Abstract
We have previously identified Cia10 as an arthritis severity and articular damage quantitative trait locus. In this study we used Illumina RatRef-12 microarrays to analyze the expression of 21,922 genes in synovial tissues from arthritis-susceptible DA and arthritis-protected DA.ACI(Cia10) congenics with pristane-induced arthritis. 310 genes had significantly different expression. The genes up-regulated in DA, and reciprocally down-regulated in DA.ACI(Cia10) included IL-11, Ccl12 and Cxcl10, as well as genes implicated in Th17 responses such as IL-17A, IL-6, Ccr6, Cxcr3 and Stat4. Suppressors of immune responses Tgfb and Vdr, and inhibitors of oxidative stress were up-regulated in congenics. There was an over-representation of genes implicated in cancer and cancer-related phenotypes such as tumor growth and invasion among the differentially expressed genes. Cancer-favoring genes like Ctsd, Ikbke, and Kras were expressed in increased levels in DA, while inhibitors of cancer phenotypes such as Timp2, Reck and Tgfbr3 were increased in DA.ACI(Cia10). These results suggest that Cia10 may control arthritis severity, synovial hyperplasia and joint damage via the regulation of the expression of cancer-related genes, inflammatory mediators and Th17-related markers. These new findings have the potential to generate new targets for therapies aimed at reducing arthritis severity and joint damage in rheumatoid arthritis.
Keywords: rheumatoid, autoimmunity, synovial, erosions, animal model, genetic
INTRODUCTION
Rheumatoid arthritis (RA) is a complex autoimmune disease characterized by chronic joint inflammation that commonly leads to joint destructive changes, disability and reduced quality of living. RA affects 1% of the population and is regulated by both genetic and environmental factors 1. Most of the genetic studies have focused in the identification of susceptibility genes. The HLA-DRB1 shared-epitope association with RA susceptibility has been known for over two decades, and in recent years several non-MHC susceptibility alleles have been identified, including PTPN22 2, CTLA4 3, TNFAIP3 and TRAF1 4, 5. Yet, little is known about the genetic regulation of disease severity and joint damage, which are among the best predictors of disease outcome and disability.
We have identified the non-MHC arthritis severity and joint damage quantitative trait locus (QTL) Cia10 on rat chromosome 2, studying an intercross between MHC-identical DA (severe arthritis) and ACI (arthritis-resistant) rats 6. DA.ACI(Cia10) congenic rats, which are identical to DA except for the presence of ACI alleles at the Cia10 interval, were generated using a genotype-guided strategy and found to develop a significantly milder form of pristane-induced arthritis (PIA), and to preserve normal joint architecture compared to DA rats. DA.ACI(Cia10) congenics also had significantly reduced synovial tissue levels of mRNA of pro-inflammatory cytokines central to RA pathogenesis 7. During the refining of the congenic interval towards positional cloning of the Cia10 gene we aimed to identify molecular pathways and potential candidate genes involved in protecting DA.ACI(Cia10) congenics, using genome-wide microarray gene expression profiling of synovial tissues. In the present study we report the presence of ACI alleles at the Cia10 interval correlates with reduced expression of genes implicated in Th17 responses, including IL-17A. Additionally, congenics had reduced expression of genes involved in the regulation of cancer growth and invasion, which may suggest new targets for therapies aimed at reducing synovial pannus growth and cartilage and bone invasion and destruction.
RESULTS
DA.ACI (Cia10) congenics are protected and developed significantly milder PIA compared with DA
DA.ACI(Cia10) congenic rats developed significantly milder arthritis compared with DA, with a median ASI of 12.3 compared with 30.4, respectively (P=0.002 Mann-Whitney test) (Figure 1B). These observations were in agreement with our previous studies 7.
Figure 1. A map of the Cia10 interval in DA.ACI(Cia10) congenics and arthritis severity index (ASI) scores.
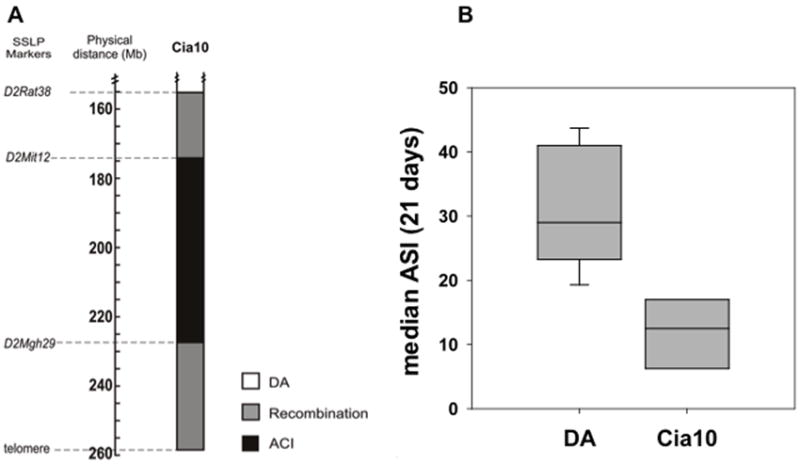
A. Map of the DA.ACI(Cia10) congenic interval on rat chromosome 2 showing DA alleles (white), ACI alleles (black) and recombination interval (grey). Key SSLP markers used in the congenic breeding are shown (Mb=position in megabases on the rat genome assembly v.3.4). B. Cummulative median ASI scores of DA (ASI=30.4) compared with DA.ACI(Cia10) (ASI=12.3) during 21 days following PIA induction (P=0.002 Mann-Whitney test) (DA n=12 and DA.ACI(Cia10) n=6). Boxes represent the 25%-75% confidence interval, and error bars are the 5%-95% confidence interval.
Gene expression analysis of synovial tissues differentiates DA from DA.ACI(Cia10) congenics
7,593 (34.6%) of the 21,922 genes present in the array were expressed by all 18 synovium samples and were used for analysis. 310 of the 7,593 genes (4.08%) expressed by all synovial tissues met the criteria for differential expression. 120 of these 310 genes (38.7%) were expressed in significantly higher levels in DA.ACI(Cia10) compared with DA rats. 190 of the 310 genes (61.3%) were expressed in significantly increased levels in DA synovium, and were reciprocally reduced in DA.ACI(Cia10).
The genes with the most significantly increased expression in the DA.ACI(Cia10) congenics and reduced expression in DA included Mup4 (4.88-fold, P=0.0067), Aldh1a1 (3.7-fold P=0.009), Rpesp (3.56-fold, P=0.0082), Igfbp6, (3.56-fold, P=0.0068), Gstm1 (2.58-fold, P=0.0029), Timp2 (2.15-fold, P=0.0024) and Reck (2.1-fold, P=0.005) (table 2).
Table 2.
Most significantly differentially expressed genes between DA.ACI(Cia10) congenics and DA.
| Gene symbol | Gene name | Accession number | Fold difference | P-value |
|---|---|---|---|---|
| Up-regulated in DA | DA/Cia10 | |||
| LOC499078 | similar to glycoprotein 49b | GI_62638899-S | 5.39 | 0.0023 |
| Cd53 | cluster of differentiation 53 | NM_012523.1 | 3.28 | 0.0040 |
| RGD1566002 | similar to RIKEN cDNA 3110001N18 | XM_221003.3 | 3.09 | 0.0038 |
| Fcgr1a | Fc fragment of IgG, high affinity Ia, receptor (CD64) | XM_001062370.1 | 2.94 | 0.0073 |
| ccdc109b | coiled-coil domain containing 109B | XM_001076863.1 | 2.89 | 0.0036 |
| LOC499136 | hypothetical gene | XM_574429.1 | 2.85 | 0.0014 |
| Rangap1 | RAN GTPase activating protein 1 | XM_576313.1 | 2.84 | 0.0051 |
| RGD1565520 | similar to 60S ribosomal protein L7a | XR_009158.1 | 2.81 | 0.0025 |
| Emr1 | egf-like module containing mucin like hormone receptor | NM_001007557.1 | 2.78 | 0.0000 |
| LOC308350 | similar to PIRB1 | XM_218261.3 | 2.73 | 0.0074 |
| RGD1564866 | similar to Heterogeneous nuclear ribonucleoprotein A1 | XR_007647.1 | 2.72 | 0.0038 |
| LOC687849 | hypothetical gene | XM_001080339.1 | 2.69 | 0.0012 |
| Rbbp7 | Retinoblastoma-binding protein 7 | NM_031816.1 | 2.68 | 0.0052 |
| RGD1563503 | similar to ribosomal protein L6 | XR_007388.1 | 2.67 | 0.0010 |
| RGD1563124 | similar to 40S ribosomal protein S20 | XM_576204.2 | 2.66 | 0.0013 |
| RGD1559935 | similar to Selenoprotein H | XR_008210.1 | 2.66 | 0.0027 |
| Ppid | peptidylprolyl isomerase D | NM_001004279.1 | 2.66 | 0.0003 |
| RGD1560979 | hypothetical gene | XR_008629.1 | 2.65 | 0.0031 |
| Acsl4 | acyl-CoA synthetase long-chain family member 4 | NM_053623.1 | 2.62 | 0.0033 |
| St14 | suppression of tumorigenicity 14 | NM_053635.2 | 2.55 | 0.0004 |
| Shc1 | Src homology 2 domain-containing transforming protein C1 | NM_053517.1 | 2.55 | 0.0001 |
| LOC306428 | similar to Chain A, T13s Mutant Of Bovine 70 Kilodalton Heat Shock Protein | XM_224824.3 | 2.55 | 0.0007 |
| LOC316373 | similar to high-mobility group (nonhistone chromosomal) protein 1-like 1 | XR_007934.1 | 2.54 | 0.0064 |
| Fcgr3a | Fc receptor, IgG 3a | NM_207603.1 | 2.51 | 0.0021 |
| Rsl1d1 | ribosomal L1 domain containing 1 | NM_001008876.1 | 2.50 | 0.0032 |
| Matk | megakaryocyte-associated tyrosine kinase | NM_021859.2 | 2.49 | 0.0044 |
| RGD1559682 | similar to peptidylprolyl isomerase A | XM_001075526.1 | 2.47 | 0.0042 |
| Fcgr2b | Fc fragment of IgG, low affinity IIb, receptor (CD32) | NM_175756.1 | 2.45 | 0.0090 |
| Smc4l1 | structural maintenance of chromosomes 4 | XM_001066172.1 | 2.43 | 0.0059 |
| Tnfaip8l2 | Tumor necrosis factor, alpha-induced protein 8-like 2 | NM_001014039.1 | 2.42 | 0.0095 |
| Psmb9 | proteasome subunit beta-1i | NM_012708.1 | 2.41 | 0.0002 |
| RGD620382 | Nucleoside 2-deoxyribosyltransferase domain containing protein | NM_133525.1 | 2.38 | 0.0037 |
| RGD1565238 | similar to Gapdh | XM_001081651.1 | 2.37 | 0.0097 |
| RGD1565306 | similar to 60S ribosomal protein L29 | XM_576023.2 | 2.35 | 0.0064 |
| Up-regulated in DA.ACI(Cia10) | Cia10/DA | |||
| LOC259244 | alpha-2u globulin PGCL3 | NM_147212.1 | 5.01 | 0.0067 |
| Mup4 | alpha-2u globulin PGCL3 | NM_198784.1 | 4.88 | 0.0067 |
| LOC682605 | alpha-2u globulin PGCL3 | XM_001062261.1 | 4.32 | 0.0074 |
| MGC72973 | beta-globulin | NM_198776.1 | 3.71 | 0.0095 |
| Aldh1a1 | aldehyde dehydrogenase 1 family, member A1 | NM_022407.3 | 3.70 | 0.0090 |
| Rpesp | RPE-spondin | XM_001063197.1 | 3.56 | 0.0082 |
| Igfbp6 | insulin-like growth factor binding protein 6 | NM_013104.2 | 3.56 | 0.0068 |
| Ccdc3 | colied coil domain containing 3 | XM_574081.1 | 3.44 | 0.0097 |
| Mt3 | metallothionein 3 | NM_053968.2 | 3.34 | 0.0089 |
| RGD1310507 | similar to RIKEN cDNA 1300017J02 | XM_236574.4 | 3.27 | 0.0072 |
| Fxyd6 | FXYD domain-containing ion transport regulator 6 | NM_022005.1 | 3.26 | 0.0090 |
| Cav2 | caveolin 2 | NM_131914.2 | 3.02 | 0.0058 |
| Fhl1 | four and a half LIM domains protein 1 | NM_145669.2 | 3.01 | 0.0086 |
| Nov | nephroblastoma overexpressed gene | NM_030868.2 | 2.86 | 0.0070 |
| Per1 | period 1 | XM_340822.2 | 2.78 | 0.0011 |
| Myh11 | myosin, heavy chain 11, smooth muscle | XM_573030.2 | 2.77 | 0.0057 |
| Gstm2 | glutathione S-transferase, mu 2 | NM_177426.1 | 2.71 | 0.0028 |
| Klhl23 | similar to 60S ribosomal protein L7a | XM_001059088.1 | 2.60 | 0.0007 |
| Pcsk1n | proprotein convertase subtilisin/kexin type 1 inhibitor | NM_019279.1 | 2.58 | 0.0072 |
| Gstm1 | glutathione S-transferase, mu 1 | NM_017014.1 | 2.58 | 0.0029 |
| Hbb | hemoglobin, beta | NM_033234.1 | 2.55 | 0.0063 |
| Etl4 | enhancer trap locus 4 | XM_001069190.1 | 2.48 | 0.0069 |
| Ctnnal1 | catenin (cadherin associated protein), alpha-like 1 | XM_001059679.1 | 2.36 | 0.0077 |
| Sptbn1 | spectrin, beta, non-erythrocytic 1 | NM_001013130.1 | 2.24 | 0.0041 |
| Cyb5r1 | cytochrome b5 reductase 1 | NM_001013126.1 | 2.20 | 0.0010 |
| Timp2 | tissue inhibitor of metalloproteinases-2 | NM_021989.2 | 2.15 | 0.0024 |
| LOC500248 | similar to 1300014I06Rik protein | XM_580052.1 | 2.14 | 0.0078 |
| Reck | reversion-inducing-cysteine-rich protein with kazal motifs | XM_001070551.1 | 2.10 | 0.0050 |
| Cldn22 | claudin 22 | XM_224843.2 | 2.10 | 0.0019 |
| Ddit4 | DNA-damage-inducible transcript 4 | NM_080906.1 | 2.09 | 0.0090 |
| RGD1311307 | similar to 1300014I06Rik protein | NM_001025719.1 | 2.08 | 0.0074 |
| Cat | catalase | NM_012520.1 | 2.07 | 0.0076 |
| Tmem117 | transmembrane protein 117 | XM_576330.2 | 2.06 | 0.0023 |
| Nes | nestin | NM_012987.1 | 2.06 | 0.0060 |
| Rgs4 | regulator of G-protein signaling 4 | NM_017214.1 | 2.05 | 0.0079 |
| Hexim1 | hexamethylene bis-acetamide inducible 1 | XM_573204.1 | 2.04 | 0.0022 |
| Axl | AXL receptor tyrosine kinase | NM_001013147.1 | 2.03 | 0.0046 |
| Tspan2 | tetraspanin-2 | NM_022589.1 | 2.02 | 0.0057 |
Among the 190 genes with significantly reduced expression levels in the synovial tissues of DA.ACI(Cia10) congenics and increased expression in DA was Gp49b (5.39-fold, P=0.0023), Cd53 (3.28-fold, P=0.004), Fcgr1a (2.94-fold, P=0.007), Emr1 (2.78-fold, P=0.00002) and Rbbp7 (2.68-fold, P=0.0051) (table 2). Seven of the differentially expressed genes were confirmed with qPCR (figure 2).
Figure 2. qPCR confirmation of the microarray results.
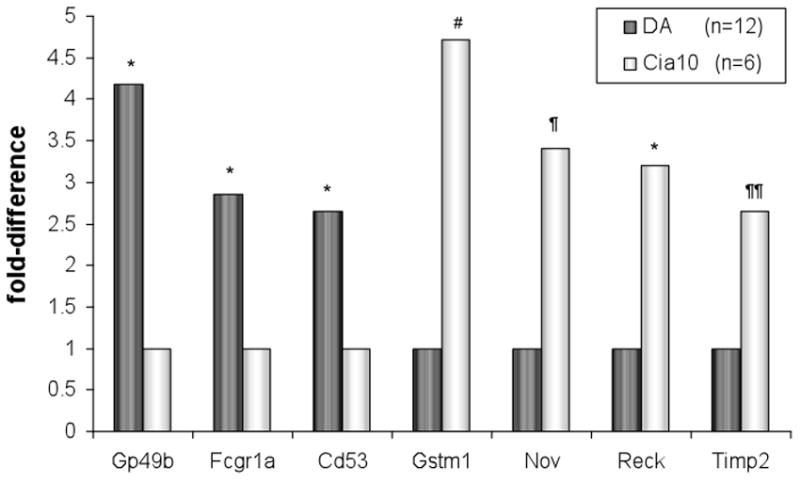
Three of the genes most significantly up-regulated in DA (Gp49b, Cd53 and Fcrg1) and four of the most up-regulated genes in DA.ACI(Cia10) (Timp2, Reck, Gstm1 and Nov) were tested with qPCR. [DA n=12 and DA.ACI(Cia10) n=6; Reck was tested in 7 DA and 4 DA.ACI(Cia10)]. * P<0.004; # P=0.016; ¶ P=0.089; ¶¶ P=0.25 (t-test using ΔCt values).
The differentially expressed genes grouped into specific “disease categories”, “cellular functions” and “gene networks”
Analyses of the 310 differentially expressed genes revealed an over-representation of ten disease categories and/or molecular and cellular functions as detected by the IPA pathway analyses (table 3). These categories included cancer (89 genes), cell death (68 genes), reproductive system disease (this category predominantly included endometriosis and genes implicated in cancer of the reproductive system or breast) (57 genes), inflammatory disease (53 genes), hematological disease (37 genes) and others (table 3).
Table 3.
Diseases and cellular functions overrepresented among the differentially expressed genes.
| Disease or cellular function | Number of Genes | p-value |
|---|---|---|
| Cancer | 89 | 0.04-0.0005 |
| Cell Death | 68 | 0.04-0.00004 |
| Reproductive System Disease* | 57 | 0.04-0.0002 |
| Inflammatory disease | 53 | 0.04-0.0005 |
| Hematological disease | 37 | 0.04-0.0001 |
| Cellular Assembly and Organization | 27 | 0.04-0.0001 |
| Tissue Development | 26 | 0.04-0.0001 |
| Molecular Transport | 25 | 0.04-0.0002 |
| Cellular Function and Maintenance | 21 | 0.04-0.0001 |
| Immunological Disease | 19 | 0.04-0.0002 |
predominantly reproductive and breast cancer phenotypes and endometriosis.
The IPA network/pathway detection analyses identified seven additional functional groups over-represented among the differentially expressed genes (supplemental table 1). These included “Embryonic Development, Tissue Development, Cell Death” (27 genes), “Inflammatory Response, Connective Tissue Disorders, Tissue Morphology” (20 genes) and “Cell Cycle, Connective Tissue Development and Function, Organ Morphology” (19 genes), all with significant scores (supplemental table 1).
Inflammatory Cytokines, Chemokines and related genes
We used three approaches to identify differentially expressed cytokines, chemokines and other known inflammatory mediators or receptors: a) genes expressed by all 18 synovial samples and meeting the criteria for significance described in the Methods section (P≤0.01 plus ≥1.5-fold-difference; b) genes expressed by only one of the strains and not the other; c) genes predominantly expressed in one strain and not in the other. Using these combined parameters we identify 21 cytokines, chemokines, receptors or related genes of relevance (table 4).
Table 4.
Cytokine and chemokine-related genes, and oxidative stress inhibitors differentially or predominantly expressed in DA.ACI(Cia10) or DA.
| Gene symbol | Gene name | Accession number | Expressed in all samples
|
Preferential strain expression
|
|||
|---|---|---|---|---|---|---|---|
| Fold-difference | P-value** | DA positive (%) | Cia10 positive (%) | P-value** | |||
| Genes up-regulated in DA | |||||||
| Ccl21b§ | chemokine (C-C motif) ligand 21b | NM_001008513.1 | 1.61 | 0.002 | |||
| Csk | c-src tyrosine kinase | XM_236290.3 | 2.02 | 0.002 | |||
| IL-11 | interleukin-11 | XM_346531.2 | 1.66 | 0.007 | |||
| IL-3ra | interleukin-3 receptor subunit alpha | NM_139260.1 | 1.52 | 0.009 | |||
| Genes predominantly expressed in DA | |||||||
| Ccl12 | chemokine (C-C motif) ligand 12 | XM_213425.2 | 11 (92) | 2 (33) | 0.021 | ||
| Ccr6§ | chemokine (C-C motif) receptor | NM_001013145.1 | 12 (100) | 3 (50) | 0.024 | ||
| Cx3cr1 | chemokine (C-X3-C motif) receptor 1 | NM_133534.1 | 10 (83) | 1 (16) | 0.012 | ||
| Cxcl10 | chemokine (C-X-C motif) ligand 10 | NM_139089.1 | 12 (100) | 3 (50) | 0.024 | ||
| Cxcr3 | chemokine (C-X-C motif) receptor 3 | NM_053415.1 | 12 (100) | 3 (50) | 0.024 | ||
| Ifi47 | interferon gamma inducible protein 47 | NM_172019.1 | 9 (75) | 0 (0) | 0.009 | ||
| IL-10 | interleukin 10 | NM_012854.1 | 8 (67) | 0 (0) | 0.012 | ||
| IL-1rl1 (IL-33R) | interleukin 1 receptor-like 1 | NM_013037.1 | 12 (100) | 2 (33) | 0.005 | ||
| IL-6 | interleukin-6 | NM_012589.1 | 11 (92) | 2 (33) | 0.021 | ||
| IL-17A | interleukin-17A | NM_001106897 | 13 (93) | 3 (43) | 0.025 | ||
| Stat4§ | signal transducer and activator of transcription 4 | NM_001012226.1 | 11 (92) | 2 (33) | 0.021 | ||
| Genes up-regulated in DA.ACI(Cia10) | |||||||
| Ccl11 | chemokine (C-C motif) ligand 11 (eotaxin) | NM_019205.1 | 1.93 | 0.003 | |||
| Tgfa | transforming growth factor alpha | NM_012671.1 | 4.20 | 0.006 | |||
| Tgfb2 | transforming growth factor beta 2 | NM_031131.1 | 1.93 | 0.0001 | |||
| Tnfsf12 | tumor necrosis factor ligand superfamily member 12 (TWEAK) | NM_001001513.2 | 2.21 | 0.003 | |||
| Cat* | catalase | NM_012520.1 | 2.07 | 0.008 | |||
| Gstm1* # | glutathione S-transferase mu 1 | NM_017014.1 | 2.71 | 0.003 | |||
| Mt3* | Metallothionein-3; growth inhibitory factor (GIF) | NM_053968.2 | 3.34 | 0.009 | |||
| Nfe2l1* | nuclear factor, erythroid derived 2,-like | XM_340886.3 | 1.76 | 0.009 | |||
| Sqstm1* | sequestosome 1 | NM_175843.3 | 1.66 | 0.002 | |||
| Genes predominantly expressed in DA.ACI(Cia10) | |||||||
| Tgfbr3 | transforming growth factor, beta receptor III | NM_017256.1 | 6 (50) | 6 (100) | 0.053 | ||
| Vdr | vitamin D (1,25- dihydroxyvitamin D3) receptor | NM_017058.1 | 4 (33) | 4 (80) | 0.321 | ||
genes associated with genetic susceptibility in rheumatoid arthritis;
gene associated with disease severity in rheumatoid arthritis. Grey box: gene implicated in Th17 cell responses.
Inhibitors of oxidative stress;
Genes expressed in all samples were compared with t-test, and preferecial strain expression with Fisher Exact Test.
IL-17A results are shown in BOLD and reflect qPCR data of 14 DA and 7 DA.ACI(Cia10) rats.
Inflammatory mediators and receptors up-regulated in DA
IL-6 was preferentially expressed in 92% of DA (n=11) and only in 33% (n=2) congenic’s synovial tissues (table 4). IL-11 was expressed in significantly increased levels in DA, compared with congenics (1.66-fold, P=0.007). These two pleiotropic cytokines are also expressed in increased levels in RA synovium, and are known to activate NFκB and osteoclasts, and to increase cell tolerance to oxidative stress, all phenomena that take place in the arthritic synovial tissues 8-11.
Cxcl10, Ccl12, and Ccl21b among others, were preferentially expressed, or expressed in significantly increased levels in DA, compared with congenics. Chemokine receptors Ccr6, Cx3cr1 and Cxcr3, as well as cytokine receptors, IL-1rl1 (IL-33r) and IL-3ra were also up-regulated in the synovial tissues from DA rats. Interestingly, Ccl21b, Ccr6 and Stat4 have been associated with RA susceptibility 12.
Additional important mediators of the inflammatory response up-regulated in DA included Csk, a key signaling gene that regulates T and B cell proliferation, activation, and migration that interacts with the RA susceptibility gene Ptpn22 2.
Four of the above genes, including IL-6, Stat4, Ccr6 and Cxcl10, have been implicated in the differentiation, activity or chemotaxis of Th17 cells, a cell type central to autoimmune disease pathogenesis. We next looked for the three genes more specifically related to Th17 cells: IL-17A, IL-17F and Rorc (RORgt). The Illumina RatRef microarray does not contain probes for IL-17A and IL-1F. Therefore we used qPCR to study the expression of these two genes. IL-17A was preferentially expressed in DA synovial tissues (93%), compared with DA.ACI(Cia10) congenics (43%), and this difference was statistically significant (p=0.025; Fisher Exact test, table 4). IL-17F was not expressed in either strain’s synovial tissues. Rorc (RORgt) expression was not significantly different between the DA and DA.ACI(Cia10) both in the microarray and qPCR (data not shown). Therefore, our results suggest that the Cia10 gene in involved in the regulation of differentiation or synovial tissue homing of Th17 cells.
Inflammatory mediators and receptors up-regulated in DA.ACI(Cia10) congenics
Tgfa, Tgfb2 and the receptor Tgfbr3 were up-regulated or predominantly expressed in DA.ACI(Cia10) congenics, compared with DA (table 4). Tgfb and its receptors are known to suppress immune responses and to induce the differentiation of Treg cells 13. Several genes up-regulated in DA.ACI(Cia10) congenics are known to interact with, or to be regulated by Tgfb, further suggesting a role for this gene in mediating protection in the congenics (figure 3).
Figure 3. Network analyses of differentially expressed genes suggest a central role for Tgfβ and NFκB.
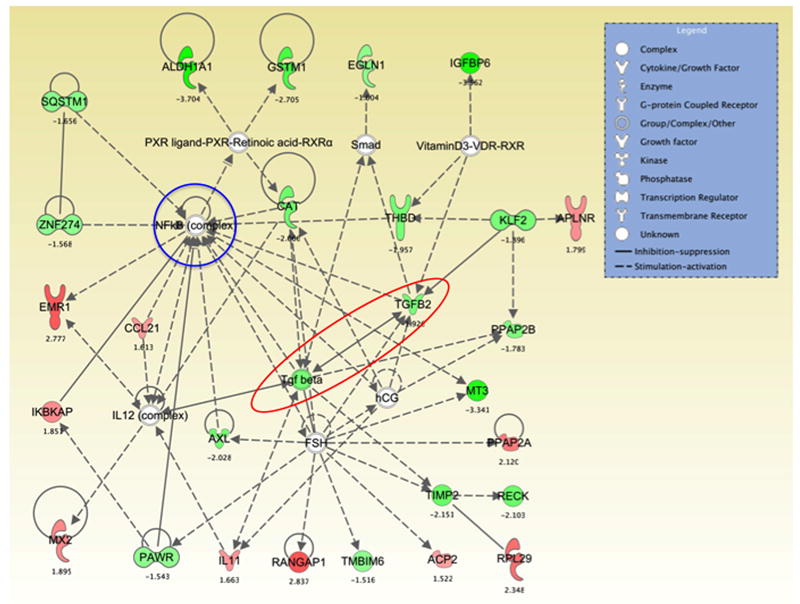
Ingenuity pathway analyses determined that several of the most significantly differentially expressed, or preferentially expressed genes interact with, induce or are induced by Tgfβ (red elipse) and NFκB (blue circle). These genes included Cat, Igfbp6, Reck, Timp2 and Vdr in the case of Tgfβ, and Ccl21, Emr1, IκBkap, IL-11 and Rangap1 in the case of NFκB.
The Vdr gene was predominantly expressed in congenics (80%) compared with DA (33%) (table 4). IPA pathway and gene interaction analyses revealed that several of the most significantly differentially expressed genes were induced by or interacted with the Vdr, including Tgfb2 (figure 3). The Vdr is known to inhibit the expression of cytokines like IL-6, which is in agreement with our observations of an inverse correlation between Vdr-Tgfb2 and IL-6.
Ccl11 (Eotaxin) expression was increased in the protected DA.ACI(Cia10) congenics (1.93-fold, P=0.003). Interestingly, Ccl11 levels have been shown to correlate with reduced radiographic damage in RA via yet unknown mechanisms 14. Ccl11 is a chemoattractant to eosinophils and Th2 cells, and modulates monocyte activity.
Genes implicated in cancer and cancer-related phenotypes accounted for the greatest percentage of the differentially expressed genes
89 genes of the 310 (28.7%) differentially expressed genes are implicated in cancer and cancer-related phenotypes. These included oncogenes, cell-cycle regulators, tumor-suppressor genes, proteases and others. The 27 genes with the strongest literature support for a role in cancer are listed on table 5. Among the cancer-favoring genes up-regulated in DA there were eight oncogenes or tumorigenesis genes, including Rbm3, Ikbke, Rcl, Kras, four cell proliferation enhancing genes (Mcts1, Aplnr, Ppap2a, and Ier3) and three invasion-associated genes (Ctsd, Cd53 and Loxl1). Two additional mediators of cancer invasion and metastasis, Cxcl10 and Cxcr3, were preferentially expressed by DA synovial tissues [DA=12 (100%), DA.ACI(Cia10)=3 (50%)] (table 4). IL-11, which as described above was expressed in increased levels in DA (table 4), has also been implicated in the pathogenesis of carcinomas 15. Histone deacetylases (Hdac1, Hdac2 and Rbbp7) had increased expression in DA, and are also commonly up-regulated in cancers and considered to regulate cell differentiation and proliferation 16.
Table 5.
Differentially expressed genes related to cancer*
| Gene symbol | Accession number | Gene name | Fold difference | P-value |
|---|---|---|---|---|
| Up-regulated in DA | ||||
| Oncogenes and tumorigenesis | DA/Cia10 | |||
| Rcl (C6ORF108) | NM_133525.1 | Nucleoside 2-deoxyribosyltransferase domain containing protein (RGD620382) | 2.38 | 0.004 |
| Kras | NM_031515.1 | v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog | 1.93 | 0.005 |
| Rbm3 | XM_001063211.1 | RNA binding motif (RNP1, RRM) protein 3 | 1.57 | 0.009 |
| Ikbke | XM_001058036.1 | Inhibitor of nuclear factor kappa-B kinase subunit epsilon | 1.51 | 0.007 |
| St14 | NM_053635.2 | suppression of tumorigenicity 14 | 2.55 | 0.0004 |
| Cdc25a | NM_133571.1 | cell division cycle 25 homolog A | 1.67 | 0.007 |
| Hdac1 | XM_216349.3 | histone deacetylase 1 | 1.52 | 0.006 |
| Hdac2 | XM_342149.3 | histone deacetylase 2 | 1.53 | 0.0007 |
| Cell proliferation | ||||
| Mcts1 | XM_001064848.1 | malignant T cell amplified sequence 1 | 2.20 | 0.001 |
| Ppap2a | NM_022538.1 | phosphatidic acid phosphatase type 2A | 2.12 | 0.002 |
| Ier3 | NM_212505.1 | immediate early response 3 | 1.93 | 0.008 |
| Aplnr | NM_031349.2 | apelin receptor | 1.80 | 0.009 |
| Cell invasion | ||||
| Cd53 | NM_012523.1 | cluster of differentiation 53 | 3.28 | 0.004 |
| Loxl1 | NM_001012125.1 | lysyl oxidase-like 1 | 1.92 | 0.005 |
| Ctsd | NM_134334.2 | cathepsin D | 2.27 | 0.001 |
| Histone deacetylation | ||||
| Rbbp7 | NM_031816.1 | retinoblastoma binding protein 7 | 2.68 | 0.005 |
| Hdac2 | XM_342149.3 | histone deacetylase 2 | 1.53 | 0.001 |
| Hdac1 | XM_216349.3 | histone deacetylase 1 | 1.52 | 0.007 |
| Up-regulated in DA.ACI(Cia10) | ||||
| Inhibitors of cell proliferation and growth | Cia10/DA | |||
| Igfbp6 | NM_013104.2 | insulin-like growth factor binding protein 6 | 3.56 | 0.007 |
| Per1 | XM_340822.2 | period 1 | 2.78 | 0.001 |
| Thbd | NM_031771.2 | thrombomodulin | 1.96 | 0.002 |
| Pawr | NM_033485.2 | PRKC, apoptosis, WT1, regulator | 1.54 | 0.005 |
| Inhibitors of invasion | ||||
| Nov | NM_030868.2 | nephroblastoma overexpressed gene | 2.86 | 0.007 |
| Rgs4 | NM_017214.1 | regulator of G-protein signaling 4 | 2.05 | 0.008 |
| Tgfbr3# | NM_017256.1 | transforming growth factor, beta receptor III | - | - |
| Inhibitors of MMPs | ||||
| Timp2 | NM_021989.2 | tissue metallopeptidase inhibitor 2 | 2.15 | 0.002 |
| Reck | XM_001070551.1 | reversion-inducing-cysteine-rich protein with kazal motifs | 2.10 | 0.005 |
Some of these genes have more than a single cancer-related function.
Tgfbr3 was expressed by 6 (50%) DA and 6 (100%) DA.ACI(Cia10) synovial tissues. Tgfbr3 is also a suppressor of cancer growth and cell proliferation.
Nine of the genes up-regulated in DA.ACI(Cia10) are known to suppress a cancer phenotype. Specifically, four of these genes (Pawr, Per1, Igfbp6 and Thbd) negatively regulate cell proliferation and growth, and three genes (Rgs4, Nov and Tgfbr3) negatively regulate cell invasion (table 4). Matrix metalloproteinases (MMPs) are known to increase invasion of cancer and synovial cells and are key mediators of cartilage and joint destruction in RA. Timp2 (2.15-fold, P=0.0024) and Reck (2.14-fold, P=0.005), two MMP inhibitors, were also significantly up-regulated in DA.ACI(Cia10). Four of the cancer-related genes (Cd53, Nov, Reck and Timp2) were confirmed with qPCR (figure 2). Therefore, our results suggest a general increased expression of genes known to favor cell proliferation and invasion in DA, while the congenics has an increased expression of anti-proliferation and anti-invasion/anti-destruction (MMP inhibitors) genes. These observations provide new insight into the molecular processes regulating the reduced synovial hyperplasia and reduced cartilage and joint damage that we had previously reported on DA.ACI(Cia10) congenics compared with DA.
Apoptosis and Cell Survival genes are over-represented among the differentially expressed genes
68 cell death regulatory genes (table 3), including 43 apoptosis genes (13.9% of all differentially expressed) had a significantly different expression between DA and DA.ACI(Cia10) (supplemental table 2). However, a similar number of pro-apoptosis [DA=11 and DA.ACI(Cia10)=8] and anti-apoptosis [DA=12 and DA.ACI(Cia10)=10] genes was up-regulated in DA and in DA.ACI(Cia10). Therefore, while several apoptosis genes were differentially expressed, there was not a clear suggestion of an increased or reduced apoptotic gene expression pattern in either strain.
Anti-oxidant genes are increased in DA.ACI(Cia10)
Genes with anti-oxidant properties (Cat, Mt3, Nfe2l1, and Sqstm1), including Gstm1, which has been associated with disease severity and articular damage in RA 17, were amongst the most significantly up-regulated genes in DA.ACI(Cia10) (figure 3 and table 4).
NFκB pathway interacting genes are differentially regulated between DA and DA.ACI(Cia10)
Ten of the differentially expressed genes are known to interact with the NFκB pathway (supplemental table 3). These included three NFκB activators (Ikbke, Ccl21b and Ikbkap) and one inhibitor (Ucp2) up-regulated in DA. As described above, IL-6 and IL-11, which are known to activate NFκB, were also expressed in increased levels, or preferentially expressed in DA.
Among the genes up-regulated in DA.ACI(Cia10) congenics, there were those that reduce activation (Cat) and transcription (Pawr and Thbd) of NFκB. Additionally, several genes up-regulated in DA.ACI(Cia10) congenics have been reported to interfere with the NFκB pathway (Supplemental figure 1). Therefore, we looked for 244 NFκB/C-rel target genes previously reported by others 18 and determined that 78 were expressed by all of our samples (32%). Only one of these genes (Acs14) was differentially expressed (2.6-fold up-regulated in DA.ACI(Cia10), P=0.003), suggesting that NFκB was not differentially activated between the two strains.
An in vitro NFκB luciferase reporter assay performed in cultured FLS obtained from DA and DA.ACI(Cia10) rat synovial tissues and stimulated with IL-1β did not detect any significant difference in NFκB activity between the two strains, in agreement with the lack on an NFκB expression signature (supplemental figure 1).
Differentially expressed genes located within the DA.ACI(Cia10) interval on chromosome 2
204 of the 7,593 genes expressed by all samples were contained within the Cia10 QTL region on rat chromosome 2. Sixteen of these 204 (7.8%) genes were differentially expressed between DA and DA.ACI(Cia10) congenics, which was more than the 76 (1%) that would have been expected by chance (P=0.018, Chi-square test). Seven of these sixteen genes were expressed in increased levels in DA.ACI(Cia10) and included Pde5a, Palmd, Tspan2, Nes, Adamtsl4 and two antioxidant genes Gstm1 (2.58–fold, P=0.0029) and Gstm2 (2.70–fold, P=0.0028; table 6).
Table 6.
Differentially expressed genes expressed within the confirmed Cia10 region on chromosome 2.
| Gene symbol | Accession number | Gene name | Cytoband | Fold difference | P-value |
|---|---|---|---|---|---|
| Genes up-regulated in DA | DA/Cia10 | ||||
| Cd53 | NM_012523.1 | cluster of differentiation 53 | 2q34 | 3.28 | 0.0040 |
| Fcgr1 | XM_001062370.1 | Fc receptor, IgG, high affinity I | 2q34 | 2.94 | 0.0073 |
| Shc1 | NM_053517.1 | Src homology 2 domain-containing transforming protein C1 | 2q34 | 2.55 | 0.0001 |
| Tnfaip8l2 | NM_001014039.1 | Tumor necrosis factor, alpha-induced protein 8-like 2 | 2q34 | 2.42 | 0.0095 |
| RGD1560825 | XM_227659.3 | similar to glyceraldehyde-3-phosphate dehydrogenase | 2q42 | 1.99 | 0.0088 |
| Sass6 | XM_227619.4 | spindle assembly 6 homolog (C. elegans) | 2q41 | 1.73 | 0.0058 |
| Hist2h2ac | XM_574997.1 | histone cluster 2, H2ac | 2q34 | 1.57 | 0.0031 |
| RGD1303130 | NM_001004226.1 | kidney predominant protein Ncu-g1 | 2q34 | 1.54 | 0.0065 |
| RGD1560263 | XM_574989.2 | similar to nuclear receptor binding factor 2 (Nrf2) | 2q34 | 1.53 | 0.0045 |
| Genes up-regulated in DA.ACI(Cia10) | Cia10/DA | ||||
| Gstm2 | NM_177426.1 | Glutathione S-transferase, mu 2 | 2q34 | 2.71 | 0.0028 |
| Gstm1 | NM_017014.1 | Glutathione S-transferase, mu 1 | 2q34 | 2.58 | 0.0029 |
| Nes | NM_012987.1 | Nestin | 2q34 | 2.06 | 0.0060 |
| Tspan2 | NM_022589.1 | Tetraspanin 2 | 2q34 | 2.02 | 0.0057 |
| Adamtsl4 | NM_001034012.1 | ADAM metallopeptidase with thrombospondin type 1 motif like | 2q34 | 1.76 | 0.0077 |
| Palmd | NM_001025688.1 | Palmdelphin | 2q41 | 1.64 | 0.0038 |
| Pde5a | NM_133584.1 | Phosphodiesterase 5A | 2q42 | 1.58 | 0.0020 |
Nine of the above 16 differentially expressed genes were up-regulated in DA, and conversely down-regulated in DA.ACI(Cia10) congenics, and included Cd53, Fcgr1a, Shc1, Tnfaip8l2, gene similar to Gapdh, Sass6, Hist2h2ac, Ncu-g1 and gene similar to Nrf2 (table 6). Cd53 (3.28-fold, P=0.004) and Fcgr1a (2.94-fold, P=0.007) were amongst the most significantly differentially expressed genes.
The differential expression of Cd53, Fcgr1a and Gstm1 was confirmed using qPCR (figure 2). Thirteen of the differentially expressed genes located within the DA.ACI(Cia10) interval are located in the same cytogenetic band on chromosome 2 (2q34) suggesting the possibility that there could be a polymorphism within the region affecting mRNA transcription or stability of multiple genes.
None of the genes contained within the Cia10 interval were preferentially expressed in one strain versus the other.
miRNAs located within the Cia10 interval contain no sequencing polymorphisms
It was hypothesized that part of the differential gene expression pattern could be explained by polymorphisms in a miRNA located within the Cia10 interval. The polymorphism could lead to an alteration of the miRNA expression or function and modulation of target genes. Eight miRNAs are predicted to reside within the Cia10 interval on rat chromosome 2 (miR9-1, miR15b, miR16, miR92b, miR137, miR186, miR190b and miR760). While seven of these microRNAs had differentially expressed targets, none of them had more targets genes than would have been expected by chance (mir9-1: n=11, mir15b: n=16, mir16: n=16, mir92b: n=9, mir137: n=9, mir186: n=5, mir760: n=6). For further confirmation, we sequenced these seven miRNAs and their surrounding DNA (500-1000bp), which contains regulatory regions. However, no polymorphisms were detected between DA and ACI.
DISCUSSION
Increased disease severity and articular damage are associated with increased risk for disability, deformities and reduced life expectancy in patients with RA 19, 20. Yet, the genes that regulate disease severity and articular damage remain largely unknown. We have been interested in the identification of these genes and consider that they have great potential to generate new and perhaps better targets for therapies aimed at preserving joint integrity and function. In the present study we used synovial tissues from DA rats, which develop severe arthritis (PIA) with pronounced synovial hyperplasia and cartilage and bone destruction, and synovial tissues from arthritis-protected and non-erosive DA.ACI(Cia10) congenics. These two strains are genetically identical except for the presence of ACI alleles at the Cia10 interval, underscoring the magnitude of the effect of this single locus on clinical disease, histologic joint damage 7 and gene expression.
DA.ACI(Cia10) and DA had significant differences in the expression of inflammatory mediators. That difference was not broad, but instead limited to a very specific set of genes, most of which have been involved in the regulation of cartilage and bone erosions, and articular damage. IL-6 and IL-11, which belong to the IL-6 family of genes and signal through gp130, were among the cytokines with significantly reduced expression in DA.ACI(Cia10) congenics. Both IL-6 and IL-11 are expressed in increased levels in RA synovium, and while IL-6 has been clearly implicated in arthritis pathogenesis and joint damage 8, 9, the literature on IL-11 is contradictory 10, 11. The chemokines Ccl12, Ccl21b and Cxcl10, and chemokine receptors Ccr6, Cx3cr1 and Cxcr3 were also expressed in reduced levels in congenics. Ccl12 is chemotactic for Ccr2-expressing monocytes and neutrophils, which have a key role in the development of synovial hyperplasia and joint erosions and damage 21. Ccl21b is chemotactic for Ccr7-expressing T and B cells, stimulates FLS, and has been implicated in the organization of germinal center-like structures in synovial tissues of RA 22, 23. Cx3cr1 is expressed by myeloid cells, and its blockade ameliorates rodent arthritis and reduces articular damage 24. Cxcl10 and its receptor Cxcr3 was predominantly expressed in DA, and these genes mediate T cell and mast cell chemotaxis, and increase FLS invasion in an autocrine and paracrine manner (Laragione et al. unpublished observations). Furthermore, blockade of Cxcr3 ameliorates adjuvant-induced arthritis in rats, also reducing joint erosion and damage 25.
Synovial tissues from DA.ACI(Cia10) congenics had increased expression of Tgfb2 and Ccl11. Tgfbr3 was also preferentially expressed in congenics’ synovial tissues. Tgfb is considered a suppressor of T cell responses and has a central role in the differentiation of Treg cells. Furthermore, treatment with Tgfb ameliorates autoimmune arthritis in rodents while anti-Tgfb antibodies exacerbate disease 26. Ccl11, also known as Eotaxin-1, is an eosinophil chemoattractant expressed by T-cells and fibroblasts. Increased serum levels of Ccl11 have been associated with reduced radiographic damage progression in early RA 14. Lastly, the Vdr was predominantly expressed in the synovial tissues of congenics. The Vdr has known IL-6-suppressive 27, Th17 differentiation-inhibitory properties 28, and Tgfb-inducing activities (figure 3), which matches the scenario identified in this study. Vdr agonists also reduced arthritis severity 29. Together, our findings suggest that Cia10 directly or indirectly regulates the expression of genes implicated in arthritis severity and joint damage, with increased levels or preferential expression of pro-inflammatory and joint damage-favoring genes in DA, while the opposite takes place in congenics.
While IL-23, IL-17-F and RORgt (Rorc) were not differentially expressed in synovial tissues collected on day 21, four of the genes up-regulated or preferentially expressed in DA, and conversely down-regulated in DA.ACI(Cia10) congenics, suggest an increased presence of IL-17 producing or inducing cells: a) IL-17A was expressed in nearly all DA samples (93%), but only in 43% of congenics; b) Ccr6 is a cell surface marker of Th17 T cells 30; c) Stat4 mediates IL-23r signaling, which is required for the generation of Th17 cells; and d) Cxcr3 is expressed by mast cells, which have been considered the main source of IL-17 in the synovial tissue 31. Furthermore, DA synovial tissues preferentially expressed IL-6, which is a key cytokine for differentiation of Th17 cells, but lacked Vdr expression, which is an inhibitor of Th17 differentiation. These observations raise the possibility that Cia10 could be a new gene controlling arthritis severity indirectly via the regulation of the differentiation and/or homing of Th17 cells to the joint.
Four of the genes with the most significantly reduced expression in DA.ACI(Cia10) congenics are either associated with genetic susceptibility to RA (Ccr6, Ccl21b, Stat4) 12, 32, or as in the case of Csk, directly interact and regulate the activity of a RA susceptibility gene (PTPN22) 2. Furthermore, Gstm1 null alleles have been associated with RA severity and radiographic erosive damage 17, and DA synovial tissues did in fact have significantly reduced expression compared with DA.ACI(Cia10). However, sequencing of DA Gstm1 gene revealed no deletions, insertions or significant sequence variants that might explain the reduced expression (data not shown). Taken together, our results suggest that the Cia10 gene could interact with RA susceptibility genes to regulate their expression and perhaps function, raising the possibility of potential epistatic interactions.
NFκB is a central regulator of synovial hyperplasia and articular damage 33, 34, and several RA susceptibility genes are involved in this pathway 35. A subset of the differentially expressed genes in the present study, including IL-6, IL-11 and Ccl21b, are known activators of NFκB, and NFκB-interacting genes were differentially expressed, raising the possibility that Cia10 might be involved in the regulation of NFκB activity. However, we were not able to detect a differentially expressed NFκB transcriptional signature 18. For further confirmation a NFκB luciferase reporter assay was studied in FLS from DA and DA.ACI(Cia10) congenics stimulated with IL-1β, and did not detect any significant difference in transcriptional activity. Therefore, our results suggest that Cia10 does not have a major effect on the regulation of the NFκB pathway.
There are several similarities between the pathogenesis of RA synovial hyperplasia, its invasion and destruction of cartilage and bone, and the behavior of cancers 36-38. Previous studies from our group and others have identified increased expression of cancer-related genes in FLS from arthritic rats and from patients with RA 39-41. In the present study one of the predominant differentially expressed groups was related to cancer and cancer phenotypes. Genes involved in oncogenesis (Kras, Ikbke, St14), cell proliferation (Cdc25a, Ier3, Ppap2a), cancer invasion (Cd53, Ctsd, Cxcl10, Cxcr3, Loxl1) 42-46 and histone deacetylation (Hdac1, Hdac2 and Rbbp7) 16 were expressed in increased levels in DA and in reduced levels in congenics.
On the contrary of the cancer-favoring genes up-regulated in DA, genes up-regulated in DA.ACI(Cia10) were protective against a cancer phenotypes. For instance, Nov, Rgs4 and Tgfbr3, which are inhibitors of cancer cell migration and invasion 47-49, were expressed in increased levels in DA.ACI(Cia10). Two MMP inhibitors (Timp2 and Reck) were up-regulated in DA.ACI(Cia10) and down-regulated in DA. Reck negatively regulates cancer cell invasion, metastasis and angiogenesis in cancer, and its expression is reduced in cancer cells 50, 51. Like in cancers and in DA synovium, Reck is expressed in reduced levels in RA synovial tissues, compared with OA controls 52. Timp2 is a negative regulator of MMPs, angiogenesis, cancer cell growth, invasion and metastasis 53, 54. Taken together, our results show that the presence of DA alleles at Cia10 increases the expression of genes favoring cancer development, proliferation and invasion, while the opposite is seen in the presence of ACI alleles. These observations suggest that these Cia10-regulated cancer genes could be involved in the regulation of synovial hyperplasia and pannus invasion and destruction of cartilage and bone.
Survival and apoptosis abnormalities have been described in cancer, in arthritic synovial tissues and in autoimmunity. We considered that the synovial hyperplasia in DA, and the lack of in DA.ACI(Cia10) could be attributed to differences in expression of apoptosis genes. While many apoptosis and cell survival genes were differentially expressed, there was not a clear bias towards having increased numbers of pro-apoptosis versus anti-apoptosis genes in either strain.
Thirteen of the sixteen differentially expressed genes located within the DA.ACI(Cia10) interval map to the same cytogenetic band on chromosome 2 (2q34) raising the possibility that a polymorphism within the region could affect a regulatory element acting in cis to influence the transcription of these closely located genes. While cis-acting transcription factor binding site polymorphisms have been a rare explanation for autoimmunity or other complex traits, it is certainly a possibility worth considering.
We also considered that a polymorphism in a microRNA located within the Cia10 interval might explain part of the differences in synovial gene expression. The Cia10 interval contains eight predicted microRNAs but sequencing these microRNAs revealed no polymorphisms. Additionally, analyses of the expression of predicted microRNA targets revealed no significant differences between the strains, making the microRNA hypothesis unlikely to explain the Cia10 effect.
In conclusion, we have determined that Cia10 regulates the expression of a unique set of inflammatory mediators, including markers of Th17 cells, IL-6, IL-17A and IL-11, oxidative stress regulators and a cancer-associated gene expression signature. These observations suggest a mechanism of action for the Cia10 gene, and several new potential prognostic biomarkers and targets for therapies aimed at reducing disease severity and joint damage in RA.
MATERIAL AND METHODS
Rats and generation of DA.ACI(Cia10) congenic strain
DA (arthritis-susceptible) and ACI (arthritis resistant) rats were originally purchased from Harlan Sprague Dawley (Indianapolis, IN). DA.ACI(Cia10) congenic rats were generated as previously described 7. Briefly, a 52.6 Mbp region from the ACI strain containing the Cia10 interval on chromosome 2 (Figure 1A) was introgressed into DA through genotype-guided breeding. This strategy selected for ACI alleles at the Cia10 interval while at the same time excluding donor genome contamination at other loci known to regulate arthritis severity 6, 55. After five backcrosses offspring heterozygous at the Cia10 interval were intercrossed to generate Cia10 homozygotes for ACI alleles (DA.ACI(Cia10)) to be used in experiments. The experimental protocol was reviewed and approved by the FIMR Institutional Animal Care and Use Committee.
Induction of PIA and arthritis scoring
Male DA (n=12) and DA.ACI(Cia10) (n=6) congenic rats 8-12 week-old were anesthetized and injected intradermally at the base of the tail with 150 μl of pristane (MP Bio, Solon, OH) divided into two injection sites (day 0) 7, 56. Arthritis severity was assessed using a previously described 80-point scoring system that uses the sum of scores to generate an arthritis severity index (ASI) 7. Ankle synovial tissues were collected for analysis 21 days post-induction of PIA.
RNA extraction
Synovial tissue was homogenized in lysis buffer using a rotor-stator homogenizer and total RNA was extracted using RNeasy (Qiagen, Valencia, CA) and including a DNase treatment step, according to manufacturer’s instructions. RNA was quantified and assessed for purity using the NanoDrop spectrophotometer (Rockland, DE), and RNA integrity was verified using the BioAnalyzer 2100 (Agilent, Palo Alto, CA).
Microarray experiments
The microarray protocol was previously described 40. Briefly, 200 ng of total RNA was amplified and biotinylated using the TotalPrep labeling kit (Ambion, Austin, TX). RNA samples were hybridized to the RatRef-12 Expression BeadChip (Illumina, San Diego, CA), which contains 22,524 probes covering 21,922 rat genes selected primarily from the NCBI RefSeq database (Release 16). Hybridization was carried out in Illumina IntelliHyb chambers, followed by washing and staining with Cy3-streptavidin. The array was scanned on a high-resolution Illumina BeadArray reader using a two-channel 0.8 μm resolution confocal laser scanner. Protocols were previously optimized for use with the Illumina Whole-Genome Expression platform.
Microarray data analysis
Illumina Bead studio software (Version 2.0) was used to extract data, which was normalized using the cubic spline algorithm. In order to reduce the false positive rate, only genes expressed by all 18 synovial tissues were used for analyses. Mean intensity values for genes detected in all 18 samples were Log2-transformed and expression data from DA and DA.ACI(Cia10) compared using the t-test. Fold-differences were calculated by elevating 2 to the difference between the means of log2-transformed DA and log2-transformed DA.ACI(Cia10) values [2(mean Log2DA − mean Log2 Cia10)], and vice-versa for DA.ACI(Cia10) fold-differences. Genes with a P-value ≤0.01 (t-test) and a fold-difference ≥1.5 between DA and DA.ACI(Cia10) were considered significantly different. Ingenuity IPA 5.5.1 program (Ingenuity, Redwood City, CA, USA) and publicly available databases such as Pubmed and Online Mendelian Inheritance in Man (OMIM) were used for pathway discovery and analysis. Enrichment for biological functions and disease groups was determined with the IPA software and calculated using the Fisher’s exact test with a cutoff p-value of ≤0.05, using the Benjamini-Hochberg correction.
We also looked for genes a) expressed in synovial tissues of one strain and not by the other, or b) predominantly in one strain and not in the other, since those could represent even more significant differences relevant to disease pathogenesis. None of the genes met criteria “a” (expressed only by one strain and not by the other), but several met criteria “b”.
cDNA synthesis and quantitative PCR (qPCR)
cDNA was synthesized from the same RNA samples hybridized to the RatRef-12 Expression BeadChip. 2μl of total RNA was used for cDNA synthesis using the Superscript III kit (Invitrogen Carlsbad, CA). cDNA was diluted 1:10 for qPCR. Rat specific primers and probes were designed using the Universal ProbeLibrary (Roche, Indianapolis, IN) (table 1). Gapdh was used as an internal control and all samples were run in duplicate. The average threshold cycle (Ct) values were used to analyze relative gene expression of Cd53, Fcgr1a, Gp49b, Timp2 Reck, Nov and Gstm1. Probes were used at a final concentration of 100nM, 5’ ends were labeled with FAM and 3’ ends with TAMRA. Primers were used at 200 nM with Eurogentec qPCR mastermix (Eurogentec, San Diego, CA). Reactions were run on an ABI 7700 qPCR thermocycler at 50°C for 2 minutes, 95°C for 10 minutes, and 45 cycles of 95°C for 0.15 minutes and 60°C for 1 minute. Relative expression of genes was normalized to Gapdh in each sample (ΔCt), and ΔCt values were used for t-test analysis. qPCR fold-differences were calculated using the 2-ΔΔCt method 57.
Table 1.
Primers and probes used for microarray results’ validation with qPCR, and primers used for microRNA sequencing§.
| Gene Symbol | Accession Number | Forward primer | Reverse primer | Probe number |
|---|---|---|---|---|
| qPCR | ||||
| Cd53 | NM_012523.1 | GGGACTTCATCCAGTCACAAC | AACCCTGAACATCTGCACCT | 4 |
| Fcgr1a | NM_001100836.1 | CCTGTGCAGTTAGAAATCTACAGAGA | CTTATTCTGCCACGGGTGA | 110 |
| Gapdh | NM_017008 | GAACGGGAAGCTCACTGGC | GCATGTCAGATCCACAACGG | Taqman probe |
| Gstm1 | NM_017014.1 | TGTTACAACCCCGACTTTGA | TCTTCTCAGGGATGGTCTTCA | 106 |
| LOC499078* | XM_001070660.1 | GGAGAGCTGGACAGTTGGAA | GGTTACAACCTGGGTCTCTTTG | 80 |
| Nov | NM_030868.2 | CGGCCTTGTGAGCAAGAG | TTCTTGGTCCGGAGACACTT | 60 |
| Reck | NM_001107954.1 | AAAAGTTGGACACAATTGCTAAGA | TCGGAGTTGATGAGGGACTC | 38 |
| Timp2 | NM_021989.2 | CTGGACGTTGGAGGAAAGAA | ACAGAGGGTAATGTGCATCTTG | 12 |
| Rorc | RGD:1595785 | AGCAGAACTGCCCCATTG | TCCCTCTGCTTCTTGGACAT | 21 |
| Il17a | NM_001106897 | CTTCACCCTGGACTCTGAGC | CCTCAGCGTTGACACAGC | 98 |
| Il17f | NM_001015011.1 | GGCTCCTGTGAAACAACCAT | TGATGGCAAATCCCAACAT | 84 |
| Sequencing | ||||
| mir9-1 | MI0000838 | CAGCTTAGATTCCCGACCTCAGAAC | ATCTTGACCCCCAGTAAAGCTGAAG | --- |
| mir15b/16# | MI0000843/MI0000844 | TGACGTCCTTCCTAACAGCAACTTC | TGTGTACAAATTAAACCCACGCAAA | --- |
| mir92b | MI0006167 | GAGGTATGGGTGGGAAAAGTACCAA | TTAAAAACATTTCGGAGGTGCATGA | --- |
| mir137 | MI0000910 | AAGCAGAGCAAATACCAAGGAAAGC | ACCACCCGAGGAAAAGAAAACATAA | --- |
| mir186 | MI0000931 | TTCTAGCATTTGAGGGCAGTTACCA | GGTGCTACAGACAACTGAGGGACAT | --- |
| mir190b | MI0006135 | AGAGGAGATGCAAACCCCTAACCTT | AGACAATAGTCTCAGAGCCCCAGGA | --- |
| mir760 | MI0006164 | CTCTAGAAGTCCAATGCGCTATCCA | AGGAAACTGAGGACCCACTCCTTC | --- |
microRNA=miR and its accession numbers were obtained from Ensembl and the miRBase;
microRNA analyses
The miRBase.org database was used to identify the microRNAs located within the Cia10 interval, and TargetScan.org was used to predict their targets. The eight microRNAs contained within the Cia10 and with differentially expressed predicted targets were sequenced as described below.
Sequencing
Splenic genomic DNA (gDNA) was extracted using DNeasy Blood and Tissue Kit (Qiagen). Primers were designed using the Whitehead Institute Primer3 website (http://frodo.wi.mit.edu/primer3/input.htm) to amplify 500-1000bp regions surrounding eight known microRNA sequences located within the Cia10 interval (table 1). PCR products were sequenced and analysed with the DNASTAR sequencing analysis software (Madison, WI).
NFκB luciferase reporter assay
Synovial tissues were collected from a different group of DA and from DA.ACI(Cia10) joints 21 days after the induction of PIA for fibroblast-like synoviocyte (FLS) isolation. FLS were isolated as previously described 40. Cells from passage four or greater (>95% FLS purity) were used for the experiments. 4.6×104 cells per cell line per strain were co-transfected with a NFκB luciferase reporter plasmid (NFκB binding sequences were originally cloned into a Promega pGL2 construct at Dr. L. Klampfer’s laboratory, Albert Einstein College of Medicine, Bronx, NY) and a Renilla internal control plasmid (Promega, Madison, WI) using Lipofectamine 2000 (Invitrogen). Control cells were transfected with a luciferase plasmid without a promoter. Following transfections cells were cultured overnight on FBS-free medium, followed by stimulation with IL-1β 10ng/ml for 48h. The NFκB reporter activity was measured with the TD 20/20 Luminometer (Turner Designs, Sunny vale, CA). Arbitrary units of the Renilla reporter were used for internal normalization.
Statistical Analysis
The t-test was used to compare the expression of genes expressed by all samples. Chi-squared and Fisher exact tests were performed using SigmaStat 3.0 (SPSS, Chicago, IL).
Supplementary Material
FLS from DA (n=4) and DA.ACI(Cia10) (n=4) were transfected with a NFκB luciferase reporter construct and stimulated with IL-1β (10ng/ml) or control vehicle for 48h. Values represent arbitrary units of luciferase luminescence.
Acknowledgments
Funded by the National Institutes of Health grants R01-AR46213, R01-AR052439 (NIAMS) and R01-AI54348 (NIAID) to Dr. P. Gulko.
Footnotes
CONFLICT OF INTEREST
The authors have no conflict of interest to declare.
References
- 1.Gregersen PK, Plenge RM, Gulko PS. Genetics of rheumatoid arthritis. In: Firestein G, Panayi G, Wollheim FA, editors. Rheumatoid arthritis. second. Oxford University Press; New York: 2006. pp. 3–14. [Google Scholar]
- 2.Begovich AB, Carlton VE, Honigberg LA, Schrodi SJ, Chokkalingam AP, Alexander HC, et al. A missense single-nucleotide polymorphism in a gene encoding a protein tyrosine phosphatase (PTPN22) is associated with rheumatoid arthritis. Am J Hum Genet. 2004;75(2):330–337. doi: 10.1086/422827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Plenge RM, Padyukov L, Remmers EF, Purcell S, Lee AT, Karlson EW, et al. Replication of putative candidate-gene associations with rheumatoid arthritis in >4,000 samples from North America and Sweden: association of susceptibility with PTPN22, CTLA4, and PADI4. Am J Hum Genet. 2005;77(6):1044–60. doi: 10.1086/498651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kurreeman FA, Padyukov L, Marques RB, Schrodi SJ, Seddighzadeh M, Stoeken-Rijsbergen G, et al. A candidate gene approach identifies the TRAF1/C5 region as a risk factor for rheumatoid arthritis. PLoS Med. 2007;4(9):e278. doi: 10.1371/journal.pmed.0040278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Plenge RM, Seielstad M, Padyukov L, Lee AT, Remmers EF, Ding B, et al. TRAF1-C5 as a risk locus for rheumatoid arthritis--a genomewide study. N Engl J Med. 2007;357(12):1199–209. doi: 10.1056/NEJMoa073491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gulko PS, Kawahito Y, Remmers EF, Reese VR, Wang J, Dracheva SV, et al. Identification of a new non-major histocompatibility complex genetic locus on chromosome 2 that controls disease severity in collagen- induced arthritis in rats. Arthrititis Rheum. 1998;41(12):2122–31. doi: 10.1002/1529-0131(199812)41:12<2122::AID-ART7>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 7.Brenner M, Meng H, Yarlett N, Griffiths M, Remmers E, Wilder R, et al. The non-MHC quantitative trait locus Cia10 contains a major arthritis gene and regulates disease severity, pannus formation and joint damage. Arthritis Rheum. 2005;52(1):322–32. doi: 10.1002/art.20782. [DOI] [PubMed] [Google Scholar]
- 8.Alonzi T, Fattori E, Lazzaro D, Costa P, Probert L, Kollias G, et al. Interleukin 6 is required for the development of collagen-induced arthritis. J Exp Med. 1998;187(4):461–8. doi: 10.1084/jem.187.4.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nishimoto N, Yoshizaki K, Miyasaka N, Yamamoto K, Kawai S, Takeuchi T, et al. Treatment of rheumatoid arthritis with humanized anti-interleukin-6 receptor antibody: a multicenter, double-blind, placebo-controlled trial. Arthritis Rheum. 2004;50(6):1761–9. doi: 10.1002/art.20303. [DOI] [PubMed] [Google Scholar]
- 10.Wong PK, Campbell IK, Robb L, Wicks IP. Endogenous IL-11 is pro-inflammatory in acute methylated bovine serum albumin/interleukin-1-induced (mBSA/IL-1)arthritis. Cytokine. 2005;29(2):72–6. doi: 10.1016/j.cyto.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 11.Walmsley M, Butler DM, Marinova-Mutafchieva L, Feldmann M. An anti-inflammatory role for interleukin-11 in established murine collagen-induced arthritis. Immunology. 1998;95(1):31–7. doi: 10.1046/j.1365-2567.1998.00568.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stahl EA, Raychaudhuri S, Remmers EF, Xie G, Eyre S, Thomson BP, et al. Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat Genet. 2010;42(6):508–14. doi: 10.1038/ng.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Green EA, Gorelik L, McGregor CM, Tran EH, Flavell RA. CD4+CD25+ T regulatory cells control anti-islet CD8+ T cells through TGF-beta-TGF-beta receptor interactions in type 1 diabetes. Proc Natl Acad Sci U S A. 2003;100(19):10878–83. doi: 10.1073/pnas.1834400100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Syversen SW, Goll GL, Haavardsholm EA, Boyesen P, Lea T, Kvien TK. A high serum level of eotaxin (CCL 11) is associated with less radiographic progression in early rheumatoid arthritis patients. Arthritis Res Ther. 2008;10(2):R28. doi: 10.1186/ar2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Putoczki T, Ernst M. More than a sidekick: the IL-6 family cytokine IL-11 links inflammation to cancer. J Leukoc Biol. 2010;88(6):1109–17. doi: 10.1189/jlb.0410226. [DOI] [PubMed] [Google Scholar]
- 16.Lane AA, Chabner BA. Histone deacetylase inhibitors in cancer therapy. J Clin Oncol. 2009;27(32):5459–68. doi: 10.1200/JCO.2009.22.1291. [DOI] [PubMed] [Google Scholar]
- 17.Mattey DL, Hutchinson D, Dawes PT, Nixon NB, Clarke S, Fisher J, et al. Smoking and disease severity in rheumatoid arthritis: association with polymorphism at the glutathione S-transferase M1 locus. Arthritis Rheum. 2002;46(3):640–6. doi: 10.1002/art.10174. [DOI] [PubMed] [Google Scholar]
- 18.Bunting K, Rao S, Hardy K, Woltring D, Denyer GS, Wang J, et al. Genome-wide analysis of gene expression in T cells to identify targets of the NF-kappa B transcription factor c-Rel. J Immunol. 2007;178(11):7097–109. doi: 10.4049/jimmunol.178.11.7097. [DOI] [PubMed] [Google Scholar]
- 19.Krause D, Schleusser B, Herborn G, Rau R. Response to methotrexate treatment is associated with reduced mortality in patients with severe rheumatoid arthritis. Arthritis Rheum. 2000;43(1):14–21. doi: 10.1002/1529-0131(200001)43:1<14::AID-ANR3>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 20.Navarro-Cano G, Del Rincon I, Pogosian S, Roldan JF, Escalante A. Association of mortality with disease severity in rheumatoid arthritis, independent of comorbidity. Arthritis Rheum. 2003;48(9):2425–33. doi: 10.1002/art.11127. [DOI] [PubMed] [Google Scholar]
- 21.Bruhl H, Cihak J, Schneider MA, Plachy J, Rupp T, Wenzel I, et al. Dual role of CCR2 during initiation and progression of collagen-induced arthritis: evidence for regulatory activity of CCR2+ T cells. J Immunol. 2004;172(2):890–8. doi: 10.4049/jimmunol.172.2.890. [DOI] [PubMed] [Google Scholar]
- 22.Bruhl H, Mack M, Niedermeier M, Lochbaum D, Scholmerich J, Straub RH. Functional expression of the chemokine receptor CCR7 on fibroblast-like synoviocytes. Rheumatology (Oxford) 2008;47(12):1771–4. doi: 10.1093/rheumatology/ken383. [DOI] [PubMed] [Google Scholar]
- 23.Manzo A, Paoletti S, Carulli M, Blades MC, Barone F, Yanni G, et al. Systematic microanatomical analysis of CXCL13 and CCL21 in situ production and progressive lymphoid organization in rheumatoid synovitis. Eur J Immunol. 2005;35(5):1347–59. doi: 10.1002/eji.200425830. [DOI] [PubMed] [Google Scholar]
- 24.Nanki T, Urasaki Y, Imai T, Nishimura M, Muramoto K, Kubota T, et al. Inhibition of fractalkine ameliorates murine collagen-induced arthritis. J Immunol. 2004;173(11):7010–6. doi: 10.4049/jimmunol.173.11.7010. [DOI] [PubMed] [Google Scholar]
- 25.Mohan K, Issekutz TB. Blockade of chemokine receptor CXCR3 inhibits T cell recruitment to inflamed joints and decreases the severity of adjuvant arthritis. J Immunol. 2007;179(12):8463–9. doi: 10.4049/jimmunol.179.12.8463. [DOI] [PubMed] [Google Scholar]
- 26.Thorbecke GJ, Shah R, Leu CH, Kuruvilla AP, Hardison AM, Palladino MA. Involvement of endogenous tumor necrosis factor alpha and transforming growth factor beta during induction of collagen type II arthritis in mice. Proc Natl Acad Sci U S A. 1992;89(16):7375–9. doi: 10.1073/pnas.89.16.7375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Srviastava MD, DeLuca H, Ambrus JL. Inhibition of IL-6 and IL-8 production in human fibroblast cell lines by 1,25 (OH)2 vitamin D3 and two of its analogs with lower calcemic activity. Res Commun Chem Pathol Pharmacol. 1994;83(2):145–50. [PubMed] [Google Scholar]
- 28.Chang JH, Cha HR, Lee DS, Seo KY, Kweon MN. 1,25-Dihydroxyvitamin D3 inhibits the differentiation and migration of T(H)17 cells to protect against experimental autoimmune encephalomyelitis. PLoS One. 2010;5(9):e12925. doi: 10.1371/journal.pone.0012925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsuji M, Fujii K, Nakano T, Nishii Y. 1 alpha-hydroxyvitamin D3 inhibits type II collagen-induced arthritis in rats. FEBS Lett. 1994;337(3):248–50. doi: 10.1016/0014-5793(94)80201-7. [DOI] [PubMed] [Google Scholar]
- 30.Hirota K, Yoshitomi H, Hashimoto M, Maeda S, Teradaira S, Sugimoto N, et al. Preferential recruitment of CCR6-expressing Th17 cells to inflamed joints via CCL20 in rheumatoid arthritis and its animal model. J Exp Med. 2007;204(12):2803–12. doi: 10.1084/jem.20071397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hueber AJ, Asquith DL, Miller AM, Reilly J, Kerr S, Leipe J, et al. Mast cells express IL-17A in rheumatoid arthritis synovium. J Immunol. 2010;184(7):3336–40. doi: 10.4049/jimmunol.0903566. [DOI] [PubMed] [Google Scholar]
- 32.Remmers EF, Plenge RM, Lee AT, Graham RR, Hom G, Behrens TW, et al. STAT4 and Risk of Rheumatoid Arthritis and Systemic Lupus Erythematosus. N Engl J Med. 2007 doi: 10.1056/NEJMoa073003. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Han Z, Boyle DL, Manning AM, Firestein GS. AP-1 and NF-kappaB regulation in rheumatoid arthritis and murine collagen-induced arthritis. Autoimmunity. 1998;28(4):197–208. doi: 10.3109/08916939808995367. [DOI] [PubMed] [Google Scholar]
- 34.Miagkov AV, Kovalenko DV, Brown CE, Didsbury JR, Cogswell JP, Stimpson SA, et al. NF-kappaB activation provides the potential link between inflammation and hyperplasia in the arthritic joint. Proc Natl Acad Sci U S A. 1998;95(23):13859–64. doi: 10.1073/pnas.95.23.13859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raychaudhuri S, Remmers EF, Lee AT, Hackett R, Guiducci C, Burtt NP, et al. Common variants at CD40 and other loci confer risk of rheumatoid arthritis. Nat Genet. 2008;40(10):1216–23. doi: 10.1038/ng.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muller-Ladner U, Kriegsmann J, Gay RE, Gay S. Oncogenes in rheumatoid arthritis. Rheum Dis Clin North Am. 1995;21(3):675–90. [PubMed] [Google Scholar]
- 37.Franz JK, Pap T, Hummel KM, Nawrath M, Aicher WK, Shigeyama Y, et al. Expression of sentrin, a novel antiapoptotic molecule, at sites of synovial invasion in rheumatoid arthritis. Arthritis Rheum. 2000;43(3):599–607. doi: 10.1002/1529-0131(200003)43:3<599::AID-ANR17>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 38.Firestein GS, Echeverri F, Yeo M, Zvaifler NJ, Green DR. Somatic mutations in the p53 tumor suppressor gene in rheumatoid arthritis synovium. Proc Natl Acad Sci U S A. 1997;94(20):10895–900. doi: 10.1073/pnas.94.20.10895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ritchlin C, Dwyer E, Bucala R, Winchester R. Sustained and distinctive patterns of gene activation in synovial fibroblasts and whole synovial tissue obtained from inflammatory synovitis. Scand J Immunol. 1994;40(3):292–8. doi: 10.1111/j.1365-3083.1994.tb03465.x. [DOI] [PubMed] [Google Scholar]
- 40.Laragione T, Brenner M, Li W, Gulko PS. Cia5d regulates a new fibroblast-like synoviocyte invasion-associated gene expression signature. Arthritis Res Ther. 2008;10(4):R92. doi: 10.1186/ar2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pap T, Franz JK, Hummel KM, Jeisy E, Gay R, Gay S. Activation of synovial fibroblasts in rheumatoid arthritis: lack of Expression of the tumour suppressor PTEN at sites of invasive growth and destruction. Arthritis Res. 2000;2(1):59–64. doi: 10.1186/ar69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Irby RB, Malek RL, Bloom G, Tsai J, Letwin N, Frank BC, et al. Iterative microarray and RNA interference-based interrogation of the SRC-induced invasive phenotype. Cancer Res. 2005;65(5):1814–21. doi: 10.1158/0008-5472.CAN-04-3609. [DOI] [PubMed] [Google Scholar]
- 43.Benes P, Vetvicka V, Fusek M. Cathepsin D--many functions of one aspartic protease. Crit Rev Oncol Hematol. 2008;68(1):12–28. doi: 10.1016/j.critrevonc.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kirschmann DA, Seftor EA, Fong SF, Nieva DR, Sullivan CM, Edwards EM, et al. A molecular role for lysyl oxidase in breast cancer invasion. Cancer Res. 2002;62(15):4478–83. [PubMed] [Google Scholar]
- 45.Zipin-Roitman A, Meshel T, Sagi-Assif O, Shalmon B, Avivi C, Pfeffer RM, et al. CXCL10 promotes invasion-related properties in human colorectal carcinoma cells. Cancer Res. 2007;67(7):3396–405. doi: 10.1158/0008-5472.CAN-06-3087. [DOI] [PubMed] [Google Scholar]
- 46.Kawada K, Sonoshita M, Sakashita H, Takabayashi A, Yamaoka Y, Manabe T, et al. Pivotal role of CXCR3 in melanoma cell metastasis to lymph nodes. Cancer Res. 2004;64(11):4010–7. doi: 10.1158/0008-5472.CAN-03-1757. [DOI] [PubMed] [Google Scholar]
- 47.Xie Y, Wolff DW, Wei T, Wang B, Deng C, Kirui JK, et al. Breast cancer migration and invasion depend on proteasome degradation of regulator of G-protein signaling 4. Cancer Res. 2009;69(14):5743–51. doi: 10.1158/0008-5472.CAN-08-3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Benini S, Perbal B, Zambelli D, Colombo MP, Manara MC, Serra M, et al. In Ewing’s sarcoma CCN3(NOV) inhibits proliferation while promoting migration and invasion of the same cell type. Oncogene. 2005;24(27):4349–61. doi: 10.1038/sj.onc.1208620. [DOI] [PubMed] [Google Scholar]
- 49.Gatza CE, Oh SY, Blobe GC. Roles for the type III TGF-beta receptor in human cancer. Cell Signal. 2010;22(8):1163–74. doi: 10.1016/j.cellsig.2010.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oh J, Takahashi R, Kondo S, Mizoguchi A, Adachi E, Sasahara RM, et al. The membrane-anchored MMP inhibitor RECK is a key regulator of extracellular matrix integrity and angiogenesis. Cell. 2001;107(6):789–800. doi: 10.1016/s0092-8674(01)00597-9. [DOI] [PubMed] [Google Scholar]
- 51.Takahashi C, Sheng Z, Horan TP, Kitayama H, Maki M, Hitomi K, et al. Regulation of matrix metalloproteinase-9 and inhibition of tumor invasion by the membrane-anchored glycoprotein RECK. Proc Natl Acad Sci U S A. 1998;95(22):13221–6. doi: 10.1073/pnas.95.22.13221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van Lent PL, Span PN, Sloetjes AW, Radstake TR, van Lieshout AW, Heuvel JJ, et al. Expression and localisation of the new metalloproteinase inhibitor RECK (reversion inducing cysteine-rich protein with Kazal motifs) in inflamed synovial membranes of patients with rheumatoid arthritis. Ann Rheum Dis. 2005;64(3):368–74. doi: 10.1136/ard.2004.027870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.DeClerck YA, Perez N, Shimada H, Boone TC, Langley KE, Taylor SM. Inhibition of invasion and metastasis in cells transfected with an inhibitor of metalloproteinases. Cancer Res. 1992;52(3):701–8. [PubMed] [Google Scholar]
- 54.Imren S, Kohn DB, Shimada H, Blavier L, DeClerck YA. Overexpression of tissue inhibitor of metalloproteinases-2 retroviral-mediated gene transfer in vivo inhibits tumor growth and invasion. Cancer Res. 1996;56(13):2891–5. [PubMed] [Google Scholar]
- 55.Meng H, Griffiths M, Remmers E, Kawahito Y, Li W, Neisa R, et al. Identification of two novel female-specific non-MHC loci regulating collagen-induced arthritis severity and chronicity, and evidence of epistasis. Arthritis Rheum. 2004;50(8):2695–705. doi: 10.1002/art.20366. [DOI] [PubMed] [Google Scholar]
- 56.Vingsbo C, Sahlstrand P, Brun J, Jonsson R, Saxne T, Holmdahl R. Pristane-induced arthritis in rats: A new model for rheumatoid arthritis with a chronic disease course influenced by both major histocompatibility complex and non-major histocompatibility complex genes. Am J Pathol. 1996;149:1675–1683. [PMC free article] [PubMed] [Google Scholar]
- 57.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FLS from DA (n=4) and DA.ACI(Cia10) (n=4) were transfected with a NFκB luciferase reporter construct and stimulated with IL-1β (10ng/ml) or control vehicle for 48h. Values represent arbitrary units of luciferase luminescence.


