Background: Structural determinants of aspartyl aminopeptidase (DNPEP) enzymatic activity have not yet been elucidated.
Results: DNPEP contains a binuclear metal active site and forms a tetrahedral homo-dodecamer in solution.
Conclusion: Manganese binding at the active site stimulates DNPEP activity. The tetrahedral assembly restricts peptide access to the active site.
Significance: This study provides a structural and biochemical basis for understanding DNPEP physiology.
Keywords: Aminopeptidase, Angiogenesis, Retina, Retinal Degeneration, Vision
Abstract
Aminopeptidases are key enzymes involved in the regulation of signaling peptide activity. Here, we present a detailed biochemical and structural analysis of an evolutionary highly conserved aspartyl aminopeptidase called DNPEP. We show that this peptidase can cleave multiple physiologically relevant substrates, including angiotensins, and thus may play a key role in regulating neuron function. Using a combination of x-ray crystallography, x-ray absorption spectroscopy, and single particle electron microscopy analysis, we provide the first detailed structural analysis of DNPEP. We show that this enzyme possesses a binuclear zinc-active site in which one of the zinc ions is readily exchangeable with other divalent cations such as manganese, which strongly stimulates the enzymatic activity of the protein. The plasticity of this metal-binding site suggests a mechanism for regulation of DNPEP activity. We also demonstrate that DNPEP assembles into a functionally relevant tetrahedral complex that restricts access of peptide substrates to the active site. These structural data allow rationalization of the enzyme's preference for short peptide substrates with N-terminal acidic residues. This study provides a structural basis for understanding the physiology and bioinorganic chemistry of DNPEP and other M18 family aminopeptidases.
Introduction
Aminopeptidases catalyze the cleavage of the N-terminal residues from peptide or protein substrates (1). These enzymes are found in all forms of life and have important roles in a variety of biological processes involving peptide signaling such as angiogenesis, memory, tumor growth and metastasis, reproduction, and maintenance of blood pressure (2). With respect to the latter process, aminopeptidases are recognized as key players in regulating the activity of the renin-angiotensin system (RAS)3 (3).
The RAS is best known for its role in regulating blood pressure and maintaining fluid and electrolyte homeostasis (4). In addition, a number of organs in the body contain local RASs that act within the organ or tissue to modulate its function, the eye constituting one such example (5–13). The ocular RAS is involved in a number of physiological and pathological processes, including regulation of intraocular pressure (14), inflammation (15, 16), angiogenesis (17), and neuron function (18, 19), and it has been identified as a potential therapeutic target for glaucoma and retinopathies (20–22).
Production and degradation of the major bioactive angiotensin peptide, angiotensin II (Ang II), involves the action of several enzymes, including renin, angiotensin-converting enzyme, and aminopeptidases (23, 24). Renin and angiotensin-converting enzyme inhibitors as well as Ang II receptor antagonists are widely used for treatment of cardiovascular and renal diseases (25, 26). Glutamyl aminopeptidase (EC 3.4.11.7, also known as aminopeptidase A or ENPEP) is one enzyme involved in the processing of Ang II to Ang III (3, 27). Ang III is thought to play a key role in angiotensin signaling in the brain and other neuronal tissues (28). Another acidic aminopeptidase implicated in the metabolism of angiotensin peptides is a soluble enzyme termed aspartyl aminopeptidase or DNPEP (EC 3.4.11.21). A recent transcriptome study in our laboratory identified Dnpep as a highly expressed gene in the whole eye as well as the neural retina (supplemental Table S1) (29). Moreover, Dnpep expression is more than 20-fold higher relative to Enpep suggesting a key role for DNPEP in the ocular RAS.
DNPEP is an ∼55-kDa metalloenzyme that requires zinc for its enzymatic function and exists as a high molecular weight complex (30–32). The enzyme activity is enhanced by Mn(II) and bacitracin and inhibited by zinc and metal chelators (30–32). The DNPEP gene was identified by Wilk and co-workers in 1998 (32). DNPEP belongs to the M18 aminopeptidase family that is contained within the MH clan as designated in the MEROPS data base (33). Yeast aminopeptidase I, a dodecameric zinc-dependent enzyme, is the founding member of the M18 family (34). M18 peptidases are found in all kingdoms of life indicating an ancient evolutionary history. Although substrate specificity is not absolutely conserved (34), the well characterized M18 aminopeptidases Yhr113wp/Ape4 and PfM18AAP from Saccharomyces cerevisiae (35) and Plasmodium falciparum (36), respectively, also prefer N-terminal aspartyl-containing substrates. Interestingly, most vertebrate genomes encode only a single M18 family peptidase pointing to an important, highly conserved, and nonredundant physiological function for DNPEP. Although a role for DNPEP in angiotensin peptide metabolism has been proposed, its ability to metabolize other signaling peptides has not yet been tested.
Until now, only low resolution techniques have been applied to the study of DNPEP structure. His residues that are critical for DNPEP enzymatic activity have been identified, but a detailed structural analysis of the active site and metal cofactors has not yet been reported (37). DNPEP behaves as an octamer according to gel filtration analysis (31, 32). However, DNPEP is related to the M42 family archaeal aminopeptidases, which form dodecameric assemblies (38, 39). Additionally, yeast aminopeptidase I has been characterized as a dodecamer with pseudo-D3 symmetry (34, 40). Thus, DNPEP quaternary structure and stoichiometry remain unclear.
Here, we show that DNPEP can metabolize several biologically relevant signaling peptides. We demonstrate that “as-isolated” DNPEP contains a binuclear zinc center in which one site is labile and can be replaced by other metal ions. Furthermore, we elucidate the structural basis of manganese catalytic activation by showing that Mn(II) substitutes for zinc at the labile site. We also show that DNPEP assembles into a functionally relevant dodecameric tetrahedral complex that contributes to DNPEP substrate selectively.
EXPERIMENTAL PROCEDURES
Chemicals
Aspartyl-p-naphthylamide, human angiotensin I (Ang I), Ang II, cholecystokinin octapeptide (CCK-8), melanin-concentrating hormone (MCH), and neurokinin B (NKB) were purchased from Bachem (Torrance, CA). Isopropyl β-d-thiogalactopyranoside and dithiothreitol (DTT) were obtained from Anatrace (Maumee, OH). All other chemicals were purchased from Sigma.
DNPEP Activity Assay
An assay mixture containing 100 μm aspartyl-p-naphthylamide was prepared in 450 μl of 50 mm Tris-HCl, pH 7.5. Ten μl of protein sample was added to the pre-warmed assay mixture with or without the various test compounds and incubated at 37 °C with shaking at 220 rpm for 30 min. The reaction was stopped by adding an equal volume of methanol. The mixture was placed in a quartz cuvette, and fluorescence emission at 412 nm with excitation at 345 nm was recorded. The enzyme activities in different conditions were compared by fluorescence intensities with that of sample alone at pH 7.5 as a control. Each condition was performed in triplicate, and the results are presented as means ± S.D.
Molecular Biology and Protein Expression
A plasmid containing the bovine dnpep open reading frame (GenBankTM accession number BC105402) was obtained from ImaGenes (clone IRCJp5010F0111D). The DNPEP coding sequence was amplified by PCR using the following primers: 5′-ccatgggcagggcccgcaaagaggcg-3′ (forward) and 5′-aagctttcagtctaccaggaggctacggc-3′ (reverse). The resulting product encodes all but the first two residues of full-length bovine DNPEP flanked by 5′ NcoI and 3′ HindIII restriction enzymes sites. The product was cloned into a modified pET30a vector that encodes His6 tag followed by a tobacco etch virus protease recognition sequence in front of NcoI- and HindIII-cloning sites resulting in an in-frame fusion between the His6/tobacco etch virus protease site tag and the DNPEP coding sequences. The open reading frame was fully sequenced to ensure the construct was correct. The plasmid was transformed into the T7 express BL21 Escherichia coli strain (New England Biolabs, Ipswich, MA), and transformants were cultured in a shaker incubator at 37 °C. Protein expression was induced when the culture reached an A600 nm of 0.6 by addition of isopropyl β-d-thiogalactopyranoside to a final concentration of 4 μm. After overnight incubation, the cells were collected by centrifugation and either flash-frozen and stored at −80 °C or used immediately. For production of manganese-activated DNPEP, 1 g of MnCl2 was added to each liter of the bacterial culture 30 min prior to induction (41).
Protein Purification
Purification of recombinant DNPEP was performed at 4 °C. The cell pellet was resuspended in 60 ml of cold binding buffer (10 mm Tris-HCl, pH 8.0, containing 300 mm NaCl, 10 mm imidazole and 10% glycerol) and lysed by three passes through a French pressure cell. The lysate was centrifuged at 195,000 × g for 30 min, and the supernatant was loaded onto a 2-ml nickel-nitrilotriacetic acid Superflow column (Qiagen, Valencia, CA). The resin was washed with 50 ml of binding buffer followed by 50 ml of wash buffer (10 mm Tris-HCl, pH 8.0, containing 300 mm NaCl, 20 mm imidazole, and 10% glycerol). Bound protein was eluted with 20 ml of elution buffer (10 mm Tris-HCl, pH 8.0, containing 300 mm NaCl, 250 mm imidazole, and 10% glycerol). DNPEP-containing fractions were combined and loaded onto a Superdex 200 gel filtration column (GE Healthcare) equilibrated with 10 mm Tris-HCl, pH 8.0, containing 300 mm NaCl, and 10% glycerol. Eluted fractions with high activity and purity were combined, concentrated, and stored at −80 °C or used immediately for experiments.
LC-MS Activity Assay
Human Ang I, Ang II, CCK-8, MCH, and NKB were dissolved in 50 mm Tris-HCl, pH 7.5. Each substrate was prepared at five concentrations as follows: 0.05, 0.1, 0.25, 0.5, and 1 mm. 0.6 μg of DNPEP was added into 100 μl of preincubated substrate solution, and the reaction mixture was incubated at 37 °C with 550 rpm shaking. Every 5–10 min after reaction initiation, 5 μl of the mixture was added into 95 μl of 40% acetic acid to stop the reaction and reduce the peptide concentration. The peptides were analyzed by an 1100 series Agilent HPLC (Santa Clara, CA) equipped with a diode-array detector and an XBridge BEH300 C4 (3.5 μm, 2.1 × 50 mm) column (Waters, Milford, MA) equilibrated with 50% acetonitrile in water. Peptides were eluted from the column at a flow rate of 200 μl/min and directed into an LXQ linear ion trap mass spectrometer (Thermo Scientific, Waltham, MA) through an electrospray ionization source working in the positive mode. Substrate and product eluted together within 2 min. DNPEP-catalyzed hydrolysis gave rise only to des-Asp1 products. Product formation was quantified as the area ratio areaproduct/(areasubstrate + areaproduct) from substrate and product chromatograms. Product concentration was calculated as [S]initial × areaproduct/(areasubstrate + areaproduct). Vmax and Km values were obtained according to Michaelis-Menten steady-state kinetics using Sigmoidal-Hill fitting in Origin 8.1 software (OriginLab).
DNPEP Crystallization
Crystals of bovine DNPEP were obtained by the hanging drop vapor diffusion method by mixing 1 μl of purified DNPEP at 5 mg/ml with 1 μl of crystallization solution consisting of either 30% PEG 400, 100 mm Tris-HCl, pH 8.5, and 0.2 m MgCl2 (condition A), 20% 1,4-butanediol, 100 mm MES-NaOH, pH 6, and 0.2 m Li2SO4 (condition B), or 35% 2-methyl-2,4-pentanediol, 100 mm Tris-HCl, pH 7, and 0.2 m NaCl (condition C) and incubating the resulting drops at 8 °C. Cube-shaped crystals typically appeared within 1 week and grew to full size within 2 months. The largest crystals were ∼75 μm3. Mature crystals were harvested without additional cryoprotection and flash-cooled in liquid nitrogen prior to x-ray exposure.
Crystallography
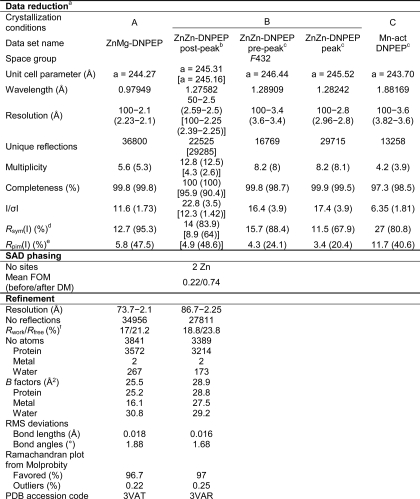
Diffraction data were collected at the Northeastern Collaborative Access Team (NECAT) 24-ID-C beamline at the Advanced Photon Source. Data sets were collected on crystals grown in condition B at the zinc K-peak and at 50 eV above and below the peak to allow anomalous scattering-based structure solution as well as to evaluate active site zinc occupancy. Datasets were also collected on a crystal grown in condition A as well as a crystal grown from manganese-activated DNPEP in condition C, the latter using an x-ray energy about 50 eV above the manganese K-edge. Data reduction was performed using either HKL2000 (42) or XDS (43) and Truncate from the CCP4 suite (44). The crystals all belong to space group F432 with one monomer in the asymmetric unit. Initial phases were obtained by zinc single wavelength anomalous diffraction (SAD) using the ZnZn-DNPEP post-peak data set. The anomalous substructure, consisting of two zinc atoms, was located using SHELXD (45) and refined using Phaser (46). The occupancy of the two zinc atoms refined to values near 1. The SAD phases were improved by density modification resulting in an overall figure of merit of 0.737 (44). The density-modified electron density maps were of sufficient quality to allow automatic building of ∼74% of the protein chain using ARP/wARP (47). This initial model was then refined against the dataset collected on the crystal grown in condition A (referred to as ZnMg-DNPEP) using REFMAC (48), and the model was completed manually using Coot (49). The geometric quality of the model was periodically evaluated using the MolProbity server (50). Iterative manual rebuilding and refinement produced a model with good stereochemistry, and Rwork and Rfree values of 17 and 21.2%, respectively. The Rfree-based overall estimated coordinate uncertainty is 0.15 Å as calculated in REFMAC. Residues 1–27, which consist mostly of the His tag and protease cleavage site, and residues 224–234 were not modeled because of weak or absent electron density. Additionally, the side chain of Glu33 was omitted as it was consistently found in strongly negative Fo − Fc electron density peaks. Six residues were modeled in split conformations. Two metal ions and 267 waters were included in the model.
The refined model was stripped of heteroatoms and refined against the ZnZn-DNPEP post-peak dataset, which was reprocessed to maximize the resolution. Notably, this model has more regions that could not be modeled because of weak electron density compared with the ZnMg-DNPEP model. Bijvoet-difference Fourier maps clearly showed that both metal-binding sites were occupied by zinc. Iterative manual rebuilding and refinement led to a model with Rwork and Rfree values of 18.8 and 23.8%, respectively, and good stereochemistry. Residues 125–128, 164–171, 193–203, 223–238, 378–384, and 427–435 were omitted from the model because of weak or absent electron density. Four residues were modeled in split conformations, and 173 waters were included in the model. Data collection, phasing, and structure refinement statistics are shown in Table 1. All figures of the crystallographic model were generated using PyMOL (Schrodinger).
TABLE 1.
Crystallographic data
a Values in parentheses are for the highest resolution shell of data.
b Values on top are for the data processed for use in SAD phasing, and values in square brackets are for the reprocessed data set used for refinement.
c Statistics were calculated with Bijvoet pairs unmerged.
d Rsym(I) = Σhkl Σi|Ii(hkl)−〈I(hkl)〉|/Σhkl|Σi Ii(hkl).
e Rpim(I) = Σhkl(1/(N − 1))1/2 Σi| Ii(hkl) − 〈I(hkl)〉|/Σhkl ΣiIi(hkl) as defined in Ref. 78 is the precision-indicating merging R factor.
f Rwork = Σhkl ‖Fobs| − |Fcalc‖/Σhkl | Fobs| Rfree is calculated in the same manner using ∼5% of reflections excluded from refinement.
Electron Microscopy
A 3-μl drop of freshly purified DNPEP at a concentration of 40 μg/ml was adsorbed to a glow-charged and carbon-coated electron microscopy (EM) grid for 1 min. The grid was then washed twice and stained with 1% uranyl acetate. Micrographs were obtained using a 200-keV FEI Tecnai F20 XT field emission transmission electron microscope at a magnification of ×67,834. In total, 105 micrographs were obtained by using a Primescan D 7100 drum scanner (Heidelberger Druckmaschinen AG) at a pixel size corresponding to 2.2 Å.
Single particle reconstruction of DNPEP was performed using the EMAN2 software package (51). About 5000 particles were boxed manually using e2boxer followed by contrast transfer function corrections. Class averages were calculated using refine2d with tetrahedral symmetry imposed. The initial three-dimensional model was built up from the 31 class averages using e2initialmodel. A refined map was obtained after six iterations with an angle step of 2.5°. The map resolution was determined according to the 0.5 criterion using a Fourier shell correlation plot generated from an even/odd test. Figures of the map were generated using Chimera (52). The crystallographic model of the DNPEP tetrahedral complex was docked into the EM map by rigid body fitting in Chimera.
X-ray Absorption Spectroscopy
X-ray absorption spectroscopy (XAS) data were collected on beamline X3B of the National Synchrotron Light Source at Brookhaven National Laboratory. Sample temperatures were maintained at 20–25 K during data collection. Samples were loaded into Kapton-wrapped Lucite cuvettes with a volume of ∼30 μl and immediately frozen in liquid nitrogen. At the zinc K-edge, datasets were collected over an energy range of 9.47–10.36 keV for both ZnZn-DNPEP (0.25 mm protein, 13 cumulative scans) and manganese-activated DNPEP (1.08 mm protein, 8 cumulative scans). At the manganese K-edge, a dataset was collected for an energy range of 6.36–7.24 keV on a freshly prepared, unexposed sample of manganese-activated DNPEP (1.08 mm protein, 13 cumulative scans). Spectra were measured in 5-eV steps in the pre-edge (1-s count time), 0.3-eV steps in the edge region (2-s count time), and 0.05k steps above the edge (count time increasing from 3 to 11 s over the scan), such that each scan required ∼45 min. XAS spectra were obtained as fluorescence excitation spectra using a 31-element solid state germanium fluorescence detector array (Canberra, Meriden, CT). Copper (for zinc K-edge) or chromium (for manganese K-edge) filters were placed between the sample and detector to keep incident count rates on individual detector elements below 100 kHz. As appropriate, zinc or manganese metal foil spectra were recorded concomitantly for energy calibration, with the first inflection point of the reference spectrum assigned an energy of 9569 eV for zinc or 6539 eV for manganese. Datasets for each sample were inspected and merged using the EXAFSPAK package (53). Following calibration and averaging, the background absorption was removed by fitting a Gaussian function to the pre-edge region and subtracting this function from the entire spectrum. A three-segment spline with fourth order components was then fit to the extended x-ray absorption fine structure (EXAFS) region of the spectrum to extract χ(k). Normalized x-ray absorption near edge spectra (XANES) were obtained using the AUTOBK algorithm implemented in Athena (54). Theoretical phase and amplitude parameters were generated using FEFF 8.40 (55) for simplified models of the binuclear DNPEP-active site containing both monodentate and bidentate carboxylates, histidine ligands, and both terminal and bridging solvent ligands. The geometry of these input models was modified as needed to better reflect the EXAFS fitting results. In all analyses, the coordination number of a given shell was a fixed parameter and was varied iteratively, whereas bond lengths (r) and Debye-Waller factors (σ2) were allowed to freely float. The amplitude reduction factor S0 was fixed at 0.9. The edge shift parameter ΔE0 was allowed to float as a single value for all shells in fits of a given complex. Therefore, in any given fit, the number of floating parameters was typically equal to (2× number of shells) + 1. The goodness of fit F was defined as (Σk6(χexptl − χcalc)2/Σk6χexptl2)1/2. For fits to Fourier-filtered data, the goodness-of-fit metric F′ was employed. F′ = (Σ(χexptl − χcalc)2)2/(NIDP − NVAR), where NVAR is the number of floated variables in the fit, and NIDP is the number of independent data points and is defined as NIDP = 2ΔkΔr/π. In the latter equation, Δk is the k-range over which the data are fit, and Δr is the back-transformation range employed in fitting Fourier-filtered data. F′ provides a useful assessment of the effect of additional shells on improving fit quality (56). The bond valence sum (BVS) for the first coordination sphere was also used to assess the appropriateness of a given fit. The BVS was calculated using Σ(exp((r0 − r)/0.37)), where r0 is an empirically derived parameter for a given pair of atoms and r is the actual bond length (57, 58). Although EXAFS cannot distinguish between atoms differing by Z ± 1, we assumed an oxygen-rich environment based upon the crystal structure data.
RESULTS
Purification and Characterization of Recombinant Bovine DNPEP
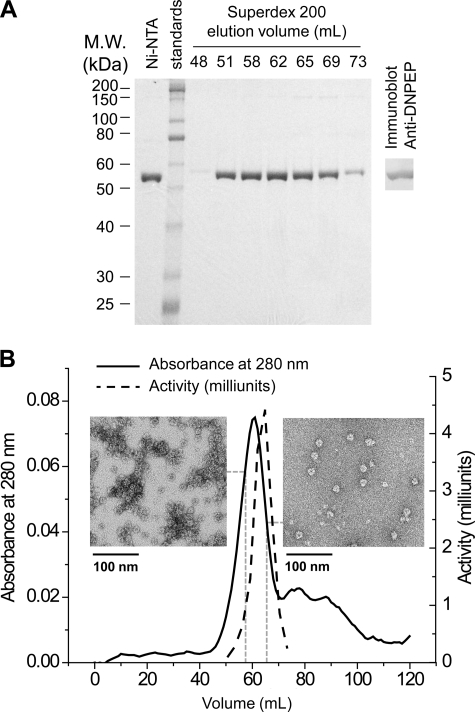
Bovine DNPEP was produced recombinantly as an N-terminal His6 fusion protein in E. coli. A two-step purification consisting of sequential metal affinity and gel filtration chromatography led to 51-fold purified DNPEP with ∼13.8% yield, equal to 5 mg of purified protein/liter of bacterial culture (Fig. 1A and supplemental Table S2). Initial gel filtration chromatograms consisted of two peaks, one minor peak corresponding to the void volume and a second dominant peak with a longer retention time that contained all of the enzymatic activity (supplemental Fig. S1A). Optimization of the purification protocol greatly reduced the size of the void peak (Fig. 1B and supplemental Fig. S1A). SDS-PAGE analysis of the purified protein showed a single 54-kDa band corresponding to the mass of the fusion protein, and the identity of the band was confirmed by immunoblotting (Fig. 1A). The elution volume of the major peak corresponded to an apparent molecular mass of 395 kDa thus giving a stoichiometry of 7–8 subunits per complex (supplemental Fig. S1B). The peak of the enzyme activity curve was shifted slightly rightward relative to the protein absorbance maximum, which we attribute to aggregation that occurred during the time before the fractions could be assayed (Fig. 1B). Indeed, micrographs of negatively stained DNPEP particles from two different fractions collected immediately after elution from the gel filtration column clearly showed more aggregation in the fraction eluted earlier compared with the one eluted later (Fig. 1B).
FIGURE 1.
Purification of recombinant DNPEP. A, SDS-PAGE (10% gel) analysis of the eluted fractions from sequential metal affinity and gel filtration chromatography steps of the purification. Proteins were visualized by Coomassie staining, and the identity of the ∼55-kDa protein as DNPEP was confirmed by immunoblotting (right). B, gel filtration chromatogram overlaid with the relative activity curve. The slight shift of the activity peak is likely due to the aggregation of DNPEP during and after elution, as shown in the inset micrographs of negatively stained particles from the fractions eluted at 58 ml (left panel) and 65 ml (right panel).
Effects of Metals and Metal Chelators on DNPEP Activity
Previous studies indicate that DNPEP uses zinc as a cofactor (31). DNPEP activity has been reported to be supported or even stimulated by manganese in some studies (30, 31) but not others (32, 37). Additionally, an inhibitory effect of EDTA on enzyme activity has been inconsistently observed. To further characterize the properties of DNPEP as a metalloenzyme, various divalent metals and reported inhibitors and activators were individually added to the activity assay mixture as shown in Table 2. We found that addition of Mn(II) to a final concentration of 4 mm increased the recombinant enzyme activity 4-fold. A dose-effect experiment revealed that the enzyme is half-maximally stimulated at a manganese concentration of ∼6 μm with the effect plateauing around 100 μm (supplemental Fig. S2). Notably, the intracellular manganese concentration in mammalian cells is in the low to middle micromolar range indicating that this activation phenomenon could be relevant in vivo (59). Consistent with these results, recombinant DNPEP purified from a culture supplemented with 5 mm MnCl2 displayed 30-fold higher catalytic efficiency toward aspartyl-p-naphthylamide compared with unsupplemented enzyme. The activity simulation was primarily the result of a 14-fold elevation in kcat, although a 2-fold lower Km value was also observed for the manganese-activated enzyme (supplemental Table S3). Co(II) also increased DNPEP activity, albeit to a lesser degree than Mn(II), whereas 0.4 mm Zn(II) almost completely abolished its activity. Excess zinc has been noted to be generally inhibitory for a number of peptidases (60). In one case, the inhibitory effect was attributed to disruption of the peptidase quaternary structure (61). Unlike glutamyl aminopeptidase (62), Ca(II) does not affect DNPEP activity. Mg(II) also did not affect DNPEP activity. Metal chelators such as EDTA and 1,10-phenanthroline reduced but did not abolish DNPEP activity. Imidazole and DTT were both inhibitory probably because of their ability to bind zinc. Although bacitracin A was previously reported to stimulate DNPEP enzymatic activity (32), we observed only 20% activation when 0.02 mm of bacitracin A was added to the assay (Table 2).
TABLE 2.
Effects of divalent metals and potential activators and inhibitors on the activity of bovine DNPEP
| Compound | Concentrationa | Activityb (percentage of control) |
|---|---|---|
| mm | ||
| Zn2+ | 0.4 | 0.9 (±0.1) |
| Mn2+ | 4 | 433 (±105) |
| Co2+ | 1 | 267 (±19) |
| 4 | 145 (±7) | |
| Ca2+ | 10 | 67 (±5) |
| Mg2+ | 4 | 96 (±18) |
| Imidazole | 20 | 60 (±2) |
| DTT | 1 | 66 (±13) |
| 0.1 | 73 (±13) | |
| Bacitracin A | 0.2 | 97 (±20) |
| 0.02 | 127 (±37) | |
| EDTA | 10 | 43 (±9) |
| 5 | 72 (±16) | |
| 1,10-Phenanthroline | 2 | 47 (±9) |
a Final concentration of each compound in the reaction solution. Divalent cations were added as chloride salts.
b Activity is shown as a percentage of the mean rate of three replicates over the control condition in which only DNPEP was added into the reaction solution, and the error in parentheses is the standard deviation of the three replicates in percentage to control. The fluorescence activity assay is described under “Experimental Procedures.”
DNPEP Activity toward Candidate Signaling Peptide Substrates
We next evaluated the ability of DNPEP to hydrolyze five candidate signaling peptide substrates that are known to be present in the eye along with their cognate receptors (supplemental Tables S1 and S4). Ang I and II are known substrates for DNPEP, whereas NKB and CCK-8 are tachykinins with N-terminal aspartyl residues and similar peptide length as Ang I and II. MCH, a 19-residue, N-aspartyl peptide with an internal disulfide bond, was tested to determine whether larger peptides with more complex structure can be cleaved by the enzyme. The hydrolysis of each potential substrate was measured by LC-MS, and the data were fit to a Michaelis-Menten kinetic model (Table 3 and supplemental Fig. S3). Among the peptides tested, Ang I and II were the most favored substrates as evidenced by their high catalytic efficiency values (Table 3). Km values were similar for the two peptides, although DNPEP catalytic efficiency was higher for Ang II because of a 3-fold higher kcat value. CCK-8 was a moderately good substrate, whereas MCH and NKB have low catalytic efficiency values. It is noteworthy that the precursor of CCK-8, cholecystokinin, is more highly expressed compared with other neural peptides (supplemental Table S4) indicating that CCK-8 could also be a physiological substrate of DNPEP. The poor activity toward MCH might be due to its larger size and disulfide-bonded nature, whereas the poor activity toward NKB was surprising given that it is a short peptide with the same number of residues as Ang II. These data confirm that angiotensins are favored substrates of this enzyme but also demonstrate that other biologically relevant peptides may also be regulated by DNPEP.
TABLE 3.
Aspartic aminopeptidase activity of DNPEP toward physiologically relevant peptides
DNPEP aspartic aminopeptidase activity was measured by LC-MS. Both product measurements and kinetic data fitting are described under “Experimental Procedures.” Errors in parentheses were obtained from Hill fitting by Origin 8.1.
| Peptide | Length (amino acids) | Relative mass | Peptide sequence (N → C) | Km | kcat | kcat/Km (×103m−1s−1) |
|---|---|---|---|---|---|---|
| Da | mm | s−1 | ||||
| Ang I | 10 | 1296.5 | Asp-Arg-Val-Tyr-Ile-His-Pro-Phe-His-Leu | 0.07 (±0.02) | 2.3 (±0.1) | 30 (±8) |
| Ang II | 8 | 1046.2 | Asp-Arg-Val-Tyr-Ile-His-Pro-Phe | 0.10 (±0.02) | 7.7 (±0.4) | 76 (±16) |
| CCK-8 | 8 | 1063.2 | Asp-Tyr-Met-Gly-Trp-Met-Asp-Phe | 0.65 (±0.19) | 5.3 (±0.8) | 8.1 (±2.6) |
| Melanin-concentrating hormone (MCH) | 19 | 2386.9 | Asp-Phe-Asp-Met-Leu-Arg-Cys-Met-Leu-Gly-Arg-Val-Tyr-Arg-Pro-Cys-Trp-Gln-Val (disulfide-bonded) | 1.4 (±0.5) | 1.4 (±0.3) | 0.99 (±0.43) |
| NKB | 10 | 1210.4 | Asp-Met-His-Asp-Phe-Phe-Val-Gly-Leu-Met | 0.47 (±0.35) | 0.18 (±0.06) | 0.38 (±0.32) |
Crystallographic Structure of DNPEP
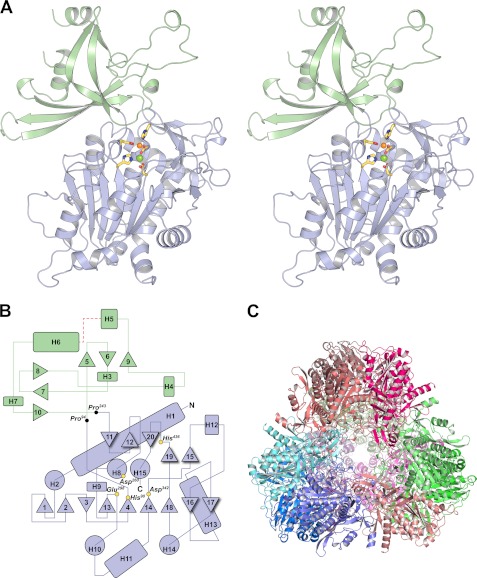
The DNPEP structure is composed of two domains, a catalytic domain that houses a binuclear metal active site and a dimerization domain that, along with portions of the catalytic domain, is involved in oligomer formation (Fig. 2, A and B). The general shape of the monomer can be likened to a flat lobed leaf or a piece of jigsaw puzzle. The prominent lobes and intervening grooves form a complex topography with self-complementary surfaces that promote homo-oligomerization. A single DNPEP monomer resides in the asymmetric unit of the crystal, but application of a subset of the crystallographic symmetry operations generates a DNPEP dodecamer with tetrahedral symmetry (Fig. 2C and supplemental Movie S1).
FIGURE 2.
Structure and topology of DNPEP. A, crystallographic structure of the DNPEP monomer. The DNPEP monomer constituting the asymmetric unit of the crystal is shown as a divergent stereo pair. The protein consists of two domains, a catalytic domain (light blue) that houses the binuclear metal active site and is common to all members of the MH clan, and a dimerization domain (pale green) that is important for oligomer formation and may also contribute to substrate selectivity and catalytic function in the context of the tetrahedral oligomeric structure. The active site metals are shown as orange (zinc) and green (magnesium) spheres together with the direct protein ligands as yellow sticks. All figures of the crystallographic model were generated using PyMOL. B, topology plot of DNPEP. Helical regions are represented by circles (parallel to viewing direction) or rounded rectangles (perpendicular to viewing direction), and β strands are indicated by triangles. The positions of the direct metal ligands are marked by yellow circles. N and C termini are indicated by black letters. A 10-residue disordered segment is indicated by a red dashed line. Residue numbering corresponds to the native amino acid sequence. This figure was prepared using Adobe Illustrator. C, DNPEP tetrahedral assembly generated by application of crystallographic symmetry operators. Each monomer is shown as a different color.
The catalytic domain, consisting of residues 1–94 and 243–471, contains a mixed parallel/antiparallel β sheet at its core with α helices packed against both faces in an αβα sandwich fold (Fig. 2, A and B). (Note that the residue numbering throughout refers to the native sequence, and to obtain the corresponding residue number for the crystallographic structure, add 25.) Two short antiparallel β sheets that sit on the top of the posterior α layer complete the description of the domain fold. All of the binuclear metal protein ligands are found within this domain. Interestingly, the Asp260–Asn261 peptide bond within helix 8 is found in a cis conformation with an ω angle of 5.8°. Non-Pro-containing cis peptide bonds are exceedingly rare in protein structures but when present are usually of high importance for the function of the protein (63). Indeed, Asp260 is a bridging ligand for the binuclear metal center, and it is likely that the higher energy conformation of the residue is somehow coupled to the metal coordination. Examination of other peptidases in the MH clan reveals that a cis peptide bond at the bridging ligand position is a feature common to this group of proteins (38, 64–66). In the case of DNPEP, it is clear that the cis conformation is critical for the proper orientation of the Asp260 side chain for metal coordination. Hydrogen bonding interactions between the Asp260 backbone carbonyl oxygen and the Ser118 hydroxyl as well as a water molecule probably help stabilize the cis conformation.
Residues 95–242 compose the dimerization domain and fold into a pair of orthogonal, three-stranded, anti-parallel β sheets with five intervening α helices. Close inspection revealed that these sheets form a β barrel-like structure on one end but are separated from each other on the other end by helix 6. The first strand in this domain, strand 5, possesses a somewhat unusual S4 β bulge motif with two protruding Arg residues that may be functionally important (67). Notably, these Arg residues along with other basic residues in this highly positively charged strand are positioned near the substrate entrance pore of the tetrahedron and through ion-ion interactions form contacts between subunits. A number of contacts exist between the two domains that confer rigidity to the overall structure.
Active Site and Identity of the Metal Cofactors
The active site binuclear metal center is located in a pocket on the top surface of the catalytic domain (Fig. 2A). All direct protein ligands of the metals are located within this domain (Fig. 2B). Based on a previously established convention for MH clan peptidases (66), we refer to the metal-binding site located furthest away from the core β sheet of the catalytic domain as site A and the other as site B. Although the active site appears to reside in an extended cleft, a major portion of the cleft is occupied by a lobe from an adjacent monomer as discussed below. Thus, the accessible active site is actually a short cavity that extends into the protein past the binuclear center and terminates in a predominantly hydrophobic pocket with the side chain of Lys370 pointing toward its base. Notably, there are no free acidic residues in proximity to the Lys370 amino group, which presumably possesses a positive charge, to act as a counterion. The strategic location of this residue at the base of the active site where the N-terminal aspartyl residue side chain is likely to bind indicates that it may strongly contribute to the preference of DNPEP for N-terminal aspartyl-containing peptides. To further evaluate features of the active site that favor N-terminal Asp-containing peptides, we carried out an electrostatics calculation on DNPEP in the context of the dodecameric assembly observed in the crystal structure. Whereas the exterior of the dodecamer is relatively neutral, its interior is highly positively charged with the highest charge density located in the active site pocket and areas immediately adjacent to the pocket (supplemental Fig. S4). It is evident from this calculation that negatively charged substrates would be drawn toward the active site through electrostatic forces.
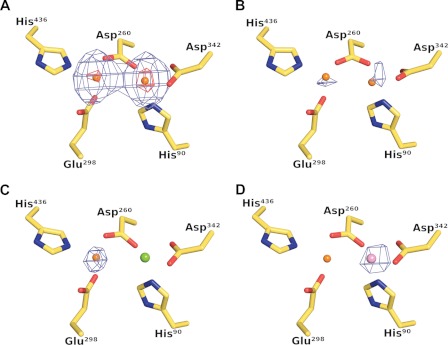
Based on the SAD phasing experiment, we expected that the metal-binding sites in as-isolated DNPEP were primarily occupied by zinc. However, they could also be occupied by other first-row transition metals close to zinc in the periodic table as such atoms would also exhibit significant anomalous scattering at the zinc K-edge. To confirm the identity of the active site metals as zinc, we collected datasets on a crystal grown in condition B using x-rays that were either 50 eV below or directly at the peak of the zinc K absorption edge, as determined by a zinc fluorescence scan on the same crystal (68, 69). Strong peaks were observed at both metal sites in the Bijvoet-difference Fourier electron density map calculated using the peak data with a somewhat higher peak height observed at site B compared with site A, whereas much weaker peaks were observed in the corresponding map calculated using the pre-edge data (Fig. 3, A and B). The sharp increase in anomalous scattering upon passing through the zinc K-edge clearly demonstrated that the enzyme, as-isolated from E. coli, contains a binuclear zinc center.
FIGURE 3.
Crystallographic identification of active site metals. The left and right metal-binding sites are referred to as site A and site B, respectively. A, Bijvoet-difference Fourier maps calculated using data collected at the zinc K absorption peak and contoured at the 5 r.m.s.d. (blue mesh) and 25 r.m.s.d. (red mesh) levels show strong anomalous scattering at the two metal sites, whereas the corresponding map calculated using pre-edge data shows only weak anomalous scattering at both sites as shown in B, thus demonstrating the presence of zinc at both sites in the as-isolated protein. C, anomalous scattering is only observed at site A in crystals grown in the presence of magnesium, suggesting that magnesium occupies site B in this crystal form. D, Bijvoet-difference Fourier map calculated using manganese K absorption peak data collected from a crystal composed of manganese-activated DNPEP. A single 9 r.m.s.d. peak is found at site B demonstrating that manganese can substitute for zinc at site B leading to an enhancement of enzymatic activity.
For the ZnMg-DNPEP dataset obtained from a crystal grown in condition A, we initially modeled both metals as zinc. However, a Bijvoet-difference Fourier map showed significant anomalous scattering only at site A, indicating that a metal other than zinc was present at site B (Fig. 3C). Because the crystal mother liquor contained a high concentration of magnesium, we tested the possibility that magnesium rather than zinc could occupy site B in this crystal by calculating element-specific combined real-imaginary SAD-log likelihood gradient maps as implemented in Phaser (46). Magnesium log likelihood gradient maps produced higher Z-scores at site B compared with the corresponding maps for zinc, which indicated that magnesium was the predominant metal at site B in this structure.
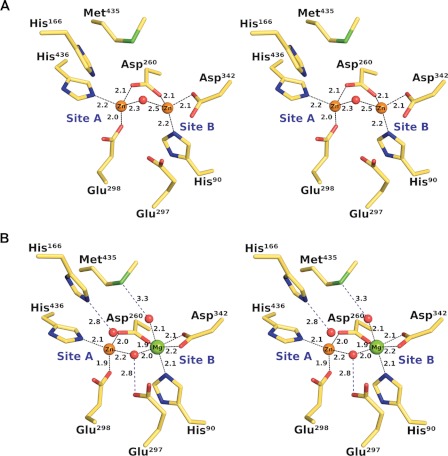
We observe different coordination geometries for the binuclear metal center in the two structures, consistent with the difference in metal content. In the ZnZn-DNPEP structure, ZnA is terminally coordinated by His436 and Glu298, whereas His90 and Asp342 serve as terminal ligands for ZnB (Fig. 4A). The metal-carboxylate interactions appear to be monodentate at both sites. The metals are bridged by Asp260 and a solvent-derived water molecule (or hydroxide ion) with a metal-metal distance of ∼3.3 Å. The ligand geometry around both zincs is distorted tetrahedral, but it appears that the solvent molecule is mobile and can move perpendicularly with respect to the metal-metal axis to form hydrogen-bonding interaction with either Glu297 or Met435 side chains. The bridging solvent ligand is located ∼2.3 and ∼2.5 Å away from ZnA and ZnB, respectively, which is most consistent with a relatively mobile, bound water molecule.
FIGURE 4.
Coordination geometry of the DNPEP binuclear center. A, stereoview of the as-isolated ZnZn-DNPEP structure showing a four-coordinate distorted tetrahedral geometry for both zinc ions. The metal-metal distance is ∼3.3 Å. The electron density maps indicate that the bridging solvent is mobile and can move perpendicularly to the metal-metal axis to form hydrogen bonding interactions with Met435 and Glu297. B, stereoview of the ZnMg-DNPEP structure showing a distorted tetrahedral geometry for the site A zinc and an octahedral geometry for the magnesium at site B. The metal-metal distance is ∼3.4 Å. The position of the bridging solvent is substantially different compared with the ZnZn-DNPEP structure, and it is seen to form a hydrogen bonding interaction with the nearby Glu297 residue.
By contrast, the higher resolution ZnMg-DNPEP structure contains a tetrahedrally coordinated zinc at site A and an octahedrally coordinated magnesium at site B, consistent with the known geometrical preferences of these cations (Fig. 4B) (70, 71). The protein ligands are arranged in much the same way as in the ZnZn-DNPEP structure with one notable difference, i.e. the terminal Asp342 carboxylate ligand binds magnesium in a bidentate fashion. The position of the bridging solvent ligand is quite different compared with the ZnZn-DNPEP structure. In the ZnMg-DNPEP structure, the bridging solvent ligand is oriented such that it forms a hydrogen-bonding interaction with Glu297. The metal-solvent bond lengths are significantly shorter with Zn-O and Mg-O distances of 2.2 and 2.0 Å, respectively. The shorter metal-solvent bond length together with the observed solvent carboxylate hydrogen bonding interaction suggests that the bridging solvent in this structure is a hydroxide ion. In addition to the direct metal ligands, a number of water molecules are located in close proximity to the metal center in the ZnMg-DNPEP structure. One of these water molecules that is likely of catalytic importance is located 2.8 Å away from ZnA. This water molecule does not appear to be directly bound to ZnA but rather is held in place near the metal center by the highly conserved His166 residue from a symmetry-related molecule. The lobe containing His166 is located in the dimerization domain and interacts extensively with the major interdomain groove in an adjacent monomer forming a highly complementary association. His166, located near the tip of the lobe, enters deeply into the active site to form a hydrogen bond with the water molecule (supplemental Movie S1). The extended conformation of the conserved lobe and the high complementarity of the surfaces strongly suggest that the interaction is physiologically relevant and that His166 plays an important role in the peptide hydrolysis reaction. Indeed, a prior study showed that Phe substitution of the corresponding His residue in human DNPEP abolishes enzymatic activity (37).
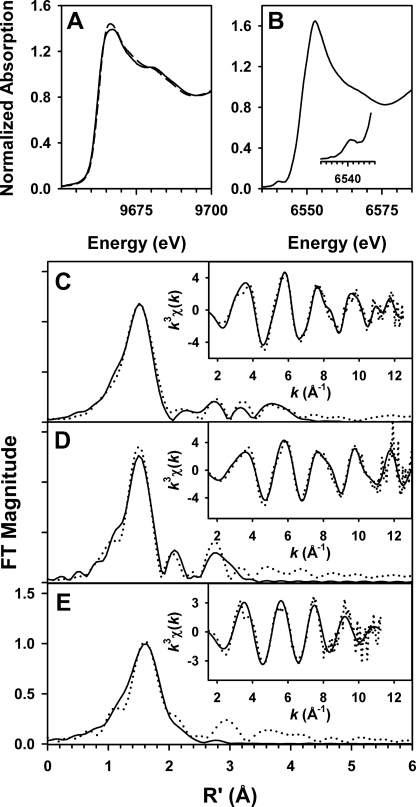
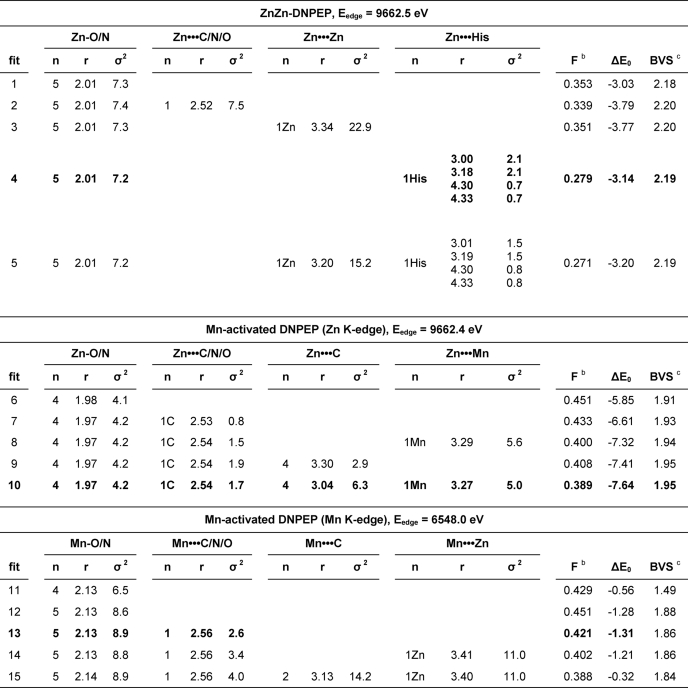
The structure of the DNPEP metal center was further evaluated by zinc K-edge XAS. Examination of metal-specific fluorescence from the as-isolated preparation revealed a significant signal only for zinc, which confirmed the homogeneity of the active site metal content. The XANES spectrum was typical for Zn(II) in an N/O-rich environment (72) with the edge inflection energy of 9662.5 eV (Fig. 5A). The best one-shell fits to the EXAFS data consisted of a fairly disordered set of five O/N scatterers at ∼2.01 Å (Fig. 5C and Table 4). A modest improvement in fit quality was obtained by splitting this shell into two subshells of O/N scatterers at 1.97 and 2.10 Å, but the difference in bond lengths between the two shells was only slightly smaller than the resolution of the data (0.143 Å), indicating that a two-shell fit is not justifiable (supplemental Table S5). The outer shell features were best modeled by a set of single-scattering and multiple-scattering paths from the imidazole of a single Zn(II)-bound His ligand, in agreement with the single His per Zn(II) site seen in the ZnZn-DNPEP crystal structure. Attempts to include a zinc-zinc interaction either in the presence or absence of His were unsuccessful, based on the lack of improvement in fit quality and the very large Debye-Waller factors obtained for the Zn···Zn shell (Table 4 and supplemental Table S6). The absence of a detectable metal-metal interaction is in accord with the mobile bridging solvent ligand observed in the ZnZn-DNPEP crystal structure, as metal-metal interactions are typically not clearly detectable by EXAFS for binuclear sites lacking a rigid single-atom bridge such as hydroxide (56, 73).
FIGURE 5.
XAS of DNPEP. A, x-ray absorption edge spectra at the zinc K-edge for both ZnZn-DNPEP and manganese-activated DNPEP. The spectra are quite similar to one another, with edge inflections at ∼9662.5 eV. B, x-ray absorption edge spectrum at the manganese K-edge for manganese-activated DNPEP. A weak pre-edge feature, magnified in the inset, is present at 6540.2 eV. C–E, Fourier transforms (FT) and unfiltered EXAFS spectra (k3χ(k), insets) of ZnZn-DNPEP at the zinc K-edge (C), manganese-activated DNPEP at the zinc K-edge (D), and manganese-activated DNPEP at the manganese K-edge (E) are shown. Experimental data are shown by dotted lines and fits by solid lines. The fit parameters correspond to those shown in boldface in Table 4.
TABLE 4.
EXAFS analysis of recombinant and Mn-substituted DNPEPa
a r is in units of Å; σ2 is in units of 10−3 Å2; ΔE0 is in units of eV. All fits are to unfiltered EXAFS data, as follows: ZnZn-DNPEP, Fourier transform range of k = 1.5–12.5 Å−1 (resolution = 0.143 Å); Mn-activated DNPEP (Zn K-edge), Fourier transform range of k = 1.5–13.0 Å−1 (resolution = 0.137 Å); Mn-activated DNPEP (Mn K-edge), Fourier transform range of k = 1.5–11.25 Å−1 (resolution = 0.161 Å).
b F = (Σk6(χexptl − χcalc)2/Σk6χexptl2)1/2.
c BVS = bond valence sum for the first-shell atoms of the fit. The BVS is defined as Σ(exp((r0 − r)/0.37)), where r0 is an empirically derived parameter for a given pair of atoms, and r is the actual bond length. The values of r0 were taken from Refs. 57, 58. Whereas EXAFS cannot distinguish between atoms differing by Z ± 1, we assumed an oxygen-rich environment based upon the crystal structure data.
Structural Basis of Manganese Activation
The strong stimulatory effect of Mn(II) on the catalytic activity of DNPEP prompted us to examine its structural basis. For these experiments, we used manganese-activated DNPEP that was produced by supplementation of the E. coli expression culture with 5 mm MnCl2 and displayed a 30-fold higher catalytic efficiency toward aspartyl-p-naphthylamide compared with unsupplemented protein. XANES and EXAFS data were recorded on the manganese-activated sample at both the zinc and manganese K-edges. Element-specific fluorescence clearly showed the presence of both manganese and zinc in the sample, which provided the rationale for collecting data at both absorption edges. The zinc K-edge XANES spectrum for manganese-activated DNPEP was similar to that of the as-isolated sample with a single edge inflection at ∼9662.4 eV. The manganese K-edge XANES spectrum exhibited a sharp rising absorption with a primary edge inflection at 6548.0 eV, a shoulder inflection at ∼6550 eV, and a weak pre-edge feature centered at ∼6540.2 eV (Fig. 5B). These features are in good agreement with those of other biological Mn(II) sites studied by XAS (73). The sharpness of the rising absorption edge and weakness of the pre-edge feature were consistent with the Mn(II) ion being in a high symmetry five- or six-coordinate environment. Zinc K-edge EXAFS analysis of the manganese-activated sample revealed some structural alteration in the Zn(II) site(s) compared with the as-isolated DNPEP. Specifically, the best one-shell fit to the data consisted of four Zn-O/N scatterers at 1.97 Å, in good agreement with the bond lengths derived from the crystallographic structure (Fig. 5D and Table 4). It is also notable that the Debye-Waller factor of this shell is smaller for manganese-activated DNPEP compared with as-isolated DNPEP, consistent with a narrower distribution of metal-ligand bond lengths. Furthermore, there is evidence for a low-Z scatterer at ∼2.53 Å, with the small magnitude of σ2 for this shell suggesting that it can be attributed to the bridging carbon of a bidentately bound carboxylate. The outer shell features were satisfactorily modeled by single scattering from low Z atoms at ∼3.04 Å, which can be attributable to carbons of coordinated His and carboxylate ligands, as well as a weak metal-metal interaction (modeled as Zn···Mn) at 3.27 Å (fit 10 in Table 4 and supplemental Table S7). Fitting the 3.3 Å feature to a set of Zn···C scatterers provided an apparent good fit to the data but exhibited a poorer visual match to the data and cannot be plausibly assigned to any zinc ligands in the active site because binuclear metalloproteins with N/O ligation generally lack any low zinc atoms at this distance. It was not possible to model scattering from coordinated histidine ligands. We note that the Zn···Mn distance of 3.27 Å is in good agreement with the metal-metal separations of 3.3 and 3.4 Å observed in the ZnZn-DNPEP and ZnMg-DNPEP crystal structures, respectively, and supports the notion of a solvent-derived hydroxide bridge in manganese-activated DNPEP.
We next analyzed the Manganese K-edge EXAFS data collected on the manganese-activated sample. As with the zinc K-edge analysis, the first coordination shell was best described by simple one-shell fits. Although a shell of four Mn-O/N scatterers provided the best fit quality, the BVS value of 1.5 for this fit was clearly too low for a Mn(II) site and is likewise unexpected given the XANES spectra, which indicated five or six coordinate Mn(II). Instead, fits consisting of a highly disordered set of five to six Mn-O/N scatterers at ∼2.13 Å are in better agreement with BVS calculations and the empirical XANES observations (Fig. 5E and Table 4). Studies of structurally well characterized Mn(II) model complexes and proteins have shown that coordination number can be significantly underestimated by EXAFS for highly disordered Mn(II) sites that have a wide distribution of bond lengths (73, 74). The significantly longer average Mn(II)-ligand bond lengths relative to the Zn(II)-ligand bond lengths are in accord with the larger ionic radius of Mn(II). Attempts to split the first shell into two sub-shells were unsuccessful (supplemental Table S4); however, as with the zinc K-edge EXAFS on this sample, the data clearly support the presence of a Mn-C/N/O scatterer at 2.56 Å that exhibits a small Debye-Waller value and can also be assigned to the bridging carbon atom of a bound carboxylate ligand. The outer shell features could be partially simulated by inclusion of a manganese-zinc interaction at 3.4 Å, a distance seemingly at odds with the metal-metal separation of 3.27 Å obtained at the zinc K-edge. However, the improvement in fit quality upon addition of this shell is smaller than was the case at the zinc K-edge, and it exhibits a fairly large Debye-Waller factor (fits 13–15, Table 4 and supplemental Table S8), indicating that a metal-metal scattering path contributes only weakly to the EXAFS. Indeed, several studies have shown that accurate detection of biological metal-metal separations longer than 3.0 Å by EXAFS is especially difficult in cases where the available k-range of the data is limited, as is the case here (56, 73, 74). We conclude that the available manganese K-edge EXAFS data for manganese-activated DNPEP does not provide convincing evidence for the presence of a metal-metal interaction. Taken together, the XAS data on manganese-activated DNPEP suggests that Mn(II) occupies site B of the binuclear center with a ligand geometry similar to that of the six-coordinate Oh Mg(II) ion in the ZnMg-DNPEP crystal structure, whereas zinc remains bound at site A with a more ordered set of ligands.
To provide further support for this idea, we collected x-ray diffraction data just above the manganese K-edge on a DNPEP crystal that was grown from the same manganese-activated DNPEP preparation used for the XAS experiments. Although the crystals displayed rapid and severe radiation damage at the long wavelength, we were able to collect a dataset that displayed reasonable anomalous signal as judged from anomalous correlation coefficients. The Bijvoet-different Fourier map calculated from these data shows a single ∼9 r.m.s.d. peak at site B clearly demonstrating that manganese does indeed substitute for zinc at site B (Fig. 3D).
DNPEP Oligomeric Structure
Examination of symmetry-related molecules in the crystal revealed that DNPEP forms a homo-dodecameric tetrahedral complex with eight such tetrahedra present in the unit cell of the crystal. To evaluate the possibility that this assembly could represent a physiological complex as opposed to a crystal packing artifact, we analyzed the extent and complementarity of self-association using the PISA server (75) and SC program (76) from the CCP4 program suite (44). Two major interfaces are responsible for the formation of the tetrahedral complex (supplemental Movie S1). The first interface between monomers related by the 2-fold symmetry element of the tetrahedron buries 3775 Å2 of solvent-accessible surface area per monomer. As mentioned above, a conserved lobe within the dimerization domain of one monomer extends into the complementary surface of a second monomer within the large groove between the catalytic and dimerization domains, resulting in the formation of an anti-parallel dimer. The dimers of DNPEP form the edges of the tetrahedron, which are ∼118 Å in length. The second interface is between monomers related by the 3-fold symmetry element of the tetrahedron and buries 2661 Å2 of solvent-accessible surface area per monomer. Helices 11, 13, and 14 and strands 16 and 17 as well as some intervening loop regions pack against the front face of the protein in the region between the two domains with the C-terminal tail of helix 15 inserted into the small groove between domains. This interaction repeats twice to form a hollow cone-shaped trimer with a closed apex and open base containing the dimerization sites. Both interfaces have large negative values for solvation-free energy gain upon complexation, indicating high thermodynamic stability. In the context of the tetrahedral assembly, ∼40% of the DNPEP solvent-accessible surface area is buried by interfacial contacts. The dimeric and trimeric interfaces have shape complementarity values of 0.709 and 0.706, respectively, comparable with well established oligomers (76). Nevertheless, in gel filtration experiments from both us and others, the protein eluted at a volume consistent with an 8-mer. This discrepancy prompted us to examine the structure and symmetry DNPEP by negative stain EM.
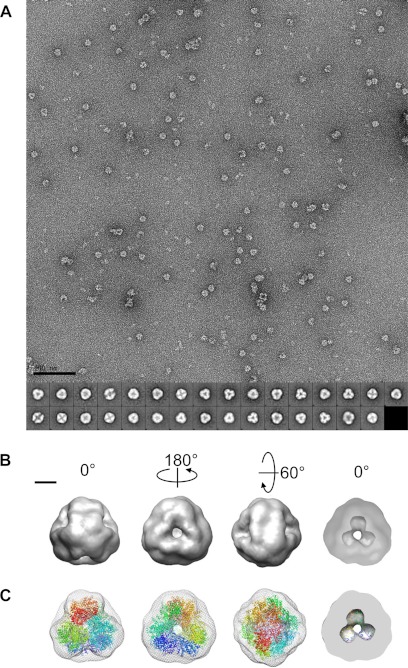
Fig. 6A shows a raw micrograph of negatively stained DNPEP particles at ×67,834 magnification. The particles are about 17 nm in diameter and are primarily triangular or square-shaped in projection. Most particles exhibit dark central staining indicative of a hollow structure. The size and projection symmetry of the particles observed in electron micrographs are fully compatible with the hypothesis that DNPEP exists as a dodecameric complex with tetrahedral symmetry. To further support this hypothesis, we performed a three-dimensional reconstruction of the DNPEP complex based on two-dimensional projections of the negatively stained particles. An initial model generated from 5000 particles without any imposed symmetry was hollow, roughly spherical with a slight elongation in one direction, and had clear features of the 2- and 3-fold symmetry axes expected for tetrahedral symmetry. Based on these agreements, tetrahedral symmetry was imposed during generation of class averages and the starting model as well as during model refinement (Fig. 6, A and B). The Euler plot demonstrates that the particle set used for refinement displayed good coverage of the unique projection space (supplemental Fig. S5A). Not unexpectedly, the particles had a tendency to sit on the EM grid with a base of the tetrahedron facing down. The three-dimensional EM map at 22 Å resolution (supplemental Fig. S5B) shows a hollow tetrahedral complex (Fig. 6B). Each face of the tetrahedron features a large pore in its center. A section through the map shows that the interior space extends toward the vertices of the tetrahedron and forms a pocket where the active sites of the complex are located. As shown in Fig. 6C, the crystallographic model docked well into the EM map. The excellent agreement between the single particle analysis and crystallographic data confirm that DNPEP exists as a dodecameric tetrahedral complex in solution and that this complex represents the active, functional form of the enzyme.
FIGURE 6.
Single particle reconstruction of DNPEP. A, micrograph of DNPEP (40 μg/ml) coated with 1% uranyl acetate at ×67,834 magnification. Aggregated and incomplete particles were omitted from the particle set. The scale bar on the bottom left indicates 100 nm. The 31 class averages derived from 5000 particles using EMAN2 are shown at the bottom of the image. B, 22-Å resolution map shown in different orientations. The cut-through section in the rightmost panel shows the hollow core of the particle with cavities extending toward the vertices of the tetrahedron. The scale bar on the top left indicates 50 Å. C, alignment of the EM map (mesh) with the crystallographic model of the DNPEP tetrahedral dodecamer (ribbons colored differently for each monomer) with the same orientations shown in B. Figures were prepared using Chimera.
DISCUSSION
In this report, we present the first detailed structural analysis of a mammalian M18 aminopeptidase member, which has provided a basis for understanding many of the biochemical features of this enzyme family. Our interest in this enzyme stems from its relatively high expression level in the eye and possible involvement in the ocular RAS, a system that has been implicated in a variety of eye diseases such as diabetic retinopathy.
Prior to this study, cytosolic aspartyl aminopeptidase activity toward angiotensin peptides had been reported (32, 77) but little was known about its ability to metabolize other physiologically relevant substrates. We have analyzed the DNPEP-catalyzed hydrolysis of five N-Asp-containing bioactive peptides expressed in the retina (Table 3 and supplemental Table S4). Although all of these peptides are hydrolyzed by DNPEP, the angiotensin peptides Ang I and Ang II are found as preferred substrates, which is consistent with the previous supposition that DNPEP acts as an angiotensinase. In addition, CCK-8 has also been found as a relatively favored substrate of DNPEP, which indicates this enzyme could be a regulator of neuropeptides in the central nervous system including the eye.
Our crystallographic and single particle EM data clearly show that DNPEP forms a tetrahedral dodecamer in contrast to the previously proposed octameric aggregation state that was based on the results of gel filtration experiments. This disagreement could potentially be due to weak binding of DNPEP to gel filtration resins with a resultant prolongation in its elution time (32, 35, 36). The preference of DNPEP toward short peptides can be rationalized by our structural data, which demonstrate that access to the active site cavity of DNPEP is greatly restricted due to its sequestration inside the tetrahedral complex. There are four ∼20-Å-wide “selectivity pores” that would likely prevent entrance of all but single-stranded peptides into the interior cavity (Figs. 2C and 6B). Mutagenesis and disulfide bond cross-linking experiments on the M42 family member TET have shown that by partially blocking the entrance pore of the tetrahedron, its enzymatic activity could be substantially reduced thus confirming the role of this pore in substrate selectivity (61).
The physiological purpose of oligomerization may extend beyond substrate size exclusion. We observed that monomers in this complex related by 2-fold symmetry extend lobe-like structural elements into each other's active sites that greatly reduces the size of the active site cleft and may contribute to substrate specificity and catalysis (supplemental Movie S1). The highly conserved His166 located near the tip of this extension is in close proximity to the binuclear metal center, and mutation of the homologous residue in human DNPEP led to loss of activity without disruption of oligomer formation (37) consistent with a role for this residue in catalysis.
Electrostatics analysis of the DNPEP complex revealed that the active site pocket is highly positively charged consistent with its substrate preference (supplemental Fig. S4). Lys370 located at the base of the active site cavity appears to be correctly positioned for interaction with the side-chain carboxylate of the N-Asp of a substrate peptide. While this study was in progress, a crystal structure of human DNPEP in complex with aspartyl-β-hydroxamate, a known DNPEP inhibitor (37), was deposited in the Protein Data Bank (3L6S). The inhibitor is bound in the active site with the carboxylate moiety forming a salt bridge with a Lys residue analogous to Lys370 in bovine DNPEP, as well as hydrogen bonds with Tyr and His side chains. These interactions would guide the scissile peptide bond to be positioned in front of the binuclear center for hydrolysis to occur. Lys370 is conserved among aspartyl aminopeptidases in the M18 family, whereas it is variable in neutral and basic aminopeptidases in this family.
Like some other MH clan peptidases, the active site of as-isolated recombinant DNPEP contains a binuclear zinc center. Interestingly, we observed that the zinc at site B is capable of exchange with other divalent cations. We have shown through XAS and x-ray crystallographic analysis of manganese-activated DNPEP that manganese substitution for zinc at site B is responsible for the strong stimulatory effect of this metal on DNPEP activity. These results suggest a critical role for site B of the binuclear center in the control of DNPEP enzymatic activity. Future studies will examine which metal ions are associated with DNPEP under physiological conditions and whether the activation phenomenon is relevant in vivo.
M42 family peptidases, which also belong to the MH clan, form tetrahedral assemblies suggesting an especially close relationship to the M18 family. M42 peptidases are found only in archaea and bacteria, whereas the M18 peptidases are more widespread. Phylogenetic analysis of crystallographically characterized M18 and M42 members shows a clear segregation into distinct families, but there are a number of commonalities between members of the two families (supplemental Fig. S6A). Both families exhibit two-domain architecture. The catalytic domain, including residues directly involved in metal binding, is structurally well conserved among the M18 and M42 peptidases. By contrast, notable structural differences in the dimerization domain are found between the two families. The dimerization domain of the M42 peptidases adopts a ferredoxin reductase-like, orthogonal, and six-stranded anti-parallel β barrel fold that is capped on both ends by α helices. The analogous domain of M18 peptidases, as represented by DNPEP, has a similar chain topology, but the β barrel fold, in comparison with M42 peptidases, appears to be unsealed and pealed apart on one end with helix 6 nestled between the two orthogonal β sheets (supplemental Fig. S6B). The expanded conformation of the M18 dimerization domain increases the contact surface area of the tetrahedron potentially increasing its stability. As seen in DNPEP, the dimerization domain of many members of both families contains a long finger-like loop insertion with a conserved His residue that extends into the active site of the dimeric partner that might complete the active site and be important for catalysis and substrate specificity.
In summary, the results presented here greatly improve our understanding of the structure, enzymology, and bioinorganic chemistry of DNPEP and the M18 aminopeptidase family in general. Equipped with this knowledge, we are now in a better position to more fully understand the physiology of this ancient family of aminopeptidases.
Supplementary Material
Acknowledgments
We thank Drs. Andreas Engel, Vera Moiseenkova-Bell, Yaroslav Tsybovsky, David Lodowski at Case Western Reserve University and Steve Ludtke at Baylor Medical Institute for their advice on electron microscopy and single particle analysis. We thank Debarshi Mustafi for sharing RNA-seq data prior to publication and Dr. Marcin Golczak for assistance with mass spectrometry. This work is based upon research conducted at the Northeastern Collaborative Access Team beam lines of the Advanced Photon Source, supported by National Institute of Health Award RR-15301 from the NCRR. Use of the Advanced Photon Source is supported by the United States Department of Energy, Office of Basic Energy Sciences, under Contract W-31-109-ENG-38. XAS experiments were carried out at beamline X3B of the National Synchrotron Light Source at Brookhaven National Laboratory. Beamline X3B is operated by the Case Center for Synchrotron Biosciences, supported by National Institutes of Health NIBIB Grant P30-EB-009998. National Synchrotron Light Source is supported by the United States Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract DE-AC02-98CH10886.
This work was supported, in whole or in part, by National Institutes of Health Grants EY009339 and P30 EY11373.

This article contains supplemental Figs. S1–S6, Tables S1–S8, Movie S1, and additional references.
The atomic coordinates and structure factors (codes 3VAR and 3VAT) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- RAS
- renin-angiotensin system
- Ang
- angiotensin
- BVS
- bond valence sum
- CCK-8
- cholecystokinin octapeptide
- DNPEP
- aspartyl aminopeptidase
- EXAFS
- extended x-ray absorption fine structure
- MCH
- melanin concentrating hormone
- NKB
- neurokinin B
- SAD
- single wavelength anomalous diffraction
- XANES
- x-ray absorption near edge spectra
- XAS
- x-ray absorption spectroscopy
- r.m.s.d.
- root mean square deviation.
REFERENCES
- 1. Taylor A. (1993) Aminopeptidases. Structure and function. FASEB J. 7, 290–298 [DOI] [PubMed] [Google Scholar]
- 2. Hooper N. M., Lendeckel U. (eds) (2004) Aminopeptidases in Biology and Disease, Kluwer Academic/Plenum Publishers, New York [Google Scholar]
- 3. Zini S., Fournie-Zaluski M. C., Chauvel E., Roques B. P., Corvol P., Llorens-Cortes C. (1996) Identification of metabolic pathways of brain angiotensin II and III using specific aminopeptidase inhibitors. Predominant role of angiotensin III in the control of vasopressin release. Proc. Natl. Acad. Sci. U.S.A. 93, 11968–11973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Guyton A. C., Hall J. E. (2000) Textbook of Medical Physiology, 10th Ed., pp. 195–209, W. B. Saunders Co., Philadelphia [Google Scholar]
- 5. Danser A. H. (1996) Local renin-angiotensin systems. Mol. Cell. Biochem. 157, 211–216 [DOI] [PubMed] [Google Scholar]
- 6. Danser A. H., Derkx F. H., Admiraal P. J., Deinum J., de Jong P. T., Schalekamp M. A. (1994) Angiotensin levels in the eye. Invest. Ophthalmol. Vis. Sci. 35, 1008–1018 [PubMed] [Google Scholar]
- 7. Deinum J., Derkx F. H., Danser A. H., Schalekamp M. A. (1989) Renin in the bovine eye. J. Hypertens. Suppl. 7, S216–S217 [DOI] [PubMed] [Google Scholar]
- 8. Ferrari-Dileo G., Ryan J. W., Rockwood E. J., Davis E. B., Anderson D. R. (1988) Angiotensin-converting enzyme in bovine, feline, and human ocular tissues. Invest. Ophthalmol. Vis. Sci. 29, 876–881 [PubMed] [Google Scholar]
- 9. Downie L. E., Vessey K., Miller A., Ward M. M., Pianta M. J., Vingrys A. J., Wilkinson-Berka J. L., Fletcher E. L. (2009) Neuronal and glial cell expression of angiotensin II type 1 (AT1) and type 2 (AT2) receptors in the rat retina. Neuroscience 161, 195–213 [DOI] [PubMed] [Google Scholar]
- 10. Berka J. L., Stubbs A. J., Wang D. Z., DiNicolantonio R., Alcorn D., Campbell D. J., Skinner S. L. (1995) Renin-containing Muller cells of the retina display endocrine features. Invest. Ophthalmol. Vis. Sci. 36, 1450–1458 [PubMed] [Google Scholar]
- 11. Milenkovic V. M., Brockmann M., Meyer C., Desch M., Schweda F., Kurtz A., Todorov V., Strauss O. (2010) Regulation of the renin expression in the retinal pigment epithelium by systemic stimuli. Am. J. Physiol. Renal Physiol. 299, F396–F403 [DOI] [PubMed] [Google Scholar]
- 12. Wagner J., Jan Danser A. H., Derkx F. H., de Jong T. V., Paul M., Mullins J. J., Schalekamp M. A., Ganten D. (1996) Demonstration of renin mRNA, angiotensinogen mRNA, and angiotensin-converting enzyme mRNA expression in the human eye. Evidence for an intraocular renin-angiotensin system. Br. J. Ophthalmol. 80, 159–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Paul M., Poyan Mehr A., Kreutz R. (2006) Physiology of local renin-angiotensin systems. Physiol. Rev. 86, 747–803 [DOI] [PubMed] [Google Scholar]
- 14. Costagliola C., Di Benedetto R., De Caprio L., Verde R., Mastropasqua L. (1995) Effect of oral captopril (SQ 14225) on intraocular pressure in man. Eur. J. Ophthalmol. 5, 19–25 [DOI] [PubMed] [Google Scholar]
- 15. Nagai N., Oike Y., Noda K., Urano T., Kubota Y., Ozawa Y., Shinoda H., Koto T., Shinoda K., Inoue M., Tsubota K., Yamashiro K., Suda T., Ishida S. (2005) Suppression of ocular inflammation in endotoxin-induced uveitis by blocking the angiotensin II type 1 receptor. Invest. Ophthalmol. Vis. Sci. 46, 2925–2931 [DOI] [PubMed] [Google Scholar]
- 16. Kurihara T., Ozawa Y., Shinoda K., Nagai N., Inoue M., Oike Y., Tsubota K., Ishida S., Okano H. (2006) Neuroprotective effects of angiotensin II type 1 receptor (AT1R) blocker, telmisartan, via modulating AT1R and AT2R signaling in retinal inflammation. Invest. Ophthalmol. Vis. Sci. 47, 5545–5552 [DOI] [PubMed] [Google Scholar]
- 17. Otani A., Takagi H., Suzuma K., Honda Y. (1998) Angiotensin II potentiates vascular endothelial growth factor-induced angiogenic activity in retinal microcapillary endothelial cells. Circ. Res. 82, 619–628 [DOI] [PubMed] [Google Scholar]
- 18. Jacobi P. C., Osswald H., Jurklies B., Zrenner E. (1994) Neuromodulatory effects of the renin-angiotensin system on the cat electroretinogram. Invest. Ophthalmol. Vis. Sci. 35, 973–980 [PubMed] [Google Scholar]
- 19. Fletcher E. L., Phipps J. A., Ward M. M., Vessey K. A., Wilkinson-Berka J. L. (2010) The renin-angiotensin system in retinal health and disease: Its influence on neurons, glia and the vasculature. Prog. Retin. Eye Res. 29, 284–311 [DOI] [PubMed] [Google Scholar]
- 20. Vaajanen A., Vapaatalo H. (2011) Local ocular renin-angiotensin system. A target for glaucoma therapy? Basic Clin. Pharmacol. Toxicol. 109, 217–224 [DOI] [PubMed] [Google Scholar]
- 21. Mauer M., Zinman B., Gardiner R., Suissa S., Sinaiko A., Strand T., Drummond K., Donnelly S., Goodyer P., Gubler M. C., Klein R. (2009) Renal and retinal effects of enalapril and losartan in type 1 diabetes. N. Engl. J. Med. 361, 40–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Silva K. C., Rosales M. A., Biswas S. K., Lopes de Faria J. B., Lopes de Faria J. M. (2009) Diabetic retinal neurodegeneration is associated with mitochondrial oxidative stress and is improved by an angiotensin receptor blocker in a model combining hypertension and diabetes. Diabetes 58, 1382–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lavoie J. L., Sigmund C. D. (2003) Minireview. Overview of the renin-angiotensin system. An endocrine and paracrine system. Endocrinology 144, 2179–2183 [DOI] [PubMed] [Google Scholar]
- 24. Reid I. A., Morris B. J., Ganong W. F. (1978) The renin-angiotensin system. Annu. Rev. Physiol. 40, 377–410 [DOI] [PubMed] [Google Scholar]
- 25. Unger T. (2002) The role of the renin-angiotensin system in the development of cardiovascular disease. Am. J. Cardiol. 89, 3A–9A [DOI] [PubMed] [Google Scholar]
- 26. Krause M. W., Fonseca V. A., Shah S. V. (2011) Combination inhibition of the renin-angiotensin system. Is more better? Kidney Int. 80, 245–255 [DOI] [PubMed] [Google Scholar]
- 27. Jensen L. L., Harding J. W., Wright J. W. (1989) Increased blood pressure induced by central application of aminopeptidase inhibitors is angiotensinergic-dependent in normotensive and hypertensive rat strains. Brain Res. 490, 48–55 [DOI] [PubMed] [Google Scholar]
- 28. Reaux A., Fournie-Zaluski M. C., David C., Zini S., Roques B. P., Corvol P., Llorens-Cortes C. (1999) Aminopeptidase A inhibitors as potential central antihypertensive agents. Proc. Natl. Acad. Sci. U.S.A. 96, 13415–13420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mustafi D., Kevany B. M., Genoud C., Okano K., Cideciyan A. V., Sumaroka A., Roman A. J., Jacobson S. G., Engel A., Adams M. D., Palczewski K. (2011) Defective photoreceptor phagocytosis in a mouse model of enhanced S-cone syndrome causes progressive retinal degeneration. FASEB J. 25, 3157–3176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cheung H. S., Cushman D. W. (1971) A soluble aspartate aminopeptidase from dog kidney. Biochim. Biophys. Acta 242, 190–193 [DOI] [PubMed] [Google Scholar]
- 31. Kelly J. A., Neidle E. L., Neidle A. (1983) An aminopeptidase from mouse brain cytosol that cleaves N-terminal acidic amino acid residues. J. Neurochem. 40, 1727–1734 [DOI] [PubMed] [Google Scholar]
- 32. Wilk S., Wilk E., Magnusson R. P. (1998) Purification, characterization, and cloning of a cytosolic aspartyl aminopeptidase. J. Biol. Chem. 273, 15961–15970 [DOI] [PubMed] [Google Scholar]
- 33. Rawlings N. D., Barrett A. J., Bateman A. (2012) MEROPS. The database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 40, D343–D350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Metz G., Röhm K. H. (1976) Yeast aminopeptidase I. Chemical composition and catalytic properties. Biochim. Biophys. Acta 429, 933–949 [DOI] [PubMed] [Google Scholar]
- 35. Yokoyama R., Kawasaki H., Hirano H. (2006) Identification of yeast aspartyl aminopeptidase gene by purifying and characterizing its product from yeast cells. FEBS J. 273, 192–198 [DOI] [PubMed] [Google Scholar]
- 36. Teuscher F., Lowther J., Skinner-Adams T. S., Spielmann T., Dixon M. W., Stack C. M., Donnelly S., Mucha A., Kafarski P., Vassiliou S., Gardiner D. L., Dalton J. P., Trenholme K. R. (2007) The M18 aspartyl aminopeptidase of the human malaria parasite Plasmodium falciparum. J. Biol. Chem. 282, 30817–30826 [DOI] [PubMed] [Google Scholar]
- 37. Wilk S., Wilk E., Magnusson R. P. (2002) Identification of histidine residues important in the catalysis and structure of aspartyl aminopeptidase. Arch. Biochem. Biophys. 407, 176–183 [DOI] [PubMed] [Google Scholar]
- 38. Russo S., Baumann U. (2004) Crystal structure of a dodecameric tetrahedral-shaped aminopeptidase. J. Biol. Chem. 279, 51275–51281 [DOI] [PubMed] [Google Scholar]
- 39. Franzetti B., Schoehn G., Hernandez J. F., Jaquinod M., Ruigrok R. W., Zaccai G. (2002) Tetrahedral aminopeptidase. A novel large protease complex from archaea. EMBO J. 21, 2132–2138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schu P. (2008) Aminopeptidase I enzymatic activity. Methods Enzymol. 451, 67–78 [DOI] [PubMed] [Google Scholar]
- 41. Yoshimoto T., Tanaka N., Kanada N., Inoue T., Nakajima Y., Haratake M., Nakamura K. T., Xu Y., Ito K. (2004) Crystal structures of creatininase reveal the substrate-binding site and provide an insight into the catalytic mechanism. J. Mol. Biol. 337, 399–416 [DOI] [PubMed] [Google Scholar]
- 42. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 43. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Collaborative Computational Project No. 4 (1994) The CCP4 Suite. Programs for Protein Crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 45. Sheldrick G. M. (2008) A short history of SHELX. Acta Crystallogr. A Found. Crystallogr. 64, 112–122 [DOI] [PubMed] [Google Scholar]
- 46. Read R. J., McCoy A. J. (2011) Using SAD data in Phaser. Acta Crystallogr. D Biol. Crystallogr. 67, 338–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Langer G., Cohen S. X., Lamzin V. S., Perrakis A. (2008) Automated macromolecular model building for x-ray crystallography using ARP/wARP version 7. Nat. Protoc. 3, 1171–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Murshudov G. N., Skubák P., Lebedev A. A., Pannu N. S., Steiner R. A., Nicholls R. A., Winn M. D., Long F., Vagin A. A. (2011) REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Emsley P., Cowtan K. (2004) Coot. Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 50. Chen V. B., Arendall W. B., 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C. (2010) MolProbity. All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tang G., Peng L., Baldwin P. R., Mann D. S., Jiang W., Rees I., Ludtke S. J. (2007) EMAN2. An extensible image processing suite for electron microscopy. J. Struct. Biol. 157, 38–46 [DOI] [PubMed] [Google Scholar]
- 52. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) UCSF Chimera. A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 53. George G. N. (2000) EXAFSPAK. Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory, Stanford, CA [Google Scholar]
- 54. Ravel B., Newville M. (2005) ATHENA, ARTEMIS, HEPHAESTUS: data analysis for X-ray absorption spectroscopy using IFEFFIT. J. Synchrotron Radiat. 12, 537–541 [DOI] [PubMed] [Google Scholar]
- 55. Ankudinov A. L., Ravel B., Rehr J. J., Conradson S. D. (1998) Real space multiple-scattering calculation and interpretation of x-ray-absorption near-edge structure. Phys. Rev. B 58, 7565 [Google Scholar]
- 56. Riggs-Gelasco P. J., Stemmler T. L., Penner-Hahn J. E. (1995) XAFS of dinuclear metal sites in proteins and model compounds. Coord. Chem. Rev. 144, 245–286 [Google Scholar]
- 57. Brown I. D., Altermatt D. (1985) Bond valence parameters obtained from a systematic analysis of the Inorganic Crystal Structure Database. Acta Crystallogr. B Struct. Sci. 41, 244–247 [Google Scholar]
- 58. Liu W., Thorp H. H. (1993) Bond valence sum analysis of metal-ligand bond lengths in metalloenzymes and model complexes. 2. Refined distances and other enzymes. Inorg. Chem. 32, 4102–4105 [Google Scholar]
- 59. Vettori M. V., Gatti R., Orlandini G., Belletti S., Alinovi R., Smargiassi A., Mutti A. (1999) An in vitro model for the assessment of manganese neurotoxicity. Toxicol. In Vitro 13, 931–938 [DOI] [PubMed] [Google Scholar]
- 60. Barrett A. J. (1994) Classification of peptidases. Methods Enzymol. 244, 1–15 [DOI] [PubMed] [Google Scholar]
- 61. Borissenko L., Groll M. (2005) Crystal structure of TET protease reveals complementary protein degradation pathways in prokaryotes. J. Mol. Biol. 346, 1207–1219 [DOI] [PubMed] [Google Scholar]
- 62. Goto Y., Hattori A., Ishii Y., Mizutani S., Tsujimoto M. (2006) Enzymatic properties of human aminopeptidase A. Regulation of its enzymatic activity by calcium and angiotensin IV. J. Biol. Chem. 281, 23503–23513 [DOI] [PubMed] [Google Scholar]
- 63. Jabs A., Weiss M. S., Hilgenfeld R. (1999) Non-proline cis peptide bonds in proteins. J. Mol. Biol. 286, 291–304 [DOI] [PubMed] [Google Scholar]
- 64. Greenblatt H. M., Almog O., Maras B., Spungin-Bialik A., Barra D., Blumberg S., Shoham G. (1997) Streptomyces griseus aminopeptidase. X-ray crystallographic structure at 1.75 Å resolution. J. Mol. Biol. 265, 620–636 [DOI] [PubMed] [Google Scholar]
- 65. Rowsell S., Pauptit R. A., Tucker A. D., Melton R. G., Blow D. M., Brick P. (1997) Crystal structure of carboxypeptidase G2, a bacterial enzyme with applications in cancer therapy. Structure 5, 337–347 [DOI] [PubMed] [Google Scholar]
- 66. Chevrier B., Schalk C., D'Orchymont H., Rondeau J. M., Moras D., Tarnus C. (1994) Crystal structure of Aeromonas proteolytica aminopeptidase. A prototypical member of the co-catalytic zinc enzyme family. Structure 2, 283–291 [DOI] [PubMed] [Google Scholar]
- 67. Chan A. W., Hutchinson E. G., Harris D., Thornton J. M. (1993) Identification, classification, and analysis of β-bulges in proteins. Protein Sci. 2, 1574–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Sukumar N., Choi M., Davidson V. L. (2011) Replacement of the axial copper ligand methionine with lysine in amicyanin converts it to a zinc-binding protein that no longer binds copper. J. Inorg. Biochem. 105, 1638–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Evans G., Pettifer R. F. (2001) CHOOCH. A program for deriving anomalous-scattering factors from x-ray fluorescence spectra. J. Appl. Crystallogr. 34, 82–86 [Google Scholar]
- 70. Dokmaniæ I., Sikiæ M., Tomiæ S. (2008) Metals in proteins. Correlation between the metal-ion type, coordination number, and the amino acid residues involved in the coordination. Acta Crystallogr. D Biol. Crystallogr. 64, 257–263 [DOI] [PubMed] [Google Scholar]
- 71. Holm R. H., Kennepohl P., Solomon E. I. (1996) Structural and functional aspects of metal sites in biology. Chem. Rev. 96, 2239–2314 [DOI] [PubMed] [Google Scholar]
- 72. Jacquamet L., Aberdam D., Adrait A., Hazemann J. L., Latour J. M., Michaud-Soret I. (1998) X-ray absorption spectroscopy of a new zinc site in the fur protein from Escherichia coli. Biochemistry 37, 2564–2571 [DOI] [PubMed] [Google Scholar]
- 73. Stemmler T. L., Sossong T. M., Jr., Goldstein J. I., Ash D. E., Elgren T. E., Kurtz D. M., Jr., Penner-Hahn J. E. (1997) EXAFS comparison of the dimanganese core structures of manganese catalase, arginase, and manganese-substituted ribonucleotide reductase and hemerythrin. Biochemistry 36, 9847–9858 [DOI] [PubMed] [Google Scholar]
- 74. Riggs-Gelasco P. (1995) Structural characterization of the manganese cluster of the oxygen evolving complex of photosystem II using x-ray absorption spectroscopy. Ph.D. dissertation, University of Michigan [Google Scholar]
- 75. Krissinel E., Henrick K. (2007) Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 76. Lawrence M. C., Colman P. M. (1993) Shape complementarity at protein/protein interfaces. J. Mol. Biol. 234, 946–950 [DOI] [PubMed] [Google Scholar]
- 77. Sim M. K., Qiu X. S. (1994) Formation of des-Asp-angiotensin I in the hypothalamic extract of normo- and hypertensive rats. Blood Press. 3, 260–264 [DOI] [PubMed] [Google Scholar]
- 78. Weiss M. S. (2001) Global indicators of x-ray data quality. J. Appl. Crystallogr. 34, 130–135 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.