4q22.1 rs6532023[G] correlated with reduced SPP1 (osteopontin) expression in adipose tissue and at 17q21.31 rs227584[A] correlated with increased C17orf65 expression in monocytes, adipose tissue, whole blood and lymphoblasts.
GRAIL analysis
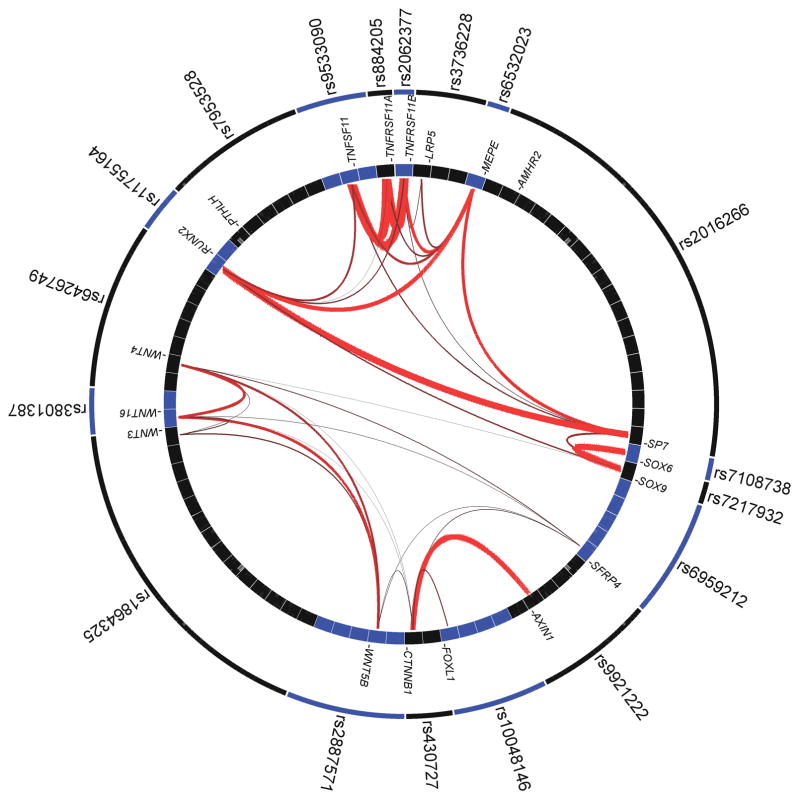
We applied the GRAIL text-mining algorithm19 to investigate connections between genes in the 55 autosomal BMD loci. This analysis revealed significant (GRAIL SNP P<0.01) connections between genes in 18 of the 55 input loci (Fig. 4 and Supplementary Table 16). The strongest connections were seen for members of three key biologic pathways: RANK-RANKL-OPG pathway (TNFRSF11A-TNFSF11-TNFRSF11B); mesenchymal stem cell differentiation (RUNX2, SP7, SOX9); and Wnt-signalling (LRP5, CTNNB1, SFRP4, WNT3, WNT4, WNT5B, WNT16, AXIN1) with the ten most frequently connecting terms being: ‘bone’, ‘catenin’, ‘signaling’, ‘differentiation’, ’rank’, ‘osteoblast’, ‘diacylglycerol’, ‘kappab’, ‘development’, and ‘osteoclast’. To assess the significance of this “biological” gene connection enrichment we applied GRAIL to 2000 random matched sets of 55 SNPs (See Supplementary note for details) and we did not observe any set with 15 or more loci with significant enriched connectivity (Supplementary Fig. 9) providing strong statistical evidence of the significant clustering of our BMD loci (P < 0.0005).
Figure 4.
Graphic representation of GRAIL connections between SNPs and corresponding genes for the 18 SNPs as determined with GRAIL P<0.01. The top 10 keywords linking the genes were: ‘bone’, ‘catenin’, ‘signaling’, ‘differentiation’, ’rank’, ‘osteoblast’, ‘diacylglycerol’, ‘kappab’, ‘development’, and ‘osteoclast’. Thicker redder lines imply stronger literature-based connectivity. Blue and black boxes depict loci boundaries represented per top-associated marker (outer circle) and per gene in the region (inner circle).
Discussion
In this, the largest GWAS for osteoporosis traits to date, we identified 32 novel genomic loci bringing to 56 the number of loci robustly associated with BMD variation. Furthermore, we report for the first time that six of these BMD loci are associated with low-trauma fractures at P<5×10−8. As for other complex traits, our results indicate hundreds of variants with small effects may be contributing to the genetic architecture of BMD and fracture risk.20 Our hypothesis-free assessment of common variants of the genome provides novel insights into biology, implicating several factors clustering in bone-active pathways.
Our results highlight the highly polygenic nature underlying BMD variation and the critical role of several biological pathways influencing osteoporosis and fracture susceptibility (Supplementary Fig. 10). On top of the Wnt factors known to be associated with BMD (CTNNB1, SOST, LRP4, LRP5, WLS, WNT4, MEF2C) several of the newly discovered loci also implicate additional Wnt signalling factors (including WNT5B, WNT16, DKK1, PTHLH, SFRP4 and AXIN1). Another clearly delineated pathway is that involved in mesenchymal cell differentiation, including the newly identified RUNX2, SOX4 and SOX9 BMD loci along with the previously known SP7. Another bone-relevant pathway includes that of “Endochondral Ossification” which involves essential processes during the fetal development of the mammalian skeleton and which implicated several of our identified BMD loci including: SPP1, MEF2C, RUNX2, SOX6, PTHLH, SP7 and SOX9. In addition, the biological relevance of our associations is accentuated by the identification of genes underlying rare monogenetic forms of osteoporosis and/or high bone mass such as SOST, CLCN7, LRP5 21–23 (Supplementary Table 17) which also contain common variants involved in normal BMD variation at the population level.11,14,16 This is supportive of a genetic architecture where both common and rare genetic variation may reside in the same locus.24 Other genes have not been reported to be associated with monogenic forms for osteoporosis but have clear involvement in bone development in animal models. For example, SNPs in the 16q12.1 BMD locus map near CYLD. Human mutations in this gene have been described to cause familial cylindromatosis a condition without phenotypic skeletal manifestations. However, it has been shown that Cyld knock-out mice have significant bone loss leading to a severe osteoporosis phenotype25 and also that CYLD regulates osteoclastogenesis.26 Moreover, evidence from the GWAS and eQTL analyses also suggests some loci contain more than one common variant with independent effects on BMD and fracture risk. On the other hand, when no correlation is observed between gene expression and a particular SNP, it is difficult to draw conclusions. A correlation might be missed if the expression of the transcript was not measured in a relevant tissue or if the expression of a particular splice-variant was not measured.27
BMD and fracture genetic effects correlate to some extent, but some important fracture risk variants may have minimal impact on BMD and vice versa. This is the case for the 18p11.21 signal (Fig. 2B) mapping to a gene coding for a protein of unknown function, which despite a modest effect on BMD (0.02% variance explained) displayed the most significant association with fracture risk (OR=1.08, 95%CI[1.06–1.10], P=8.8×10−13). This is in contrast to variants with known stronger effects on BMD which were not significantly associated with fracture risk. For example, variants in the RANK-RANKL-OPG pathway, known to play a critical role in osteoclastogenesis, had clear associations with BMD but not fracture risk (Fig. 2A). Even though loci discovery was based on the BMD phenotype, these findings reflect the heterogeneous and complex nature of the mechanistic pathways leading to fracture. Therefore, given our study design, we cannot rule out the possibility that yet unidentified genetic loci are influencing risk of fracture independently of BMD. Future well-powered GWAS meta-analyses on fracture risk will address this question while corroborating the associations with fracture that we report for some of the BMD loci (particularly those not associated with fracture at P<5×10−8).
Our study also provides indication that there is sex- and site-specificity underlying BMD variation. One of the GWAS signals (Xp22.31) was only significant in the sex-stratified analysis in men and displayed significant sex heterogeneity (Phet=1.62×10−8). This is expected considering the sexual dimorphism of bone.28,29 In fact, in a recent GWAS, the rs5934507 SNP mapping to Xp22.31, which is associated with BMD in the current study, has been previously associated with male serum testosterone levels.30 Thus, it is likely that rs5934507 affects serum testosterone, which in turn regulates BMD. In line with the different types of bone composition at the different skeletal sites (predominantly trabecular at the lumbar spine while predominantly cortical at the femoral neck) we observed some indication of site specificity in 10 of the 56 BMD loci, suggesting differential genetic influences on BMD determination across skeletal sites. As has been previously shown31, we did not find in our results major differences in effect sizes between individuals of European and East Asian ancestry (Supplementary Fig. 7). However, this may be due to reduced power given the smaller number of individuals of East Asian ancestry. We tested a genetic risk score to identify individuals at risk of osteoporosis and fracture and showed that cumulatively, the identified variants generate a gradient of risk. These gradients reach ORs of 1.56 for osteoporosis and 1.60 for fractures when comparing participants with the highest risk scores with those reflecting the mean score. Yet, at present there is limited clinical utility in using this score as evidenced by the non-significant contribution to case discrimination after considering clinical risk factors with strong effects on osteoporosis and fracture risk (like age and weight). This is not unexpected given the small fraction of genetic risk for either BMD or fracture that has been identified thus far.
Our study has limitations. The identified SNPs are probably not the causal variants; it is more likely that these markers are in LD with the underlying causal variants. Additional analyses on potential functional SNPs identified in this study will be required to determine if they are causal to these relationships with BMD. Moreover, the causal genes underlying the GWAS signals may be different from the candidate genes we describe, considering that our understanding of their role in bone biology is limited. Further exploration of these loci with more detailed sequencing, gene expression, and translational studies will be required. Such studies can also disentangle the diverse types of complex relationships we currently cannot distinguish in the BMD loci with secondary signals, i.e., if these are the result of true allelic heterogeneity or if they are driven by a second gene in the same region.32 Similarly, despite our large sample size, power limitations still play a role for detecting additional associations with smaller effect sizes and/or arising from rarer variants. Finally, given the different levels of data availability and the difficulties for standardization across studies, we did not evaluate the effect of additional risk factors for osteoporosis, such as menopausal status and smoking, which can influence the genetic associations with BMD. Nonetheless, despite these limitations we have identified many novel and previously unsuspected associations with BMD variation and fracture risk.
Finally, the relatively weak effects of the variants discovered by GWAS do not undermine the biological relevance of the genes identified, as exemplified by the identification of genetic signals at the location of genes coding for proteins currently targeted by novel osteoporosis treatments (Supplementary Fig. 10). The novel genes identified in our study may represent new candidates to target for osteoporosis drug discovery. Most established treatments for osteoporosis currently focus on curtailing bone resorption (eg. bisphosphonates, RANKL inhibitors) while only few anabolic treatments are currently approved for the treatment of osteoporosis (i.e. recombinant truncated or altered PTH). Other anabolic compounds under Phase II development include PTHrP fragments and Wnt-signaling enhancers such as anti-Sclerostin antibodies.33 Several of the variants robustly associated with BMD map in or close to genes of proteins involved in these pharmacologic pathways, namely osteoprotegerin (TNFRSF11B), RANK (TNFRSF11A), RANKL (TNFSF11), PTHrP (PTHLH), Low-density lipoprotein receptor-related protein 5 (LRP5), Sclerostin (SOST), and Dickkopf-1 (DKK1).
In conclusion, these findings highlight the highly polygenic and complex nature underlying BMD variation, shedding light on the pathophysiological mechanisms underlying fracture susceptibility and harbouring potential for the future identification of drug targets for the treatment of osteoporosis.
ONLINE METHODS
Study design
This study is part of the GEnetic Factors for OSteoporosis consortium (GEFOS), a coalition of teams of investigators dedicated to identify the genetic determinants of osteoporosis. The discovery samples comprised 17 GWA studies (n=32,961) from populations across North America, Europe, East Asia and Australia, with a variety of epidemiological designs (Supplementary Table 18A) and patient characteristics (Supplementary Table 18B); a subset of which had fracture information (Supplementary Table 18C). Subjects from 34 additional studies with BMD data (n=50,933) were used for replication while association with fracture was tested across 50 studies with fracture information, most of them also used for the BMD analysis (n=31,016 cases and 102,444 controls) (Figure 1 and Supplementary Tables 19A-C and 20A-C). All studies were approved by their institutional ethics review committees and all participants provided written informed consent.
BMD measurements and fracture definition
BMD of the lumbar spine (LS-BMD) and femoral neck (FN-BMD) was measured in all cohorts using dual-energy X-ray absorptiometry following standard manufacturer protocols (Supplementary Tables 18B, 19B and 20B). Three clinically-distinct fracture definitions were used: 1) Any type, consisting of low-trauma fractures at any skeletal site (except fingers, toes and skull) occurring after age 18 years assessed by X-ray, radiographic report, clinical record, clinical interview and/or questionnaire; 2) Validated non-vertebral, consisting of fractures occurring after age 50 years with diagnosis confirmed by hospital records and/or radiographs; and 3) Radiographic vertebral fractures, from lateral morphometry scored on X-rays. The first is most-inclusive, while the latter two are more stringent fracture definitions commonly used in randomized trials.35,36 Controls were defined as individuals without a history of fracture using for each fracture type the same age limit categories of the cases.
Stage 1 Genome-wide association analysis
Genotyping and Imputation
GWAS genotyping was done by each study following standard manufacturer protocols followed by imputation to ~2.5 million SNPs from HapMap37 Phase II release 22 using Genome Build 36. Quality control was performed independently for each study. To facilitate meta-analysis, each group performed genotype imputation with BIMBAM38, IMPUTE39, or MACH40 software using genotypes from the HapMapPhase II release 22 (CEU or CHB/JPT as appropriate). HapMap release 21 was used as reference for SNPs residing on the X chromosome and IMPUTE software was used for imputation. Overall imputation quality scores for each SNP were obtained from IMPUTE (proper_info) and MACH (rsq_hat) statistics. Details on the genotyping platform used, genotype quality control procedures and software for imputation employed for each study are presented in the Supplementary Tables 18D and 19D.
Association analysis with BMD
each study performed genome-wide association analysis for FN-BMD and LS-BMD using sex-specific, age- weight- and principal components-adjusted standardized residuals analyzed under an additive (per allele) genetic model. Analyses of autosomal and chromosome X markers were done separately. Analysis of imputed genotype data accounted for uncertainty in each genotype prediction by using either the dosage information from MACH or the genotype probabilities from IMPUTE and BIM-BAM. Studies used MACH2QTL40 directly or via GRIMP41 (which uses genotype dosage value as a predictor in a linear regression framework), SNPTEST39, Merlin42, BIM-BAM or the linear mixed effects model of the Kinship and ProbABEL43 (Supplementary Tables 18D and 19D). For analysis of the X-chromosome either SNPTEST or R package was used in each participating study. We coded “effect allele homozygous genotype” as “2” and “other allele homozygous genotype” as “0” in the genotyped SNPs in men on the X chromosome. The imputed genotypes were coded as continuous variables from 0 to 2 to take into account imputation uncertainty. The genomic control method44 was used to correct the standard error (SE) by the square root of the genomic inflation factor (lambda): SEcorrected = SE × √lambda.
Meta-analysis of the GWA studies
before performing meta-analysis on the genome-wide association data, SNPs with poor imputation quality scores (rsq_hat < 0.3 in MACH, proper_info < 0.4 in IMPUTE or the ratio of observed to expected dosage variance < 0.3 in BIMBAM) and markers with a minor allele frequency < 1% were excluded for each study. All individual GWAS were genomic control corrected before meta-analysis.44 Individual study-specific genomic control values ranged from 0.98 to 1.08. (Supplementary Table 18D). A total of 2,483,766 autosomal SNPs were meta-analyzed across: 17, 16 and 13 studies for FN-BMD (Pooled, women-only, and men-only analyses, respectively) and 16, 13 and 12 studies for LS-BMD (Pooled, women-only, and men-only analyses, respectively). A total of 76,253 X-linked SNPs were meta-analyzed across: 14, 13 and 10 studies for LS and FN-BMD (Pooled, women-only, and men-only analyses, respectively). In our discovery analysis, we chose to implement a fixed effects models approach as it is generally preferable for the purposes of initial discovery, where the aim is to screen and identify as many of the true variants as possible.45,46 SNPs present in less than three studies were removed from the meta-analysis yielding ~ 2.2 million SNPs in the final results. Genomic inflation factors (λ) were 1.11, 1.09, 1.06 for FN-BMD BMD (Pooled, women-only, and men-only analyses, respectively) and 1.13, 1.09, 1.06 for LS-BMD (Pooled, women-only, and men-only analyses, respectively). A second GC correction was applied to the overall meta-analysis results, although such second correction is considered overly conservative.47 Significance for BMD association was set at P<5×10−8 while a Bonferroni correction was used for the association with fracture.48
Selection of SNPs for follow up
we took forward the most significant 96 SNPs for replication. Based on power estimations, after adding 30,000 samples in stage 2 these variants had a priori Power >=85% to reach P= 5 × 10−8 in the meta-analysis. Loci were considered independent when separated by at least 1 Mb down and upstream of the top GWAS signal. The 96 variants included the 82 index SNPs representing each of the 82 loci reaching P<5×10−6 in Stage 1, 9 SNPs that lie within the same 2Mb windows as the 82 but which were independent from the main signal (secondary signals), and the top-five most associated SNPs of the X-chromosome (with P <5×10−5).
Association analyses with fracture risk
effect estimates (odds ratio) for association of allele dosage of the top hits with fracture risk were obtained from logistic regression models adjusted for age, age2 weight, sex, height and four principal components. The proportion of the fracture risk explained by FN-BMD was calculated from the regression coefficients as (βunadjusted - βBMDadjusted)/βunadjusted in a subset of replication samples for which both FN-BMD and complete fracture information was available.
Stage 2 follow up
Samples and genotyping
fracture association results were also obtained for the 82 most significant SNPs from 54,244 individuals of European ancestry from 7 GWAS (in-silico genotyping) that had not been included in the stage 1 analyses (Supplementary Tables 19A, 19B and 19C). Subjects from 34 studies of the GENOMOS consortium with BMD and/or fracture information were studied for replication (Supplementary Tables 3A, 3B and 3C). De-novo replication genotyping was done in the UK (Kbiosciences), Iceland (deCODE Genetics), Australia (University of Queensland Diamantina Institute) and the USA (WHI GeCHIP) using KASPar, Centaurus, OpenArray and iSelect assays respectively (Supplementary Note). Minimum genotyping quality control criteria were defined as: Sample call rate > 80%, SNP call rate > 90%, HWE P > 1×10−4, MAF > 1%.
Association analyses and meta-analysis
We tested the association between the 96 SNPs and BMD and fracture risk in each in-silico and de-novo “Stage 2” study separately as described for the “Stage 1” studies. We subsequently meta-analyzed effects and standard errors from the “Stage 2” studies, followed by a meta-analysis of the summary statistics of both “Stage 1” and “Stage 2” using the inverse-variance method in METAL. At this replication stage, where more than 30 studies were synthesized, we chose to first assess the underlying heterogeneity considering both the Cochran’s Q statistic and the I2 metric. If the heterogeneity was not significant fixed effects models were applied. If the Cochrane Q P-value<0.0005 and the I2 was > 50% we used the more conservative random effects models.
Additional analyses
Further analyses were performed for the SNPs carried forward for replication. Each of these analyses is described in detail in the “Supplementary Note”. In brief, we performed: 1) a conditional genome-wide association analysis to examine whether any of the 82 BMD loci harbored additional independent signals; 2) tested gene-by-gene pair-wise interactions between these BMD loci; 3) assessed within the independent setting of the PERF study (for details on study design see Supplementary Tables 20A, 20B & 20C) the predictive ability derived from the cumulative effect of the 63 genome-wide significant autosomal BMD SNPs in relation to BMD levels and osteoporosis risk; and that of the 16 BMD SNPs also associated with fracture risk in relation to fracture risk; 4) identified SNPs having r2 ≥ 0.80 with the lead SNP that were potentially functional (nonsense, nonconservative non-synonymous, synonymous, exonic splicing, transcription factor binding sites, etc) using regional imputation with the 1000 Genomes data (June 2010 release); 5) tested the relationship between gene expression profiles from a) trans-iliacal bone biopsies and BMD in 84 unrelated postmenopausal women49 and b) also examined cis- associations between each of the 55 significant BMD SNPs and expression of nearby genes in different tissues including lymphoblastoid cell lines50–52, primary human fibroblasts and osteoblasts53, adipose tissue54, whole blood54 and circulating monocytes55; and finally 6) evaluated the connectivity and relationships between identified loci using the literature-based annotation with Gene Relationships across Implicated Loci (GRAIL19) statistical strategy.
Supplementary Material
Acknowledgments
We thank all study participants for making this work possible. This research and the Genetic Factors for Osteoporosis (GEFOS) consortium have been funded by the European Commission (HEALTH-F2-2008-201865-GEFOS). We acknowledge funding from the following organizations: NIH research grants: R01 AG18728, R01HL088119, R01AR046838, U01 HL084756, P30DK072488, T32AG000262, F32AR059469, P01 AG-18397, R01AG041517 and M01 RR-00750. NIH contract N01-AG-12100, the NIA Intramural Research Program, Hjartavernd (the Icelandic Heart Association), and the Althingi (the Icelandic Parliament). Icelandic Heart Association. National Health and Medical Research Council (Australia) (grant reference 511132). Australian Cancer Research Foundation and Rebecca Cooper Foundation (Australia). National Health and Medical Research Council (Australia). National Health and Medical Research Council (Australia) Career Development Award (569807). Medical Research Council New Investigator Award (MRC G0800582). Health Research Council of New Zealand. Sanofi-Aventis, Eli Lilly, Novartis, Pfizer, Proctor&Gamble Pharmaceuticals and Roche. National Health and Medical Research Council, Australia. Australian National Health and Medical Research Council, MBF Living Well foundation, the Ernst Heine Family Foundation and from untied educational grants from Amgen, Eli Lilly International, GE-Lunar, Merck Australia, Novartis, Sanofi-Aventis Australia and Servier. Medical research Council UK. Arthritis Research UK (17539, 15389). The Victorian Health Promotion Foundation and the Geelong Region Medical Research Foundation, and the National Health and Medical Research Council, Australia (project grant 628582). Action Research UK. European Commission (QLRT-2001-02629) and the UK Food Standards Agency. BioPersMed (COMET K-project 825329), Austrian Federal Ministry of Transport, Innovation and Technology (BMVIT) and the Austrian Federal Ministry of Economics and Labour/the Federal Ministry of Economy, Family and Youth (BMWA/BMWFJ) and the Styrian Business Promotion Agency (SFG)”Red de Envejecimiento y fragilidad (RETICEF), Instituto Carlos III. Spanish Ministry of Education and Science (SAF2010-15707). Catalan Government (2009SGR971, 2009SGR818). Instituto de Salud Carlos III-Fondo de Investigaciones Sanitarias (PI 06/0034, PI08/0183). Healthway Health Promotion Foundation of Western Australia, Australasian Menopause Society and the Australian National Health and Medical Research Council Project (254627, 303169 and 572604). Academy of Finland and Finnish Ministry of Education. Merck Frosst Canada Ltd.; Eli Lilly Canada Inc.; Novartis Pharmaceuticals Inc.; The Alliance: sanofi-aventis & Procter and Gamble Pharmaceuticals Canada Inc.; Servier Canada Inc.; Amgen Canada Inc.; The Dairy Farmers of Canada; and The Arthritis Society. NHLBI contracts N01-HC-85239, N01-HC-85079 through N01-HC-85086; N01-HC-35129, N01 HC-15103, N01 HC-55222, N01-HC-75150, N01-HC-45133, HL080295, HL075366, HL087652, HL105756 NINDS. Additional support was provided through AG-023629, AG-15928, AG-20098, and AG-027058 from the NIA. National Center for Research Resources grant M01-RR00425 to the Cedars-Sinai General Clinical Research Center Genotyping core and National Institute of Diabetes and Digestive and Kidney Diseases grant DK063491 to the Southern California Diabetes Endocrinology Research Center. deCODE Genetics. The UK’s NIMR Biomedical Research Centre Grant. Cancer Research Campaign; Medical Research Council; Stroke Association; British Heart Foundation; Department of Health; Europe Against Cancer Programme Commission of the European Union and the Ministry of Agriculture, Fisheries and Food. EU Biomed 1 (BMHICT920182, CIPDCT925012, ERBC1PDCT 940229, ERBC1PDCT930105), Medical Research Council G9321536 and G9800062, Wellcome Trust collaborative Research Initiative 1995, MAFF AN0523, EU FP5 (QLK6-CT-2002-02629), Food Standards Agency N05046. The UK’s NIMR Biomedical Research Centre Grant to Cambridge. Netherlands Organisation for Scientific Research (NWO), Erasmus University Medical Center, the Centre for Medical Systems Biology (CMSB1 and CMSB2) of the Netherlands Genomics Initiative (NGI). F.I.R.M.O. Fondazione Raffaella Becagli. National Institute for Arthritis, Musculoskeletal and Skin Diseases and National Institute on Aging (R01 AR/AG 41398, DPK N01AG62101, N01AG62103, N01AG62106, 1R01AG032098 and R01 AR 050066; DK National Heart, Lung, and Blood Institute’s Framingham Heart Study (N01-HC-25195, N02-HL-6-4278). Canadian Institutes for health research operating grant funding reference #86748. Federal program of Ministry of Education and Science of Russian Federation “Scientific and pedagogical staff of innovative Russia” in 2009-2013 (state contract P-601) and Federal program “Research and development of prior directions of scientific-technological complex of Russia in 2007-2012” (state contract 16.512.11.2032). Swedish Research Council (K2010-54X-09894-19-3, 2006-3832 and K2010-52X-20229-05-3), the Swedish Foundation for Strategic Research, the ALF/LUA research grant in Gothenburg, the Lundberg Foundation, the Torsten and Ragnar Söderberg’s Foundation, the Västra Götaland Foundation, the Göteborg Medical Society, the Novo Nordisk foundation. University of Athens, Greece (Kapodistrias 2009). Medical Research Council; NIHR Musculoskeletal BRU Oxford; NIHR Nutrition BRU Southampton. The Center for Inherited Disease Research (CIDR). National Institutes of Health contract number HHSN268200782096C. Hong Kong Research Grant Council (HKU 768610M); The Bone Health Fund of HKU Foundation; The KC Wong Education Foundation; Small Project Funding (201007176237); Matching Grant, CRCG Grant and Osteoporosis and Endocrine Research Fund, and the Genomics Strategic Research Theme of The University of Hong Kong. Direct grant, Chinese University of Hong Kong. Korea Health 21 R&D Project, Ministry of Health & Welfare, Republic of Korea (Project No.: A010252); Korea Healthcare technology R&D Project, Ministry for Health, Welfare and Family Affairs (Project No.: A110536). Netherlands Ministry of Health Welfare and Sports, Directorate of Long-Term Care. World Anti-Doping Agency, the Danish Ministry of Culture, Institute of Clinical Research of the University of Southern Denmark. Chief Scientist Office of the Scottish Government, the Royal Society, and the European Union framework program 6 EUROSPAN project (LSHG-CT-2006-018947). European Union’s Seventh Framework Programme (FP7/2007-2013) grant HEALTH-F2-2009-223004 PHASE. Netherlands Organisation of Scientific Research NWO Investments (nr. 175.010.2005.011, 911-03-012); Research Institute for Diseases in the Elderly (014-93-015; RIDE2); Netherlands Genomics Initiative/Netherlands Consortium for Healthy Aging (050-060-810); German Bundesministerium fuer Forschung und Technology (#01 AK 803 A-H and # 01 IG 07015 G); NIHR Biomedical Research Centre (grant to Guys’ and St. Thomas’ Hospitals and King’s College London); the Chronic Disease Research Foundation; Wellcome Trust; Canadian Institutes of Health Research, the Canadian Foundation for Innovation, the Fonds de la Recherche en Santé Québec, The Lady Davis Institute, the Jewish General Hospital and Ministère du Développement économique, de l’Innovation et de l’Exportation du Quebec. Swedish Research Council (K20006-72X-20155013). The Swedish Sports Research Council (87/06), the Swedish Society of Medicine, the Kempe-Foundation (JCK-1021), Medical Faculty of Umeå Unviersity (ALFVLL:968:22-2005, ALFVL:-937-2006, ALFVLL:223:11-2007, ALFVLL:78151-2009) and from the county council of Västerbotten (Spjutspetsanslag VLL:159:33-2007)HL 043851 and HL69757 from the National Heart, Lung, and Blood Institute and CA 047988 from the National Cancer Institute, the Donald W. Reynolds Foundation and the Fondation Leducq. Amgen. Academy of Finland: grants 126925, 121584, 124282, 129378 (Salve), 117787 (Gendi), and 41071 (Skidi), the Social Insurance Institution of Finland, Kuopio, Tampere and Turku University Hospital Medical Funds (grant 9M048 for TeLeht), Juho Vainio Foundation, Paavo Nurmi Foundation, Finnish Foundation of Cardiovascular Research and Finnish Cultural Foundation, Tampere Tuberculosis Foundation and Emil Aaltonen Foundation (T.L). K08AR055688. A detailed list of acknowledgements by study is given in the Supplementary Note.
Footnotes
Author Contributions
This work was done under the auspices of the European Commission sponsored Genetic Factors for Osteoporosis (GEFOS) consortium.
Study-specific design and management: U.S., M.A., L.M., J.P., S.B., M.B., B.M.B., C. Christiansen, C. Cooper, G.D., I.F., M.F., D.G., J.G-M., M. Kähönen, M. Karlsson, J-M.K., P.K., B.L.L., W.D.L., P.L., Ö.L., R.S.L., J.M., D.M., J.M.O., U.P., J.A.R., P.M.R., F. Rousseau, P.E.S., N.L.T., R.U., W.V., J.V., M.T.Z., K.M.G., T.P., D.I.C., S.R.C., R.E., J.A.E., V.G., A.H., R.D.J., G.J., J.W.J., K-T.K., T.L., M. Lorentzon, E.M., B.D.M., G.C.N., M.P., H.A.P., R.L.P., O.R., I.R.R., P.N.S., P.C.S., A.R.S., F.A.T., C.M.D., N.J.W., L.A.C., M.J.E., T.B.H., A.W-C.K., B.M.P., J. Reeve, T.D.S., E.A.S., M.Z., U.T., C.O., J.B.R., M.A.B., K. Stefansson, A.G.U., S.H.R., J.P.I., D.P.K. and F. Rivadeneira.
Study-specific Genotyping: K.E., U.S., E.L.D., L.O., L.V., S.X., A.K.A., D.J.D., S.G., R.K., C.K., A.Z.L., J.R.L., S.M., S.S., S.T., O.T., S.C., E.K., J.M., B.O-P., Y.S.A., E.G., L.H., H.J., T. Kwan, R. Luben, C.M., S.T.P., S. Reppe, J.I.R., J.B.v., D.V., K.M.G., D.I.C., G.R.C., P.D., R.D.J., T.L., Y.L., M. Lorentzon, R.L.P., N.J.W., L.A.C., C.O., M.A.B., A.G.U. and F. Rivadeneira.
Study-specific Phenotyping: U.S., E.L.D., O.M.A., A.M., S.X., N. Alonso, S.K.K., S.G.W., A.K.A., T.A., J.R.C., Z.D., N.G-G., S.G., G.H., L.B.H., K.A.J., G.K., C.K., T. Koromila, M. Kruk, M. Laaksonen, A.Z.L., S.L., P.C.L., L.M., X.N., J.P., L.M.R., K. Siggeirsdottir, O.S., N.M.v., J.W., K.Z., M.B., C. Christiansen, M.F., M. Kähönen, M. Karlsson, J-M.K., Ö.L., J.M., D.M., B.O-P., J.M.O., U.P., D.M.R., J.A.R., P.M.R., F. Rousseau, W.V., J.V., M.C-B., E.G., T.I., R. Luben, S. Reppe, G.S., J.B.v., D.V., F.M.W., K.M.G., J.A.C., D.I.C., E.M.D., R.E., J.A.E., V.G., A.H., R.D.J., G.J., Y.L., M. Lorentzon, E.M., G.C.N., B.A.O., M.P., H.A.P., R.L.P., O.R., I.R.R., J. Robbins, P.N.S., C.M.D., M.J.E., J. Reeve, E.A.S., M.Z., C.O., M.A.B., A.G.U., D.P.K. and F. Rivadeneira.
Study-specific data analysis: K.E., U.S., E.E., Y.H., E.L.D., E.E.N., L.O., O.M.A., N. Amin, J.P.K., D.L.K., G.L., C.L., R.L.M., A.M., L.V., D.W., S.X., L.M.Y-A., H.Z., J.E., C.M.K., S.K.K., P.J.L., G.T., J.F.W., V.A., A.K.A., T.A., J.R.C., G.H., L.J.H., C.K., T. Koromila, A.Z.L., S.M., T.V.N., M.S.P., J.P., L.M.R., A.V.S., O.S., S.T., S.C., J.M., B.O-P., U.P., R. Li, R. Luben, S. Reppe, J.I.R., A.R.W., Y.Z., S. Raychaudhuri, D.I.C., J.A.E., R.D.J., T.L., K.N., O.R., D.M.E., D.K., J.B.R., M.A.B., J.P.I., D.P.K. and F. Rivadeneira.
Analysis plan design: K.E., E.E., U.S. D.K., D.P.K, J.P.I. and F. Rivadeneira.
Meta-analyses: K.E., E.E., Y.H. and E.E.N.
Gene by Gene interaction: K.E., E.E. and A.R.W.
Risk modeling and secondary signals: K.E. and F. Rivadeneira.
Expression QTL: U.S., G.T., E.G., S. Reppe, K.M.G. and T.P.
Functional SNP prediction: Y.H.
Gene relationships across implicated loci (GRAIL): K.E., E.L.D., D.W. and S. Raychaudhuri.
Standardization of phenotype and genotype replication datasets: K.E., U.S., E.E., E.L.D., L.O., G.T., L.H. and C.M.
Interpretation of results (BMD working group): K.E., U.S., E.E., Y.H., E.L.D., E.E.N., L.O., O.M.A., N. Amin, D.L.K., C.L., R.L.M., A.M., L.V., D.W., S.X., L.M.Y-A., J.E., C.M.K., S.K.K., A.W-C.K., J. Reeve, M.Z., C.O., D.K., J.B.R., M.A.B., A.G.U., S.H.R., J.P.I., D.P.K. and F. Rivadeneira.
Manuscript draft (BMD writing group): K.E., U.S., E.E., Y.H., E.L.D., E.E.N., L.O., O.M.A., A.M., C.O., D.K., J.B.R., M.A.B., A.G.U., S.H.R., J.P.I., D.P.K. and F. Rivadeneira.
GEFOS Steering committee: U.S., E.E., U.T., A.G.U., S.H.R., J.P.I. and F. Rivadeneira.
Competing financial interests
The coauthors affiliated with deCODE Genetics in Reykjavík Iceland withhold stock options in that company.
URLs
GEFOS Consortium, http://www.gefos.org/;
GENOMOS Consortium, http://www.genomos.eu/;
HapMap Project, http://hapmap.ncbi.nlm.nih.gov/;
1000 Genomes Project, http://www.1000genomes.org/;
LocusZoom, http://csg.sph.umich.edu/locuszoom/;
References
- 1.Burge R, et al. Incidence and economic burden of osteoporosis-related fractures in the United States, 2005–2025. J Bone Miner Res. 2007;22:465–75. doi: 10.1359/jbmr.061113. [DOI] [PubMed] [Google Scholar]
- 2.Johnell O, et al. Predictive value of BMD for hip and other fractures. J Bone Miner Res. 2005;20:1185–94. doi: 10.1359/JBMR.050304. [DOI] [PubMed] [Google Scholar]
- 3.Kanis JA, et al. The use of clinical risk factors enhances the performance of BMD in the prediction of hip and osteoporotic fractures in men and women. Osteoporos Int. 2007;18:1033–46. doi: 10.1007/s00198-007-0343-y. [DOI] [PubMed] [Google Scholar]
- 4.Peacock M, Turner CH, Econs MJ, Foroud T. Genetics of osteoporosis. Endocr Rev. 2002;23:303–26. doi: 10.1210/edrv.23.3.0464. [DOI] [PubMed] [Google Scholar]
- 5.Ralston SH, Uitterlinden AG. Genetics of osteoporosis. Endocr Rev. 2010;31:629–62. doi: 10.1210/er.2009-0044. [DOI] [PubMed] [Google Scholar]
- 6.Hardy J, Singleton A. Genomewide association studies and human disease. N Engl J Med. 2009;360:1759–68. doi: 10.1056/NEJMra0808700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Manolio TA. Genomewide association studies and assessment of the risk of disease. N Engl J Med. 2010;363:166–76. doi: 10.1056/NEJMra0905980. [DOI] [PubMed] [Google Scholar]
- 8.Richards JB, et al. Bone mineral density, osteoporosis, and osteoporotic fractures: a genome-wide association study. Lancet. 2008;371:1505–12. doi: 10.1016/S0140-6736(08)60599-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Styrkarsdottir U, et al. Multiple genetic loci for bone mineral density and fractures. N Engl J Med. 2008;358:2355–65. doi: 10.1056/NEJMoa0801197. [DOI] [PubMed] [Google Scholar]
- 10.Rivadeneira F, et al. Twenty bone-mineral-density loci identified by large-scale meta-analysis of genome-wide association studies. Nat Genet. 2009;41:1199–206. doi: 10.1038/ng.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Styrkarsdottir U, et al. New sequence variants associated with bone mineral density. Nat Genet. 2009;41:15–7. doi: 10.1038/ng.284. [DOI] [PubMed] [Google Scholar]
- 12.Hsu YH, et al. An integration of genome-wide association study and gene expression profiling to prioritize the discovery of novel susceptibility Loci for osteoporosis-related traits. PLoS Genet. 2010;6:e1000977. doi: 10.1371/journal.pgen.1000977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kung AW, et al. Association of JAG1 with bone mineral density and osteoporotic fractures: a genome-wide association study and follow-up replication studies. Am J Hum Genet. 2010;86:229–39. doi: 10.1016/j.ajhg.2009.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duncan EL, et al. Genome-wide association study using extreme truncate selection identifies novel genes affecting bone mineral density and fracture risk. PLoS Genet. 2011;7:e1001372. doi: 10.1371/journal.pgen.1001372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Richards JB, et al. Collaborative meta-analysis: associations of 150 candidate genes with osteoporosis and osteoporotic fracture. Ann Intern Med. 2009;151:528–37. doi: 10.7326/0003-4819-151-8-200910200-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Meurs JB, et al. Large-scale analysis of association between LRP5 and LRP6 variants and osteoporosis. Jama. 2008;299:1277–90. doi: 10.1001/jama.299.11.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bagger YZ, et al. Links between cardiovascular disease and osteoporosis in postmenopausal women: serum lipids or atherosclerosis per se? Osteoporos Int. 2007;18:505–12. doi: 10.1007/s00198-006-0255-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kiel DP, et al. Genetic variation at the low-density lipoprotein receptor-related protein 5 (LRP5) locus modulates Wnt signaling and the relationship of physical activity with bone mineral density in men. Bone. 2007;40:587–96. doi: 10.1016/j.bone.2006.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raychaudhuri S, et al. Identifying relationships among genomic disease regions: predicting genes at pathogenic SNP associations and rare deletions. PLoS Genet. 2009;5:e1000534. doi: 10.1371/journal.pgen.1000534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lango Allen H, et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467:832–8. doi: 10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brunkow ME, et al. Bone dysplasia sclerosteosis results from loss of the SOST gene product, a novel cystine knot-containing protein. Am J Hum Genet. 2001;68:577–89. doi: 10.1086/318811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo YF, et al. Polymorphisms of the low-density lipoprotein receptor-related protein 5 (LRP5) gene are associated with obesity phenotypes in a large family-based association study. J Med Genet. 2006;43:798–803. doi: 10.1136/jmg.2006.041715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kornak U, et al. Loss of the ClC-7 chloride channel leads to osteopetrosis in mice and man. Cell. 2001;104:205–15. doi: 10.1016/s0092-8674(01)00206-9. [DOI] [PubMed] [Google Scholar]
- 24.Panagiotou OA, Evangelou E, Ioannidis JP. Genome-wide significant associations for variants with minor allele frequency of 5% or less--an overview: A HuGE review. Am J Epidemiol. 2010;172:869–89. doi: 10.1093/aje/kwq234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jin W, et al. Deubiquitinating enzyme CYLD negatively regulates RANK signaling and osteoclastogenesis in mice. J Clin Invest. 2008;118:1858–66. doi: 10.1172/JCI34257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sundaram K, Shanmugarajan S, Rao DS, Reddy SV. Mutant p62P392L stimulation of osteoclast differentiation in Paget’s disease of bone. Endocrinology. 2011;152:4180–9. doi: 10.1210/en.2011-1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grundberg E, et al. Population genomics in a disease targeted primary cell model. Genome Res. 2009;19:1942–52. doi: 10.1101/gr.095224.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duan Y, Beck TJ, Wang XF, Seeman E. Structural and biomechanical basis of sexual dimorphism in femoral neck fragility has its origins in growth and aging. J Bone Miner Res. 2003;18:1766–74. doi: 10.1359/jbmr.2003.18.10.1766. [DOI] [PubMed] [Google Scholar]
- 29.Karasik D, Ferrari SL. Contribution of gender-specific genetic factors to osteoporosis risk. Ann Hum Genet. 2008;72:696–714. doi: 10.1111/j.1469-1809.2008.00447.x. [DOI] [PubMed] [Google Scholar]
- 30.Ohlsson C, et al. Genetic determinants of serum testosterone concentrations in men. PLoS Genet. 2011;7:e1002313. doi: 10.1371/journal.pgen.1002313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Styrkarsdottir U, et al. European bone mineral density loci are also associated with BMD in East-Asian populations. PLoS One. 2010;5:e13217. doi: 10.1371/journal.pone.0013217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wood AR, et al. Allelic heterogeneity and more detailed analyses of known loci explain additional phenotypic variation and reveal complex patterns of association. Human molecular genetics. 2011;20:4082–92. doi: 10.1093/hmg/ddr328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mosekilde L, Torring O, Rejnmark L. Emerging anabolic treatments in osteoporosis. Curr Drug Saf. 2011;6:62–74. doi: 10.2174/157488611795684712. [DOI] [PubMed] [Google Scholar]
- 34.Hasselblad V, Hedges LV. Meta-analysis of screening and diagnostic tests. Psychol Bull. 1995;117:167–78. doi: 10.1037/0033-2909.117.1.167. [DOI] [PubMed] [Google Scholar]
- 35.Harris ST, et al. Effects of risedronate treatment on vertebral and nonvertebral fractures in women with postmenopausal osteoporosis: a randomized controlled trial. Vertebral Efficacy With Risedronate Therapy (VERT) Study Group. JAMA. 1999;282:1344–52. doi: 10.1001/jama.282.14.1344. [DOI] [PubMed] [Google Scholar]
- 36.Jackson RD, et al. Calcium plus vitamin D supplementation and the risk of fractures. N Engl J Med. 2006;354:669–83. doi: 10.1056/NEJMoa055218. [DOI] [PubMed] [Google Scholar]
- 37.Frazer KA, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–61. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Servin B, Stephens M. Imputation-based analysis of association studies: candidate regions and quantitative traits. PLoS Genet. 2007;3:e114. doi: 10.1371/journal.pgen.0030114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–13. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 40.Li Y, Willer C, Sanna S, Abecasis G. Genotype imputation. Annu Rev Genomics Hum Genet. 2009;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Estrada K, et al. GRIMP: a web-and grid-based tool for high-speed analysis of large-scale genome-wide association using imputed data. Bioinformatics. 2009;25:2750–2. doi: 10.1093/bioinformatics/btp497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin--rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 43.Aulchenko YS, Struchalin MV, van Duijn CM. ProbABEL package for genome-wide association analysis of imputed data. BMC Bioinformatics. 2010;11:134. doi: 10.1186/1471-2105-11-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Devlin B, Roeder K, Wasserman L. Genomic control, a new approach togenetic-based association studies. Theor Popul Biol. 2001;60:155–66. doi: 10.1006/tpbi.2001.1542. [DOI] [PubMed] [Google Scholar]
- 45.Ioannidis JP, Thomas G, Daly MJ. Validating, augmenting and refining genome-wide association signals. Nature reviews. Genetics. 2009;10:318–29. doi: 10.1038/nrg2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pereira TV, Patsopoulos NA, Salanti G, Ioannidis JP. Discovery properties of genome-wide association signals from cumulatively combined data sets. Am J Epidemiol. 2009;170:1197–206. doi: 10.1093/aje/kwp262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang J, et al. Genomic inflation factors under polygenic inheritance. Eur J Hum Genet. 2011;19:807–12. doi: 10.1038/ejhg.2011.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pe’er I, Yelensky R, Altshuler D, Daly MJ. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet Epidemiol. 2008;32:381–5. doi: 10.1002/gepi.20303. [DOI] [PubMed] [Google Scholar]
- 49.Reppe S, et al. Eight genes are highly associated with BMD variation in postmenopausal Caucasian women. Bone. 2010;46:604–12. doi: 10.1016/j.bone.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 50.Ge B, et al. Global patterns of cis variation in human cells revealed by high-density allelic expression analysis. Nat Genet. 2009;41:1216–22. doi: 10.1038/ng.473. [DOI] [PubMed] [Google Scholar]
- 51.Montgomery SB, et al. Transcriptome genetics using second generation sequencing in a Caucasian population. Nature. 2010;464:773–7. doi: 10.1038/nature08903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stranger BE, et al. Population genomics of human gene expression. Nat Genet. 2007;39:1217–24. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grundberg E, et al. Global analysis of the impact of environmental perturbation on cis-regulation of gene expression. PLoS Genet. 2011;7:e1001279. doi: 10.1371/journal.pgen.1001279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Emilsson V, et al. Genetics of gene expression and its effect on disease. Nature. 2008;452:423–8. doi: 10.1038/nature06758. [DOI] [PubMed] [Google Scholar]
- 55.Zeller T, et al. Genetics and beyond--the transcriptome of human monocytes and disease susceptibility. PLoS One. 2010;5:e10693. doi: 10.1371/journal.pone.0010693. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.