Background: The Sec machinery transports proteins from the bacterial cytoplasm to the periplasm.
Results: Protein translocation kinetics are single exponential when multiple polylysine sites are simultaneously introduced into precursor proteins.
Conclusion: Translocation through the SecYEG pore is not the rate-limiting step of transport.
Significance: These data argue against the SecA motor model.
Keywords: Bacteria, Fluorescence, Kinetics, Protein Translocation, Trafficking
Abstract
The bacterial Sec protein translocation system catalyzes the transport of unfolded precursor proteins across the cytoplasmic membrane. Using a recently developed real time fluorescence-based transport assay, the effects of the number and distribution of positive charges on the transport time and transport efficiency of proOmpA were examined. As expected, an increase in the number of lysine residues generally increased transport time and decreased transport efficiency. However, the observed effects were highly dependent on the polylysine position in the mature domain. In addition, a string of consecutive positive charges generally had a more significant effect on transport time and efficiency than separating the charges into two or more charged segments. Thirty positive charges distributed throughout the mature domain resulted in effects similar to 10 consecutive charges near the N terminus of the mature domain. These data support a model in which the local effects of positive charge on the translocation kinetics dominate over total thermodynamic constraints. The rapid translocation kinetics of some highly charged proOmpA mutants suggest that the charge is partially shielded from the electric field gradient during transport, possibly by the co-migration of counter ions. The transport times of precursors with multiple positively charged sequences, or “pause sites,” were fairly well predicted by a local effect model. However, the kinetic profile predicted by this local effect model was not observed. Instead, the transport kinetics observed for precursors with multiple polylysine segments support a model in which translocation through the SecYEG pore is not the rate-limiting step of transport.
Introduction
In Gram-negative bacteria such as Escherichia coli, all periplasmic and outer membrane proteins are synthesized in the cytosol, and they therefore need to be transported across the cytoplasmic membrane to reach their final destination. The major route for protein translocation across or into the cytoplasmic membrane is through the Sec pathway. The core structure of the Sec machinery consists of the protein-conducting channel, SecYEG, and a peripheral ATPase, SecA. When a typical exported precursor protein emerges from the ribosome, the molecular chaperone SecB associates with the nascent chain, maintaining it in a translocation-competent state. SecA recognizes the signal sequence and targets the nascent chain to the SecYEG complex (for reviews, see Refs. 1–6). Based on x-ray crystal structure data, the ∼20–25-Å-long SecYEG channel is hourglass-shaped with a ring of hydrophobic amino acid residues at its central constriction, which is ∼5–8 Å wide (7). The pore is blocked on the periplasmic side by a plug formed by a short α-helix, which is displaced during precursor translocation (7–10).
Considerable effort has focused on explaining exactly how precursor proteins migrate through the SecYEG pore. There are two potential sources of energetic input that could influence translocation speed and directionality, ATP and the protonmotive force (PMF),3 and both are essential for efficient protein translocation (11–13). The need for ATP is outlined first. Based on numerous protease and chemical reagent accessibility studies, SecA undergoes significant conformational changes upon ATP hydrolysis (11, 13–18). The general picture that has been favored over approximately 2 decades is that ATP hydrolysis results in the translocation of a discrete segment of the polypeptide chain and that multiple similar SecA ATPase cycles are required for full precursor translocation (11, 19). In the context of the x-ray structure of the SecA-SecYEG complex, a “two-helix finger” of SecA has been proposed to be involved in driving precursor translocation through the SecYEG pore upon ATP hydrolysis (20, 21). A SecA “clamp” appears to be properly positioned to hold the preprotein at some step of the translocation process (22). Precursor length dependence studies support a model in which translocation through the pore is the slow step of transport (23, 24). Consequently, the implication of the SecA conformational cycling model is that the kinetics of translocation should be reasonably well described by a model consisting of a series of first-order reaction steps. Surprisingly, the transport kinetics observed with our recently developed real time fluorescence-based assay appear inconsistent with this stepwise translocation model (24).
The PMF is composed of the transmembrane pH and electric field gradients, the ΔpH and Δψ, respectively (25). At some point, the electric field gradient, which is typically positive in the periplasm (13, 26, 27), likely has a significant effect on precursor translocation rate due to electrophoretic effects on charge mobility. However, the influence of the electric field on the translocation of secretory proteins has been controversial. Some data are consistent with an electrophoretic explanation (28–30), whereas other data support an alternative hypothesis (31, 32). The “positive-inside” rule arises from the observation that, in the case of membrane proteins, the more positive end of a transmembrane segment is typically found on the cytoplasmic side on the membrane (33, 34). Although the influence of the basic physical constraints behind membrane protein integration and secretory protein translocation may not be completely identical due to the different results and the use of a different complement of protein complexes for these two activities (35, 36), the SecYEG channel is utilized for both, and therefore it seems reasonable that the electric field gradient could be felt similarly by membrane and secretory pre-proteins. Although an obvious explanation for the positive-inside rule is that the electric field simply orients the transmembrane span so that the charges near the end of the span are aligned in the thermodynamically most favorable configuration with respect to the field, it is clear that the explanation is not so simple (34, 37). In acidophiles, the positive-inside rule still holds, and yet the electric field gradient is reversed (32).
Conformational and kinetic influences on translocation are most likely felt locally by the precursor polypeptide, i.e. while discrete segments are interacting with elements of the Sec machinery. For example, the PMF affects the conformation and conformational stability of the SecYEG complex (38–40) and the SecA affinity for SecYEG (13) and ADP (41). These effects can in turn influence the rate at which a precursor transports through the channel or how it interacts with the channel and thus would constitute indirect effects of the PMF on precursor translocation. Alternatively, local structures within the precursor protein likely affect the translocation of polypeptide segments. For example, changing the spacing between two hydrophobic segments in proOmpA resulted in an altered digestion pattern when a protease was added during transport (42), suggesting that a hydrophobic segment is transported more slowly or less efficiently, possibly because a decision needs to be made whether the segment is a transmembrane span (43). A Δψ-dependent effect on charged segments is only expected to be felt during translocation of the charged region through the polypeptide pore, where local transient secondary motifs and/or interactions with the translocation system could modulate the electric field effect. Such local kinetic effects could significantly complicate overall translocation kinetics and cause significant deviation from the most thermodynamically favored result.
Here, we report the first methodical examination of the effects of the location and distribution of positive charges within the mature domain of a precursor protein. Our recently developed fluorescence-based assay with 1 s time resolution (24) provided transport efficiencies and detailed kinetic profiles of the translocation process for numerous charge mutants of proOmpA. The effect of charge was clearly dependent on where in the mature domain they were introduced. Charges distributed throughout the mature domain had significantly lower effects on translocation than consecutive charges, indicating the higher importance of local kinetic influences rather than overall thermodynamics. The transport times of precursors with multiple polylysine sites were largely predicted from the transport times of precursors with single polylysine sites. Surprisingly, however, the kinetic profiles of these mutants do not support a multistep translocation model.
EXPERIMENTAL PROCEDURES
Proteins
Wild type proOmpA-HisC was described previously (24). Lysine residues were introduced at the locations indicated in the figures via inverse PCR (44, 45). The coding region of all plasmid constructs was confirmed by DNA sequencing. Precursor proteins were purified as described earlier (24), with the exception that protein expression was initiated when the A600 reached ∼2. Fluorescent proteins were labeled with Atto565 maleimide (24). Biotinylated proOmpA was made similarly using N-(3-maleimidylpropionyl)biocytin (Invitrogen) but without gel purification of the precursor length protein. SecA and SecB were purified and quantified as described previously (24).
IMV Preparation
Inverted membrane vesicles (IMVs) were prepared from E. coli strain MC4100 using a modified form of pET610 (a gift from Arnold Driessen) (46). To eliminate any possible effects of Ni2+ binding to SecY, the His6 tag on SecY encoded by pET610 was removed by inverse PCR yielding pΔHis-SecY. SecYEG was induced with 0.5 mm isopropyl 1-thio-β-d-galactopyranoside at 37 °C for 2 h when the A600 reached ∼2, and IMVs were purified as described earlier (24), with the exception that 10 mm EDTA was added during lysozyme treatment.
Transport Reactions
Import reactions were performed essentially as described previously (24), except that an ATP-regenerating system was used. In short, import reactions consisted of precursor protein (10 nm, unless otherwise noted), IMVs (A280 = 1.0), 8 μm SecB, and 200 nm SecA (concentrations based on monomeric forms) in import buffer (10 mm KH2PO4, 5 mm MgSO4, 10 mm NaCl, 250 mm sucrose, 0.4 mg/ml bovine serum albumin, 1 mm β-mercaptoethanol, pH 7.5). A constant ATP concentration (1 mm) was maintained with an ATP-regenerating system (5 mm creatine phosphate, 0.2 mg/ml creatine phosphokinase). Succinate (5 mm) was added 30 s before ATP addition to induce a PMF, unless otherwise indicated. Transport of Atto565-labeled precursor proteins was assayed with an SLM-8100 spectrofluorometer, as described previously (24). Excitation and emission wavelengths were 565 nm (4 nm slits) and 590 nm (8 nm slits), respectively. Reactions were initiated by ATP addition, unless otherwise indicated. NeutrAvidin and NeutrAvidin-horseradish peroxidase conjugate (NeutrAvidin-HRP) were from Invitrogen.
Transport Time and Transport Efficiency Calculations
In general, the transport kinetics for the mutant proteins exhibited a small upward base-line drift, for unknown reasons. Therefore, the transport kinetics were fit to an exponential plus a linear base-line, i.e. fluorescence = a + be−kt + mt, where a (base-line), b (pre-exponential factor), k (rate constant), and m (slope) are constants, and t is time. Because the base-line drift was linear and not exponential, as would be expected for a slow component of transport, it is unclear if the base-line drift reflected instability of the sample or slow transport. Therefore, the transport time was defined as the time constant of the exponential (τ = 1/k), and the fluorescence changes due to the base-line drift were not included in transport efficiency calculations. Thus, the transport efficiency = A/(A + B + C), where A = the total fluorescence change explained by the exponential component of the fitted data (i.e. given by the absolute value of the pre-exponential factor), B = the total fluorescence change due to the base-line drift, and C = the total fluorescence change resulting from EDTA addition, which was used to estimate the amount of nontransported precursor protein (24). This is considered a conservative estimate of the transport efficiency, which in some cases is significantly higher when the base-line drift is included (see supplemental material).
Linear Transport Velocity Model
During translocation through the SecYEG pore, polylysine segments may locally affect the translocation of the lysines and the surrounding peptide. More precisely, proOmpA appears to translocate at a relatively constant rate (23, 24), and thus, a polylysine sequence may alter the translocation rate of a segment of the polypeptide, due, for example, to some combination of electrophoretic, electrostatic, and conformational effects. If the effects are local, then the translocation of nK (n lysines) at one site should not have any influence on the translocation of a distant nK segment. We assumed that a 5K sequence locally affects the translocation of a polypeptide segment ∼20 residues long. This includes the 5K segment, as well as an extended chain length on each side of the lysines capable of extending across the bilayer. With these basic principles, the transport time, τ, is predicted as shown in Equation 1,
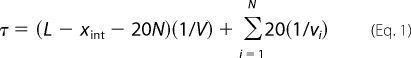 |
where L is the total precursor length (which is 356 for proOmpA-HisC); xint is the x-intercept of a plot of transport time against precursor length (which is 61 for proOmpA-HisC) and suggests that an early segment of the polypeptide chain is transported very quickly through the SecYEG channel (24); N is the number of 5K segments; V is the average linear transport velocity of the entire polypeptide chain (calculated from the wild type transport time); and vi is the average linear transport velocity of the 20-residue segment influenced by the ith 5K segment. Values for vi were calculated from the single 5K mutant data in Fig. 1B (summarized in supplemental Table S1).
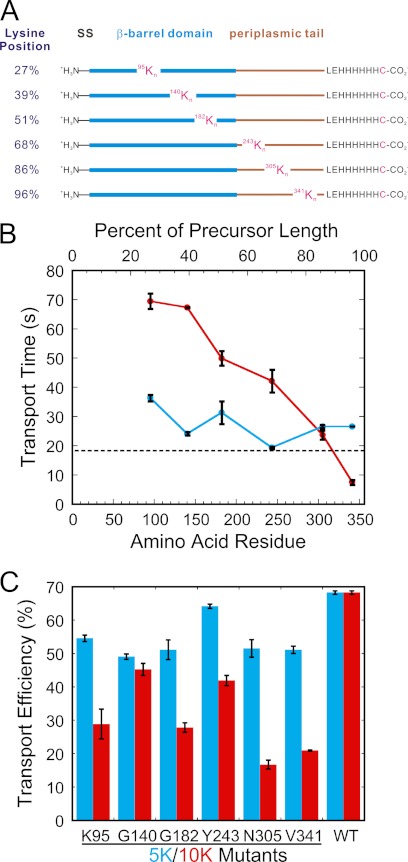
FIGURE 1.
Design of the 5K and 10K polylysine mutants, and summary of their transport times and transport efficiencies. A, positions of mutations. Segments of wild type proOmpA-HisC were replaced with n = 5 or 10 consecutive lysines. Mutation sites are identified by residue number and percent of total precursor length from the N terminus. SS, signal sequence. B, transport times. The transport times (τ) obtained from the fits in supplemental Fig. S2 are plotted against the position of the mutated segment (5K, blue; 10K, red). The wild type transport time is indicated by a dashed line. C, transport efficiencies. Transport efficiencies of the 5K (blue) and 10K (red) mutants are compared with that of the wild type protein (WT). Transport efficiencies were calculated as described under “Experimental Procedures.” The wild type controls were identical for the 5K and 10K mutants because the same batch of IMVs was used for all data.
Errors
Error bars are reported as means ± S.E. Transport times and translocation efficiencies varied for different IMV preparations, although only high translocation efficiency IMV preparations were used for the reported experiments. The same batch of IMVs was used for each mutant data set within each figure panel, and a wild type control is provided for each IMV preparation used. Values compared between panels may be different due to different IMV preparations.
RESULTS
Mutant Design and Experimental Approach
To probe how positive charge affects transport time and efficiency, we introduced lysines at six approximately equally spaced locations within the mature domain of proOmpA. We varied both the number of lysines introduced at a particular site, as well as the number of sites at which lysines were introduced. In all cases, the wild type residue was mutated to lysine, so that the total length of each precursor protein was identical (356 residues). Sites are identified both by the residue number of the first residue that was mutated and the percent of the total length that this position represents (Fig. 1A).
Precursor transport rates and transport efficiencies were measured by a recently described real time fluorescence-based transport assay (24). In short, overexpressed and purified proOmpA mutants were tagged at the C terminus with the Atto565 dye. The dye's fluorescence was initially quenched by Ni2+, which bound to a His6 tag adjacent to the dye labeling site. During translocation into IMVs, the Ni2+ was removed, presumably stripped off during translocation through the SecYEG pore, resulting in an increase in fluorescence. A control experiment performed in the absence of Ni2+ was used to correct for dye self-quenching that occurs upon transfer into the vesicle lumen. Dequenching the fluorescence of the untransported protein with EDTA, which removes/chelates the Ni2+, allowed estimation of the transport efficiency, as described previously (24). With this approach, transport efficiency is defined as the percentage of the precursor protein that was added to the assay that ultimately transported across the IMV membrane into the lumen.
Transport of 5K and 10K Mutants
It was previously concluded that the insertion of a stretch of five consecutive lysine residues (5K) at position 249 had essentially no effect on proOmpA transport but that 10 lysines (10K) almost completely blocked transport (31). We first sought to determine whether similar results would be obtained no matter where the mutations were made in the mature domain of proOmpA. Mutations were made at the six positions discussed earlier (Fig. 1A). In principle, a significant number of consecutive lysine residues could provide a substantial barrier to translocation as the transfer of positively charged resides through the SecYEG pore occurs against the electric field gradient. To illustrate the local thermodynamic barrier relative to the total thermodynamic barrier, an estimate of the amount of total charge translocated is plotted against the amino acid position for each of the 5K and 10K mutants tested (supplemental Fig. S1).
Raw data for the 5K and 10K mutants is shown in supplemental Fig. S2, and summarized in Fig. 1, B and C. At first glance, the transport time versus position plots for the 5K and 10K mutants reveals approximately linear correlations, with a significantly steeper slope for the 10K mutants (Fig. 1B). However, this is a substantially oversimplified picture as the error bars are well outside a strict linear correlation. For example, the transport times of Lys95-5K-proOmpA, Tyr243-5K-proOmpA, and Val341-5K-proOmpA clearly do not lie on a straight line (Fig. 1B). These data therefore indicate a nonlinear position dependence of 5K on transport time. The position dependence of 10K on transport time was quite dramatic, with a transport time range from ∼10 to ∼70 s. A 10K segment significantly slowed transport time when located near the N-terminal end of the mature domain (Fig. 1B). All of the 5K mutants exhibited a slightly decreased transport efficiency (∼70–95% of wild type, Fig. 1C and supplemental Fig. S3A). Intriguingly, the 10K mutants with the shortest transport times yielded the lowest transport efficiencies (∼20–30% of wild type, Fig. 1C and supplemental Fig. S3B).
Transport of 2x5K Mutants
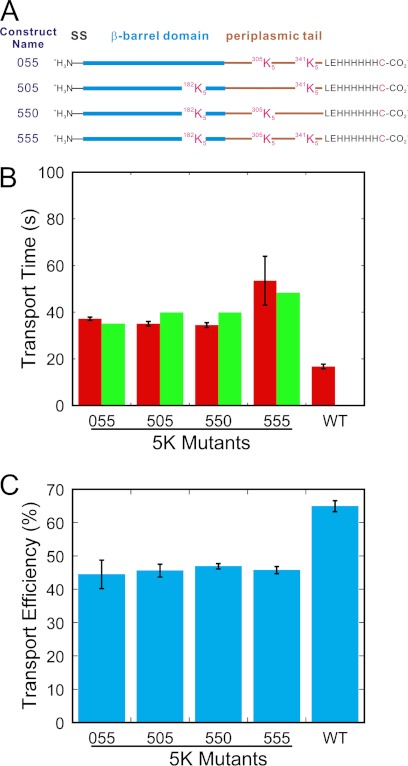
The total thermodynamic barrier to translocation of a precursor protein with 10 consecutive charged residues against the Δψ is identical to that for transporting the same protein with these 10 charged residues distributed nonconsecutively within the mature domain. To investigate the role of overall thermodynamics in influencing transport time and transport efficiency, we constructed proOmpA mutants with two 5K sequences. We chose three positions, Gly182, Asn305, and Val341, for which the single 5K mutants exhibited similar transport times and translocation efficiencies (Fig. 1, B and C). Using these three amino acid positions, we constructed the three possible 2x5K mutants, and, for completeness, the 3x5K mutant. All three 2x5K mutants yielded similar transport times (∼40 s) and transport efficiencies (∼70% of wild type). The 3x5K mutant yielded a similar translocation efficiency but a slower transport time (Fig. 2 and supplemental Fig. S4). The properties of the 10K mutants do not appear to be useful for predicting the properties of the 2x5K mutants. For example, the Asn305 and Val341 10K mutants had relatively low transport efficiencies and short transport times. In contrast, the 055-proOmpA mutant, which has a 5K segment beginning at each of these positions, had a higher transport efficiency and longer transport time. These data therefore support the hypothesis that the overall thermodynamics of transporting against a Δψ is not the major influence on the transport time and transport efficiency of these mutants.
FIGURE 2.
Transport kinetics of the 2x5K and 3x5K mutants. A, design of the mutants. SS, signal sequence. B, transport times. Shown in red are the transport times (τ) obtained from the fits in supplemental Fig. S4. Transport times predicted based on the linear transport velocity model (see text) are shown in green. C, transport efficiencies.
Although our data do not support a picture wherein the overall thermodynamics is the major determining factor for transport time, the introduction of a 5K sequence within proOmpA does yield a position-dependent effect on transport time. Previous investigators found that the length of a polylysine segment influences translocation rate (31), presumably by a partial or full arrest during translocation of the polylysine segment through the SecYEG pore. We therefore tested whether the effects of 5K mutations on transport time were additive using a linear transport velocity model (see “Experimental Procedures”). The transport times predicted by this linear transport velocity model are indicated by the green bars in Fig. 2B. All the 2x5K and 3x5K transport time data agree with this model.
Transport of Nx3K Mutants
According to the linear transport velocity model, a translocation pause occurs at each polylysine segment due to the difficulty of translocating such sequences through the SecYEG pore. This agrees with the conclusion that long polylysine segments can cause the precursor to become trapped within the translocon at (or near) the position of the polylysine (31). Whereas longer polylysine segments could completely block transport, shorter such segments may simply reduce transport rate and efficiency. A pause in the translocation process should produce a lag phase at the beginning of the bulk translocation kinetics. A series of multiple pauses should give a longer lag phase and therefore should be more easily detected. The basic principle here is that the kinetics should follow a model consisting of a series of first-order steps, the more steps, the longer the lag phase as shown in Scheme 1,
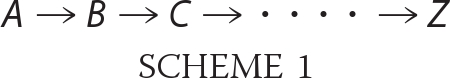 |
In Scheme 1, the number of intermediates is determined by the number of pause sites. Thus, the transport of wild type proOmpA, which yields single exponential kinetics, is well described by a single step model. One pause is predicted to require a two-step model, if the pause is of an appropriate duration. Similarly, n pauses should require an (n + 1)-step model.
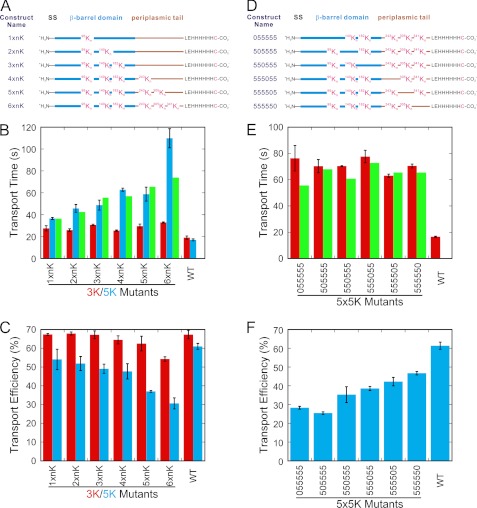
Although the 2x5K transport time data agrees with this linear transport velocity model, we further tested these ideas by creating a series of precursor proteins with an increasing number of polylysine sites. To introduce a large number of potential pause sites with a minimal effect on transport efficiency, we introduced three consecutive lysine residues (3K) as potential pause sites. Using the same six positions in the mature domain identified in Fig. 1A, we introduced up to six 3K sites (Fig. 3A). All of the 3K mutants exhibited a moderately longer transport time (∼25–30 s versus 19 s for wild type), indicating that the introduced lysines had an effect on transport (red bars in Fig. 3B). Transport efficiency was essentially unaffected for all except the 6x3K mutant, for which transport efficiency was only moderately reduced (red bars in Fig. 3C). The transport kinetics for all the 3K mutants were well fit by a single exponential. If discrete pauses were introduced by the 3K sites, they were insufficient to yield a detectable lag phase (supplemental Fig. S5).
FIGURE 3.
Transport characteristics of proOmpA mutants with multiple polylysine segments. A, design of the Nx3K and Nx5K polylysine mutants. Segments of wild type proOmpA-HisC were replaced with n = 3 or 5 consecutive lysines. SS, signal sequence. B, transport times (τ) of the Nx3K (red) and Nx5K (blue) mutants obtained from the fits in supplemental Figs. S5 and S6. Predicted transport times for the Nx5K mutants based on the linear transport velocity model (see text) are shown in green. C, transport efficiencies of the Nx3K (red) and Nx5K (blue) mutants. For B and C, the different values for the wild type controls are a consequence of the different batches of IMVs used for the 3K and 5K data sets. D, design of the 5x5K mutants. E, transport times (τ) of the 5x5K mutants (red) obtained from the fits in supplemental Fig. S8. Predicted transport times based on the linear transport velocity model (see text) are shown in green. F, transport efficiencies of the 5x5K mutants.
Transport of Nx5K Mutants
We next tested whether a series of 5K sites would be more effective in producing a lag phase in the transport kinetics. We introduced up to six 5K sites, as we did earlier for the 3K sites (Fig. 3A). Transport time generally increased as the number of 5K sites increased, supporting the hypothesis that the 5K sites are more effective at introducing translocation pauses than the 3K sites (compare red and blue bars in Fig. 3B). With the exception of the 6x5K mutant, the transport times are well predicted by the linear transport velocity model discussed earlier (green bars in Fig. 3B). Transport efficiencies decreased in the presence of a higher number of 5K sites (blue bars in Fig. 3C).
The Nx5K mutants sometimes exhibited an initial lag phase in their transport kinetics (supplemental Fig. S6). This was expected, according to Scheme 1. Therefore, the transport kinetics were fit to a Scheme 1 kinetic model with n identical rate constants (n = 1–7; supplemental Fig. S7). The best fit was typically n = 1–2. This was surprising, because the transport time predictions of the linear transport velocity model fit the data reasonably well (Figs. 2B and 3B). Therefore, the kinetic profiles indicate that the polylysine mutants do not follow a single turnover stepwise translocation model in which polylysine segments cause rate-limiting pauses during translocation through the pore. More succinctly, although transport times largely agree with the linear transport velocity model, the kinetic profiles do not support this model.
Transport of 5x5K Mutants
Because the data in Fig. 1B indicated that individual polylysine segments had position-dependent effects, we next sought to further investigate whether multiple polylysine segments exhibited such effects. The data in Fig. 2 suggested that this was not the case. Therefore, we constructed a series of six 5x5K mutants using the same six mature domain positions as before, with a 5K site in all but one position (Fig. 3D). The transport times of these six 5x5K mutants were all fairly similar (∼60–80 s) and were reasonably well predicted by the linear transport velocity model (Fig. 3E). The transport efficiency decreased as the 5K sites were moved toward the C terminus (Fig. 3F), suggesting that C-terminal positive charges have a more detrimental effect on transport efficiency. Lag phases were small, if observed, even for the same 5x5K mutant tested earlier that did exhibit a lag phase (compare supplemental Figs. S7E and S9F), despite the fact that similar transport efficiencies and transport times were observed in both experiments. We attribute these differences to IMV batch dependences, based on nine different measurements using four different IMV preparations. For completeness, we fit the data to the Scheme 1 kinetic model (supplemental Fig. S9). In all cases, the best fit was for n = 1–2, again indicating that the Scheme 1 kinetics predicted by the linear transport velocity model are not supported by the data.
Correlation between Transport Time and Transport Efficiency
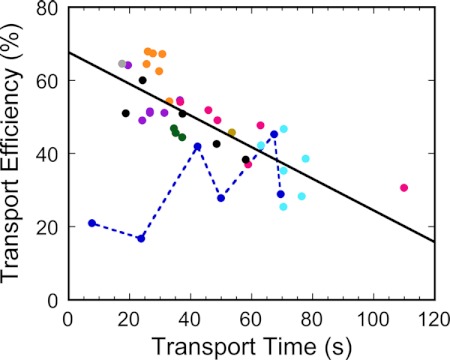
There is a moderate negative correlation (R2 = 0.60) between transport time and transport efficiency (Fig. 4). One possibility is that a precursor protein has a cumulatively increased probability to abort transport the longer it interacts with the transport machinery. There are two significant outliers, the Asn305-10K and Val341-10K mutants. In fact, for the 10K mutants, transport time and transport efficiency appear positively correlated (dashed line in Fig. 4). Transport time and transport efficiency are moderately correlated with the total charge added to proOmpA-HisC (R2 = 0.63 and 0.27, respectively; supplemental Fig. S10).
FIGURE 4.
Correlation between transport time and transport efficiency for all of the proOmpA mutants reported here and for precursors of different lengths (see “Discussion”), as reported earlier (24). The solid line is the linear fit (R2 = 0.60) not including the two outliers (Asn305-10K and Val341-10K mutants). If all points are included, R2 = 0.25. The slope and R2 values are virtually identical for the polylysine mutants and for the precursors of different lengths. Data from the 10K mutants are connected by the dashed line. Gray, wild type; purple, single 5K mutants; dark blue, 10K mutants; green, 2x5K mutants; brown, 3x5K mutant; orange, Nx3K mutants; pink, Nx5K mutants; light blue, 5x5K mutants; and black, precursors of different lengths. Error bars are omitted for clarity.
Polylysine Mutants Do Not Get Trapped in the Translocon
Nouwen et al. (31) obtained a low transport efficiency for a 10K precursor protein and concluded that the polylysine segment caused the precursor protein to become trapped in the translocon. We therefore tested whether our highly charged polylysine mutants form stable translocation intermediates that block transport. We addressed this possibility as follows. A mutant unlabeled precursor protein was incubated with IMVs under transport conditions at a concentration (500 nm or 3 μm) exceeding that of functional translocons (24). If the polylysine-containing precursor protein became trapped in the translocon, further transport activity should have been blocked. However, the transport of wild type fluorescent proOmpA-HisC was not blocked by the unlabeled Nx5K mutants or the Gly182-10K mutant, even after a short (2 min) incubation time, long enough for the polylysine precursors to be transported but short enough to minimize the possibility for spontaneous clearance (Fig. 5, A and B). These data indicate that under fully energized transport conditions, the polylysine mutants do not become trapped within the translocation channel. Moreover, each of the polylysine mutants exhibited fluorescence self-quenching, as was described earlier for the wild type precursor protein (24), consistent with transport into the IMV lumen. Thus, we conclude that all of our polylysine mutants were able to fully translocate through the SecYEG translocon.
FIGURE 5.
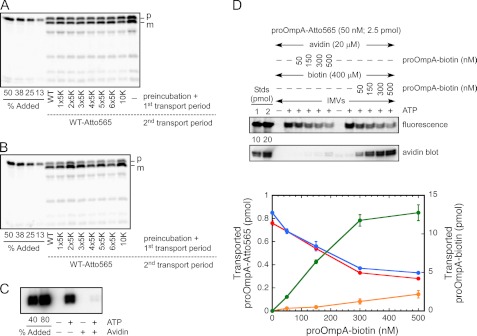
Effect of polylysine mutations and bound avidin on the translocation of wild type proOmpA. A, preincubation with 500 nm polylysine mutants. IMVs were preincubated with the indicated precursor protein (500 nm, nonfluorescent) for 5 min at 37 °C. Transport was initiated by the addition of 1 mm ATP and 5 mm succinate. After 2 min at 37 °C (first transport period), 50 nm wild type proOmpA-HisC-Atto565 (WT-Atto565) was added, and transport was continued for another 2 min (second transport period). The fluorescent translocated protein was analyzed after proteinase K treatment by SDS-PAGE and direct in-gel fluorescence using a phosphorimager. The leftmost 4 lanes are standards calibrated to the total amount of proOmpA-HisC-Atto565 added, as indicated. The rightmost lane is a control indicating the amount of transport when no precursor was present in the preincubation period and first transport period. Transport reactions in the middle lanes contained the indicated precursor protein in the preincubation period and the first transport period. These data demonstrate that the Gly182-10K mutant and the Nx5K mutants had no significantly different effect on transport than wild type proOmpA. Thus, they did not block the SecYEG translocons. B, preincubation with 3 μm polylysine mutants. Earlier work indicated that the functional SecYEG concentration at the IMV concentration used in A was ∼100–400 nm (24). To test for the possibility that transport blockage was not observed in A due to a significant underestimate of the SecYEG and endogenous SecA concentrations, the experiment was repeated by preincubating with 3 μm of the indicated precursor proteins. Other experimental conditions were the same as in A, except that both transport reaction periods were 15 min instead of 2 min. These data clearly demonstrate that the tested precursor proteins do not become trapped in the SecYEG translocation channel under these conditions. Precursor (p) and mature (m) length bands are indicated. C, effect of avidin on the transport of biotinylated proOmpA. Transport reactions were performed with proOmpA-HisC-biotin (500 nm) in the presence or absence of NeutrAvidin (20 μm) and ATP (1 mm) as indicated. Reactions were performed at 37 °C for 10 min. Samples were treated with proteinase K (1.3 μg/ml) at RT for 45 min and quenched with PMSF (20 mm). D, competition between biotinylated and unbiotinylated proOmpA in the presence and absence of avidin. The fluorescent precursor (proOmpA-HisC-Atto565) concentration was fixed at 50 nm, and the biotinylated precursor concentration (proOmpA-HisC-biotin) was varied from 0 to 500 nm. Transport of the two proteins was determined by in-gel fluorescence imaging or by probing a blot with avidin-HRP conjugate, respectively. Reagents were added in the order indicated from top to bottom. Biotin was added before or after the biotinylated precursor. Due to the high biotin-avidin affinity, the precursor-avidin complex could only form in the latter situation. Unless otherwise indicated, conditions were identical as in C. The graph shows the average transport yields from four independent experiments under “avidin-bound” and “avidin not bound” conditions, which reflect whether biotinylated precursor was added before or after free biotin, respectively, and hence whether proOmpA-biotin was bound or not bound to avidin. The left axis corresponds to the quantification of fluorescence scans reflecting the transported proOmpA-Atto565 under avidin-bound (red) and avidin not bound (blue) conditions. The right axis corresponds to the quantification of avidin blots reflecting the transported proOmpA-biotin under avidin-bound (orange) and avidin not bound (green) conditions.
SecYEG Translocons Are Not Blocked by a Translocation-incompetent Precursor
As a further test to determine whether the SecYEG pore could be blocked under our transport conditions, we attached a biotin moiety to wild type proOmpA, and then blocked transport with avidin (Fig. 5C). The basic principle of this experiment is that due to the strong biotin-avidin interaction, proOmpA is not translocated due to the large unfoldable avidin domain attached to the C terminus of the precursor. The expectation was that the precursor-avidin construct would get trapped within the SecYEG pore. To investigate whether pore blockage occurred, we examined the transport of a translocation competent precursor, e.g. one that does not have avidin attached, under conditions in which the translocation-incompetent precursor is expected to block the pore. However, there are potentially two inhibitory effects, simple competition (for binding sites) and pore blockage, that need to be distinguished. We examined this by determining the transport of fluorescent precursor as a function of increasing concentrations of biotinylated precursor in the presence and absence of bound avidin (Fig. 5D). We found that transport is inhibited by the precursor-avidin complex, but the inhibition can be entirely explained by competition; pore blockage does not seem to occur. This result was unexpected. However, this is exactly the result that was obtained earlier for the twin arginine translocation (Tat) machinery, for which the interpretation was that transport could be aborted (47, 48).
DISCUSSION
This study reports the application of a fluorescence-based transport assay to examine the influence of a positive charge on the transport time and transport efficiency of proOmpA by the Sec protein export machinery. This is the first report that systematically investigates the influence of the number and distribution of positive charges throughout the mature domain of a precursor protein. The high time resolution of a novel real time kinetic assay is essential for testing the predictions of mechanistic models. The data support the following major conclusions. First, multiple lysine residues inhibit precursor translocation, and more lysine residues are more inhibitory (Fig. 1C). Second, consecutive lysine residues are more inhibitory than an equivalent number of lysine residues distributed over multiple locations (compare Figs. 1C and 2C). Third, the effect of polylysine sequences is dependent on position within the mature domain (Fig. 1, B and C). Fourth, overall the transport efficiency is negatively correlated with transport time (Fig. 4). Fifth, the effects on transport time resulting from 5K mutations at individual sites are largely additive, assuming that preprotein segments transport at position-dependent linear rates (Fig. 3, B and E). Sixth, the translocation kinetics are typically approximately exponential even in the presence of multiple rate-influencing polylysine segments (supplemental Figs. S2 and S4–S9). Seventh, under fully energized conditions, precursors do not get trapped within the SecYEG translocation pore, they either translocate or abort transport (Fig. 5). The implications of all of these conclusions are now discussed.
The movement of positive charge against a Δψ is thermodynamically unfavorable. The data presented here indicate, however, that the total electrophoretic interaction of positive charges in the precursor protein with the Δψ is not the major factor controlling the translocation behavior of the precursor protein. For example, the 10K mutants had widely varying transport times and transport efficiencies (Fig. 1), the 6x3K mutant behaved similarly to the 5K mutants, which have less than one-third the total positive charge (Figs. 1 and 3), and the 6x5K mutant behaved similar to the Lys95-10K mutant (Figs. 1 and 3). One interpretation is that the Δψ thermodynamic barrier is not that imposing for the process of moving a precursor protein across a membrane. However, an alternative and more likely possibility is that counter ions within the hydration shell of a precursor protein can migrate with charged residues through the translocation channel. This picture is consistent with recent work that demonstrated the facile transport of 18-Å rigid tags, indicative of a large channel (49), and it agrees with experiments that demonstrated ion and water flux through an open channel (50). Note that a narrow channel that requires dehydration would be energetically expensive. If counter ions do indeed move with charged residues through the translocation channel, sliding of the precursor protein in both directions could be relatively facile. Backsliding has been demonstrated experimentally (51). Note that the total charge translocated does not have to be entirely compensated, on average, by co-migrating counter ions. Thus, electrophoretic influences could still be felt by the precursor protein, although not as strongly as might be predicted based on the charge distribution in the linear sequence of the precursor.
Considering that the cause of the observed effects of polylysine cannot be simply accounted for by electrophoretic influences, a different mechanism is required to explain our results. The data in supplemental Fig. S10 support a role for electrostatics in influencing transport time and transport efficiency because about half of the variability in these parameters is explained by total positive charge. One possibility is that a lysine side chain exhibits a stronger interaction with elements of the translocation system than other side chains. This interaction could be electrostatic (e.g. the amino group interacts with one or more negatively charged domains) or through a hydrophobic face or pocket (e.g. via the aliphatic carbons). Polylysine sites of a translocating preprotein could also interact electrostatically with the negatively charged membrane lipids. Such lipid interactions have been found to explain the “positive inside” rule for membrane proteins (52, 53). To explore the effects of electrostatics, we examined the transport kinetics of the Nx5K mutants in the presence of a high salt (200 mm NaCl) concentration (supplemental Fig. S11). Under such conditions, electrostatic interactions should be largely shielded. Although the transport of wild type proOmpA was about 2-fold slower under these high salt conditions, the effects of 5K segments on transport time and transport efficiency were largely eliminated. These data therefore suggest that the effects of polylysine segments are largely determined by electrostatics.
The electrostatic effects of polylysine segments on precursor transport could be manifested locally by influencing polypeptide translocation through the SecYEG pore. Such local effects could potentially be additive, such that multiple distant polylysine sites could produce independent translocation pauses. This is the basis behind the linear transport velocity model, which predicts the observed transport times fairly well (Figs. 2B and 3, B and E). In only two cases did the transport time predictions fail, for the 6x5K mutant (Fig. 3B) and, to a lesser extent, for the 055555 mutant (Fig. 3E). The problem is that this model also predicts that the kinetics should follow Scheme 1. We did not observe the extended lag phase predicted by Scheme 1. Thus, the linear transport velocity model appears incorrect.
One explanation for the approximately single exponential kinetics observed is that the transport time could be primarily determined by the precursor release rate from SecA and/or SecB. Because the polylysine sites introduced here are at least 74 residues from the signal sequence, it seems unlikely that the polylysine mutations affect the interaction of SecA with the signal sequence. However, covalently attached signal sequences are not required for binding to the SecA-SecYEG complex (54), suggesting direct interaction of SecA with mature domains. Thus, mature domain polylysine stretches could interfere with SecA binding affinity. While the interactions of SecA with the proOmpA mature domain are uncertain, SecB clearly interacts directly with the OmpA mature domain. Compared with other amino acids, lysine residues have a moderate affinity for SecB (55). Thus, the polylysine mutants could have a higher affinity for SecB, and a slower release from SecB could lead to a slower transport rate. A rate-limiting slow release from SecB dependent on the number and length of polylysine sites would yield the approximately single exponential kinetics that we observed. A protease protection assay revealed that a 10K peptide has an affinity of ∼1 μm for SecB, whereas polylysines of ≤7 residues interact significantly weaker (56, 57). The significantly stronger affinity of SecB for 10K versus 5K polypeptides could potentially explain the differences in transport times and transport efficiencies observed for our 5K and 10K proOmpA mutants (Fig. 1). However, considering that precursor affinities for SecB are typically in the 10–50 nm range (58, 59), a 10K affinity of 1 μm may not have a significant effect on precursor off-rate. Nonetheless, multiple 3K or 5K sites could yield significantly stronger SecB affinities due to avidity, especially if the binding motifs include nearby aromatic residues (55). We addressed this issue directly for some of the Nx5K mutants by performing transport reactions in the presence and absence of SecB. We found that transport is not substantially faster without SecB (supplemental Fig. S12A). Thus, SecB release does not appear to be the rate-limiting step of transport. Because SecB is required for efficient transport of proOmpA (supplemental Fig. S12B), we conclude that the observed kinetics are dominated by the behavior of the SecB-bound form of proOmpA.
Transport efficiencies could potentially be affected by the solubility of proOmpA mutants. The solubility of wild type proOmpA and all of the Nx5K and 10K mutants in the presence of SecA and SecB is ∼40–50% (supplemental Fig. S13). These data suggest that a lower solubility is not responsible for the lower transport efficiency of the 5x5K and 6x5K mutants (Fig. 3C) or the wide range of transport efficiencies of the 10K mutants (Fig. 1C). Moreover, Fig. 4 indicates that transport efficiency is generally negatively correlated with transport time, i.e. a longer transport time generally results in a lower transport efficiency. Lower solubility in and of itself is not expected to affect transport time. Thus, an alternative explanation for lower transport efficiencies is needed. An important clue comes from Fig. 5D, which indicates that a translocation incompetent precursor can be competitive for transport, and yet not block the translocation pore. This can happen if the proOmpA-avidin complex binds to the Sec translocation system but then aborts transport. A high probability to abort transport implies a low transport efficiency of a single attempt, and it predicts a slower overall transport rate. If precursors that abort transport do not always return for another translocation attempt, a lower overall transport efficiency will be observed. This picture therefore explains the negative correlation of transport time with transport efficiency.
This discussion therefore leads us to propose an alternate transport model, which we term the abortive diffusion model. Brownian diffusion on the nanoscale is quite fast, and it is physically reasonable that translocation through the SecYEG channel occurs on the sub-second time scale, as we discussed earlier (24) and as was originally proposed by others (60). We therefore suggest that the major reason why translocation is observed as a slow process is that the probability of aborted transport is high. For example, if transport can occur in ∼5 s (including diffusion to the SecYEG complex, binding, and translocation), and yet is only successful ∼25% of the time, the average transport time would be about ∼20 s, as we observed for wild type proOmpA. The predicted kinetics of such a transport process is approximately exponential, as we observed. In principle, the presence of polylysine segments may increase the abortive probability due to aberrant interactions with SecA, SecB, and/or the SecYEG complex, thereby leading to a slower overall transport rate that still remains approximately exponential, again in agreement with our data. Note, however, that we concluded earlier that SecB release from the precursor protein was not rate-limiting, and therefore it is not expected to influence abortive probability under our conditions. A more reasonable explanation is that polylysine segments inhibit translocation through the SecYEG channel for electrostatic reasons, e.g. either due to interactions with the SecA-SecYEG complex or due to formation of a translocation incompetent intermediate (partially folded precursor). Each time a precursor protein aborts transport, it likely can rebind to SecA/SecB and try again or achieve a transport-incompetent conformation. The latter possibility is consistent with the recent finding that a transport-incompetent form of proOmpA is trypsin-resistant and is therefore partially folded (61). Thus, an increase in abortive transport is predicted to result in slower net transport and a lower net transport efficiency, in agreement with Fig. 4. This model also predicts that ATP will be required throughout the observed kinetics because ATP will be needed for each new translocation attempt, in agreement with our earlier results (24). Although a stable SecA-SecB-precursor-SecYEG complex can be obtained in the absence of ATP and a PMF (11, 62), SecA is rapidly released from the membrane in the presence of a PMF (13), indicating complex disassembly. Thus, the lifetime of the SecA-SecB-precursor-SecYEG complex is likely quite short under fully energized conditions, as is required for the abortive diffusion model. The scatter in Fig. 4 can be explained by positive or negative effects on the abortive efficiency depending on the manner in which the precursor protein interacts with the translocation system. We note that the proposed rapid aborted transport is predicted to occur only under conditions in which the membrane remains energized. When the PMF is reduced or collapses (e.g. during sample work-up), the precursor likely becomes trapped within the SecYEG channel (13, 14, 31).
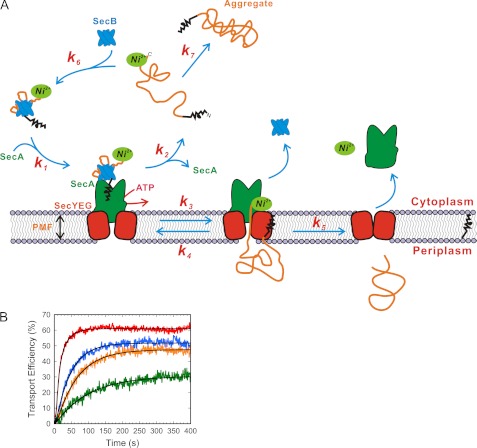
The abortive diffusion model is outlined in Fig. 6A. There are a few important features of this model. First, this model does not formally rule out stepwise transport, e.g. catalyzed by conformational cycling of the SecA ATPase. However, these conformational cycling steps would have to occur very rapidly such that the entire process is complete within a few seconds (summarized by the k3 step in Fig. 6A). Polylysine segments could inhibit the interaction of SecA with the translocating polypeptide leading to an increase in the abortive probability. Second, the model predicts that precursors with low transport efficiency do not become trapped in the translocation pore but rather are cleared from the translocon. Such rejected precursor molecules may eventually become transport-incompetent (e.g. aggregated or misfolded) and no longer able to interact with the transport system. Such translocon clearing is expected to be highly useful in vivo to remove precursor molecules that achieve transport-incompetent (partially folded) configurations during transport. Third, because the translocation step is postulated to occur primarily through random diffusion through the pore, longer precursor proteins are less likely to transport successfully on any given attempt. Thus, longer precursor proteins are expected to abort transport more frequently leading to lower transport rates, as has been observed (24, 31). Finally, the model is consistent with the kinetic profiles of our polylysine mutants. To illustrate this point, Fig. 6B shows a global fit to the translocation kinetics of wild type proOmpA and three Nx5K mutants. Although there are many fit parameters and thus they are highly underdetermined, this figure shows that the general kinetic profile of the model is consistent with our data and that different transport times and transport efficiencies are easily accommodated. Single molecule experiments would allow direct determination of the rate constants k2 and k3 and therefore would provide very useful constraints to test the model.
FIGURE 6.
Abortive diffusion model. A, kinetic scheme of the model. The precursor protein and translocation system bind to form a receptor-substrate complex. Precursor translocation occurs during the k3 step, and retrotranslocation is possible (k4). The step defined by k5 is the fluorescence dequenching step, after which the precursor is converted to the mature protein. Precursor that has aborted transport can aggregate or be converted back into translocation competent precursor (e.g. by the action/binding of SecA and/or SecB). B, global fit of the model to Nx5K mutant data. Due to the large number of kinetic constants, the fits are highly underdetermined (many sets of values yield similar fits), and thus the output values are unreliable. The main message is that an increase in abortive transport (an increased k2/k3 ratio) is sufficient to explain the reduced transport efficiencies and the apparently longer transport times for the Nx5K mutants. Note that the actual transport time of a single attempt (1/k3) does not need to change. For the fits shown (black), it was assumed that k3 = k4, but different for each precursor protein, and that k1 was the same for all precursor proteins. Neither of these assumptions is necessary to obtain similar quality fits. Data from supplemental Fig. S6 were corrected by subtracting the linear base line (see “Experimental Procedures”). The origin was determined by first individually fitting the data to the model: red, wild type; blue, 2x5K mutant; orange, 4x5K mutant; green, 6x5K mutant.
In summary, we have investigated the effect of polylysine segments on precursor transport by the Sec translocation machinery. We found that both location and charge distribution are important, demonstrating that any electrophoretic effect is substantially modulated by other structural, electrostatic, and/or interactive effects. Most importantly, the observed kinetic profiles do not support a model in which distinct polylysine segments individually produce rate-limiting pauses during translocation through the pore. Consequently, the data argue that translocation through the pore is not the rate-limiting step of transport. Although this contradicts our earlier interpretation (24), we have provided an alternative explanation that is consistent with our current and previous data. We expect that mathematical modeling and single molecule approaches will be instrumental for more precisely deciphering the molecular interactions influencing translocation behavior.
Acknowledgments
We thank Arnold Driessen for the SecYEG and SecB overexpression plasmids; Donald Oliver for the SecA overexpression plasmid; Timothy Yahr and William Wickner for the proOmpA overexpression plasmid, and Anton Zilman for informative discussions.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 GM065534 (to S. M. M.). This work was also supported by Welch Foundation Grant BE-1541 (to S. M. M.).

This article contains supplemental Table S1 and Figs. S1–S13.
- PMF
- protonmotive force
- OmpA
- outer membrane protein A
- IMV
- inverted membrane vesicle.
REFERENCES
- 1. Rapoport T. A. (2007) Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature 450, 663–669 [DOI] [PubMed] [Google Scholar]
- 2. Driessen A. J., Nouwen N. (2008) Protein translocation across the bacterial cytoplasmic membrane. Annu. Rev. Biochem. 77, 643–667 [DOI] [PubMed] [Google Scholar]
- 3. Xie K., Dalbey R. E. (2008) Inserting proteins into the bacterial cytoplasmic membrane using the Sec and YidC translocases. Nat. Rev. Microbiol. 6, 234–244 [DOI] [PubMed] [Google Scholar]
- 4. Cross B. C., Sinning I., Luirink J., High S. (2009) Delivering proteins for export from the cytosol. Nat. Rev. Mol. Cell Biol. 10, 255–264 [DOI] [PubMed] [Google Scholar]
- 5. Sardis M. F., Economou A. (2010) SecA. A tale of two protomers. Mol. Microbiol. 76, 1070–1081 [DOI] [PubMed] [Google Scholar]
- 6. du Plessis D. J., Nouwen N., Driessen A. J. (2011) The Sec translocase. Biochim. Biophys. Acta 1808, 851–865 [DOI] [PubMed] [Google Scholar]
- 7. Van den Berg B., Clemons W. M., Jr., Collinson I., Modis Y., Hartmann E., Harrison S. C., Rapoport T. A. (2004) X-ray structure of a protein-conducting channel. Nature 427, 36–44 [DOI] [PubMed] [Google Scholar]
- 8. Harris C. R., Silhavy T. J. (1999) Mapping an interface of SecY (PrlA) and SecE (PrlG) by using synthetic phenotypes and in vivo cross-linking. J. Bacteriol. 181, 3438–3444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tam P. C., Maillard A. P., Chan K. K., Duong F. (2005) Investigating the SecY plug movement at the SecYEG translocation channel. EMBO J. 24, 3380–3388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lycklama A., Nijeholt J. A., Bulacu M., Marrink S. J., Driessen A. J. (2010) Immobilization of the plug domain inside the SecY channel allows unrestricted protein translocation. J. Biol. Chem. 285, 23747–23754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schiebel E., Driessen A. J., Hartl F. U., Wickner W. (1991) ΔμH+ and ATP function at different steps of the catalytic cycle of preprotein translocase. Cell 64, 927–939 [DOI] [PubMed] [Google Scholar]
- 12. Driessen A. J. (1992) Precursor protein translocation by the Escherichia coli translocase is directed by the protonmotive force. EMBO J. 11, 847–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nishiyama K., Fukuda A., Morita K., Tokuda H. (1999) Membrane deinsertion of SecA underlying proton-motive force-dependent stimulation of protein translocation. EMBO J. 18, 1049–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Economou A., Wickner W. (1994) SecA promotes preprotein translocation by undergoing ATP-driven cycles of membrane insertion and deinsertion. Cell 78, 835–843 [DOI] [PubMed] [Google Scholar]
- 15. Economou A., Pogliano J. A., Beckwith J., Oliver D. B., Wickner W. (1995) SecA membrane cycling at SecYEG is driven by distinct ATP binding and hydrolysis events and is regulated by SecD and SecF. Cell 83, 1171–1181 [DOI] [PubMed] [Google Scholar]
- 16. Kim Y. J., Rajapandi T., Oliver D. (1994) SecA protein is exposed to the periplasmic surface of the E. coli inner membrane in its active state. Cell 78, 845–853 [DOI] [PubMed] [Google Scholar]
- 17. Ramamurthy V., Oliver D. (1997) Topology of the integral membrane form of Escherichia coli SecA protein reveals multiple periplasmically exposed regions and modulation by ATP binding. J. Biol. Chem. 272, 23239–23246 [DOI] [PubMed] [Google Scholar]
- 18. Jilaveanu L. B., Oliver D. B. (2007) In vivo membrane topology of Escherichia coli SecA ATPase reveals extensive periplasmic exposure of multiple functionally important domains clustering on one face of SecA. J. Biol. Chem. 282, 4661–4668 [DOI] [PubMed] [Google Scholar]
- 19. van der Wolk J. P., de Wit J. G., Driessen A. J. (1997) The catalytic cycle of the Escherichia coli SecA ATPase comprises two distinct preprotein translocation events. EMBO J. 16, 7297–7304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zimmer J., Nam Y., Rapoport T. A. (2008) Structure of a complex of the ATPase SecA and the protein-translocation channel. Nature 455, 936–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Erlandson K. J., Miller S. B., Nam Y., Osborne A. R., Zimmer J., Rapoport T. A. (2008) A role for the two-helix finger of the SecA ATPase in protein translocation. Nature 455, 984–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bauer B. W., Rapoport T. A. (2009) Mapping polypeptide interactions of the SecA ATPase during translocation. Proc. Natl. Acad. Sci. U.S.A. 106, 20800–20805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tomkiewicz D., Nouwen N., van Leeuwen R., Tans S., Driessen A. J. (2006) SecA supports a constant rate of preprotein translocation. J. Biol. Chem. 281, 15709–15713 [DOI] [PubMed] [Google Scholar]
- 24. Liang F. C., Bageshwar U. K., Musser S. M. (2009) Bacterial Sec protein transport is rate-limited by precursor length. A single turnover study. Mol. Biol. Cell 20, 4256–4266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Azzone G., Benz R., Bertl A., Colombini M., Crofts A., Dilley R., Dimroth P., Dutton P. L., Felle H., Harold F., Junge W., Kaback H. R., Knaff D., Krulwich T., Lodish H., Malmstrom B., Maloney P., Mannella C., Padan E., Papa S., Rottenberg H., Rudnick G., Rydstrom J., Silverstein T., Skulachev V., Slayman C., Tedeschi H., Wikstrom M., Wilson T. H. (1993) Transmembrane measurements across bioenergetic membranes. Biochim. Biophys. Acta 1183, 1–3 [Google Scholar]
- 26. Riondet C., Cachon R., Waché Y., Alcaraz G., Diviès C. (1999) Changes in the proton-motive force in Escherichia coli in response to external oxidoreduction potential. Eur. J. Biochem. 262, 595–599 [DOI] [PubMed] [Google Scholar]
- 27. Rottenberg H. (1979) The measurement of membrane potential and ΔpH in cells, organelles, and vesicles. Methods Enzymol. 55, 547–569 [DOI] [PubMed] [Google Scholar]
- 28. Geller B., Zhu H. Y., Cheng S., Kuhn A., Dalbey R. E. (1993) Charged residues render pro-OmpA potential dependent for initiation of membrane translocation. J. Biol. Chem. 268, 9442–9447 [PubMed] [Google Scholar]
- 29. Li P., Beckwith J., Inouye H. (1988) Alteration of the amino terminus of the mature sequence of a periplasmic protein can severely affect protein export in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 85, 7685–7689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cao G., Kuhn A., Dalbey R. E. (1995) The translocation of negatively charged residues across the membrane is driven by the electrochemical potential. Evidence for an electrophoresis-like membrane transfer mechanism. EMBO J. 14, 866–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nouwen N., Berrelkamp G., Driessen A. J. (2009) Charged amino acids in a preprotein inhibit SecA-dependent protein translocation. J. Mol. Biol. 386, 1000–1010 [DOI] [PubMed] [Google Scholar]
- 32. van de Vossenberg J. L., Albers S. V., van der Does C., Driessen A. J., van Klompenburg W. (1998) The positive inside rule is not determined by the polarity of the Δpsi (transmembrane electrical potential) Mol. Microbiol. 29, 1125–1127 [DOI] [PubMed] [Google Scholar]
- 33. von Heijne G., Gavel Y. (1988) Topogenic signals in integral membrane proteins. Eur. J. Biochem. 174, 671–678 [DOI] [PubMed] [Google Scholar]
- 34. Seppälä S., Slusky J. S., Lloris-Garcerá P., Rapp M., von Heijne G. (2010) Control of membrane protein topology by a single C-terminal residue. Science 328, 1698–1700 [DOI] [PubMed] [Google Scholar]
- 35. Beck K., Wu L. F., Brunner J., Müller M. (2000) Discrimination between SRP- and SecA/SecB-dependent substrates involves selective recognition of nascent chains by SRP and trigger factor. EMBO J. 19, 134–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Koch H. G., Hengelage T., Neumann-Haefelin C., MacFarlane J., Hoffschulte H. K., Schimz K. L., Mechler B., Müller M. (1999) In vitro studies with purified components reveal signal recognition particle (SRP) and SecA/SecB as constituents of two independent protein-targeting pathways of Escherichia coli. Mol. Biol. Cell 10, 2163–2173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Andersson H., von Heijne G. (1994) Membrane protein topology. Effects of ΔμH+ on the translocation of charged residues explain the “positive inside” rule. EMBO J. 13, 2267–2272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nouwen N., de Kruijff B., Tommassen J. (1996) prlA suppressors in Escherichia coli relieve the proton electrochemical gradient dependency of translocation of wild type precursors. Proc. Natl. Acad. Sci. U.S.A. 93, 5953–5957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tani K., Tokuda H., Mizushima S. (1990) Translocation of ProOmpA possessing an intramolecular disulfide bridge into membrane vesicles of Escherichia coli. Effect of membrane energization. J. Biol. Chem. 265, 17341–17347 [PubMed] [Google Scholar]
- 40. Duong F., Wickner W. (1999) The PrlA and PrlG phenotypes are caused by a loosened association among the translocase SecYEG subunits. EMBO J. 18, 3263–3270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shiozuka K., Tani K., Mizushima S., Tokuda H. (1990) The proton-motive force lowers the level of ATP required for the in vitro translocation of a secretory protein in Escherichia coli. J. Biol. Chem. 265, 18843–18847 [PubMed] [Google Scholar]
- 42. Sato K., Mori H., Yoshida M., Tagaya M., Mizushima S. (1997) Short hydrophobic segments in the mature domain of ProOmpA determine its stepwise movement during translocation across the cytoplasmic membrane of Escherichia coli. J. Biol. Chem. 272, 5880–5886 [DOI] [PubMed] [Google Scholar]
- 43. Duong F., Wickner W. (1998) Sec-dependent membrane protein biogenesis. SecYEG, preprotein hydrophobicity and translocation kinetics control the stop-transfer function. EMBO J. 17, 696–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ochman H., Gerber A. S., Hartl D. L. (1988) Genetic applications of an inverse polymerase chain reaction. Genetics 120, 621–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Dominy C. N., Andrews D. W. (2003) Site-directed mutagenesis by inverse PCR. Methods Mol. Biol. 235, 209–223 [DOI] [PubMed] [Google Scholar]
- 46. Kaufmann A., Manting E. H., Veenendaal A. K., Driessen A. J., van der Does C. (1999) Cysteine-directed cross-linking demonstrates that helix 3 of SecE is close to helix 2 of SecY and helix 3 of a neighboring SecE. Biochemistry 38, 9115–9125 [DOI] [PubMed] [Google Scholar]
- 47. Bageshwar U. K., Whitaker N., Liang F. C., Musser S. M. (2009) Interconvertibility of lipid- and translocon-bound forms of the bacterial Tat precursor pre-SufI. Mol. Microbiol. 74, 209–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Musser S. M., Theg S. M. (2000) Characterization of the early steps of OE17 precursor transport by the thylakoid ΔpH/Tat machinery. Eur. J. Biochem. 267, 2588–2598 [DOI] [PubMed] [Google Scholar]
- 49. Bonardi F., Halza E., Walko M., Du Plessis F., Nouwen N., Feringa B. L., Driessen A. J. (2011) Probing the SecYEG translocation pore size with preproteins conjugated with sizable rigid spherical molecules. Proc. Natl. Acad. Sci. U.S.A. 108, 7775–7780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Saparov S. M., Erlandson K., Cannon K., Schaletzky J., Schulman S., Rapoport T. A., Pohl P. (2007) Determining the conductance of the SecY protein translocation channel for small molecules. Mol. Cell 26, 501–509 [DOI] [PubMed] [Google Scholar]
- 51. Erlandson K. J., Or E., Osborne A. R., Rapoport T. A. (2008) Analysis of polypeptide movement in the SecY channel during SecA-mediated protein translocation. J. Biol. Chem. 283, 15709–15715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bogdanov M., Xie J., Heacock P., Dowhan W. (2008) To flip or not to flip. Lipid-protein charge interactions are a determinant of final membrane protein topology. J. Cell Biol. 182, 925–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Dowhan W., Bogdanov M. (2009) Lipid-dependent membrane protein topogenesis. Annu. Rev. Biochem. 78, 515–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gouridis G., Karamanou S., Gelis I., Kalodimos C. G., Economou A. (2009) Signal peptides are allosteric activators of the protein translocase. Nature 462, 363–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Knoblauch N. T., Rüdiger S., Schönfeld H. J., Driessen A. J., Schneider-Mergener J., Bukau B. (1999) Substrate specificity of the SecB chaperone. J. Biol. Chem. 274, 34219–34225 [DOI] [PubMed] [Google Scholar]
- 56. Randall L. L. (1992) Peptide binding by chaperone SecB. Implications for recognition of nonnative structure. Science 257, 241–245 [DOI] [PubMed] [Google Scholar]
- 57. Diamond D. L., Randall L. L. (1997) Kinetic partitioning. Poising SecB to favor association with a rapidly folding ligand. J. Biol. Chem. 272, 28994–28998 [DOI] [PubMed] [Google Scholar]
- 58. Hardy S. J., Randall L. L. (1991) A kinetic partitioning model of selective binding of nonnative proteins by the bacterial chaperone SecB. Science 251, 439–443 [DOI] [PubMed] [Google Scholar]
- 59. Randall L. L., Topping T. B., Suciu D., Hardy S. J. (1998) Calorimetric analyses of the interaction between SecB and its ligands. Protein Sci. 7, 1195–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Simon S. M., Peskin C. S., Oster G. F. (1992) What drives the translocation of proteins? Proc. Natl. Acad. Sci. U.S.A. 89, 3770–3774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Nishiyama K., Tokuda H. (2010) Preparation of a highly translocation-competent proOmpA/SecB complex. Protein Sci. 19, 2402–2408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Driessen A. J., Wickner W. (1991) Proton transfer is rate-limiting for translocation of precursor proteins by the Escherichia coli translocase. Proc. Natl. Acad. Sci. U.S.A. 88, 2471–2475 [DOI] [PMC free article] [PubMed] [Google Scholar]