Abstract
Bats are the only mammals that use highly developed laryngeal echolocation, a sensory mechanism based on the ability to emit laryngeal sounds and interpret the returning echoes to identify objects. Although this capability allows bats to orientate and hunt in complete darkness, endowing them with great survival advantages, the genetic bases underlying the evolution of bat echolocation are still largely unknown. Echolocation requires high-frequency hearing that in mammals is largely dependent on somatic electromotility of outer hair cells. Then, understanding the molecular evolution of outer hair cell genes might help to unravel the evolutionary history of echolocation. In this work, we analyzed the molecular evolution of two key outer hair cell genes: the voltage-gated potassium channel gene KCNQ4 and CHRNA10, the gene encoding the α10 nicotinic acetylcholine receptor subunit. We reconstructed the phylogeny of bats based on KCNQ4 and CHRNA10 protein and nucleotide sequences. A phylogenetic tree built using KCNQ4 amino acid sequences showed that two paraphyletic clades of laryngeal echolocating bats grouped together, with eight shared substitutions among particular lineages. In addition, our analyses indicated that two of these parallel substitutions, M388I and P406S, were probably fixed under positive selection and could have had a strong functional impact on KCNQ4. Moreover, our results indicated that KCNQ4 evolved under positive selection in the ancestral lineage leading to mammals, suggesting that this gene might have been important for the evolution of mammalian hearing. On the other hand, we found that CHRNA10, a gene that evolved adaptively in the mammalian lineage, was under strong purifying selection in bats. Thus, the CHRNA10 amino acid tree did not show echolocating bat monophyly and reproduced the bat species tree. These results suggest that only a subset of hearing genes could underlie the evolution of echolocation. The present work continues to delineate the genetic bases of echolocation and ultrasonic hearing in bats.
Keywords: Chiroptera, CHRNA10, hearing, KCNQ4, natural selection, parallel evolution
Introduction
During the evolution of mammals, the inner ear underwent many important morphological and functional variations different from other vertebrates. Some of the changes are noteworthy, like the elongation of the papilla, the division of hair cells into two subtypes with differential functions (inner hair cells [IHCs] and outer hair cells [OHCs]), and the appearance of somatic electromotility for active amplification of sounds (Manley 2000; Manley et al. 2004). The somatic electromotility is a mammalian novelty (Brownell et al. 1985; Dallos 2008) and is based on the unique properties of the motor protein prestin (Zheng et al. 2000; Liberman et al. 2002). Prestin is the fifth member of the SLC26 family of multifunctional anion exchangers (Mount and Romero 2004), which had been under positive selection in the lineage leading to mammals (Franchini and Elgoyhen 2006; Elgoyhen and Franchini 2011) most probably to acquire new functional capacities. The evolution of the inner ear has endowed mammals with the capability of hearing a wide range of sound frequencies (Manley et al. 2004), including the highest (more than 200 kHz) among vertebrates (Fenton and Bell 1981).
Bats are one of the most specialized and diversified mammalian groups. In addition to being the only mammals that can fly, bats exploit advanced echolocation to perceive the surroundings. The combination of flight and echolocation has most probably allowed bats to conquer a high variety of habitats on Earth and become a very diverse and successful group of mammals (Altringham 1996; Jones and Teeling 2006). In recent years, several molecular studies have addressed the evolution of echolocation in bats (Teeling et al. 2000, 2002; Eick et al. 2005). A large amount of molecular data support a phylogeny of bats in which the family Pteropodidae (mainly nonecholocating Old World fruit bats with the exception of tongue-clicking in one genus) groups together with the echolocating superfamily Rhinolophoidea in the suborder Yinpterochiroptera, whereas all other echolocating bats belong to the monophyletic suborder Yangochiroptera (Teeling et al. 2005). This topology suggests that either laryngeal echolocation was gained once in the ancestor of bats and then lost in the Old World fruit bats or that laryngeal echolocation evolved independently multiple times in bats (Teeling et al. 2005). However, a phylogenetic reconstruction of bats using PRESTIN coding sequences unites echolocating bats into a monophyletic clade, with the pteropodids in a basal position (Li et al. 2008), suggesting that this gene evolved convergently in laryngeal echolocating bats. Moreover, natural selection has driven convergent prestin substitutions in echolocating dolphins, which group with echolocating bats in a phylogenetic tree (Li et al. 2010; Liu et al. 2010). Altogether, these results suggest that the emergence of echolocation in mammals is correlated with the evolution of PRESTIN, a key gene for high-frequency hearing, a requirement for echolocation (Vater and Kössl 2004). We hypothesize that the evolution of echolocation required more genes to be shaped by natural selection. In this regard, we propose that in order to understand the genetic bases underlying echolocation evolution, more genes and pathways involved in any of the necessary abilities that allow bats to echolocate and hear ultrasonic sound need to be studied.
OHC somatic electromotility makes a major contribution to mechanical amplification, hearing sensitivity, and frequency selectivity (Brownell et al. 2001; Dallos et al. 2008; Mellado Lagarde et al. 2008) and most likely contributes to the extended high-frequency sensitivity of mammals (Manley 2000; Manley et al. 2004). Thus, genes expressed in OHCs, which underlie or modulate somatic electromotility, become candidates to study in order to further understand the genetic bases of echolocation and its evolution. In this work, we sequenced and analyzed the evolution in bats of two key genes expressed in OHCs: the voltage-gated potassium channel subfamily KQT member 4 (KCNQ4) and the gene coding for the α10 nicotinic acetylcholine receptor (nAChR) subunit (CHRNA10). KCNQ4 is expressed exclusively in the basal membrane of OHCs (Kharkovets et al. 2000) and produces a major K+ conductance (IK,n) at resting potential of the cells (Housley and Ashmore 1992), which facilitate the maturation of OHCs (Marcotti and Kros 1999). Furthermore, mutations in KCNQ4 can lead to slowly progressive hearing loss in autosomal dominant nonsyndromic deafness (DFNA2) patients (Coucke et al. 1999; Kubisch et al. 1999). On the other hand, the α10 nAChR subunit assembles with α9 to form the receptor that mediates synaptic transmission between medial olivocochlear fibers and OHCs (Elgoyhen et al. 1994, 2001), which serves inhibition of somatic electromotility (Guinan 1996). Since KCNQ4 and CHRNA10 contribute to the modulation of somatic electromotility in OHCs and because CHRNA10 is the only inner ear gene, apart from PRESTIN, where signatures of positive selection in the mammalian ancestor have been found so far (Franchini and Elgoyhen 2006; Elgoyhen and Franchini 2011), both genes are attractive candidates to analyze, in order to further uncover the evolution of echolocation. In the present work, multiple phylogenetic and molecular evolutionary analyses were used to investigate whether the evolution of KCNQ4 and CHRNA10 is associated with bat echolocation.
Materials and Methods
Taxonomic Coverage, Gene Cloning, and Sequencing
In order to obtain a wide taxonomic coverage of the order Chiroptera, we included representative echolocating and nonecholocating bat species in this study. For KCNQ4, we collected samples from 41 bat species covering 11 families, and for CHRNA10, we collected and sequenced eight bat species. We included three species of Old World fruit bats of the suborder Yinpterochiroptera (Cynopterus sphinx, Pteropus vampyrus, and Rousettus leschenaultii). Although R. leschenaultii uses tongue-clicking echolocation, the other pteropodid species included do not have laryngeal echolocation or associated high-frequency sensitivity. We also included laryngeal echolocating bats from both Yinpterochiroptera and Yangochiroptera. Our species sampling covered both constant-frequency (Rhinolophus ferrumequinum and Hipposideros armiger) and frequency-modulated echolocating species (Myotis lucifugus, Tadarida plicata, and Rhinopoma hardwickii). All bats were collected in the wild, and a small piece of skin from the wing was taken. Then, the animal was freed into the wild as soon as possible. Some individuals were sacrificed and bat tissues, such as brain, were kept at −80 °C for RNA preservation. Parts of the brain samples were collected during the surveillance program for coronaviruses (Li, Wang, et al. 2007). The sample collection of bats followed ethical principles by the Animal Ethics Committee of East China Normal University, and all the samples were held at the School of Life Sciences, East China Normal University.
Genomic DNA was extracted from bat wing membrane biopsy using the DNeasy kit (Qiagen). Total RNA was isolated from bat brain sample using the RNAiso kit (TaKaRa), and cDNA was synthesized for cloning with Superscript III reverse transcriptase (Invitrogen). Based on KCNQ4 sequences from public databases, we designed two degenerate primers to amplify coding regions from 19 bat cDNA samples (F-general and R-general). The polymerase chain reaction (PCR) protocol was 95 °C for 5 min, 30 cycles at 95 °C for 30 s, 57 °C for 30 s and 72 °C for 2 min, and a final extension at 72 °C for 10 min. We also sequenced KCNQ4 missing exons in P. vampyrus, Tursiops truncatus, and Felis catus from genomic DNAs, in order to complete the sequence information obtained from Ensembl (www.ensembl.org). To further determine the evolutionary patterns of amino acids at positions 388 and 406 of KCNQ4, we sequenced partial exon 12 from additional 21 bat species using genomic DNA (F-exon 12 and R-exon 12). To sequence whole coding regions of CHRNA10, we divided the gene into three sections and performed independent PCRs. The primers were designed based on conserved regions of mammalian CHRNA10 sequences from public databases. The first gene segment covered exons 2 and 3, the second ranged from exon 3 to 4 and the last from exon 4 to 5. We used the following PCR protocol: 95 °C for 5 min, 10–15 touch-down cycles with annealing temperature from 68 °C to 52°C (decreasing 1 °C per cycle), 15–20 cycles using the final annealing temperature, and then extension at 72 °C for 10 min. We also designed a primer set to sequence the missing part of exon 4 in the M. lucifugus genome. For detailed information on primers used in this study, see supplementary table S1 (Supplementary Material online). All the PCR products were cloned using pGEM-T Easy vector (Promega) and sequenced on an ABI 3730 sequencer (Applied Biosystems). Sequences of KCNQ4 and CHRNA10 from our experiments were deposited in the European Nucleotide Archive EMBL-Bank database, and the supplementary tables S2 and S3 (Supplementary Material online) contain detailed information of the new sequences (accession nos. HE608266–HE608319).
Sequence Acquisition from Public Databases
To study the molecular evolution of KCNQ4 and CHRNA10 in bats and in mammals, we downloaded coding sequences from either NCBI (www.ncbi.nlm.nih.gov) or Ensembl (www.ensembl.org). We used BLAT to search Ensembl with the human KCNQ4 and CHRNA10 sequences as queries and only sequences containing whole coding regions were kept. For phylogenetic reconstruction of the KCNQ4 gene tree, we used 13 mammalian coding sequences, including two bat genes (P. vampyrus and M. lucifugus). To study the evolution of the KCNQ4 exon 12 in detail, we retrieved additional sequences from another 11 mammalian species. For the reconstruction of the CHRNA10 phylogenetic tree, we used 15 mammalian coding sequences from the public databases. For branch-site tests of positive selection, further vertebrate sequences were also downloaded directly from Ensembl. Accession numbers of all sequences downloaded from public databases were also given in supplementary tables S2 and S3 (Supplementary Material online).
Phylogenetic Reconstruction
The coding sequences obtained were aligned using CLUSTAL W (Thompson et al. 1994) implemented in MEGA 4 software (Tamura et al. 2007). The programs ModelTest (Posada and Crandall 1998; Posada 2006) and ProtTest (Abascal et al. 2005) were used to evaluate nucleotide and amino acid substitution models individually, according to Akaike information criterion (Akaike 1974). GTR + I + Γ and JTT + I + Γ + F models were respectively used for nucleotide and amino acid tree reconstructions in KCNQ4 analyses. GTR + I + Γ and JTT + Γ + F models were used for CHRNA10 data. Bayesian phylogeny reconstructions of amino acid sequences were performed using MrBayes 3.1.2 (Ronquist and Huelsenbeck 2003). A parameter of 5 million generations was set in the Markov chain, including a burn-in step corresponding to the first 2 million generations. Markov chain convergence after burn-in was confirmed by AWTY graphical analyses (Nylander et al. 2008). Bayesian phylogenetic inferences were also used for nucleotide sequences of both genes. The program RAxML 7.0.4 was implemented to reconstruct the maximum-likelihood (ML) phylogeny (Stamatakis 2006). Maximum parsimony (MP), and neighbor joining (NJ) trees were reconstructed from MEGA 4, both with 2,000 bootstrap replicates. In addition, synonymous and nonsynonymous substitution trees of KCNQ4 were also carried out in MEGA 4, using Nei-Gojobori p distance (Nei and Gojobori 1986; Tamura et al. 2007).
Evolutionary Analysis in Bats
To test selective pressure of the hearing genes in bat lineages, we performed two-ratio model using CODEML program in PAML 4 package and compared it with one-ratio model through a likelihood ratio test (LRT) (Yang 1998, 2007). The two-ratio model assumes focal lineage(s) have a different ω value (rate of nonsynonymous substitutions/rate of synonymous substitutions; dN/dS) than others lineages; whereas, the one-ratio model supposes ω value is the same across all branches. The statistic 2Δℓ (twice the log likelihood difference between the nested models) was compared with the chi-square distribution. First, we tested the ancestors leading to all bats and Old World fruit bats individually. Then we tested whether the ω value in six ancestral branches leading to echolocating bats was significantly higher than background lineages. We also performed free-ratio model to infer ancestral states of KCNQ4 and CHRNA10 (Yang 1998). A species topology based on published phylogenetic studies was used in the analyses (Murphy et al. 2001; Giannini and Simmons 2003; Hoofer and Van Den Bussche 2003; Teeling et al. 2005; Steiper and Young 2006; Miller-Butterworth et al. 2007; McGowen et al. 2009; Churakov et al. 2010).
Then we searched for parallel amino acid substitutions along paraphyletic echolocating bat lineages. The software CONVERG 2 was used to test whether the observed parallel substitutions in our focal echolocating branches were fixed randomly or by natural selection (Zhang and Kumar 1997). For the expanded KCNQ4 exon 12 dataset, Mesquite 2.72 was used to trace ancestral states of the 388 and 406 residues in mammals under parsimony criterion (Maddison WP and Maddison DR 2009). The expanded species tree was based on additional reported phylogenetic data (Li, Liang, et al. 2007; Thabah et al. 2007; Datzmann et al. 2010). Multivariate analysis of protein polymorphism (MAPP) was used to determine the physicochemical impact of parallel amino acid substitutions in echolocating bats (Stone and Sidow 2005). Physicochemical constraint for each site was estimated using an alignment of KCNQ4 excluding both echolocating bat and mouse sequences in order to prevent the influence of Ile and Ser at positions 388 and 406.
To carry out in-depth adaptive evolutionary analysis, the modified branch-site model A (test 2) of positive selection was carried out in vertebrate sequences (Zhang et al. 2005). The focal branches in this test were the ancestors of Yangochiroptera, yinpterochiropteran echolocating bats, and Old World fruit bats. Additionally, we also analyzed the ancestors of mammals and placentals. Two nested hypotheses were tested for each focal branch. In the alternative hypothesis, three ω values (ω0, ω1, and ω2) were calculated representing the codons under negative, null, and positive selection, respectively. However, in the null hypothesis, only two ω values (ω0 and ω1) were estimated and no positively selected sites were allowed. In the case where positive selection was found in our focal branch, particular sites with positive selection will be detected by the Bayes empirical Bayes method (Yang et al. 2005).
Results
Genetic Data from Bats
We obtained over 80% KCNQ4 coding sequences from 19 bat species, including three species from the fruit bat family Pteropodidae and 16 species from nine echolocating bat families (Rhinolophidae, Hipposideridae, Rhinopomatidae, Megadermatidae, Vespertilionidae, Miniopteridae, Molossidae, Mormoopidae, and Phyllostomidae). Several KCNQ4 variants were cloned randomly from different species. The splice variants were conserved in bats and were not included in the analyses (see supplementary fig. S1, Supplementary Material online). The numbering of KCNQ4 amino acid sites was based on splice variant four of human KCNQ4. We also obtained exon 12 sequence data from another 21 bats, in order to investigate the evolution of positions 388 and 406 more thoroughly. Coupled with exon 12 sequences from additional 11 mammals, we totally collected data for KCNQ4 exon 12 in 64 mammalian species. For CHRNA10, we sequenced and collected the whole coding regions of the gene from nine bats, without the signal peptide sequences. Two of them (R. leschenaultii and P. vampyrus) are from family Pteropodidae and the other seven species belong to echolocating bat families (Rhinolophidae, Hipposideridae, Rhinopomatidae, Vespertilionidae, Molossidae, and Phyllostomidae). We analyzed in total 24 mammalian CHRNA10 sequences.
Phylogenetic Conflict between KCNQ4 Gene Tree and Bat Species Tree
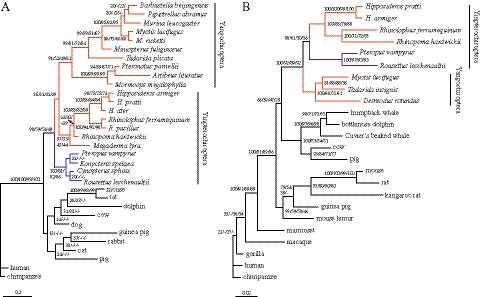
Our results showed that in a Bayesian phylogenetic tree of KCNQ4 amino acid sequences, echolocating bats from the two suborders grouped together with a Bayesian posterior probability of 36% (fig. 1A). In addition, amino acid trees performed using ML, MP, and NJ methods also supported the monophyly of echolocating bats (fig. 1A). In contrast, the nucleotide tree (supplementary fig. S2A, Supplementary Material online) showed similar topology to the bat species tree (Teeling et al. 2005). Moreover, we found that synonymous and nonsynonymous substitution trees also support the topology conflict. A tree based on synonymous substitutions showed echolocating bat paraphyly, whereas the nonsynonymous tree revealed monophyly of echolocating bats (supplementary fig. S3, Supplementary Material online). Our phylogenetic analyses indicated that nonsynonymous nucleotide substitutions and amino acid substitutions led to the topology difference and as expected, synonymous nucleotide substitutions evolved more neutrally.
Fig. 1.
KCNQ4 (A) and CHRNA10 (B) protein trees. Different phylogenetic methods were used to reconstruct the amino acid trees of both genes. Values on the branches indicate statistical supports from Bayesian, ML, MP, and NJ, respectively, and the negative sign (−) indicates lack of support for a specific method. Echolocating bats (orange) and Old World fruit bats (blue) are indicated in both trees. The human sequences were used to root the trees and the scale bars indicate amino acid substitutions per amino acid site.
Tests of Positive Selection on Bat KCNQ4
The results obtained from the two-ratio model indicated that the ancestor of all bats was under purifying selection (ω = 0.027, 2Δℓ = 0.00044, df = 1, P = 0.983; ω from one-ratio model is 0.027) and did not show evidence of episodic positive selection. This result suggested that there was not a correlation between adaptive molecular changes on KCNQ4 and the acquisition of functional capacities underlying echolocation in the ancestor of bats. The results also indicated that the ancestral branch leading to Old World fruit bats was under purifying selection (ω = 0.015, 2Δℓ = 0.69, df = 1, P = 0.406). This finding did not support the hypothesis that the clustering of echolocating bats observed was due to accelerated evolution of the fruit bat lineage. In addition, we tested six focal branches leading to echolocating bat ancestors (dotted lineages in fig. 2A) because substitutions in these branches may have fundamental influences on the evolution of bats. Our results showed that the six focal echolocating bat ancestor branches had significantly higher ω value compared with other branches (0.053 and 0.025, respectively, 2Δℓ = 5.92, df = 1, P = 0.015), suggesting the potential action of positive selection during early stages of the evolution of echolocating bats. However, the LRT is not significant after the amino acid sites at positions 388 and 406 were removed (2Δℓ = 2.89, df = 1, P = 0.089). The free-ratio model was significantly better than the one-ratio model (2Δℓ = 96.32, df = 60, P = 0.002). This result indicated that a model of different selective pressures acting on KCNQ4 lineages adjusted better to the evolutionary history of this gene than the one-ratio model. Hence, we used the ancestral sequence reconstructions from the free-ratio model.
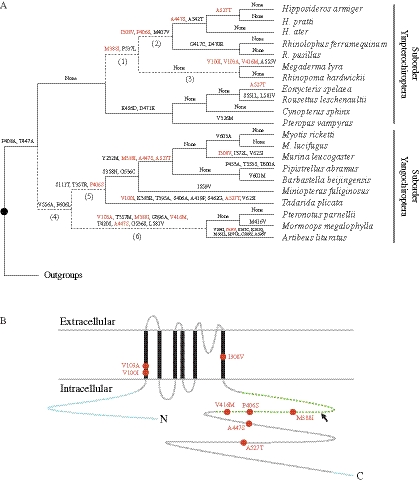
Fig. 2.
Parallel substitutions in KCNQ4. (A) KCNQ4 amino acid substitutions among different lineages of bats. The eight substitutions that were fixed independently in yinpterochiropteran and yangochiropteran echolocating bats are indicated with orange letters. Dotted lines and numbers in parentheses indicate the focal echolocating bat branches that were tested by the two-ratio model and the parallel evolution test (table 1). Outgroup species are: mouse, rat, guinea pig, rabbit, dolphin, cow, pig, cat, dog, chimpanzee, and human. (B) Schematic protein model of KCNQ4 was based on prediction from the Universal Protein Resource (http://www.uniprot.org/uniprot/P56696). The parallel amino acid substitutions are indicated with orange circles, the six transmembrane domains are indicated with black bars, and the intracellular membrane proximal region is indicated by a green dotted line. The splice variants of KCNQ4 take place in the membrane proximal region (indicated by an arrow). The missing sequences of KCNQ4 in the study are shown in blue.
Parallel Evolution of KCNQ4 among Echolocating Bats
There were a total of eight parallel amino acid substitutions identified in the two echolocating bat clades (fig. 2A), and the parallel changes were highlighted in a schematic model of KCNQ4 (fig. 2B). Two of them (M388I and P406S) appeared early on during the evolution of echolocating bats, and the two parallel substitutions deviated significantly from random expectation at a significance level of 0.05 (table 1). Therefore, we proposed that the parallel evolution of KCNQ4 in echolocating bats was driven by natural selection. Interestingly, when the 388 site was eliminated, the amino acid phylogeny of KCNQ4 showed paraphyly of echolocating bats again (supplementary fig. S4, Supplementary Material online). The ancestral states of residues at positions 388 and 406 were also reconstructed from the expanded exon 12 data. With the exception of four species from the families Miniopteridae and Molossidae, we observed that all echolocating bats shared the amino acid Ile at position 388. Surprisingly, mice, which are also sensitive to high-frequency sounds, fixed Ile independently, whereas the remaining mammalian species conserved a Met residue at position 388 (fig. 3A). On the other hand, the derived Ser residue at position 406 was exclusively shared by two bat families (Rhinolophidae and Hipposideridae) from suborder Yinpterochiroptera and two (Vespertilionidae and Miniopteridae) from suborder Yangochiroptera (fig. 3B). MAPP scores were calculated for the M388I and P406S amino acid replacements and the results indicated that these two replacements had an important functional effect (score = 21.99, P < 0.0001 and 9.76, P = 0.0025, respectively) on KCNQ4.
Table 1.
Statistical Tests for Parallel Evolution.
| Gene | Parallel Substitution | Branch Paira | Observed Number | Expected Number | P value (after correction for multiple testingb) |
| KCNQ4 | M388I | 1 versus 6 | 1 | 0.002 | 0.019 |
| P406S | 2 versus 5 | 1 | 0.002 | 0.015 | |
| CHRNA10 | A100T | 9 versus 10 | 1 | 0.013 | 0.116 |
The numbers correspond to focal echolocating branches in figure 2A and supplementary figure S5, Supplementary Material online.
The P value of each test was multiplied by nine (total branch pairs among focal echolocating lineages) to correct for multiple testing.
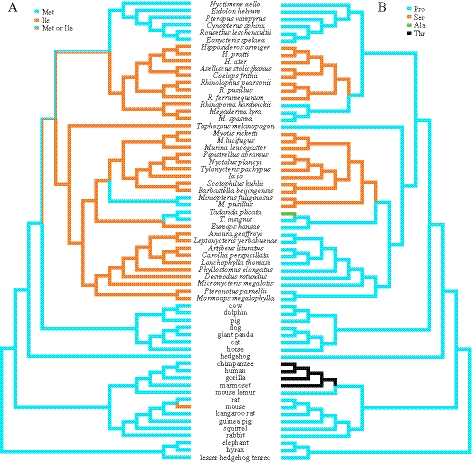
Fig. 3.
Evolution of the amino acid positions 388 (A) and 406 (B) in KCNQ4 by parsimony criterion. Derived states of the two amino acids in echolocating bats are shown in orange and ancestral states are shown in blue. Amino acid states at position 388 are not resolved in branches leading to yinpterochiropterans and all bats. The bat Tadarida plicata and some primates show an Ala (green) and a Thr (black) at position 406.
Phylogeny and Molecular Evolution of CHRNA10
The amino acid trees of CHRNA10 did not support the monophyly of echolocating bats. Moreover, the CHRNA10 trees, either at the amino acid or nucleotide level, were similar with the bat species tree (fig. 1B and supplementary fig. S2B, Supplementary Material online). The free-ratio model suited the data better than the one-ratio model (2Δℓ = 65.05, df = 44, P = 0.021). The ω values from the two-ratio tests for bat ancestor (ω = 0.108, 2Δℓ = 0.268, df = 1, P = 0.605) and Old World fruit bat ancestor (ω = 0.055, 2Δℓ = 1.13, df = 1, P = 0.288) were not significantly better than the one-ratio model (ω from one-ratio model is 0.08). The test for the echolocating bat ancestors was also not significantly better than the one-ratio model (ω = 0.106, 2Δℓ = 2.33, df = 1, P = 0.127). Interestingly, there was one mutation in CHRNA10 (A100T) that occurred in two focal echolocating ancestors (supplementary fig. S5, Supplementary Material online). The test for parallel amino acid substitutions was not significant after the correction for multiple testing, so we could not exclude the possibility that the parallel changes were fixed randomly in echolocating bats (table 1).
In-Depth Branch-Site Analyses
Our branch-site model results obtained with CHRNA10 and KCNQ4 demonstrated that none of the analyzed bat lineages in the species tree showed a significant probability to fit an adaptive evolutionary hypothesis better than a neutral evolutionary hypothesis. On the other hand, the mammalian and placental lineages showed strong signals of adaptive evolution (supplementary table S4, Supplementary Material online). The results obtained with CHRNA10 were in agreement with previously published data (Franchini and Elgoyhen 2006). The strong signals of adaptive evolution detected in KCNQ4, in the lineage leading to mammals and in the placental lineage, suggested that this gene might have acquired novel functional capacities. However, more functional studies are necessary to probe this hypothesis.
Discussion
Our results indicated that both KCNQ4 and CHRNA10 genes were under purifying selection in most mammals and suggested that these two OHC genes are functionally important. In addition, our branch-site evolutionary studies revealed that KCNQ4 evolved under positive selection in the common ancestor of mammals, suggesting that this gene might be important for the evolution of mammalian hearing. On the other hand, our results showed that amino acid phylogenies of KCNQ4 based on various reconstruction methods, consistently grouped together all echolocating bats, whereas CHRNA10 amino acid trees did not support the monophyly of echolocating bats. In contrast, phylogenetic trees based on nucleotide sequences for both genes were consistent with the bat species tree. Several alternative explanations can account for the phylogeny discordance between the KCNQ4 gene tree and the bat species tree, such as gene duplication following subsequent loss, long-branch attraction (LBA), and parallel sequence evolution. The topology discordance observed is unlikely to be the result of an ancient gene gain and loss process in bats, since there is only one known KCNQ4 gene in mammals. In addition, the nucleotide gene tree supported the paraphyly of echolocating bats, which is in agreement with the reported bat species tree (Teeling et al. 2005). We can also exclude the possibility that LBA led to the topology difference, because the two ancestral branches leading to yinpterochiropteran and yangochiropteran echolocating bats are not obviously long (0.08 and 0.09 amino acid substitutions per site) compared with the ancestral branch of Old World fruit bats (0.08 amino acid substitutions per site). Cases of convergent molecular evolution that cause phylogenetic conflict with well-established species trees have been reported in the past. Among these cases, it is noteworthy to mention the finding that prestin evolved convergently in echolocating bats and cetaceans (Li et al. 2010; Liu et al. 2010), and also the case of convergent evolution of mitochondrial genes in agamid lizards and snakes (Castoe et al. 2009). Our data showed that only trees based on amino acid sequences and nonsynonymous substitutions led to the topology difference, indicating that parallel amino acid changes were responsible for the clustering of echolocating bats. Furthermore, the higher ω value observed in echolocating ancestors was consistent with potential positive selection acting on these branches during the evolution of KCNQ4 in bats. In contrast, the ancestor of Old World fruit bats showed signatures of purifying selection. Altogether, these results suggest that there is no relaxed selective pressure during the evolution of Old World fruit bats.
Among the eight parallel substitutions in KCNQ4 identified in echolocating bats, two of them, the 388 and 406 residues, occurred in basal ancestors, and the 388 residue exerted a substantial contribution to the KCNQ4 phylogeny difference. Both parallel amino acid substitutions showed probabilities that deviated significantly from neutral evolution, which indicated that the acquisitions of the two identical residues in echolocating bat lineages were most probably driven by natural selection. Moreover, when the two sites are removed, the ω value of focal echolocating ancestors was not significantly higher than background lineages. This result gave further support to the hypothesis that these two sites underlay the selective pressure change in echolocating bat ancestors. The detailed evolutionary study of the 388 position in mammalian KCNQ4 revealed that Ile was fixed almost exclusively in echolocating taxa. Among the 36 echolocating bat species analyzed at this position, 32 possess the Ile residue and the 4 species of the families Molossidae and Miniopteridae that do not have this change seem to have evolved from an ancestor that did possess Ile at this position. The result suggests that Ile was selected independently early on during the evolution of echolochating bats of the two suborders. An alternative explanation is that pteropodids evolved from an ancestor having Ile at this position and mutated it back to the ancestral state in mammals. There are two hypotheses concerning the origin of bat echolocation: one hypothesis suggests that laryngeal echolocation evolved once in bats, and it was subsequently lost in pteropodids (Springer et al. 2001), whereas the second hypothesis suggests that echolocation arose independently in the two bat suborders (Eick et al. 2005). In any case, the fact that the majority of echolocating bat species possess the Ile residue suggests that this amino acid might be important for the function of KCNQ4 in high-frequency hearing and echolocation. On the other hand, the substitution P406S of KCNQ4 is exclusively shared by echolocating bats from two yinpterochiropteran families (Rhinolophidae and Hipposideridae) and two yangochiropteran families (Vespertilionidae and Miniopteridae), indicating that the substitutions were fixed by parallel evolution.
Although it would be tempting to correlate these findings with the hearing capacity in each one of these species, the characterization of bat hearing is still incomplete. In the absence of a clear knowledge concerning the best hearing frequency for each echolocating bat species, it is difficult to conclude whether these amino acid substitutions have association with the acquisition of high-frequency sensitivity. Among all mammalian species analyzed, the only species other than bats displaying an Ile residue at position 388 of KCNQ4 is the domestic mouse (Mus musculus), which can also use ultrasonic sound in some conditions, and whose upper hearing limit is around 90 kHz, with the best hearing frequency at 16 kHz (Heffner and Masterton 1980). The MAPP predictions indicated that M388I and P406S parallel substitutions would exert a significant functional impact on KCNQ4. These two residues are located in the predicted membrane proximal region of the C terminus, which has been reported to be responsible for the targeting of KCNQ channels to the membrane (Chung et al. 2006; Xu et al. 2007). Due to the cellular membrane distribution of KCNQ channels influencing their physiological functions significantly (Chung et al. 2006), KCNQ4 is expressed exclusively in the basal pole of OHCs, and the expression level is higher in basal turns of cochlea, where high-frequency sounds are detected (Kharkovets et al. 2000). Further studies are needed in order to establish the consequences of amino acid substitutions at positions 388 and 406 of KCNQ4 in the functional properties and targeting of this protein in OHCs, including how this affects somatic electromotility and how it relates to echolocation and/or ultrasonic hearing.
Different to that described for KCNQ4, CHRNA10 does not show parallel evolution driven by natural selection in echolocating bats. Although the A100T substitutions were independently fixed in two echolocating bat clades, the statistical analysis did not support the action of natural selection on the fixation of this residue. In addition, our analysis of the CHRNA10 gene showed that the ancestor of Old World fruit bats was under purifying selection. Even though there were nine amino acid substitutions that occurred in this branch, in comparison to only three substitutions in the two lineages leading to echolocating bat ancestors, the higher number of substitutions was more likely correlated with a longer evolutionary time (Teeling et al. 2005). It has been previously shown that the genes encoding prestin and the α10 nAChR subunit evolved adaptively in the ancestral branch leading to mammals (Franchini and Elgoyhen 2006; Elgoyhen and Franchini 2011), and convergent prestin substitutions occurred in echolocating bats and whales (Li et al. 2008; Li et al. 2010; Liu et al. 2010). Thus, lack of parallel evolution driven by natural selection in echolocating bats for CHRNA10 is in contrast to the previously described evolutionary history of PRESTIN in this group of mammals. Altogether, these data indicate that after a process of positive selection in the branch leading to mammals shared by PRESTIN and CHRNA10, the two genes seem to have followed different evolutionary pathways: whereas PRESTIN continued to evolve adaptively in echolocating mammals, CHRNA10 has been under strong purifying selection in all groups of mammals. It has been suggested that the evolution of OHC mechanical amplification coevolved with the efferent system, which specialized to modulate the amplifier (Manley and Köppl 1998; Köppl 2011). Taken together, the present results might indicate that whereas some key OHC proteins necessary for mammalian mechanical amplification acquired amino acid substitutions to best fit echolocation and ultrasonic hearing, the efferent system as known in mammals has all the capabilities to fine-tune amplification and frequency selectivity even in the highest frequency hearing mammals.
In summary, our data showed topology differences in KCNQ4, but not in CHRNA10 phylogeny due to parallel amino acid substitutions in the two lineages of echolocating bats. These acquired changes had been probably important for the evolution of echolocation and ultrasonic hearing. Thus, the present work contributes toward the identification of genes that were targeted by selective forces during the evolution of bat ultrasonic hearing and echolocation and further help understand the intimacies of the mammalian cochlea at work.
Supplementary Material
Supplementary tables S1–S4 and figures S1–S5 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
We thank Gang Li for valuable advice in early stages of this work and Yi-Hsuan Pan for helpful comments. This work was supported by the Key Construction Program of the National “985” Project and “211” Project to S.Z., the Ministry Reward for Excellent Doctors in Academics (MXRZZ2010010) to Y.L., Agencia Nacional de Promoción Científica y Tecnológica (ANPCyT) and Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET) Research Grants to L.F.F., an International Research Scholar Grant from the Howard Hughes Medical Institute, ANPCyT, and CONICET to A.B.E., a CONICET doctoral fellowship to F.P. and Department of Science and Technology (SR/FT/L-97/2005) financial support to K.E.R.
References
- Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- Akaike H. A new look at statistical model identification. IEEE Trans Automat Contr. 1974;19:716–723. [Google Scholar]
- Altringham JD. Bats: biology and behaviour. Oxford: Oxford University Press; 1996. [Google Scholar]
- Brownell WE, Bader CR, Bertrand D, de Ribaupierre Y. Evoked mechanical responses of isolated cochlear outer hair cells. Science. 1985;227:194–196. doi: 10.1126/science.3966153. [DOI] [PubMed] [Google Scholar]
- Brownell WE, Spector AA, Raphael RM, Popel AS. Micro- and nanomechanics of the cochlear outer hair cell. Annu Rev Biomed Eng. 2001;3:169–194. doi: 10.1146/annurev.bioeng.3.1.169. [DOI] [PubMed] [Google Scholar]
- Castoe TA, de Koning AP, Kim HM, Gu W, Noonan BP, Naylor G, Jiang ZJ, Parkinson CL, Pollock DD. Evidence for an ancient adaptive episode of convergent molecular evolution. Proc Natl Acad Sci U S A. 2009;106:8986–8991. doi: 10.1073/pnas.0900233106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung HJ, Jan YN, Jan LY. Polarized axonal surface expression of neuronal KCNQ channels is mediated by multiple signals in the KCNQ2 and KCNQ3 C-terminal domains. Proc Natl Acad Sci U S A. 2006;103:8870–8875. doi: 10.1073/pnas.0603376103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churakov G, Sadasivuni MK, Rosenbloom KR, Huchon D, Brosius J, Schmitz J. Rodent evolution: back to the root. Mol Biol Evol. 2010;27:1315–1326. doi: 10.1093/molbev/msq019. [DOI] [PubMed] [Google Scholar]
- Coucke PJ, Van Hauwe P, Kelley PM, et al. (20 co-authors) Mutations in the KCNQ4 gene are responsible for autosomal dominant deafness in four DFNA2 families. Hum Mol Genet. 1999;8:1321–1328. doi: 10.1093/hmg/8.7.1321. [DOI] [PubMed] [Google Scholar]
- Dallos P. Cochlear amplification, outer hair cells and prestin. Curr Opin Neurobiol. 2008;18:370–376. doi: 10.1016/j.conb.2008.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallos P, Wu X, Cheatham MA, et al. (12 co-authors) Prestin-based outer hair cell motility is necessary for mammalian cochlear amplification. Neuron. 2008;58:333–339. doi: 10.1016/j.neuron.2008.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datzmann T, von Helversen O, Mayer F. Evolution of nectarivory in phyllostomid bats (Phyllostomidae Gray, 1825, Chiroptera: Mammalia) BMC Evol Biol. 2010;10:165. doi: 10.1186/1471-2148-10-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eick GN, Jacobs DS, Matthee CA. A nuclear DNA phylogenetic perspective on the evolution of echolocation and historical biogeography of extant bats (Chiroptera) Mol Biol Evol. 2005;22:1869–1886. doi: 10.1093/molbev/msi180. [DOI] [PubMed] [Google Scholar]
- Elgoyhen AB, Franchini LF. Prestin and the cholinergic receptor of hair cells: positively-selected proteins in mammals. Hear Res. 2011;273:100–108. doi: 10.1016/j.heares.2009.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgoyhen AB, Johnson DS, Boulter J, Vetter DE, Heinemann S. α9: an acetylcholine receptor with novel pharmacological properties expressed in rat cochlear hair cells. Cell. 1994;79:705–715. doi: 10.1016/0092-8674(94)90555-x. [DOI] [PubMed] [Google Scholar]
- Elgoyhen AB, Vetter DE, Katz E, Rothlin CV, Heinemann SF, Boulter J. α10: a determinant of nicotinic cholinergic receptor function in mammalian vestibular and cochlear mechanosensory hair cells. Proc Natl Acad Sci U S A. 2001;98:3501–3506. doi: 10.1073/pnas.051622798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenton MB, Bell GP. Recognition of species of insectivorous bats by their echolocation calls. J Mammal. 1981;62:233–243. [Google Scholar]
- Franchini LF, Elgoyhen AB. Adaptive evolution in mammalian proteins involved in cochlear outer hair cell electromotility. Mol Phylogenet Evol. 2006;41:622–635. doi: 10.1016/j.ympev.2006.05.042. [DOI] [PubMed] [Google Scholar]
- Giannini NP, Simmons NB. A phylogeny of megachiropteran bats (Mammalia: Chiroptera: Pteropodidae) based on direct optimization analysis of one nuclear and four mitochondrial genes. Cladistics. 2003;19:496–511. doi: 10.1111/j.1096-0031.2003.tb00385.x. [DOI] [PubMed] [Google Scholar]
- Guinan JJ. The physiology of olivocochlear efferents. In: Dallos P, Popper AN, Fay RR, editors. The cochlea. New York: Springer; 1996. pp. 435–502. [Google Scholar]
- Heffner H, Masterton B. Hearing in Glires: domestic rabbit, cotton rat, feral house mouse, and kangaroo rat. J Acoust Soc Am. 1980;68:1584–1599. [Google Scholar]
- Hoofer SR, Van Den Bussche RA. Molecular phylogenetics of the chiropteran family Vespertilionidae. Acta Chiropt. 2003;5(Suppl):1–63. [Google Scholar]
- Housley GD, Ashmore JF. Ionic currents of outer hair cells isolated from the guinea-pig cochlea. J Physiol. 1992;448:73–98. doi: 10.1113/jphysiol.1992.sp019030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones G, Teeling EC. The evolution of echolocation in bats. Trends Ecol Evol. 2006;21:149–156. doi: 10.1016/j.tree.2006.01.001. [DOI] [PubMed] [Google Scholar]
- Kharkovets T, Hardelin JP, Safieddine S, Schweizer M, El-Amraoui A, Petit C, Jentsch TJ. KCNQ4, a K+ channel mutated in a form of dominant deafness, is expressed in the inner ear and the central auditory pathway. Proc Natl Acad Sci U S A. 2000;97:4333–4338. doi: 10.1073/pnas.97.8.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köppl C. Birds—same thing, but different? Convergent evolution in the avian and mammalian auditory systems provides informative comparative models. Hear Res. 2011;273:65–71. doi: 10.1016/j.heares.2010.03.095. [DOI] [PubMed] [Google Scholar]
- Kubisch C, Schroeder BC, Friedrich T, Lütjohann B, El-Amraoui A, Marlin S, Petit C, Jentsch TJ. KCNQ4, a novel potassium channel expressed in sensory outer hair cells, is mutated in dominant deafness. Cell. 1999;96:437–446. doi: 10.1016/s0092-8674(00)80556-5. [DOI] [PubMed] [Google Scholar]
- Li G, Liang B, Wang Y, Zhao H, Helgen KM, Lin L, Jones G, Zhang S. Echolocation calls, diet, and phylogenetic relationships of Stoliczka's trident bat, Aselliscus stoliczkanus (Hipposideridae) J Mammal. 2007;88:736–744. [Google Scholar]
- Li G, Wang J, Rossiter SJ, Jones G, Cotton JA, Zhang S. The hearing gene Prestin reunites echolocating bats. Proc Natl Acad Sci U S A. 2008;105:13959–13964. doi: 10.1073/pnas.0802097105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Wang J, Rossiter SJ, Jones G, Zhang S. Accelerated FoxP2 evolution in echolocating bats. PLoS One. 2007;2:e900. doi: 10.1371/journal.pone.0000900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Liu Z, Shi P, Zhang J. The hearing gene Prestin unites echolocating bats and whales. Curr Biol. 2010;20:R55–56. doi: 10.1016/j.cub.2009.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberman MC, Gao J, He DZ, Wu X, Jia S, Zuo J. Prestin is required for electromotility of the outer hair cell and for the cochlear amplifier. Nature. 2002;419:300–304. doi: 10.1038/nature01059. [DOI] [PubMed] [Google Scholar]
- Liu Y, Cotton JA, Shen B, Han X, Rossiter SJ, Zhang S. Convergent sequence evolution between echolocating bats and dolphins. Curr Biol. 2010;20:R53–R54. doi: 10.1016/j.cub.2009.11.058. [DOI] [PubMed] [Google Scholar]
- Maddison WP, Maddison DR. Mesquite: a modular system for evolutionary analysis. Version 2.72. 2009. Available from: http://mesquiteproject.org. [Google Scholar]
- Manley GA. Cochlear mechanisms from a phylogenetic viewpoint. Proc Natl Acad Sci U S A. 2000;97:11736–11743. doi: 10.1073/pnas.97.22.11736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manley GA, Köppl C. Phylogenetic development of the cochlea and its innervation. Curr Opin Neurobiol. 1998;8:468–474. doi: 10.1016/s0959-4388(98)80033-0. [DOI] [PubMed] [Google Scholar]
- Manley GA, Popper AN, Fay RR. Evolution of the vertebrate auditory system. New York: Springer; 2004. [Google Scholar]
- Marcotti W, Kros CJ. Developmental expression of the potassium current IK,n contributes to maturation of mouse outer hair cells. J Physiol. 1999;520:653–660. doi: 10.1111/j.1469-7793.1999.00653.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGowen MR, Spaulding M, Gatesy J. Divergence date estimation and a comprehensive molecular tree of extant cetaceans. Mol Phylogenet Evol. 2009;53:891–906. doi: 10.1016/j.ympev.2009.08.018. [DOI] [PubMed] [Google Scholar]
- Mellado Lagarde MM, Drexl M, Lukashkina VA, Lukashkin AN, Russell IJ. Outer hair cell somatic, not hair bundle, motility is the basis of the cochlear amplifier. Nat Neurosci. 2008;11:746–748. doi: 10.1038/nn.2129. [DOI] [PubMed] [Google Scholar]
- Miller-Butterworth CM, Murphy WJ, O'Brien SJ, Jacobs DS, Springer MS, Teeling EC. A family matter: conclusive resolution of the taxonomic position of the long-fingered bats, Miniopterus. Mol Biol Evol. 2007;24:1553–1561. doi: 10.1093/molbev/msm076. [DOI] [PubMed] [Google Scholar]
- Mount DB, Romero MF. The SLC26 gene family of multifunctional anion exchangers. Pflugers Arch. 2004;447:710–721. doi: 10.1007/s00424-003-1090-3. [DOI] [PubMed] [Google Scholar]
- Murphy WJ, Eizirik E, O'Brien SJ, et al. (11 co-authors) Resolution of the early placental mammal radiation using Bayesian phylogenetics. Science. 2001;294:2348–2351. doi: 10.1126/science.1067179. [DOI] [PubMed] [Google Scholar]
- Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- Nylander JA, Wilgenbusch JC, Warren DL, Swofford DL. AWTY (are we there yet?): a system for graphical exploration of MCMC convergence in Bayesian phylogenetics. Bioinformatics. 2008;24:581–583. doi: 10.1093/bioinformatics/btm388. [DOI] [PubMed] [Google Scholar]
- Posada D. ModelTest Server: a web-based tool for the statistical selection of models of nucleotide substitution online. Nucleic Acids Res. 2006;34:W700–W703. doi: 10.1093/nar/gkl042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D, Crandall KA. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Springer MS, Teeling EC, Madsen O, Stanhope MJ, de Jong WW. Integrated fossil and molecular data reconstruct bat echolocation. Proc Natl Acad Sci U S A. 2001;98:6241–6246. doi: 10.1073/pnas.111551998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Steiper ME, Young NM. Primate molecular divergence dates. Mol Phylogenet Evol. 2006;41:384–394. doi: 10.1016/j.ympev.2006.05.021. [DOI] [PubMed] [Google Scholar]
- Stone EA, Sidow A. Physicochemical constraint violation by missense substitutions mediates impairment of protein function and disease severity. Genome Res. 2005;15:978–986. doi: 10.1101/gr.3804205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Teeling EC, Madsen O, Van Den Bussche RA, de Jong WW, Stanhope MJ, Springer MS. Microbat paraphyly and the convergent evolution of a key innovation in Old World rhinolophoid microbats. Proc Natl Acad Sci U S A. 2002;99:1431–1436. doi: 10.1073/pnas.022477199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teeling EC, Scally M, Kao DJ, Romagnoli ML, Springer MS, Stanhope MJ. Molecular evidence regarding the origin of echolocation and flight in bats. Nature. 2000;403:188–192. doi: 10.1038/35003188. [DOI] [PubMed] [Google Scholar]
- Teeling EC, Springer MS, Madsen O, Bates P, O'Brien SJ, Murphy WJ. A molecular phylogeny for bats illuminates biogeography and the fossil record. Science. 2005;307:580–584. doi: 10.1126/science.1105113. [DOI] [PubMed] [Google Scholar]
- Thabah A, Li G, Wang Y, Liang B, Hu K, Zhang S, Jones G. Diet, echolocation calls, and phylogenetic affinities of the great evening bat (Ia io; Vespertilionidae): another carnivorous bat. J Mammal. 2007;88:728–735. [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vater M, Kössl M. The ears of whales and bats. In: Thomas JA, Moss CF, Vater M, editors. Echolocation in bats and dolphins. Chicago: The University of Chicago Press; 2004. pp. 89–99. [Google Scholar]
- Xu T, Nie L, Zhang Y, et al. (12 co-authors) Roles of alternative splicing in the functional properties of inner ear-specific KCNQ4 channels. J Biol Chem. 2007;282:23899–23909. doi: 10.1074/jbc.M702108200. [DOI] [PubMed] [Google Scholar]
- Yang Z. Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol Biol Evol. 1998;15:568–573. doi: 10.1093/oxfordjournals.molbev.a025957. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Yang Z, Wong WS, Nielsen R. Bayes empirical Bayes inference of amino acid sites under positive selection. Mol Biol Evol. 2005;22:1107–1118. doi: 10.1093/molbev/msi097. [DOI] [PubMed] [Google Scholar]
- Zhang J, Kumar S. Detection of convergent and parallel evolution at the amino acid sequence level. Mol Biol Evol. 1997;14:527–536. doi: 10.1093/oxfordjournals.molbev.a025789. [DOI] [PubMed] [Google Scholar]
- Zhang J, Nielsen R, Yang Z. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol Biol Evol. 2005;22:2472–2479. doi: 10.1093/molbev/msi237. [DOI] [PubMed] [Google Scholar]
- Zheng J, Shen W, He DZ, Long KB, Madison LD, Dallos P. Prestin is the motor protein of cochlear outer hair cells. Nature. 2000;405:149–155. doi: 10.1038/35012009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.