Background: Tdp1 is a DNA repair enzyme conserved across eukaryotes.
Results: Tdp1 repairs not only 3′-tyrosyl-DNA bonds and 3′-phosphoglycolates but also 5′-tyrosyl-DNA bonds and 3′-deoxyribose phosphates.
Conclusion: The end processing functions of Tdp1 extend to the repair of Top2-DNA adducts and DNA breaks from base alkylation.
Significance: Tdp1 has a broad range of DNA repair activities and is a potential drug target in anticancer therapy.
Keywords: Anticancer Drug, Cancer Therapy, DNA Damage, DNA Repair, DNA Topoisomerase, Tyrosyl-DNA Phosphodiesterase, Base Alkylation, Topoisomerase DNA Cleavage Complex
Abstract
Tyrosyl-DNA phosphodiesterase 1 (Tdp1) repairs topoisomerase I cleavage complexes (Top1cc) by hydrolyzing their 3′-phosphotyrosyl DNA bonds and repairs bleomycin-induced DNA damage by hydrolyzing 3′-phosphoglycolates. Yeast Tdp1 has also been implicated in the repair of topoisomerase II-DNA cleavage complexes (Top2cc). To determine whether vertebrate Tdp1 is involved in the repair of various DNA end-blocking lesions, we generated Tdp1 knock-out cells in chicken DT40 cells (Tdp1−/−) and Tdp1-complemented DT40 cells with human TDP1. We found that Tdp1−/− cells were not only hypersensitive to camptothecin and bleomycin but also to etoposide, methyl methanesulfonate (MMS), H2O2, and ionizing radiation. We also show they were deficient in mitochondrial Tdp1 activity. In biochemical assays, recombinant human TDP1 was found to process 5′-phosphotyrosyl DNA ends when they mimic the 5′-overhangs of Top2cc. Tdp1 also processes 3′-deoxyribose phosphates generated from hydrolysis of abasic sites, which is consistent with the hypersensitivity of Tdp1−/− cells to MMS and H2O2. Because recent studies established that CtIP together with BRCA1 also repairs topoisomerase-mediated DNA damage, we generated dual Tdp1-CtIP-deficient DT40 cells. Our results show that Tdp1 and CtIP act in parallel pathways for the repair of Top1cc and MMS-induced lesions but are epistatic for Top2cc. Together, our findings reveal a broad involvement of Tdp1 in DNA repair and clarify the role of human TDP1 in the repair of Top2-induced DNA damage.
Introduction
DNA topoisomerase I (Top1)3 is an essential eukaryotic enzyme that regulates DNA topology by relaxing both positive and negative DNA supercoiling generated during replication and transcription (1–3). To untwist the DNA, Top1 nicks one DNA strand by covalently linking its catalytic tyrosine residue to a 3′-phosphate, which allows the controlled rotation of the broken strand around the intact strand (4, 5). Once supercoiling is removed, Top1 is released by religation of the DNA 3′-phosphate with the 5′-hydroxyl end (2, 6). Camptothecin (CPT) derivatives, such as topotecan and irinotecan, are Top1 inhibitors widely used in cancer chemotherapy (7). The indenoisoquinolines, another class of non-CPT Top1 inhibitors, are in clinical development (8). These agents kill cancer cells by stabilizing Top1 cleavage complex (Top1cc) (9) and inducing the formation of DNA double strand breaks (DSBs) upon replication fork collisions with Top1cc (3, 10–13). Transcription has also been shown to contribute to the formation of DSBs (14–16).
Tyrosyl-DNA phosphodiesterase 1 (Tdp1) removes Top1cc (17–23), while Top1cc-induced DSBs resulting from replication fork collisions are primarily repaired by homologous recombination (24–28). Mutation and genetic inactivation of Tdp1 cause the neurodegenerative disease spinocerebellar ataxia neuropathy 1 (29). Spinocerebellar ataxia neuropathy 1 lymphoblastoid cells, Tdp1 knock-out mice, and murine cells derived from these mice all exhibit hypersensitivity to CPT (20, 21, 30–36). The recent finding that Tdp1 localizes to mitochondria and contributes to efficient repair of oxidative damage in mitochondrial DNA revealed a novel role for Tdp1 in mitochondrial DNA repair (6).
In addition to 3′-phosphotyrosyl bonds, Tdp1 hydrolyzes other adducts that block DNA ends, including 3′-phosphoglycolates (3′-PGs) (37) and AP sites (19, 38) albeit less effectively (18, 39). Endogenous oxidative damage or anticancer treatments (i.e. chemotherapy or ionizing radiations) can lead to such 3′-DNA blocking lesions. Ionizing radiation (IR) induces a variety of DNA lesions, including a small number of lethal DSBs and many more single strand breaks with 3′-PG ends (40, 41). Bleomycin induces DSBs with 3′-PG ends (42). The involvement of Tdp1 in the repair of such lesions is significant given that several of the agents listed above are clinically used for cancer treatment.
To better understand the role of Tdp1 in vertebrate cells, we took advantage of the relative ease to delete specific genes in chicken DT40 cells (43), which have well characterized repair pathways (44). We generated Tdp1-deficient chicken DT40 (Tdp1−/−) cell lines and Tdp1−/− cells complemented with human TDP1. Notably, there is a high degree of amino acid sequence identity and similarity between human and chicken Tdp1 (69 and 11%, respectively) with all catalytic residues conserved (45) (Fig. 1). We then examined the role of Tdp1 in repairing a broad range of DNA damages using the Tdp1−/− cells in cell survival assays and biochemical assays.
FIGURE 1.
Alignment of human and chicken Tdp1 amino acid sequences. The sequences share 69% identity (asterisks) and 11% similarity (dots). Conserved catalytic residues are shown with bold letters and are indicated by arrowheads.
The role of Tdp1 in topoisomerase II cleavage complex (Top2cc) removal has been controversial. Etoposide, a clinically used Top2 inhibitor, stabilizes Top2cc, while the Top2 homodimers are covalently linked to extruding 4-nucleotide overhangs on the 5′-end of DNA (3, 9, 46). Some reports have suggested the involvement of Tdp1 in Top2cc repair (47, 48), whereas others have provided contradictory data (18, 30, 32, 49). The results of the present study clarify the involvement of Tdp1 in resolving Top2cc-mediated DNA damage. Finally, because CtIP (RBBP8) has recently emerged as a critical factor for the repair of topoisomerase-DNA complexes (26, 27, 50), we generated double mutant DT40 cells for Tdp1 and CtIP and investigated the relative contribution of the Tdp1- and CtIP-dependent pathways in the cellular responses to Top1- and Top2-targeting drugs as well as methyl methanesulfonate (MMS).
EXPERIMENTAL PROCEDURES
Cell Culture
DT40 cells were cultured at 37 °C with 5% CO2 in RPMI 1640 medium supplemented with 1% chicken serum (Invitrogen), 10−5 m β-mercaptoethanol, penicillin, streptomycin, and 10% fetal calf serum.
Generation of Tdp1−/− DT40 Cells
Tdp1 gene disruption construct 1 (Tdp1-1-puro) was generated from genomic PCR products combined with puromycin-resistant selection cassettes flanked by loxP sites using MultiSite Gateway technology (Invitrogen). All procedures were performed according to the manufacturer's instructions. Genomic DNA sequences of wild type cells were amplified using primers 5′-ttttgggacccttgtgtcttctgctc-3′ and 5′-gttccaaatgcaatatccagtttggc-3′ for the 5′-arm and 5′-gtaagtaacaatgtctagcagg-3′ and 5′-cttccagactttgcctcacattgctc-3′ for the 3′-arm. To generate the 5′- and 3′-arm entry clones, 2.7 kb from the 5′- and 3.7 kb from the 3′-arm were subcloned by BP recombination into the donor vectors pDONRTM P4-P1R and pDONR P2R-P3, respectively. To generate the targeting vector by LR recombination, we used the 5′-arm clone, 3′-arm clone, pDEST DTA-MLS, and Puro entry clones (51). To generate Tdp1+/− cells, the Tdp1-1-puro construct was linearized by the AscI restriction enzyme and transfected by electroporation (Bio-Rad).
To generate Tdp1 gene disruption construct 2 (Tdp1-2-hyg), genomic DNA sequences of Tdp1+/− cells were amplified using primers 5′-agtcactcgaggggcccgcacaagcacgcccttttgagaacattagc-3′ and 5′-agtcactcgagcattccttgagcacaggagaactgtaagcc-3′ for the 5′-arm and 5′-gcctgttgtgggacagttctcaagcattgg-3′ and 5′-cacagctgtttctgtgcggtctgtatgctg-3′ for the 3′-arm. The amplified 3′-arm PCR product (2.2 kb) was subcloned into pCR2.1-TOPO vector (Invitrogen). The hygromycin resistance gene that harbors NotI sites at both ends (51) was cloned into the NotI site of the pCR2.1-TOPO vector carrying the 3′-arm. The 5′-arm PCR product (3.2 kb) harboring ApaI and XhoI sites at 5′- and 3′-sites, respectively, was cloned into the ApaI and XhoI sites of the pCR-2.1-TOPO vector carrying the 3′-arm and hygromycin resistance gene. To generate Tdp1−/− cells, Tdp1-2-hyg was linearized using the SacI restriction enzyme and transfected by electroporation (Bio-Rad).
The 0.7-kb fragment generated by PCR of genomic DNA using the primers 5′-tgtaagggtaagaagcagtgtgcacctg-3′ and 5′-ctaatacagacagcagtcctagagctcc-3′ was used as a probe for Southern blot analysis. The genomic DNA of the transfectants was digested with NotI and NcoI, and the targeted clones were confirmed by Southern blotting. To confirm the gene disruption by RT-PCR, the following primers were used: 5′-gcacaagcacgcccttttgagaacattagc-3′ (primer a), 5′-cacagctgtttctgtgcggtctgtatgctg-3′ (primer b), 5′-aggacagcactgaggagaattctgacagtg-3′ (primer c), and 5′-gctaatgttctcaaaagggcgtgcttgtgc-3′ (primer d). To establish cell lines that stably express the human TDP1 transgene in Tdp1−/− cells (Tdp1−/−;hTDP1 cells), pCMV-Tag2-FLAG-hTDP1 expression vector (Stratagene, La Jolla, CA) (33) was transfected in Tdp1−/− cells.
Generation of CtIP S332A/−/−;Tdp1−/− DT40 Cells
To generate the double knock-out CtIP S332A/−/−;Tdp1−/− cells, the linearized Tdp1 disruption constructs Tdp1-1-puro and Tdp1-2-hyg were transfected sequentially by electroporation. Gene disruption was confirmed by Southern blot and RT-PCR (see above).
Immunoblotting and Antibodies
To prepare whole cell lysates, cells were lysed by CelLyticTM M lysis reagent (C2978, Sigma-Aldrich). After thorough mixing and incubation at 4 °C for 30 min, lysates were then centrifuged at 12,000 × g at 4 °C for 20 min. Supernatants were collected, aliquoted, and stored at −80 °C. Preparation of mitochondrial and nuclear extracts was performed as described (6). Lysates were prepared in the same manner as whole cell lysates. Immunoblotting was carried out using standard procedures. Rabbit polyclonal anti-Tdp1 antibody was obtained from Abcam (Ab4166; Cambridge, MA). Mouse monoclonal anti-γH2AX antibody was purchased from Upstate Biotechnology (Lake Placid, NY). Actin antibodies were purchased from Sigma. Mouse monoclonal anti-Top1 antibody was purchased from BD Biosciences (556597). Rabbit polyclonal anti-Porin (AB-5; voltage-dependent anion channel) antibody was purchased from EMD Millipore (PC548T-5UG). Secondary antibodies were horseradish peroxidase (HRP)-conjugated antibodies to mouse or rabbit Ig (GE Healthcare).
Preparation of Radiolabeled Oligonucleotides and Substrates
Oligonucleotides with 5′- and 3′-phosphotyrosine linkages were synthesized by Midland Certified Reagent Co., Inc. (Midland, TX). All other oligonucleotides were synthesized by Integrated DNA Technologies (Coralville, IA). T4 polynucleotide kinase (New England Biolabs, Cambridge, MA) and [γ-32P]ATP (PerkinElmer Life Sciences) were used for 5′-end labeling, and terminal deoxynucleotidyl transferase (Invitrogen) and [α-32P]cordycepin 5′-[α-32P]triphosphate (PerkinElmer Life Sciences) were used for 3′-end labeling. For the preparation of internally labeled DNA oligonucleotides, a 22-nt DNA (5′-gcgcagctagcggcggatggca-3′) with a 3′-phosphate was labeled with 32P at the 5′-end. An 18-nt DNA (5′-tccgttgaagcctgcttt-3′) harboring the phosphotyrosine, hydroxyl, or phosphate at the 5′-end was mixed with 5′-labeled 22-nt DNA before annealing to a 36-nt DNA with complementary sequence. The nicks were sealed with T4 DNA ligase (New England Biolabs). The resulting internally labeled 40-nt product harboring 5′-phosphotyrosine, -hydroxyl, or -phosphate was then gel-purified and eluted for use (named Y40, OH40, and P40, respectively). Y40 was then annealed to a complementary 40-nt DNA (5′-tgccatccgccgctagctgcgcaaagcaggcttcaacgga-3′) or to shorter complementary DNA strands (missing 2, 4, or 6 nt from the 3′-end) to generate Y40/40, Y40/38,Y40/36, or Y40/34, respectively (see Fig. 4). Double-stranded OH40/36 and P40/36 were generated in the same manner. For the 3′-deoxyribose phosphate (3′-dRP) substrate, 5′-labeled 25-nt DNA carrying uracil at the 15th nt from the 5′-end was annealed to a complementary 25-nt DNA harboring adenine opposite the uracil (supplemental Fig. S4). Annealed DNA was incubated with uracil-DNA glycosylase for 1 h at 37 °C, and then Endonuclease III (New England BioLabs) was added for 1 h at 37 °C to generate the 3′-dRP at a nicked DNA site. Unincorporated radioactive nucleotides were removed using a mini Quick Spin Oligo column (Roche Diagnostics).
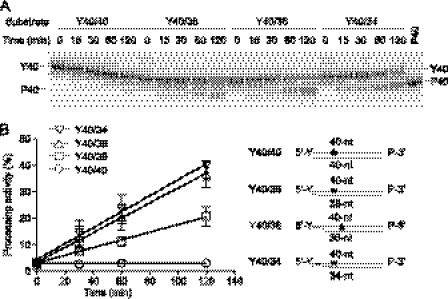
FIGURE 4.
Processing activity of recombinant human TDP1 on double-stranded substrate harboring 5′-phosphotyrosyl linkage with blunt end (Y40/40) or 2- (Y40/38), 4- (Y40/36), or 6-base (Y40/34) 5′-overhangs. A, the substrates were incubated with 1 μm TDP1 for the indicated time. P40 was used as marker. A gel representative of consistent results in independent experiments is shown. B, quantification of TDP1 processing activity from A (left). The error bars represent the S.E. (n = 3). A scheme of the DNA substrates used in A is shown (right). An asterisk indicates the internal radiolabeled site.
Complementary oligodeoxynucleotides were annealed in equimolar amounts by heating a solution to 95 °C for 3 min followed by slow cooling to room temperature. The other sequences of the oligonucleotides used in the experiments are as follows: 14Y, 5′-gatctaaaagactt-pY-3′; 14P, 5′-gatctaaaagactt-p-3′; 14OH, 5′-gatctaaaagactt-OH-3′; Y19, 5′-Yp-tccgttgaagcctgcttta-3′; Y19/19, 5′-Yp-tccgttgaagcctgcttta-3′ and 3′-aggcaacttcggacgaaat-5′; P19, 5′-p-tccgttgaagcctgcttta-3′; OH19, 5′-OH-tccgttgaagcctgcttta-3′; and 6-FAM19, 5′-FAM-tccgttgaagcctgcttta-3′. The bold “a” indicates the added adenosine through the 3′-end labeling using [α-32P]cordycepin 5′-triphosphate (Yp, tyrosylphospho linkage; p, phosphate linkage; pY, phosphotyrosyl linkage).
DNA Reactions and Gel Analyses
Preparation of cell lysate was carried out as same as for immunoblotting described above. One nanomolar labeled DNA substrates in a 10-μl reaction volume were incubated with the indicated concentration of recombinant human TDP1 (52) or cell lysate for the indicated time at 25 °C in a buffer containing 80 mm KCl, 2 mm EDTA, 1 mm dithiothreitol (DTT), 40 μg/ml bovine serum albumin, 50 mm Tris-HCl, pH 7.5, and 0.01% Tween 20. Reactions were terminated by adding 1 volume of gel loading buffer (96% (v/v) formamide, 10 mm EDTA, 1% (w/v) xylene cyanol, and 1% (w/v) bromphenol blue). Double-stranded substrates were heated at 95 °C for 3 min before loading. Samples were subjected to 16% denaturing PAGE. Gels were dried and exposed on PhosphorImager screens. Imaging and quantification were done using a Typhoon 8600 and ImageQuant software (GE Healthcare).
Measurement of Cellular Sensitivity to DNA-damaging Agents
To assess IR sensitivity, 3 × 105 cells in 10 ml of medium were irradiated with a 137Cs source. To measure the sensitivity of cells to CPT, etoposide, MMS, bleomycin, and cisplatin, cells were continuously exposed to various concentrations of the drugs. For exposure of cells to hydrogen peroxide (H2O2), 3 × 105 cells were treated in 1 ml of medium containing H2O2 for 30 min and then washed with PBS. Two hundred cells were seeded into a 384-well white plate (6007680, PerkinElmer Life Sciences) with 40 μl of medium/well. Plates were incubated at 37 °C for 72 h. Cell survival was determined using the ATPlite 1step kit (PerkinElmer Life Sciences). Briefly, 40 μl of ATPlite solution was added to each well. After 5 min, luminescence was measured by an Envision 2104 Multilabel Reader (PerkinElmer Life Sciences).
Immunostaining
Cells were treated with or without 10 nm CPT for 2 h. After cytospin, cells were fixed with 4% paraformaldehyde for 10 min at room temperature. Primary antibody against γH2AX was detected using anti-mouse IgG secondary antibodies labeled with Alexa Fluor 488/568 (Invitrogen). Cells were mounted in antifade solution with DAPI (Vector Laboratories, Burlingame, CA) and examined using a laser-scanning confocal microscope (Zeiss LSM510) with a ×63 oil objective. Images were collected and processed using the Zeiss AIM software and sized in Adobe Photoshop 7.0.
RESULTS
Generation of Tdp1−/− DT40 Cells
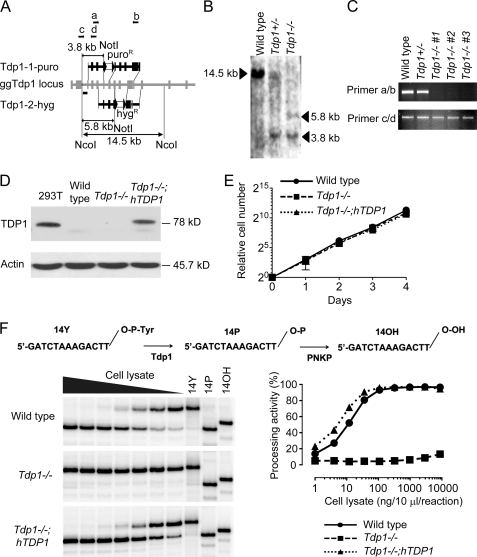
We disrupted one of two Tdp1 alleles using targeting construct 1 (Tdp1-1-puro) carrying a puromycin resistance gene (Fig. 2A) to generate Tdp1+/− cells. Gene targeting was confirmed by Southern blotting (Fig. 2B). However, disruption of the other allele was not successful using targeting construct 1 carrying a hygromycin resistance gene. This indicated the possibility of an allele-specific mutation(s) between the two Tdp1 gene alleles. Therefore, we generated targeting construct 2 (Tdp1-2-hyg; Fig. 2A) in which the homology arms were amplified using the genomic DNA from the Tdp1+/− cells as template. Targeting construct 2 successfully disrupted the other Tdp1 allele (Fig. 2B). We confirmed Tdp1 gene disruption by RT-PCR using paired primers a/b that were designed to flank both resistance genes containing stop codons. Other paired primers c/d that were designed from the 5′-side of the targeted sites were used as a control. As expected, primers a/b amplified the cDNA of wild type and Tdp1+/− cells, whereas none of three independent Tdp1−/− cell clones yielded detectable product (Fig. 2C). To complement the Tdp1−/− cells, we transfected the FLAG-tagged human TDP1 cDNA in Tdp1−/− cells. The chicken Tdp1−/− cells complemented with human TDP1 (hereafter referred to as Tdp1−/−;hTDP1 cells) showed a similar amount of TDP1 protein expression compared with human 293T cells (Fig. 2D). Several available antibodies raised against human TDP1 did not react with the chicken Tdp1 (Fig. 2D and data not shown). The proliferative properties of generated cells were indistinguishable from those of wild type cells as monitored by growth curves (Fig. 2E) and by cell cycle analysis (data not shown).
FIGURE 2.
Targeted disruption of Tdp1 in DT40 cells and characterization of Tdp1−/− cells. A, schematic representation of the chicken Tdp1 locus and configuration of the targeted allele. Black squares indicate the exons. Two targeting plasmids, Tdp1-1-puro and Tdp1-2-hyg, with different homology arms and resistance genes were used. The restriction enzyme sites used for the Southern blot analysis are indicated. Primers (a–d) used for the RT-PCR in C are indicated by arrows. The probe used in the Southern blot analysis is indicated as a horizontal bar. loxP sites are indicated as black triangles flanking each resistance gene (white square). gg, gallus gallus. B, Southern blot analysis of NcoI- and NotI-digested genomic DNA from cells with the indicated genotypes using a flanking probe as shown in A. The arrowheads indicate the position of the detected band. C, representative RT-PCR analysis with the indicated genotypes using the paired primers shown in A. D, Western blot analysis of whole cell lysates prepared from the indicated genotypes. Blots were probed with anti-human TDP1 and anti-actin. E, growth curves of cells of the indicated genotypes. The error bars represent S.D. (n = 3). F, Tdp1 is the primary 3′-tyrosyl phosphodiesterase in DT40 cells. Top, scheme for the conversion of the 14-nt 3′-phosphotyrosyl substrate (14Y) to the 14-nt 3′-hydroxyl product (14OH) by sequential action of Tdp1 and polynucleotide kinase (PNKP). Bottom left, representative gel images of cleavage assays from the indicated cell lines. 5′-32P-Labeled 14Y (1 nm) was incubated with serially diluted (1:3) whole cell lysates. The highest concentration was 8.8 μg in a 10-μl reaction volume. Bottom right, quantification of processing activity for each cell lysate.
To examine the tyrosyl-DNA phosphodiesterase activity of the mutant cells, we performed gel-based assays using a 14-nt single-stranded DNA oligonucleotide bearing a 3′-phosphotyrosine (14Y) labeled at the 5′ terminus with 32P (19, 23, 49). Tdp1 activity was measured by the extent of substrate conversion into a 3′-phosphate DNA product (14P). The 14P product can be readily converted further to a 3′-hydroxyl DNA product (14OH) by polynucleotide kinase 3′-phosphatase (Fig. 2F, upper panel) (53). The 32P-labeled 14Y substrate was incubated with serially diluted whole cell lysates from wild type, Tdp1−/−, and Tdp1−/−;hTDP1 cells in buffer without magnesium. Wild type and Tdp1−/−;hTDP1 cells showed similar processing activity that was detectable with as little as 0.1 ng/μl whole cell lysate. By contrast, no processing activity was observed in Tdp1−/− cells extract even at 8,800-fold excess (880 ng/μl) (Fig. 2F, lower panel). These results show that Tdp1 is the only factor able to process the 3′-phosphotyrosyl-DNA bond under divalent cation-free conditions and that human TDP1 can complement chicken Tdp1 for the 3′-phosphotyrosyl-DNA bond processing activity.
To determine whether Tdp1 was also functioning in DT40 cell mitochondria, we examined the Tdp1 activity of mitochondrial extracts by fractionating wild type and Tdp1−/− cells into nuclear and mitochondrial fractions and then performed the gel-based assays using 14Y substrates (supplemental Fig. S1). The mitochondrial fraction from wild type cells showed robust 3′-phosphotyrosyl diesterase activity, whereas no detectable processing activities were observed for the mitochondrial fraction of Tdp1−/− cells. These data demonstrate that the nuclear Tdp1 gene provides 3′-phosphodiesterase activity to both nuclear and mitochondrial DNA.
Broad Involvement of Tdp1 for DNA Repair
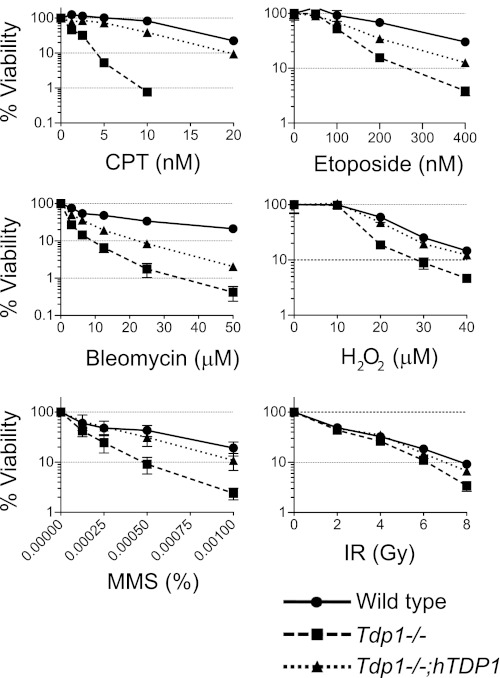
We examined the sensitivity of Tdp1−/− cells to various types of DNA-damaging agent (Fig. 3). As expected (31, 32), Tdp1−/− cells showed hypersensitivity to CPT. Low dose CPT (10 nm) also induced high γH2AX signal (12) in the Tdp1−/− cells as detected by immunostaining and Western blotting (supplemental Fig. S2, A and B) and induced the accumulation of cells at the G2 phase in Tdp1−/− cells (supplemental Fig. S2C).
FIGURE 3.
Involvement of Tdp1 for DNA repair induced by broad range of DNA-damaging agents. Survival curves of cells treated with the indicated DNA-targeting agents are shown. The indicated cells were treated with the indicated DNA-targeting agents continuously for 72 h before measuring ATP activity, which was used to measure cell viability. The survival of untreated cells was set as 100%. Error bars represent S.D. (n = 3). Gy, grays.
Tdp1−/− cells showed consistent hypersensitivity to etoposide, a selective Top2 inhibitor (3, 46), marginal sensitivity to IR, and strong sensitivity to bleomycin in agreement with the fact that both IR and bleomycin induce DSBs and/or single strand breaks with 3′-PG ends that can be processed by Tdp1 (6, 37, 39, 54). Tdp1−/− cells also showed significant sensitivity to MMS and mild sensitivity to H2O2. Complementation of the Tdp1−/− cells with human TDP1 reduced the sensitivities to these treatments. These results suggest that Tdp1 has broad involvement in the repair of lesions induced by a variety of DNA-damaging agents.
Cleavage of Top2cc by Tdp1
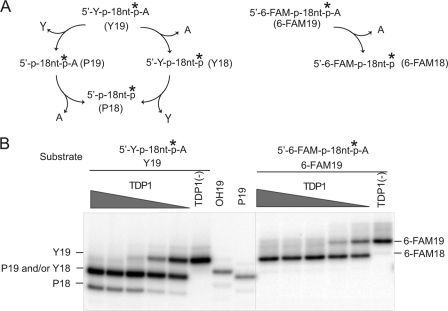
Our cell survival results showing that Tdp1−/− cells are hypersensitive to etoposide prompted us to examine the involvement of Tdp1 for the repair of Top2cc in greater detail. To test the activity of Tdp1 for Top2cc removal, we performed gel-based cleavage assays using DNA substrates with 5′-phosphotyrosyl ends. In addition to a blunt-ended double strand substrate (Y40/40), we tested substrates with 5′-overhangs of 2, 4, or 6 bases (Y40/38, Y40/36, and Y40/34, respectively; Fig. 4). The internally labeled substrates contained phosphate groups on the 3′-end to prevent the 3′-nucleosidase activity of Tdp1 from removing the last base at the 3′-end (19, 49). Use of these particular substrates eliminated the confounding issue of multiple products generated by TDP1 and allowed us to unambiguously interpret our results of 5′-phosphotyrosine processing activity by TDP1. When these substrates were incubated with recombinant human TDP1, the 5′-phosphotyrosine was removed for the 2-, 4-, and 6-base overhang substrates (Y40/38, Y40/36, and Y40/34) but not for the blunt-ended substrate (Y40/40). Notably, the 4- and 6-base overhang substrates (Y40/36 and Y40/34) were processed more efficiently than the 2-base overhang substrate (Y40/38). TDP1 also cleaved a single-stranded DNA harboring 5′-phosphotyrosine efficiently in a dose-dependent manner (Fig. 5). Note the substrates labeled on the 3′-end generated one additional product due to the 3′-nucleosidase activity of TDP1 (Fig. 5). When we tested a 5′-fluorescein (6-FAM)-labeled oligonucleotide, only one product was generated from this substrate. Because TDP1 had full 3′-nucleosidase activity under the same conditions (Fig. 5B, compare left and right panels), we attributed the product to the 3′-nucleosidase activity of TDP1 (Fig. 5A). Consistent with our previously published results (49), we conclude that TDP1 cannot process a 5′-fluorescein (6-FAM)-labeled oligonucleotide. Additional experiments revealed that TDP1 did not process 5′-phosphate or 5′-hydroxyl DNA ends to any appreciable degree (supplemental Fig. S3). Altogether, these results demonstrate that human TDP1 has specific processing activity for 5′-phosphotyrosine linkage in the context of 5′-overhangs, which are landmarks of Top2cc.
FIGURE 5.
Differential activity of recombinant human TDP1 for single-stranded DNA substrates harboring 5′-phosphotyrosine (Y19) and 5′-fluorescein (6-FAM19). A, scheme for the processing pathways of Y19 (left) and 6-FAM19 (right) by TDP1. Because of the nucleosidase activity of TDP1 (19), which removes the last 3′-end base, the Y19 substrate can be processed in two ways. One is that the 5′-tyrosine is removed first (P19) before the 3′-terminal adenine (P18) (clockwise). The other is that the 3′-adenine is removed first (Y18) before the 5′-tyrosine (P18) (counterclockwise). An asterisk indicates the radiolabeled site. B, each substrate was incubated with serial dilutions (1:3) of TDP1 at 25 °C for 30 min with the highest TDP1 concentration starting at 1 μm. Processing of 6-FAM19 by TDP1 yielded one product (right), whereas processing of Y19 yielded two distinct products (left), indicating that TDP1 processes 5′-phosphotyrosine but not 5′-fluorescein.
Involvement of Tdp1 for Repair of Abasic Sites
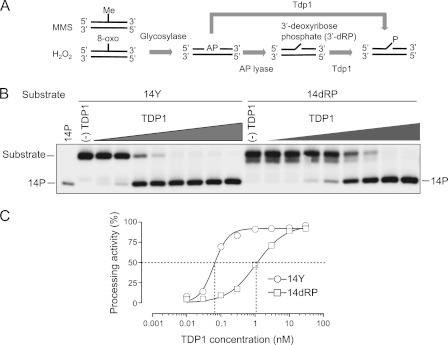
Because the hypersensitivity of Tdp1−/− cells to MMS was strong and almost fully reverted by human TDP1 (Fig. 3), we investigated the biochemical basis of this hypersensitivity. MMS is a strong electrophile that attacks the most nucleophilic centers in DNA, resulting in DNA base methylation (55). Methylated DNA bases can be converted into abasic (AP) sites by DNA N-glycosylase. H2O2 also generates modified bases such as 8-oxoguanine, which can lead to AP sites (56) (Fig. 6A). AP sites are then repaired by various pathways, including base excision repair, nucleotide excision repair, translesion synthesis, or homologous recombination. In the process of base excision repair, a 3′-dRP can form as a result of β-elimination by OGG1 and NTH1 that inhibits the gap filling by DNA polymerase β (57). Although 3′-dRP is known to be removed by AP endonuclease 1, a recent study revealed that both an AP site and a 3′-dRP are also cleaved by Tdp1 (38). Consistent with these results, we also demonstrated that recombinant human TDP1 can process a 3′-dRP substrate. The processing efficiency of 3′-dRP by TDP1 was 17-fold less than that of 3′-phosphotyrosine in terms of EC50 (the half-maximal effective concentration) (Fig. 6, B and C, and supplemental Fig. S4). Therefore, the sensitivity of Tdp1−/− cells to MMS and H2O2 is consistent with impaired repair activity of AP sites and 3′-dRP in these cells.
FIGURE 6.
Involvement of Tdp1 for repair of abasic (AP) sites. A, scheme for the conversion of MMS- and H2O2-induced DNA damage into the substrates for Tdp1. DNA adducts like methylated base and 8-oxoguanine (8-oxo) are converted into AP sites by DNA glycosylase. In one way, Tdp1 cleaves the AP site, directly generating the 3′-phosphate end. In the other way, the AP site turns out to be a single strand break with a 3′-dRP end after hydrolysis by AP lyase. Tdp1 cleaves the 3′-dRP, generating the 3′-phosphate end. B, processing activity of recombinant human TDP1 on the 14-nt 3′-phosphotyrosyl substrate (14Y) and the 14-nt 3′-dRP substrate (14dRP). Each substrate was incubated with serial dilutions (1:3) of TDP1 at 25 °C for 30 min with the highest concentration starting at 30 nm. The 14-nt 3′-phosphate (14P) was used as a marker. A gel representative of consistent results in independent experiments is shown. C, quantification of TDP1 processing activity for each substrate from B. EC50 values (the half-maximal effective concentration) for 14Y and 14dRP are 0.062 and 1.078 nm, respectively.
Parallel and Overlapping DNA Repair Functions of Tdp1 and CtIP
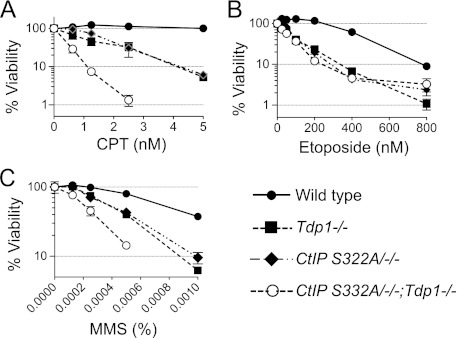
CtIP is known to be involved in homologous recombination pathways. Recent studies show that CtIP, together with BRCA1, acts in the nuclease-mediated elimination of Top1cc and Top2cc (26, 27, 50). CtIP S332A/−/− DT40 cells, which harbor an alanine substitution at residue Ser-332 of CtIP that abrogates its interaction with BRCA1, are hypersensitive to CPT, etoposide, and MMS but remain proficient in homologous recombination (26). Because this function overlaps with that of Tdp1, we generated double mutant DT40 cells (referred to as CtIP S332A/−/−;Tdp1−/−) to examine the relationship between Tdp1 and CtIP in different types of DNA repair. Both Tdp1−/− and CtIP S332A/−/− single mutant cells were hypersensitive to CPT, etoposide, and MMS, respectively (Fig. 7). CPT and MMS showed greater than an additive effect in the double mutant CtIP S332A/−/−;Tdp1−/− cells, indicating that Tdp1 and CtIP have parallel and complementary functions for the repair of Top1cc and MMS-induced DNA damage. On the contrary, CtIP S332A/−/−;Tdp1−/− cells showed similar sensitivity to etoposide compared with each Tdp1−/− and CtIP S332A/−/− single mutant, suggesting that Tdp1 and CtIP are epistatic for the repair of Top2-induced lesions.
FIGURE 7.
Interactions between CtIP and Tdp1 for repair of Top1cc, Top2cc, and MMS-induced damage. A–C, survival curves of cells treated with CPT (A), etoposide (B), or MMS (C). Sensitivity was determined as in Fig. 3. The survival of untreated cells was set as 100%. Error bars represent S.D. (n = 3).
DISCUSSION
The role of Tdp1 is well established for the repair of Top1-induced DNA damage (20, 21, 31, 32, 35, 58, 59). Because inactivation of Tdp1 augments the antiproliferative effect of Top1 inhibitors, Tdp1 inhibitors are being explored as anticancer drugs in cancer cells with preexisting Top1cc repair defects (45, 60).
Our results expand the spectrum of repair pathways that involve Tdp1, including the repair of DNA lesions produced by etoposide, IR, bleomycin, MMS, and H2O2. Biochemical assays corroborate this view given that human TDP1 can process 3′-PGs (6, 37, 39, 54), 3′-dRP (present study), 5′-phosphotyrosine (present study), and AP sites (19, 38). 3′-PG ends represent up to half of the breaks induced by IR (40) and are present at a large fraction of the DSB ends produced by bleomycin (37, 54). Accordingly, cells from Tdp1 knock-out mice are defective in the repair of IR- and H2O2-induced breaks and hypersensitive to bleomycin (31, 32). Spinocerebellar ataxia neuropathy 1 cells are also radiosensitive and unable to rapidly repair H2O2-induced single strand breaks (20). Consistent with these results, our finding that Tdp1−/− cells are hypersensitive to H2O2, bleomycin, and IR suggests a role for Tdp1 in the processing of 3′-PG and possibly AP sites. Moreover, oxidative lesions produced by IR and H2O2 are known to lead to the formation of Top1cc (61), which would also require Tdp1 for their removal.
5′-Phosphodiesterase activity has been reported for yeast Tdp1. Moreover, deletion of the Tdp1 gene in yeast confers hypersensitivity to etoposide (47), and overexpression of human TDP1 in human cells counteracts the DNA damage mediated by etoposide (48). On the other hand, Tdp1 knock-out mice and embryonic fibroblasts as well as spinocerebellar ataxia neuropathy 1 cells are not noticeably hypersensitive to etoposide (21, 32). Thus, the potential role of Tdp1 in the repair of Top2-mediated DNA damage has been a long standing controversy (18, 47). Our cell survival data with etoposide clearly indicate that Tdp1 can be involved in Top2cc repair in metazoans (Fig. 3). Why the sensitivity to etoposide is detectable in chicken Tdp1−/− cells but not in mouse and human Tdp1 mutant cells is unclear. One possibility is that chicken DT40 cells might be limited in their alternative repair pathways for Top2cc, whereas redundant pathways are present in human and murine cells. The repair protein complexes involved in the repair of Top2cc are not well understood in vertebrate cells except for the recent discovery of Tdp2 (62, 63). It is also possible that DT40 cells show a differential response to etoposide because of the rapid cell cycle of DT40 cells (∼8 h for one cycle) with most of the cells in S phase. This may also contribute to the hypersensitivity of DT40 cells to CPT. The complementation by exogenous expression of human TDP1 in Tdp1−/− cells only partially restored the resistance of Tdp1−/− cells to etoposide, whereas it almost fully restored normal responses to CPT and MMS (Fig. 3). This difference might be due to species-specific protein-protein interactions whereby human TDP1 fails to interact fully with other chicken-specific repair complexes that are critical for Top2cc and alkylation damage removal. Despite these differences, our biochemical results using recombinant human TDP1 (Figs. 4 and 5) strongly support that Tdp1 plays a role in the removal of Top2cc and contributes to the response to Top2 inhibitors.
To our knowledge, the involvement of Tdp1 in the repair of the DNA lesions induced by the classical alkylating agent MMS has not been reported. MMS produces N7-guanine methyl adducts that are readily converted to AP sites by glycosylases or/and spontaneous base elimination. Such lesions are known to trap Top1cc (64–66), which could explain the participation of Tdp1 in their repair. However, our present study also suggests a more direct involvement of Tdp1 in the repair of abasic sites by direct cleavage of the 3′-blocking dRP lesions (19, 38).
The repair of Top1- and Top2-induced DNA lesions involves redundant pathways (46, 66). A recent study revealed that CtIP, together with BRCA1, acts in the nuclease-mediated elimination of Top1cc and Top2cc (26). Deletion of Ctp1, the ortholog of CtIP in Schizosaccharomyces pombe, markedly sensitizes cells to CPT (50), and deletion of Sae2, the Saccharomyces cerevisiae ortholog of CtIP, produces a mild sensitization to CPT (36, 67). Our data show at least an additive sensitization of Tdp1−/−;CtIP S322A/−/− double mutant cells compared with the single mutants, indicating parallel (redundant) activities of CtIP and Tdp1 for Top1cc repair in DT40 cells. By contrast, we found that Tdp1 and CtIP are epistatic for the repair of Top2cc, suggesting that Tdp1 and CtIP work together for the removal of Top2cc. Further investigations are warranted to elucidate the possible interactions of Tdp1 and CtIP for the repair of Top2-induced DNA lesions.
Supplementary Material
Acknowledgments
We thank Kouji Hirota and other members at Kyoto University who contributed to the generation of the heterozygous of Tdp1 mutant chicken DT40 cells.
This work was supported, in whole or in part, by the National Institutes of Health through the Intramural Program of the NCI Center for Cancer Research.

This article contains supplemental Figs. S1–S4.
- Top1
- topoisomerase I
- Top2
- topoisomerase II
- CPT
- camptothecin
- Top1cc
- topoisomerase I DNA cleavage complex(es)
- Top2cc
- topoisomerase II DNA cleavage complex(es)
- MMS
- methyl methanesulfonate
- 3′-dRP
- 3′-deoxyribose phosphate
- TDP1
- tyrosyl-DNA phosphodiesterase 1
- DSB
- double strand break
- PG
- phosphoglycolate
- AP
- apurinic/apyrimidinic
- IR
- ionizing radiation
- nt
- nucleotide(s)
- FAM
- fluorescein.
REFERENCES
- 1. Liu L. F., Wang J. C. (1987) Supercoiling of the DNA template during transcription. Proc. Natl. Acad. Sci. U.S.A. 84, 7024–7027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Champoux J. J. (2001) DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 70, 369–413 [DOI] [PubMed] [Google Scholar]
- 3. Pommier Y., Leo E., Zhang H., Marchand C. (2010) DNA topoisomerases and their poisoning by anticancer and antibacterial drugs. Chem. Biol. 17, 421–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Stewart L., Redinbo M. R., Qiu X., Hol W. G., Champoux J. J. (1998) A model for the mechanism of human topoisomerase I. Science 279, 1534–1541 [DOI] [PubMed] [Google Scholar]
- 5. Koster D. A., Croquette V., Dekker C., Shuman S., Dekker N. H. (2005) Friction and torque govern the relaxation of DNA supercoils by eukaryotic topoisomerase IB. Nature 434, 671–674 [DOI] [PubMed] [Google Scholar]
- 6. Das B. B., Dexheimer T. S., Maddali K., Pommier Y. (2010) Role of tyrosyl-DNA phosphodiesterase (TDP1) in mitochondria. Proc. Natl. Acad. Sci. U.S.A. 107, 19790–19795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pommier Y. (2006) Topoisomerase I inhibitors: camptothecins and beyond. Nat. Rev. Cancer 6, 789–802 [DOI] [PubMed] [Google Scholar]
- 8. Pommier Y., Cushman M. (2009) The indenoisoquinoline noncamptothecin topoisomerase I inhibitors: update and perspectives. Mol. Cancer Ther. 8, 1008–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pommier Y., Marchand C. (2012) Interfacial inhibitors: targeting macromolecular complexes. Nat. Rev. Drug Discov. 11, 25–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tsao Y. P., Russo A., Nyamuswa G., Silber R., Liu L. F. (1993) Interaction between replication forks and topoisomerase I-DNA cleavable complexes: studies in a cell-free SV40 DNA replication system. Cancer Res. 53, 5908–5914 [PubMed] [Google Scholar]
- 11. Strumberg D., Pilon A. A., Smith M., Hickey R., Malkas L., Pommier Y. (2000) Conversion of topoisomerase I cleavage complexes on the leading strand of ribosomal DNA into 5′-phosphorylated DNA double-strand breaks by replication runoff. Mol. Cell. Biol. 20, 3977–3987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Furuta T., Takemura H., Liao Z. Y., Aune G. J., Redon C., Sedelnikova O. A., Pilch D. R., Rogakou E. P., Celeste A., Chen H. T., Nussenzweig A., Aladjem M. I., Bonner W. M., Pommier Y. (2003) Phosphorylation of histone H2AX and activation of Mre11, Rad50, and Nbs1 in response to replication-dependent DNA double-strand breaks induced by mammalian DNA topoisomerase I cleavage complexes. J. Biol. Chem. 278, 20303–20312 [DOI] [PubMed] [Google Scholar]
- 13. Koster D. A., Palle K., Bot E. S., Bjornsti M. A., Dekker N. H. (2007) Antitumour drugs impede DNA uncoiling by topoisomerase I. Nature 448, 213–217 [DOI] [PubMed] [Google Scholar]
- 14. Sordet O., Larochelle S., Nicolas E., Stevens E. V., Zhang C., Shokat K. M., Fisher R. P., Pommier Y. (2008) Hyperphosphorylation of RNA polymerase II in response to topoisomerase I cleavage complexes and its association with transcription- and BRCA1-dependent degradation of topoisomerase I. J. Mol. Biol. 381, 540–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tuduri S., Crabbé L., Conti C., Tourrière H., Holtgreve-Grez H., Jauch A., Pantesco V., De Vos J., Thomas A., Theillet C., Pommier Y., Tazi J., Coquelle A., Pasero P. (2009) Topoisomerase I suppresses genomic instability by preventing interference between replication and transcription. Nat. Cell Biol. 11, 1315–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sordet O., Nakamura A. J., Redon C. E., Pommier Y. (2010) DNA double-strand breaks and ATM activation by transcription-blocking DNA lesions. Cell Cycle 9, 274–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Interthal H., Champoux J. J. (2011) Effects of DNA and protein size on substrate cleavage by human tyrosyl-DNA phosphodiesterase 1. Biochem. J. 436, 559–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yang S. W., Burgin A. B., Jr., Huizenga B. N., Robertson C. A., Yao K. C., Nash H. A. (1996) A eukaryotic enzyme that can disjoin dead-end covalent complexes between DNA and type I topoisomerases. Proc. Natl. Acad. Sci. U.S.A. 93, 11534–11539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Interthal H., Chen H. J., Champoux J. J. (2005) Human Tdp1 cleaves a broad spectrum of substrates, including phosphoamide linkages. J. Biol. Chem. 280, 36518–36528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. El-Khamisy S. F., Saifi G. M., Weinfeld M., Johansson F., Helleday T., Lupski J. R., Caldecott K. W. (2005) Defective DNA single-strand break repair in spinocerebellar ataxia with axonal neuropathy-1. Nature 434, 108–113 [DOI] [PubMed] [Google Scholar]
- 21. Miao Z. H., Agama K., Sordet O., Povirk L., Kohn K. W., Pommier Y. (2006) Hereditary ataxia SCAN1 cells are defective for the repair of transcription-dependent topoisomerase I cleavage complexes. DNA Repair 5, 1489–1494 [DOI] [PubMed] [Google Scholar]
- 22. Pouliot J. J., Robertson C. A., Nash H. A. (2001) Pathways for repair of topoisomerase I covalent complexes in Saccharomyces cerevisiae. Genes Cells 6, 677–687 [DOI] [PubMed] [Google Scholar]
- 23. Debéthune L., Kohlhagen G., Grandas A., Pommier Y. (2002) Processing of nucleopeptides mimicking the topoisomerase I-DNA covalent complex by tyrosyl-DNA phosphodiesterase. Nucleic Acids Res. 30, 1198–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Eng W. K., Faucette L., Johnson R. K., Sternglanz R. (1988) Evidence that DNA topoisomerase I is necessary for the cytotoxic effects of camptothecin. Mol. Pharmacol. 34, 755–760 [PubMed] [Google Scholar]
- 25. Nitiss J., Wang J. C. (1988) DNA topoisomerase-targeting antitumor drugs can be studied in yeast. Proc. Natl. Acad. Sci. U.S.A. 85, 7501–7505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nakamura K., Kogame T., Oshiumi H., Shinohara A., Sumitomo Y., Agama K., Pommier Y., Tsutsui K. M., Tsutsui K., Hartsuiker E., Ogi T., Takeda S., Taniguchi Y. (2010) Collaborative action of Brca1 and CtIP in elimination of covalent modifications from double-strand breaks to facilitate subsequent break repair. PLoS Genet. 6, e1000828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sartori A. A., Lukas C., Coates J., Mistrik M., Fu S., Bartek J., Baer R., Lukas J., Jackson S. P. (2007) Human CtIP promotes DNA end resection. Nature 450, 509–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Regairaz M., Zhang Y. W., Fu H., Agama K. K., Tata N., Agrawal S., Aladjem M. I., Pommier Y. (2011) Mus81-mediated DNA cleavage resolves replication forks stalled by topoisomerase I-DNA complexes. J. Cell Biol. 195, 739–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Takashima H., Boerkoel C. F., John J., Saifi G. M., Salih M. A., Armstrong D., Mao Y., Quiocho F. A., Roa B. B., Nakagawa M., Stockton D. W., Lupski J. R. (2002) Mutation of TDP1, encoding a topoisomerase I-dependent DNA damage repair enzyme, in spinocerebellar ataxia with axonal neuropathy. Nat. Genet. 32, 267–272 [DOI] [PubMed] [Google Scholar]
- 30. Interthal H., Chen H. J., Kehl-Fie T. E., Zotzmann J., Leppard J. B., Champoux J. J. (2005) SCAN1 mutant Tdp1 accumulates the enzyme-DNA intermediate and causes camptothecin hypersensitivity. EMBO J. 24, 2224–2233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Katyal S., el-Khamisy S. F., Russell H. R., Li Y., Ju L., Caldecott K. W., McKinnon P. J. (2007) TDP1 facilitates chromosomal single-strand break repair in neurons and is neuroprotective in vivo. EMBO J. 26, 4720–4731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hirano R., Interthal H., Huang C., Nakamura T., Deguchi K., Choi K., Bhattacharjee M. B., Arimura K., Umehara F., Izumo S., Northrop J. L., Salih M. A., Inoue K., Armstrong D. L., Champoux J. J., Takashima H., Boerkoel C. F. (2007) Spinocerebellar ataxia with axonal neuropathy: consequence of a Tdp1 recessive neomorphic mutation? EMBO J. 26, 4732–4743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Das B. B., Antony S., Gupta S., Dexheimer T. S., Redon C. E., Garfield S., Shiloh Y., Pommier Y. (2009) Optimal function of the DNA repair enzyme TDP1 requires its phosphorylation by ATM and/or DNA-PK. EMBO J. 28, 3667–3680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hawkins A. J., Subler M. A., Akopiants K., Wiley J. L., Taylor S. M., Rice A. C., Windle J. J., Valerie K., Povirk L. F. (2009) In vitro complementation of Tdp1 deficiency indicates a stabilized enzyme-DNA adduct from tyrosyl but not glycolate lesions as a consequence of the SCAN1 mutation. DNA Repair 8, 654–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. El-Khamisy S. F., Katyal S., Patel P., Ju L., McKinnon P. J., Caldecott K. W. (2009) Synergistic decrease of DNA single-strand break repair rates in mouse neural cells lacking both Tdp1 and aprataxin. DNA Repair 8, 760–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hamilton N. K., Maizels N. (2010) MRE11 function in response to topoisomerase poisons is independent of its function in double-strand break repair in Saccharomyces cerevisiae. PLoS One 5, e15387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhou T., Lee J. W., Tatavarthi H., Lupski J. R., Valerie K., Povirk L. F. (2005) Deficiency in 3′-phosphoglycolate processing in human cells with a hereditary mutation in tyrosyl-DNA phosphodiesterase (TDP1). Nucleic Acids Res. 33, 289–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lebedeva N. A., Rechkunova N. I., Lavrik O. I. (2011) AP-site cleavage activity of tyrosyl-DNA phosphodiesterase 1. FEBS Lett. 585, 683–686 [DOI] [PubMed] [Google Scholar]
- 39. Inamdar K. V., Pouliot J. J., Zhou T., Lees-Miller S. P., Rasouli-Nia A., Povirk L. F. (2002) Conversion of phosphoglycolate to phosphate termini on 3′ overhangs of DNA double strand breaks by the human tyrosyl-DNA phosphodiesterase hTdp1. J. Biol. Chem. 277, 27162–27168 [DOI] [PubMed] [Google Scholar]
- 40. Henner W. D., Rodriguez L. O., Hecht S. M., Haseltine W. A. (1983) γ ray induced deoxyribonucleic acid strand breaks. 3′ glycolate termini. J. Biol. Chem. 258, 711–713 [PubMed] [Google Scholar]
- 41. Datta K., Purkayastha S., Neumann R. D., Pastwa E., Winters T. A. (2011) Base damage immediately upstream from double-strand break ends is a more severe impediment to nonhomologous end joining than blocked 3′-termini. Radiat. Res. 175, 97–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Povirk L. F., Zhou T., Zhou R., Cowan M. J., Yannone S. M. (2007) Processing of 3′-phosphoglycolate-terminated DNA double strand breaks by Artemis nuclease. J. Biol. Chem. 282, 3547–3558 [DOI] [PubMed] [Google Scholar]
- 43. Buerstedde J. M., Takeda S. (1991) Increased ratio of targeted to random integration after transfection of chicken B cell lines. Cell 67, 179–188 [DOI] [PubMed] [Google Scholar]
- 44. Yamazoe M., Sonoda E., Hochegger H., Takeda S. (2004) Reverse genetic studies of the DNA damage response in the chicken B lymphocyte line DT40. DNA Repair 3, 1175–1185 [DOI] [PubMed] [Google Scholar]
- 45. Dexheimer T. S., Antony S., Marchand C., Pommier Y. (2008) Tyrosyl-DNA phosphodiesterase as a target for anticancer therapy. Anticancer Agents Med. Chem. 8, 381–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nitiss J. L. (2009) Targeting DNA topoisomerase II in cancer chemotherapy. Nat. Rev. Cancer 9, 338–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nitiss K. C., Malik M., He X., White S. W., Nitiss J. L. (2006) Tyrosyl-DNA phosphodiesterase (Tdp1) participates in the repair of Top2-mediated DNA damage. Proc. Natl. Acad. Sci. U.S.A. 103, 8953–8958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Barthelmes H. U., Habermeyer M., Christensen M. O., Mielke C., Interthal H., Pouliot J. J., Boege F., Marko D. (2004) TDP1 overexpression in human cells counteracts DNA damage mediated by topoisomerases I and II. J. Biol. Chem. 279, 55618–55625 [DOI] [PubMed] [Google Scholar]
- 49. Dexheimer T. S., Stephen A. G., Fivash M. J., Fisher R. J., Pommier Y. (2010) The DNA binding and 3′-end preferential activity of human tyrosyl-DNA phosphodiesterase. Nucleic Acids Res. 38, 2444–2452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hartsuiker E., Neale M. J., Carr A. M. (2009) Distinct requirements for the Rad32(Mre11) nuclease and Ctp1(CtIP) in the removal of covalently bound topoisomerase I and II from DNA. Mol. Cell 33, 117–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Iiizumi S., Nomura Y., So S., Uegaki K., Aoki K., Shibahara K., Adachi N., Koyama H. (2006) Simple one-week method to construct gene-targeting vectors: application to production of human knockout cell lines. BioTechniques 41, 311–316 [DOI] [PubMed] [Google Scholar]
- 52. Antony S., Agama K. K., Miao Z. H., Takagi K., Wright M. H., Robles A. I., Varticovski L., Nagarajan M., Morrell A., Cushman M., Pommier Y. (2007) Novel indenoisoquinolines NSC 725776 and NSC 724998 produce persistent topoisomerase I cleavage complexes and overcome multidrug resistance. Cancer Res. 67, 10397–10405 [DOI] [PubMed] [Google Scholar]
- 53. Plo I., Liao Z. Y., Barceló J. M., Kohlhagen G., Caldecott K. W., Weinfeld M., Pommier Y. (2003) Association of XRCC1 and tyrosyl DNA phosphodiesterase (Tdp1) for the repair of topoisomerase I-mediated DNA lesions. DNA Repair 2, 1087–1100 [DOI] [PubMed] [Google Scholar]
- 54. Zhou T., Akopiants K., Mohapatra S., Lin P. S., Valerie K., Ramsden D. A., Lees-Miller S. P., Povirk L. F. (2009) Tyrosyl-DNA phosphodiesterase and the repair of 3′-phosphoglycolate-terminated DNA double-strand breaks. DNA Repair 8, 901–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Pegg A. E. (1984) Methylation of the O6 position of guanine in DNA is the most likely initiating event in carcinogenesis by methylating agents. Cancer Invest. 2, 223–231 [DOI] [PubMed] [Google Scholar]
- 56. Daroui P., Desai S. D., Li T. K., Liu A. A., Liu L. F. (2004) Hydrogen peroxide induces topoisomerase I-mediated DNA damage and cell death. J. Biol. Chem. 279, 14587–14594 [DOI] [PubMed] [Google Scholar]
- 57. Wiederhold L., Leppard J. B., Kedar P., Karimi-Busheri F., Rasouli-Nia A., Weinfeld M., Tomkinson A. E., Izumi T., Prasad R., Wilson S. H., Mitra S., Hazra T. K. (2004) AP endonuclease-independent DNA base excision repair in human cells. Mol. Cell 15, 209–220 [DOI] [PubMed] [Google Scholar]
- 58. Pouliot J. J., Yao K. C., Robertson C. A., Nash H. A. (1999) Yeast gene for a Tyr-DNA phosphodiesterase that repairs topoisomerase I complexes. Science 286, 552–555 [DOI] [PubMed] [Google Scholar]
- 59. Vance J. R., Wilson T. E. (2002) Yeast Tdp1 and Rad1-Rad10 function as redundant pathways for repairing Top1 replicative damage. Proc. Natl. Acad. Sci. U.S.A. 99, 13669–13674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Marchand C., Lea W. A., Jadhav A., Dexheimer T. S., Austin C. P., Inglese J., Pommier Y., Simeonov A. (2009) Identification of phosphotyrosine mimetic inhibitors of human tyrosyl-DNA phosphodiesterase I by a novel AlphaScreen high-throughput assay. Mol. Cancer Ther. 8, 240–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Pourquier P., Pommier Y. (2001) Topoisomerase I-mediated DNA damage. Adv. Cancer Res. 80, 189–216 [DOI] [PubMed] [Google Scholar]
- 62. Zeng Z., Cortés-Ledesma F., El Khamisy S. F., Caldecott K. W. (2011) TDP2/TTRAP is the major 5′-tyrosyl DNA phosphodiesterase activity in vertebrate cells and is critical for cellular resistance to topoisomerase II-induced DNA damage. J. Biol. Chem. 286, 403–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Cortes Ledesma F., El Khamisy S. F., Zuma M. C., Osborn K., Caldecott K. W. (2009) A human 5′-tyrosyl DNA phosphodiesterase that repairs topoisomerase-mediated DNA damage. Nature 461, 674–678 [DOI] [PubMed] [Google Scholar]
- 64. Pourquier P., Ueng L. M., Kohlhagen G., Mazumder A., Gupta M., Kohn K. W., Pommier Y. (1997) Effects of uracil incorporation, DNA mismatches, and abasic sites on cleavage and religation activities of mammalian topoisomerase I. J. Biol. Chem. 272, 7792–7796 [DOI] [PubMed] [Google Scholar]
- 65. Pourquier P., Pilon A. A., Kohlhagen G., Mazumder A., Sharma A., Pommier Y. (1997) Trapping of mammalian topoisomerase I and recombinations induced by damaged DNA containing nicks or gaps. Importance of DNA end phosphorylation and camptothecin effects. J. Biol. Chem. 272, 26441–26447 [DOI] [PubMed] [Google Scholar]
- 66. Pommier Y., Barcelo J. M., Rao V. A., Sordet O., Jobson A. G., Thibaut L., Miao Z. H., Seiler J. A., Zhang H., Marchand C., Agama K., Nitiss J. L., Redon C. (2006) Repair of topoisomerase I-mediated DNA damage. Prog. Nucleic Acid Res. Mol. Biol. 81, 179–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Deng C., Brown J. A., You D., Brown J. M. (2005) Multiple endonucleases function to repair covalent topoisomerase I complexes in Saccharomyces cerevisiae. Genetics 170, 591–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.