Background: Induced mammalian target of rapamycin (mTOR)-dependent autophagy can restrict mycobacterial growth.
Results: Mycobacterial lipids stimulate macrophage autophagy and mTOR signaling.
Conclusion: Mycobacteria naturally induce autophagy in an mTOR-independent manner.
Significance: This work will impact the development of therapeutic strategies for Mycobacteria tuberculosis infection.
Keywords: Autophagy, Lipids, mTOR, Mycobacterium, Mycobacterium tuberculosis
Abstract
The interaction of host cells with mycobacteria is complex and can lead to multiple outcomes ranging from bacterial clearance to latent infection. Although many factors are involved, the mammalian autophagy pathway is recognized as a determinant that can influence the course of infection. Intervention aimed at utilizing autophagy to clear infection requires an examination of the autophagy and signal transduction induced by mycobacteria under native conditions. With both pathogenic and non-pathogenic mycobacteria, we show that infection correlates with an increase in the mammalian target of rapamycin (mTOR) activity indicating that autophagy induction by mycobacteria occurs in an mTOR-independent manner. Analysis of Mycobacterium smegmatis and Mycobacterium bovis bacille Calmette-Guérin (BCG), which respectively induce high and low autophagy responses, indicates that lipid material is capable of inducing both autophagy and mTOR signaling. Although mycobacterial infection potently induces mTOR activity, we confirm that bacterial viability can be reduced by rapamycin treatment. In addition, our work demonstrates that BCG can reduce autophagy responses to M. smegmatis suggesting that specific mechanisms are used by BCG to minimize host cell autophagy. We conclude that autophagy induction and mTOR signaling take place concurrently during mycobacterial infection and that host autophagy responses to any given mycobacterium stem from multiple factors, including the presence of activating macromolecules and inhibitory mechanisms.
Introduction
Autophagy is a catabolic process that can occur in response to a number of diverse stimuli. Early studies focused on the role of autophagy in the cellular response to nutrient deprivation (1). Under starvation conditions, eukaryotic cells isolate and degrade unwanted cytosolic material to generate building blocks for de novo biosynthesis (1). Given the link between autophagy and metabolism, it is not surprising that the mammalian target of rapamycin (mTOR)2 would serve as a major autophagy regulator. When nutrients are plentiful, mTOR activation leads to the phosphorylation of p70-S6 kinase, which in turn leads to the phosphorylation of ribosomal protein S6 (S6). The phosphorylation of S6 is one of several mTOR targets that lead to an up-regulation of protein translation favoring cell growth and differentiation (2, 3). Conversely, nutrient starvation, or treatment with rapamycin, causes a profound reduction in both phosphorylated p70-S6 kinase and S6, resulting in reduced protein translation. The lack of mTOR signaling shuts down anabolic processes and facilitates the formation of LC3B-II-positive autophagosomes that engulf cytosolic material and fuse with lysosomes to degrade the contents. Thus, mTOR-dependent autophagy is a key and evolutionarily conserved process that supports cell viability during times of nutrient deprivation (1).
In addition to this housekeeping function, it is now well recognized that mammalian autophagy is a potent means by which intracellular microorganisms can be sensed, housed/sequestered, and destroyed (4). Although mechanistically different, the capacity of the autophagy pathway to respond to intracellular bacteria is consistent with the description of autophagy as a stress-responsive pathway (1). Interactions between the host autophagy machinery and multiple bacteria of various species have been well documented (5). Mycobacteria, including BCG and Mycobacterium tuberculosis, have also received considerable attention. It has been demonstrated that rapamycin/starvation induced autophagy is one mechanism by which mycobacterial survival within macrophages can be restricted (6, 7). There are likely other mechanisms that link the autophagy machinery to cytosolic mycobacteria, and pro-inflammatory cytokines, Toll receptors, sequestome-like receptors, and Nod-like receptors, are some of the mechanisms that participate in overall bacterial recognition that have been documented (4, 6, 8–10). Autophagic elimination of intracellular mycobacteria occurs upon fusion of bacteria housed within LC3B-II-positive autophagsomes with lysosomes, thereby exposing mycobacteria to reduced pH, proteases, and a customized formulation of neo-antimicrobial peptides (6, 9). The digestion and killing of bacteria in this fashion liberates antigenic epitopes that can be presented on the cell surface in the context of major histocompatibility class II and I protein complexes for detection by antigen-specific CD4+ and CD8+ T lymphocytes (4, 11). As expected, the infection of dendritic cells with recombinant BCG and treatment with rapamycin has been shown to enhance the immunogenicity of mycobacterial antigens (12). These findings suggest that autophagy may represent a new strategy toward developing focused and potent immune responses to traditionally difficult intracellular targets such as mycobacteria.
Utilizing autophagy to destroy mycobacteria and make epitopes available for immune system recognition will require consideration as to how the pathogen influences autophagy and the signal transduction that surrounds the overall response. For example, M. marinum recruits LC3B to form compartments within which the bacterium can reside (13). It is well known that M. tuberculosis can perturb generic phagosome-lysosome fusion and acidification along with global host cell signal transduction (14). More recently, the eis gene from M. tuberculosis has been shown to regulate host autophagy (15). Studies performed in other organisms, including Shigella, Listeria, and Staphylococcus, have elucidated a number of mechanisms by which host cell autophagy and signal transduction can be modified by the pathogen after infection (14). Consequently, the use of autophagy to destroy mycobacteria requires an understanding of the range of natural autophagy responses to the pathogen, the inducing macromolecules, and the bacterial influence upon the autophagy machinery and key signaling pathways.
In the current study we investigate the autophagy response to mycobacterial infection under natural conditions, identify mycobacterial components that elicit autophagy in macrophages, and determine the mycobacterial influence upon mTOR signaling. Our work is the first to examine autophagy induction by mycobacteria in the absence of starvation or rapamycin, and we are the first to identify mycobacterium induced autophagy as an mTOR-independent process.
EXPERIMENTAL PROCEDURES
Mycobacteria Culture
All strains of mycobacteria used in this study were grown in 7H9 media supplemented with 0.5% glycerol, 0.05% tyloxapol, and 10% oleic acid/albumin/dextrose/catalase (OADC, referred to hereafter as 7H9-OADC). M. Kanasii (ATCC # 12478), M. fortuitum (ATCC # 6841), M. tuberculosis (H37Ra, ATCC # 25177), M. tuberculosis (H37Rv, ATCC # 25618), and M. tuberculosis (Erdman, ATCC # 35801) were purchased from the ATCC. M. smegmatis (16) and BCG Danish (Statens Serum Institute) were obtained from the laboratory of Dr. William Jacobs (Albert Einstein College of Medicine). For studies involving heat-killed M. smegmatis and BCG, the mycobacteria were washed with PBS + 0.05% tyloxapol before being aliquoted in Eppendorf tubes for heat treatment. Mycobacteria were heat-killed in PBS + 0.05% tyloxapol at 95 °C for 30 min. To confirm heat killing, heat-treated M. smegmatis samples were plated on LB plates and grown at 37 °C for 3 days.
Macrophage Culture and Infection
RAW264.7 (RAW) cells were purchased from the Duke Cell Culture Facility and grown in high glucose DMEM, 10% heat-inactivated fetal bovine serum, non-essential amino acids, sodium pyruvate, and l-glutamine. Cells were thawed 24 h prior to an experiment and allowed to recover at 37 °C and 5% CO2. For instances when starvation was induced, RAW cells were washed three times with Hanks' balanced salt solution (HBSS, Sigma H9394) and incubated in HBSS as indicated (17). For primary macrophages, bone marrow from female C57BL/6 mice was cultured in IMDM + 10% HI-FBS and L929 culture supernatant. All studies involving mice were performed in accordance with procedures set forth by the Duke University Institutional Animal Care and Use Committee. RAW cells and bone marrow-derived macrophages from female C57BL/6 mice were scraped, counted, and seeded at 1 × 105, 5 × 105, and 1 × 106 cells per well in 24-, 12-, or 6-well plates, respectively. Mycobacteria to be infected were harvested by centrifugation, washed in PBS + 0.05% tyloxapol, and sonicated to disperse. The A600 was determined, and bacteria were added to the wells to achieve the MOI indicated. For some experiments, washing and resuspension in PBS + 0.05% tyloxapol was omitted to rule out the impact of tyloxapol on observed phenotypes. In those cases, the bacteria were aliquoted, rinsed in DMEM + 10% HI-FBS, and pipetted vigorously before addition into culture. Active infection was allowed to occur for 2–4 h, after which the cells were rinsed in either PBS or media and treated with gentamicin (50 μg/ml) to kill extracellular bacteria. RAW cells were harvested at the time points indicated and processed for analysis by SDS-PAGE and immunoblot. For M. smegmatis CFU determination, infected cells were lysed in water, serially diluted, and plated on LB plates. Colonies were counted after 3 days of growth at 37 °C.
RAW GFP-LC3 Cells
RAW cells were transfected with pEGFP-LC3 using Effectene (Qiagen) according to manufacturer's instructions (18). Stable transfectants were selected and maintained in G418 (1 mg/ml). Expression and function were confirmed by flow cytometry and the formation of puncta in response to rapamycin (10 mg/ml). For experiments, RAW-GFP-LC3 cells were thawed and plated in complete medium 24 h prior to an experiment in the absence of G418. On the day of an experiment, thawed RAW GFP-LC3 cells were scraped, counted, and plated on sterilized glass coverslips. The cells were allowed to adhere before being infected or treated as described. pEGFP-LC3 was obtained from Noboro Mizushima (Tokyo Medical and Dental University, Tokyo, Japan) (19, 20).
Microscopy
Glass coverslips containing treated or infected RAW GFP-LC3 cells were rinsed with 1× PBS and fixed in 4% formalin/PBS for 10 min at room temperature. The coverslips were then permeabilized with 0.1% Triton X-100/PBS for 2 min before being stained with Hoechst. The stained coverslips were then rinsed in water and mounted on slides with Aqua Polymount (Polysciences) (18). Images were taken on Nikon TE 2000 fluorescent microscopes and analyzed with NIS Elements and ImageJ software.
DQ-BSA Degradation Assays
To quantify protein degradation induced by stimuli, the self-quenched substrate DQ-BSA was utilized as described in previous studies (13, 21–23). Upon degradation, the cleaved substrate becomes highly fluorescent and can be detected by microscopy or flow cytometry. Briefly, RAW cells were seeded and loaded with 10 μg/ml DQ-BSA for 4 h at 37 °C. The cells were then washed to remove excess label followed by autophagy induction. The cells were then cultured overnight and imaged by fluorescence microscopy or analyzed by flow cytometry. Flow cytometric analysis was performed on a BD FACSCanto, and data were analyzed with FlowJo software.
Immunoblot and Chemiluminescence
Cellular protein was prepared by washing plates with PBS and adding 1× RIPA buffer (with protease and phosphatase inhibitors) directly to the plate. The cells were scraped, transferred to microcentrifuge tubes, and allowed to extract for 15–30 min on ice with frequent vortexing. Protein was quantified with the Bio-Rad protein assay. For SDS-PAGE, 5–15 μg of cellular protein was resolved on 4–20% gradient gels at 125–150 V and then transferred to 0.2-μm PVDF at 130–160 mA for 1 h (18, 24). PVDF membranes were blocked with either 5% nonfat dry milk (NFDM) or bovine serum albumin (BSA) that was dissolved in 1× TBS + 0.1% Tween 20 (TBST). Membranes were allowed to block overnight at 4 °C before probing with specific antibodies. Anti-LC3B (1:1000, BSA), anti-phospho-S6 (1:2000, BSA), anti-S6 (1:2000, NFDM), anti-P-ULK1-Ser-757 (1:1000, BSA), anti-eIF4B (1:1000, BSA), anti-P-eIF4B-Ser-422 (1:1000, BSA) anti-GFP (1:1000; BSA), and anti-rabbit-HRP (1:3000, BSA or NFDM) were all purchased from Cell Signaling (24). Anti-actin (1:2000, NFDM) and anti-HSP90 (1:2000, NFDM) were obtained from Santa Cruz Biotechnology. Anti-mouse-HRP (1:2000, NFDM) was purchased from Genscript. After probing with primary and secondary antibodies, proteins of interest were revealed by chemiluminescence using either the Lumi-LightPlus (Roche Applied Science) or the Western Lightening Plus (PerkinElmer Life Sciences) Western blotting substrates according to the manufacturer's instructions. Films were scanned, and ImageJ was used to quantify band intensities. As described in the individual figures, band intensity of LC3B-II was related to the level of a housekeeping gene such as HSP90 or Actin (25).
Preparation of Nonidet P-40 Protein Lysates from Mycobacteria
Bacteria were harvested and washed extensively in PBS to remove media components and lysed in buffer composed of 20 mm Hepes, pH 7.4, 100 mm NaCl, 0.5% Nonidet P-40, and 10% glycerol. Glass beads were added, and the bacteria were vortexed and lysed on ice for 30 min. The bacteria were then sonicated three times for 15 s at 50% power using a Branson Digital Sonifier to release protein. Multiple lysates from the same bacteria were combined and concentrated on an Amicon Ultra Centrifugal Filter with nominal molecular weight cutoff of 10,000 until a concentration of 1–2 mg/ml was achieved. Samples of each protein preparation were resolved by SDS-PAGE and stained with Coomassie stain to confirm that protein was isolated. Protein concentrations were determined BCA assay (Pierce).
Isolation and Use of Total Bacterial Lipids
M. smegmatis and BCG were grown in either Sauton media or 7H9-OADC. After washing in PBS, the bacterial pellets were mixed with chloroform:methanol:water (10:10:3, v/v) and incubated at 50 °C for 3 h. The lower organic phase was isolated and dried down. The lyophilized lipid material was then re-extracted by first mixing with chloroform:methanol (2:1, v/v). Water was added followed by vortexing and centrifugation to separate the lower organic and upper aqueous phases. The upper phases were discarded, and the lower phase containing lipid material was dried down at 50–60 °C in a tissue culture hood. Dried lipid material was weighed and dissolved in chloroform:methanol (2:1, v/v). Lipids were spotted onto glass coverslips or directly into tissue culture dishes (diluted in petroleum ether), and the solvents were evaporated prior to the addition of the cells and media.
Statistics
GraphPad Prism was used for all analyses, and samples were analyzed by analysis of variance with a Tukey post-test. A p value of ≤0.05 was deemed to be significant.
RESULTS
Autophagy Induction by Mycobacteria Differs in Magnitude and Varies by Species
Most studies performed to date have examined autophagy induction during mycobacterial infection in the context of potent inducers, i.e. stressors, such as starvation or rapamycin treatment. A comparison, however, of the range of natural autophagy induction responses that can be triggered by mycobacteria is both necessary and relevant to understanding the host-pathogen interaction that occurs in nature. This is particularly important as the immediate events that occur as host and bacterium interact can have long term biological implications. To address this issue, mouse RAW cells were either left uninfected, or infected with M. smegmatis, M. fortuitum, M. kanasii, H37Ra, or BCG for 3 h. Protein lysates were prepared, and Western blotting for either control HSP90 or β-actin and LC3B I/II was performed. Because mycobacterial infection can induce a myriad of signaling and cytoskeletal changes in infected cells, we chose to normalize LC3B-II to both HSP90 and β-actin to rule out impacts of any one control protein on our interpretation of LC3B-II. We have found that both HSP90 and β-actin behave similarly in the context of mycobacterial infection (supplemental Fig. S1).
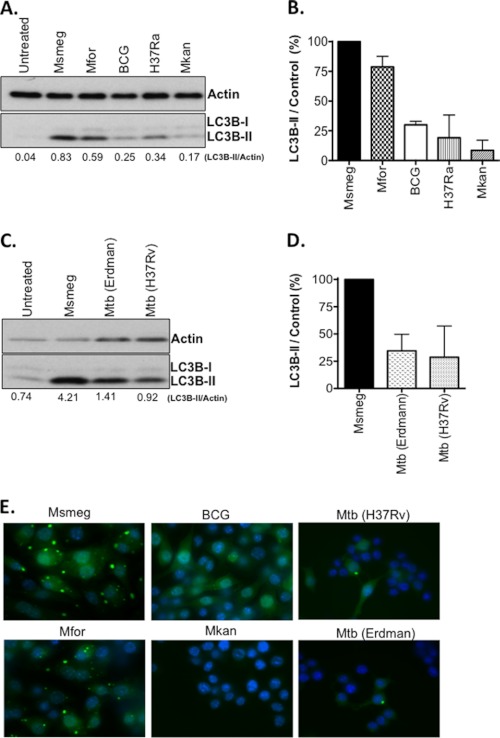
As shown in Fig. 1 (A and B), M. smegmatis and M. fortuitum exhibited consistently strong autophagy responses, as measured by an increase in lipidated LC3B-II (25). In contrast, BCG, H37Ra, and M. kanasii induced comparatively lower levels of LC3B-II in these assays (30–60% decrease). The attenuated M. tuberculosis strain H37Ra is one of the most commonly used controls for M. tuberculosis. Infection of RAW cells for 3 h with pathogenic M. tuberculosis strains H37Rv and Erdman indicate that both strains exhibit autophagy responses similar to BCG and H37Ra (Fig. 1, C and D). To confirm that differences in the levels of LC3B-II correlate with the presence of autophagosomes, RAW cells expressing GFP-LC3 were infected overnight with the same mycobacterial strains. Infection with M. smegmatis and M. fortuitum gave rise to large and numerous puncta, although infection with BCG, M. tuberculosis strains (H37Rv and Erdman), and M. kanasii yielded weaker responses (Fig. 1E). We conclude that GFP-LC3 puncta formation mirrors the results obtained by LC3B Western blot and that we observed heterogeneity under natural conditions with respect to the magnitude of autophagy induction which varies by species.
FIGURE 1.
Comparison of mycobacterial species for autophagy induction. A, the mycobacterial species indicated were used to infect RAW cells for 3–4 h at MOI 10, and Western blots were performed. Shown is the ratio of LC3B-II/Actin for each sample. B, shown is mean ± range for two experiments as performed in A. After subtracting the ratio for untreated samples, the LC3B-II/control ratio for M. smegmatis was set to 100%, and the relative responses of the indicated strains (mean ± range) is shown. C, M. smegmatis and two strains of M. tuberculosis (H37Rv and Erdman) were used to infect RAW cells for 3 h, and LC3B-I/II plus actin was analyzed by Western blot. D, shown is the mean ± range of two experiments performed as in C. E, RAW cells expressing GFP-LC3 were infected overnight (MOI 10) with the indicated mycobacteria. Images were taken at 100× magnification. Shown are merged images of GFP-LC3 (green) and Hoechst (blue). M. tuberculosis strains (H37Rv and Erdman) were evaluated independently of other species in adherence of the Biosafety Level 3 protocol along with a control M. smegmatis infection. Msmeg, M. smegmatis; Mfor, M. fortuitum; Mkan, M. kanasii; and Mtb, M. tuberculosis.
M. smegmatis Is a More Potent Inducer of Autophagy Than BCG
Our analysis of various mycobacterial species yielded a range of autophagy induction responses. This made it possible to choose high and low autophagy inducers (comparatively) and investigate bacterial products that induce autophagy in a native infection setting or have an influence upon signaling. We selected M. smegmatis and BCG as representative high and low autophagy inducers, respectively, and confirmed our original observation with these two mycobacteria. M. smegmatis infection induced substantial levels of autophagy at both 5 and 24 h, as measured by increased LC3B-II protein (Fig. 2, A, and B). In contrast, BCG induced LC3B-II levels that were at best 60% of that observed after infection with M. smegmatis. Significantly lower induction of autophagy by M. tuberculosis strains was also detected at 24-h post-infection when compared with M. smegmatis infection-induced autophagy (supplemental Fig. S2).
FIGURE 2.
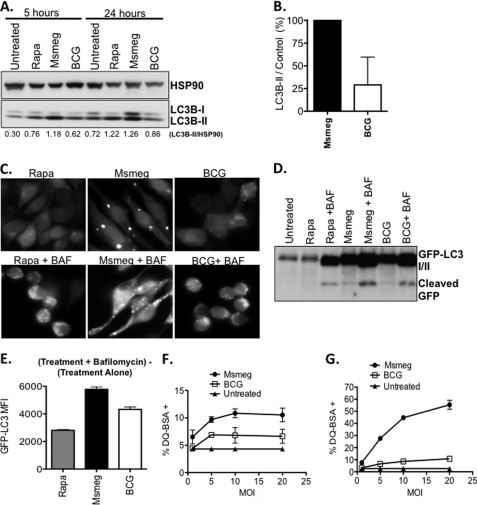
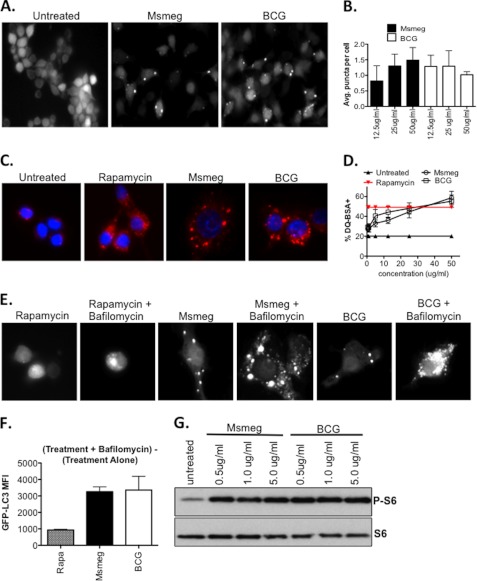
M. smegmatis is a potent inducer of autophagy. A, RAW cells were infected with either M. smegmatis or BCG (MOI: 10) or treated with rapamycin (positive control). Shown is the ratio of LC3B-II/HSP90 for each lane. B, RAW cells were infected for 24 h, and LC3B-II/HSP90 ratios were determined. After subtracting the ratio for untreated samples, the LC3B-II/control ratio for M. smegmatis alone was set to 100%, and the same ratio for BCG-infected cells is shown as the relative percentage ± S.D. Data shown are pooled from three independent assays. C, RAW GFP-LC3 cells were treated with rapamycin or infected with mycobacteria with or without bafilomycin A1 and imaged by fluorescence microscopy. D, RAW GFP-LC3 cells were treated as C, and protein lysates were analyzed by Western blot with anti-GFP antibody. E, RAW-GFP-LC3 cells were treated as in C, and flow cytometry was used to quantify the GFP rescued by bafilomycin treatment. Shown is the absolute MFI calculated as the MFI of (treatment + bafilomycin) − MFI of (treatment alone). F, DQ-BSA-loaded cells were infected with M. smegmatis and BCG for 5 h, and the percentage of DQ-BSA+ cells was determined by flow cytometry. G, assay was performed as described in F except that the cells were infected for 24 h. Msmeg, M. smegmatis.
To confirm that the autophagy induction culminates with autophagic degradation, infections were performed in the presence of bafilomycin A1, an inhibitor autophagic degradation (26, 27). As shown in Fig. 2C, overnight treatment with RAW-GFP-LC3 cells with rapamycin leads to weakly fluorescent cells with sparse puncta. However, co-treatment with bafilomycin A1 rescues the degraded GFP-LC3 signal to reveal bright cells with visible puncta. An overnight infection with M. smegmatis leads to robust puncta formation, which is enhanced by bafilomycin A1 treatment (Fig. 2C). In contrast, BCG infection of RAW-GFP-LC3 cells leads to weak puncta formation, and bafilomycin A1 treatment does recover GFP-LC3 signal. Because monitoring the cleaved GFP from GFP-LC3 allows us to examine the total flux of autophagy, Western blot for cleaved GFP was performed (25). Overnight treatment with rapamycin leads to substantial degradation such that the cleaved GFP was barely visible (Fig. 2D). However, co-treatment with bafilomycin A1 rescues cleaved GFP prior to complete degradation. Infection with both M. smegmatis and BCG led to the release of cleaved GFP, which could be recovered by bafilomycin A1 treatment (Fig. 2D). Microscopy experiments also demonstrated that more GFP signal was recovered from M. smegmatis-infected cells than BCG-infected cells. Flow cytometry was used to quantify the GFP mean fluorescent intensity (MFI). As shown in Fig. 2E, bafilomycin A1 rescued GFP fluorescence from cells treated overnight with rapamycin. Similarly, bafilomycin A1 treatment rescued GFP fluorescence from both M. smegmatis- and BCG-treated cells. As expected, we recover more GFP from M. smegmatis-infected cells consistent with the ability of M. smegmatis to induce higher levels of autophagy than BCG. Rapamycin in these assays served only as a positive control. This was especially true for assays using overnight time points that exceeded the typical time frame for maximum rapamycin-induced responses, which occurred within hours after treatment. As a result, the magnitude of the responses induced by mycobacteria should be compared among species.
Lastly, an increase in autophagy should coincide with enhanced lysosomal activity and general proteolysis. Thus, we predicted that M. smegmatis should elicit higher levels of lysosomal activity and proteolysis in comparison to BCG. We utilized the self-quenched reporter substrate, DQ-BSA, to quantify this activity. Upon proteolytic cleavage, DQ-BSA becomes highly fluorescent and provides a measure of the overall proteolytic/lysosomal activity within the cell. RAW cells were loaded with DQ-BSA and infected with M. smegmatis and BCG for 5 and 24 h, and flow cytometry was used to measure the percentage of DQ-BSA+ cells. Infection with M. smegmatis induced larger percentages of DQ-BSA+ cells at both 5 h (Fig. 2F) and 24 h (Fig. 2G) post-infection, indicating that M. smegmatis induced higher levels of cellular proteolysis. We conclude that M. smegmatis is more potent than BCG at inducing autophagy and proteolytic activity.
Autophagy Induction by M. smegmatis and BCG Occurs Independently of mTOR Inhibition
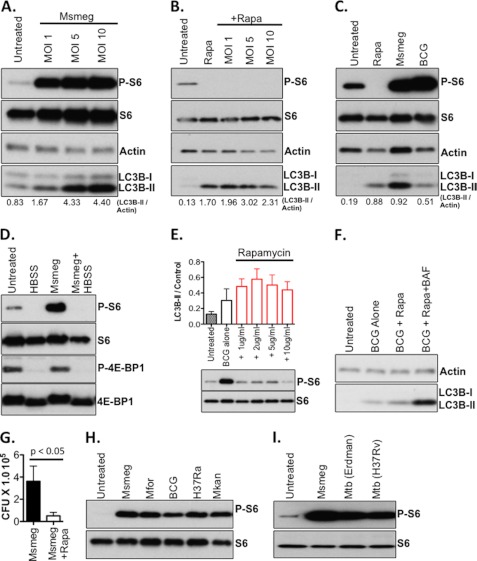
Cells lacking nutrients and/or growth factors exhibit reduced mTOR activity, thereby activating the autophagy machinery to recycle macromolecules for future use (28). A reduction in mTOR activity is known to result in reduced levels of phosphorylated ribosomal S6 kinase, leading to a reduction in phosphorylated ribosomal S6, and reduced protein translation (2, 17). However, it has not been demonstrated whether mycobacterial infection has an impact on mTOR signaling and if mTOR signaling correlates with autophagy induction. To address these questions, RAW cells were infected with M. smegmatis, and the phosphorylated S6 was determined as a read-out for mTOR activity. To our surprise, infection with M. smegmatis induced the formation of phosphorylated S6 (Fig. 3A). A significant level of autophagy (based on LC3B-II signal) was induced by M. smegmatis infection at the same MOI (Fig. 3A). To confirm that the increase in phosphorylated S6 protein induced by M. smegmatis represents the product of mTOR signaling, macrophages were pre-treated with rapamycin prior to infection. As anticipated, blockade of mTOR signaling by rapamycin abolished the increase in phosphorylated S6 protein that accompanies infection with M. smegmatis (Fig. 3B). Despite our finding that M. smegmatis and BCG induce markedly different levels of autophagy, both species induced significant levels of phosphorylated S6 protein in comparison to uninfected cells (Fig. 3C). Examination of other targets downstream of mTOR indicated that the phosphorylation of 4E-BP1 was unaffected by mycobacterial infection (Fig. 3D), but phosphorylation of eIF4B was induced by infection (supplemental Fig. S3). Phosphorylation of ULK1 was also not impacted by mycobacterial infection (supplemental Fig. S3). Because mTOR-dependent autophagy elicited by rapamycin or starvation would decrease mTOR activity, we concluded that autophagy induced by mycobacterial infection occurs independently of mTOR.
FIGURE 3.
Autophagy induction by mycobacteria occurs independently of mTOR inhibition. A, RAW cells were infected with M. smegmatis for 3 h at the MOI indicated. Western blots for phosphorylated ribosomal S6 (P-S6), total ribosomal S6 (S6), LC3B-I/II, and actin were performed. B, RAW cells were either left untreated, treated with rapamycin (25 μg/ml), or treated with rapamycin and infected with M. smegmatis at the MOI indicated for 6 h. C, RAW cells were infected with either M. smegmatis or BCG for 6 h at MOI 10. D, RAW cells were left untreated, starved in HBSS, infected with M. smegmatis (Msmeg; MOI 10), or starved and infected with M. smegmatis for 5 h. E, RAW cells were infected with BCG (MOI 5) in the absence or presence of increasing doses of rapamycin for 5 h. Shown is the LC3B-II/actin ratio (±S.E.). Western blots for P-S6 and total S6 are shown to confirm the inhibitory effects of rapamycin upon mTOR activity in these assays. F, RAW cells were infected with BCG at MOI 10, infected, and treated with rapamycin (Rapa), or infected and co-treated with rapamycin and bafilomycin A1 (BAF). G, RAW cells were infected with M. smegmatis with or without treatment with rapamycin. Shown are the mean CFU and standard deviation of 12 replicates per group pooled from four independent assays. H and I, lysates from RAW cells as described in Fig. 1A were re-analyzed for both phosphorylated ribosomal S6 (P-S6) and total ribosomal S6. Msmeg, M. smegmatis; Mfor, M. fortuitum; Mkan, M. kanasii; and Mtb, M. tuberculosis.
Because the observation that mycobacterial infection increases mTOR activity is novel, we evaluated how inhibition of mTOR activity with rapamycin would impact mycobacterium-induced autophagy by examining the levels of LC3B-II in infected cells with or without increasing doses of rapamycin. Whereas we could see trends toward higher levels of LC3B-II in rapamycin-treated cells, no statistical significance was found (Fig. 3E). Similar results were obtained with cells infected with M. smegmatis (data not shown). We also noticed that LC3B-II levels decreased, at higher doses of rapamycin. This result suggested that rapamycin treatment may enhance autophagic degradation and thus make it difficult to observe the entire pool of LC3B-II protein involved in the authophagic response. To test this hypothesis, RAW cells were infected with BCG and treated with either rapamycin alone or co-treated with rapamycin and bafilomycin A1. The addition of bafilomycin A1 to cells infected and treated with rapamycin rescued a substantial quantity of LC3B-II protein (Fig. 3F), suggesting that rapamycin enhanced the autophagic response in mycobacteria-infected cells.
Because we are the first group to observe the induction of mTOR activity (as measured by P-S6) in the context of mycobacterial infection, we decided to confirm rapamycin-induced bacterial killing. RAW cells were infected with M. smegmatis for 1 h followed by treatment with either plain media or media containing rapamycin. Consistent with published reports (6, 9), treatment with rapamycin negatively impacted bacterial viability by an average of 7-fold (Fig. 3G), indicating that bacterial stimulation of mTOR is not sufficient to render mycobacteria resistant to mTOR-dependent autophagy. In addition, it reflects our observation that rapamycin treatment enhances autophagic responses in mycobacterium-infected cells by inhibiting the mTOR pathway through which the bacteria are potently stimulated under natural infection conditions.
To evaluate whether the ability of mycobacteria to induce mTOR activity and the phosphorylation of S6 are a consistent feature among mycobacteria, we analyzed the presence of phosphorylated S6 in cells infected with both pathogenic and non-pathogenic mycobacteria. We found that all non-pathogenic (Fig. 3H) and pathogenic mycobacteria (Fig. 3I) tested induced phosphorylation of S6. We concluded that mTOR signaling, as measured by the presence of phosphorylated S6, is a consistent feature of mycobacterial infection that occurs simultaneously with autophagy induction.
The Lipid Component of Mycobacteria, but Not DNA or Protein, Induces Autophagy and mTOR Activity
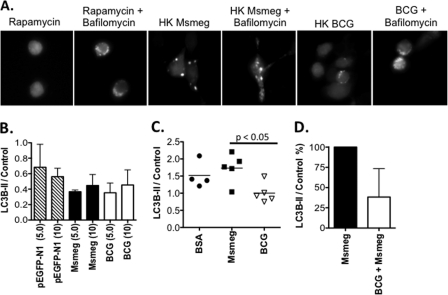
Given that M. smegmatis infection induced a potent autophagy response, we questioned whether the bacterium accomplished this exclusively through active mechanisms, or whether individual macromolecules had the ability to induce autophagy. To address this question, we tested the ability of heat-killed M. smegmatis and BCG to induce GFP-LC3B puncta formation. Although both heat-killed M. smegmatis and BCG induced puncta formation, heat-killed M. smegmatis induced larger responses in comparison to heat-killed BCG (Fig. 4A). Bafilomycin A1 treatment of cells exposed to heat-killed mycobacteria enhanced the appearance of GFP-LC3 puncta indicting the heat-killed mycobacteria induced a complete autophagy response (Fig. 4A). We also observed that heat-killed M. smegmatis induced higher levels of DQ-BSA fluorescence indicating higher levels of cellular proteolysis (supplemental Fig. S4). Based upon these results, we systematically investigated both mycobacterial species to define components that could influence the autophagy response. To test the ability of mycobacterial DNA to induce autophagy, RAW cells were treated with either vector DNA (pEGFP-N1, control) or chromosomal DNA extracted from M. smegmatis and BCG. Treatment of RAW cells with 10 μg/ml mycobacterial DNA did not induce an autophagy response above vector-control DNA-treated cells (Fig. 4B). Additionally, protein lysates of M. smegmatis and BCG were tested for their ability to induce autophagy. Although necessary for extraction, the detergent Nonidet P-40 did limit the amount of protein that could be added to cells, with 100 ng/ml being the upper limit. As a control, we prepared BSA in the same extraction buffer and concentrated it similarly. Treatment with either mycobacterial protein extract did not induce an autophagy response above cells treated with the same concentration of BSA (Fig. 4C). However, the basal level of lipidated LC3B-II was consistently reduced in RAW cells treated with BCG-derived protein extracts versus cells treated with M. smegmatis-derived protein extracts. These observations led to the novel hypothesis that BCG could reduce mycobacterial infection-induced autophagy. To test this hypothesis, RAW cells were pre-infected overnight with BCG at MOI 10. Cells were then infected for 4 h with M. smegmatis at MOI 10. Protein lysates were prepared, and Western blots for LC3B-II were performed. As shown in Fig. 4D, pre-infection with BCG reduced autophagy responses to M. smegmatis when compared with cells infected with M. smegmatis alone. It has also been observed that pre-infection with M. tuberculosis (H37Rv) can also reduce the autophagy response to M. smegmatis (data not shown). We conclude that BCG has the capacity to reduce autophagy induction in response to M. smegmatis infection, a bacterial autophagy inducer.
FIGURE 4.
Innate factors influence the induction of autophagy by mycobacteria. A, RAW-GFP-LC3 cells were treated with heat-killed (HK) M. smegmatis or BCG overnight with or without bafilomycin A1 co-treatment. B, RAW cells were treated overnight with control DNA (pEGFP-N1), or chromosomal DNA extracted from M. smegmatis and BCG. Shown is the ratio of LC3B-II/actin at 5 μg/ml and 10 μg/ml doses (mean ± range) from two independent experiments. C, RAW cells were treated with BSA or Nonidet P-40 protein lysates (100 ng/ml) prepared from either M. smegmatis or BCG. Shown is the ratio of LC3B-II/actin (mean ± individual samples) of data pooled from three independent experiments. D, RAW cells were infected with M. smegmatis (MOI 10) ± pre-infection with BCG overnight (MOI 10). Western blots for LC3B-I/II and either actin or HSP90 were performed. After subtracting the ratio for untreated samples, the LC3B-II/control ratio for M. smegmatis alone was set to 100%, and the impact of BCG pre-infection is shown as the relative percentage (mean ± S.D.). Data shown are pooled from three independent assays. Msmeg, M. smegmatis.
The last mycobacterial macromolecule that we considered in our analysis was lipid material. To test whether mycobacterial lipid material could induce autophagy, total cell wall lipids were extracted from both M. smegmatis and BCG. Surprisingly, we observed that cell wall lipids from both mycobacteria could induce GFP-LC3 puncta formation (Fig. 5A). To determine if lipid material derived from M. smegmatis and BCG differed from each other in autophagy induction, the average GFP-LC3 puncta per cell induced by various concentrations of lipid material was estimated. Overall, there were no statistical differences observed between the M. smegmatis- and BCG-derived lipids, and both appeared to be equally capable of inducing GFP-LC3 puncta (Fig. 5B). Similarly, lipid material from both M. smegmatis and BCG could induce equivalent levels of DQ-BSA hydrolysis (Fig. 5, C and D). To confirm our finding that lipid material induces proteolysis and autophagic degradation, bafilomycin A1 was used for the assay. Addition of bafilomycin A1 to lipid-treated cells revealed substantial puncta that would have been lost to degradation (Fig. 5E). Quantitation by flow cytometry confirms that bafilomycin A1 treatment indeed rescues GFP fluorescence (Fig. 5F). Therefore, we conclude that lipids derived from BCG and M. smegmatis induce similar amounts of GFP-LC3 puncta and cellular proteolysis in RAW cells. Because we observed mTOR activation during mycobacterial infection, we examined whether lipid material from mycobacteria could induce mTOR activity. The lipid material from both M. smegmatis and BCG induced the formation of phosphorylated S6, indicating mTOR signaling (Fig. 5G). We concluded that M. smegmatis- and BCG-derived lipids induced mTOR signaling, autophagic degradation, and proteolysis.
FIGURE 5.
Mycobacterial lipids stimulate autophagy. A, total lipids extracted from M. smegmatis and BCG were spotted on glass coverslips. RAW-GFP-LC3 cells were added, cultured overnight, and imaged at 100× magnification. B, RAW GFP-LC3 cells were cultured in the presence of mycobacterial lipids, and ImageJ was used to estimate the average GFP-LC3 puncta per cell (±S.D.). Between 250 and 350 cells treated with three independently prepared batches of mycobacterial lipids were evaluated. C, RAW cells loaded with DQ-BSA were cultured overnight in the presence of lipid or 10 μg/ml rapamycin and imaged at 100× magnification. D, RAW cells were loaded with DQ-BSA and treated overnight with the indicated concentrations of mycobacterial lipids or 10 μg/ml rapamycin as a positive control. Flow cytometry was used to measure the percentage of DQ-BSA+ cells. Shown are combined results (mean ± S.D.) from cells treated with four independently prepared batches of lipid (n = 9–12 per group). E, RAW GFP-LC3 cells were treated with either rapamycin (10 μg/ml) or mycobacterial lipid with or without bafilomycin (100 nm) and imaged at 100× magnification. F, RAW-GFP-LC3 cells were treated as in E, and flow cytometry was used to quantify the GFP rescued by bafilomycin treatment. Shown is the absolute MFI calculated as the MFI of (treatment + bafilomycin) − MFI of (treatment alone). G, RAW cells were treated with the indicated concentrations of mycobacterial lipids for 24 h. Western blots for phosphorylated ribosomal S6 and total ribosomal S6 were performed. Msmeg, M. smegmatis.
DISCUSSION
Autophagy has recently been described as a mechanism by which mammalian cells can limit mycobacterial infection. Originally, autophagy was believed to be exclusively a housekeeping response to nutrient deprivation and reduced mTOR signaling (1). Under conditions of amino acid and growth factor deprivation, cytosolic contents could be targeted to lysosomes to liberate nutrients for de novo biosynthesis (1, 29). More recently, a new class of autophagy termed “xenophagy” (30) was identified as a means by which invading microorganisms could be sequestered and subjected to lysosomal degradation. BCG and pathogenic M. tuberculosis (H37Rv) were among the first bacteria shown to be destroyed by starvation or treatment with rapamycin (mTOR-dependent autophagy) (1, 6, 31). These seminal studies have been extended by Jagannath et al. who demonstrated that rapamycin-induced (mTOR-dependent) autophagy could enhance adaptive immune responses to BCG (12), thus linking mTOR-dependent autophagy to immune protection. Based upon these examples, mTOR-dependent autophagy represents a powerful means by which numerous bacteria, including mycobacteria, can be identified, destroyed, and made available for recognition by the immune system (4, 30).
Fully harnessing the autophagy pathway to elicit protective responses against mycobacteria requires understanding the range of responses present under natural conditions, the inducing macromolecules, and the signaling events that occur concurrently. Toward that goal, we have studied several species of mycobacteria to evaluate the range of autophagy responses that can be induced by mycobacterial infection in the absence of autophagy-inducing agents. Although M. smegmatis and M. fortuitum gave robust responses, BCG, M. kanasii, and H37Ra gave comparably weaker responses. Infection of RAW cells for 3 and 24 h with M. tuberculosis strains H37Rv and Erdman indicated that both strains of pathogenic mycobacteria yielded autophagy responses similar to BCG and H37Ra. Similar results were obtained with clinical isolates of M. tuberculosis as well (data not shown). The differences observed between mycobacterial strains could reflect a number of factors, including differences in infectivity, proliferation, the concentration of activating macromolecules, and the activity of inhibitory mechanisms. For example, it is known that M. smegmatis and M. fortuitum grow more rapidly than other strains utilized in this study, although the autophagy induction at 3-h post-infection indicated that the difference in autophagy is not due to the mycobacterial growth rate. Further studies will be required to understand why the autophagy response is not completely uniform across species especially among slow growing species such as BCG and M. kanasii. In addition to enhancing our understanding of mycobacteria, we anticipate this work will shed light on the general mechanisms by which the autophagy machinery functions within the macrophage.
To the best of our knowledge, our work is the first to examine the impact of mycobacterial infection upon mTOR signaling. It is well known that inhibition of mTOR signaling by starvation or rapamycin treatment is a potent inducer of autophagy. Previous studies have often infected cells with mycobacteria, and simultaneously induced starvation or employed rapamycin treatment to inhibit mTOR, which obscures the impact of the mycobacteria upon mTOR signaling. Given the critical role for mTOR in regulating the autophagy response, we sought to determine if there was a correlation between mTOR signaling and autophagy in the context of mycobacterial infection. Infection with both M. smegmatis and BCG led to substantial mTOR activity, indicating that mycobacterium-induced autophagy is mTOR-independent. Both starvation and rapamycin treatment abolished this response, confirming that the increase in phosphorylated S6 is the bona fide product of mTOR activity. Similar results were observed with phosphorylated eIF4B, but phosphorylated 4E-BP1 remained unchanged during infection. An examination of the complete panel of non-pathogenic and pathogenic mycobacteria used in this study indicated that all species tested could induce the phosphorylation of S6. These results are especially interesting in light of recent work showing that Toxoplama gondii infection can induce autophagy without a change in the levels of phosphorylated S6 or 4E-BP1 (32).
Given the potency with which mTOR-dependent autophagy can decrease mycobacterial viability, these findings call into question whether mycobacteria specifically stimulates mTOR to avoid an mTOR-dependant autophagy response that can reduce bacterial burden. Our data demonstrate that rapamycin does enhance autophagic responses in infected cells that were best observed in the context of bafilomycin A1 co-treatment. Thus, it may suggest that the degradative component of the autophagy response might be the most impacted by mTOR stimulation in the context of mycobacterial infection. This correlated well with our data showing that rapamycin treatment of RAW cells decreased the viability of intracellular M. smegmatis. It is also possible that mTOR stimulation delays the kinetics of mycobacterium-induced autophagy, which can be enhanced by rapamycin treatment to affect observable bacterial killing. Additional studies will be necessary to test these hypotheses and further identify exactly how the inhibition of bacterially induced mTOR activity alters the autophagic response.
We should point out that it is equally plausible that the increase in mTOR signaling is indicative of the increase in cytokine synthesis and other effector functions that utilize nutrients and take place when macrophages become infected with mycobacteria. Both hypotheses are valid and additional experiments will be necessary to determine if the increase in mTOR signaling that occurs during mycobacterial infection favors either the pathogen or the host.
We also sought to understand the features of both M. smegmatis and BCG that could induce autophagy and mTOR signaling. Because heat-killed M. smegmatis and BCG could induce autophagy and cellular proteolysis, albeit to differing degrees, we postulated that innate features of the mycobacteria may be responsible for autophagy induction and mTOR signaling. Treatment of macrophages with DNA and protein extracted from either M. smegmatis or BCG did not yield autophagy responses above background. Moreover, protein extracts from BCG had a tendency to reduce the basal levels of LC3B-II in RAW cells. This suggested that BCG and related pathogenic mycobacteria have mechanisms to limit or reduce their autophagy signature. Consistent with this idea, we observed that pre-infection of RAW cells overnight with BCG and M. tuberculosis (not shown) reduced the autophagy response to M. smegmatis the following day. It is unclear whether pre-infection with BCG can limit the autophagy response to other inducing agents. The notion that BCG and other pathogenic mycobacteria could modify host autophagy is consistent with findings that pathogenic mycobacteria alter other host cellular responses, including apoptosis (33, 34). Our findings are especially relevant in the context of BCG vaccination, which is well documented in exhibiting variable effectiveness. We expect that additional work utilizing genetic approaches will identify mycobacterial gene products that affect or limit host autophagy responses. Because extracted protein and DNA from both mycobacteria failed to induce autophagy, we suspected a lipid component might be involved. Lipid material from both M. smegmatis and BCG induced substantial GFP-LC3 puncta and DQ-BSA proteolysis. Despite the fact that it has been shown that Escherichia coli LPS can stimulate mTOR signaling (35), our work is the first to implicate mycobacterial lipid material as an activator of mTOR signaling. As lipid components would be among the first molecules to interact with the host upon encountering a mycobacterium, bacterial stimulation of mTOR may be intentional. Once again, genetic approaches will be invaluable in identifying specific lipids and molecules that stimulate mTOR signaling and autophagy in macrophages.
The observation, that lipid material from both M. smegmatis and BCG induces similar levels of autophagy, indicates that additional factors govern the overall autophagy response to the intact pathogens. For example, it has been shown that the exposed lipids on the surface of mycobacteria vary by species (36). It is possible that lipids capable of inducing autophagy are more exposed on M. smegmatis, thereby permitting greater responses to be observed when using intact bacteria. Because we used total lipid preparations from both organisms, that spatial distinction would have been lost. Also, it has been noted that M. smegmatis infects cells with greater ease than BCG. This difference could expose macrophages to greater quantities of M. smegmatis lipid material. The fact that we can observe potent phenotypes in as little as 3 h after the addition of bacteria would suggest that additional factors beyond infectivity are involved. Studies with heat-killed mycobacteria yielded the same results, further suggesting a limited impact of infectivity upon the observed phenotype. Once inside the cell, M. smegmatis divides much more rapidly than BCG and allows for higher concentrations of stimulatory lipids inside the macrophage, which is relevant for assays designed to look at later time points. Further work testing fractionated lipids will aid in comparing specific classes of lipids and allow more precise structure-function and dosage determinations to be made. Our observation that BCG can reduce autophagy induction in response to M. smegmatis infection indicates that inhibitory mechanisms present in BCG can reduce the autophagy response to mycobacterial lipid material. Our work demonstrates that the overall autophagy response to any mycobacterium should be viewed as the summation of activating macromolecules and inhibitory mechanisms.
In sum, our work is the first to scrutinize mycobacterium autophagy without the added influence of inducing agents, and we have identified mycobacterium autophagy as a truly mTOR-independent process. Mycobacterial lipids have been shown by us to induce autophagy and activate mTOR signaling. Additionally, our study showed that BCG has an innate ability to reduce the macrophage autophagy response. Additional studies that attempt to emulate natural conditions of infection will be used to further dissect how mycobacteria and specific macromolecules interact with the host autophagy machinery. Understanding such interactions will allow for the development of strategies to impede or block mycobacterial activity and consequently improve immune responses.
Supplementary Material
Acknowledgments
We thank Holly Alley and Kristen Jurcic Smith for technical assistance. We also thank Gregory Sempowski of the Duke Human Vaccine Institute for helpful discussions and manuscript review. Special thanks to Noboro Mizushima (Tokyo Medical and Dental University, Tokyo, Japan) for providing GFP-LC3 expression plasmids.
This work was supported, in whole or in part, by National Institutes of Health Grants R21 AI095723-01 and U54 AI057157 (Southeast Regional Center of Excellence for Emerging Infections and Biodefense). Tuberculosis research was performed in the NIAID Regional Biocontainment Laboratory at Duke (UC6 AI058607). Flow cytometry was performed in the Duke Human Vaccine Institute Research Flow Cytometry Shared Resource Facility (Durham, NC) under the direction of Dr. John F. Whitesides.

This paper contains supplemental Appendix S1.
- mTOR
- mammalian target of rapamycin
- S6
- ribosomal protein S6
- BCG
- bacille Calmette-Guérin
- MOI
- multiplicity of infection
- NFDM
- nonfat dry milk
- MFI
- mean fluorescent intensity.
REFERENCES
- 1. He C., Klionsky D. J. (2009) Regulation mechanisms and signaling pathways of autophagy. Annu. Rev. Genet. 43, 67–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fingar D. C., Salama S., Tsou C., Harlow E., Blenis J. (2002) Mammalian cell size is controlled by mTOR and its downstream targets S6K1 and 4EBP1/eIF4E. Genes Dev. 16, 1472–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhou H., Huang S. (2010) The complexes of mammalian target of rapamycin. Curr. Protein Pept. Sci. 11, 409–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Deretic V. (2011) Autophagy in immunity and cell-autonomous defense against intracellular microbes. Immunol. Rev. 240, 92–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ogawa M., Mimuro H., Yoshikawa Y., Ashida H., Sasakawa C. (2011) Manipulation of autophagy by bacteria for their own benefit. Microbiol. Immunol. 55, 459–471 [DOI] [PubMed] [Google Scholar]
- 6. Gutierrez M. G., Master S. S., Singh S. B., Taylor G. A., Colombo M. I., Deretic V. (2004) Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell 119, 753–766 [DOI] [PubMed] [Google Scholar]
- 7. Kumar D., Nath L., Kamal M. A., Varshney A., Jain A., Singh S., Rao K. V. (2010) Genome-wide analysis of the host intracellular network that regulates survival of Mycobacterium tuberculosis. Cell 140, 731–743 [DOI] [PubMed] [Google Scholar]
- 8. Sanjuan M. A., Dillon C. P., Tait S. W., Moshiach S., Dorsey F., Connell S., Komatsu M., Tanaka K., Cleveland J. L., Withoff S., Green D. R. (2007) Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature 450, 1253–1257 [DOI] [PubMed] [Google Scholar]
- 9. Ponpuak M., Deretic V. (2011) Autophagy and p62/sequestosome 1 generate neo-antimicrobial peptides (cryptides) from cytosolic proteins. Autophagy 7, 336–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cooney R., Baker J., Brain O., Danis B., Pichulik T., Allan P., Ferguson D. J., Campbell B. J., Jewell D., Simmons A. (2010) NOD2 stimulation induces autophagy in dendritic cells influencing bacterial handling and antigen presentation. Nat. Med. 16, 90–97 [DOI] [PubMed] [Google Scholar]
- 11. Crotzer V. L., Blum J. S. (2010) Autophagy and adaptive immunity. Immunology 131, 9–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jagannath C., Lindsey D. R., Dhandayuthapani S., Xu Y., Hunter R. L., Jr., Eissa N. T. (2009) Autophagy enhances the efficacy of BCG vaccine by increasing peptide presentation in mouse dendritic cells. Nat. Med. 15, 267–276 [DOI] [PubMed] [Google Scholar]
- 13. Lerena M. C., Colombo M. I. (2011) Mycobacterium marinum induces a marked LC3 recruitment to its containing phagosome that depends on a functional ESX-1 secretion system. Cell Microbiol. 13, 814–835 [DOI] [PubMed] [Google Scholar]
- 14. Meena L. S., Rajni (2010) Survival mechanisms of pathogenic Mycobacterium tuberculosis H37Rv. FEBS J. 277, 2416–2427 [DOI] [PubMed] [Google Scholar]
- 15. Shin D. M., Jeon B. Y., Lee H. M., Jin H. S., Yuk J. M., Song C. H., Lee S. H., Lee Z. W., Cho S. N., Kim J. M., Friedman R. L., Jo E. K. (2010) Mycobacterium tuberculosis eis regulates autophagy, inflammation, and cell death through redox-dependent signaling. PLoS Pathog. 6, e1001230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Snapper S. B., Melton R. E., Mustafa S., Kieser T., Jacobs W. R., Jr. (1990) Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol. Microbiol. 4, 1911–1919 [DOI] [PubMed] [Google Scholar]
- 17. Korolchuk V. I., Saiki S., Lichtenberg M., Siddiqi F. H., Roberts E. A., Imarisio S., Jahreiss L., Sarkar S., Futter M., Menzies F. M., O'Kane C. J., Deretic V., Rubinsztein D. C. (2011) Lysosomal positioning coordinates cellular nutrient responses. Nat. Cell Biol. 13, 453–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zullo A. J., Michaud M., Zhang W., Grusby M. J. (2009) Identification of the small protein rich in arginine and glycine (SRAG). A newly identified nucleolar protein that can regulate cell proliferation. J. Biol. Chem. 284, 12504–12511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kimura S., Noda T., Yoshimori T. (2007) Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy 3, 452–460 [DOI] [PubMed] [Google Scholar]
- 20. Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T. (2000) LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 19, 5720–5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liang C., Lee J. S., Inn K. S., Gack M. U., Li Q., Roberts E. A., Vergne I., Deretic V., Feng P., Akazawa C., Jung J. U. (2008) Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat. Cell Biol. 10, 776–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ha S. D., Ham B., Mogridge J., Saftig P., Lin S., Kim S. O. (2010) Cathepsin B-mediated autophagy flux facilitates the anthrax toxin receptor 2-mediated delivery of anthrax lethal factor into the cytoplasm. J. Biol. Chem. 285, 2120–2129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tamai K., Tanaka N., Nara A., Yamamoto A., Nakagawa I., Yoshimori T., Ueno Y., Shimosegawa T., Sugamura K. (2007) Role of Hrs in maturation of autophagosomes in mammalian cells. Biochem. Biophys. Res. Commun. 360, 721–727 [DOI] [PubMed] [Google Scholar]
- 24. Barth S., Glick D., Macleod K. F. (2010) Autophagy. Assays and artifacts. J. Pathol. 221, 117–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Klionsky D. J., Abeliovich H., Agostinis P., Agrawal D. K., Aliev G., Askew D. S., Baba M., Baehrecke E. H., Bahr B. A., Ballabio A., Bamber B. A., Bassham D. C., Bergamini E., Bi X., Biard-Piechaczyk M., Blum J. S., Bredesen D. E., Brodsky J. L., Brumell J. H., Brunk U. T., Bursch W., Camougrand N., Cebollero E., Cecconi F., Chen Y., Chin L. S., Choi A., Chu C. T., Chung J., Clarke P. G., Clark R. S., Clarke S. G., Clavé C., Cleveland J. L., Codogno P., Colombo M. I., Coto-Montes A., Cregg J. M., Cuervo A. M., Debnath J., Demarchi F., Dennis P. B., Dennis P. A., Deretic V., Devenish R. J., Di Sano F., Dice J. F., Difiglia M., Dinesh-Kumar S., Distelhorst C. W., Djavaheri-Mergny M., Dorsey F. C., Droge W., Dron M., Dunn W. A., Jr., Duszenko M., Eissa N. T., Elazar Z., Esclatine A., Eskelinen E. L., Fesus L., Finley K. D., Fuentes J. M., Fueyo J., Fujisaki K., Galliot B., Gao F. B., Gewirtz D. A., Gibson S. B., Gohla A., Goldberg A. L., Gonzalez R., Gonzalez-Estevez C., Gorski S., Gottlieb R. A., Haussinger D., He Y. W., Heidenreich K., Hill J. A., Hoyer-Hansen M., Hu X., Huang W. P., Iwasaki A., Jaattela M., Jackson W. T., Jiang X., Jin S., Johansen T., Jung J. U., Kadowaki M., Kang C., Kelekar A., Kessel D. H., Kiel J. A., Kim H. P., Kimchi A., Kinsella T. J., Kiselyov K., Kitamoto K., Knecht E., Komatsu M., Kominami E., Kondo S., Kovacs A. L., Kroemer G., Kuan C. Y., Kumar R., Kundu M., Landry J., Laporte M., Le W., Lei H. Y., Lenardo M. J., Levine B., Lieberman A., Lim K. L., Lin F. C., Liou W., Liu L. F., Lopez-Berestein G., Lopez-Otin C., Lu B., Macleod K. F., Malorni W., Martinet W., Matsuoka K., Mautner J., Meijer A. J., Melendez A., Michels P., Miotto G., Mistiaen W. P., Mizushima N., Mograbi B., Monastyrska I., Moore M. N., Moreira P. I., Moriyasu Y., Motyl T., Munz C., Murphy L. O., Naqvi N. I., Neufeld T. P., Nishino I., Nixon R. A., Noda T., Nurnberg B., Ogawa M., Oleinick N. L., Olsen L. J., Ozpolat B., Paglin S., Palmer G. E., Papassideri I., Parkes M., Perlmutter D. H., Perry G., Piacentini M., Pinkas-Kramarski R., Prescott M., Proikas-Cezanne T., Raben N., Rami A., Reggiori F., Rohrer B., Rubinsztein D. C., Ryan K. M., Sadoshima J., Sakagami H., Sakai Y., Sandri M., Sasakawa C., Sass M., Schneider C., Seglen P. O., Seleverstov O., Settleman J., Shacka J. J., Shapiro I. M., Sibirny A., Silva-Zacarin E. C., Simon H. U., Simone C., Simonsen A., Smith M. A., Spanel-Borowski K., Srinivas V., Steeves M., Stenmark H., Stromhaug P. E., Subauste C. S., Sugimoto S., Sulzer D., Suzuki T., Swanson M. S., Tabas I., Takeshita F., Talbot N. J., Talloczy Z., Tanaka K., Tanida I., Taylor G. S., Taylor J. P., Terman A., Tettamanti G., Thompson C. B., Thumm M., Tolkovsky A. M., Tooze S. A., Truant R., Tumanovska L. V., Uchiyama Y., Ueno T., Uzcategui N. L., van der Klei I., Vaquero E. C., Vellai T., Vogel M. W., Wang H. G., Webster P., Wiley J. W., Xi Z., Xiao G., Yahalom J., Yang J. M., Yap G., Yin X. M., Yoshimori T., Yu L., Yue Z., Yuzaki M., Zabirnyk O., Zheng X., Zhu X., Deter R. L. (2008) Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy 4, 151–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kyei G. B., Dinkins C., Davis A. S., Roberts E., Singh S. B., Dong C., Wu L., Kominami E., Ueno T., Yamamoto A., Federico M., Panganiban A., Vergne I., Deretic V. (2009) Autophagy pathway intersects with HIV-1 biosynthesis and regulates viral yields in macrophages. J. Cell Biol. 186, 255–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mizushima N., Yoshimori T. (2007) How to interpret LC3 immunoblotting. Autophagy 3, 542–545 [DOI] [PubMed] [Google Scholar]
- 28. Mehrpour M., Esclatine A., Beau I., Codogno P. (2010) Overview of macroautophagy regulation in mammalian cells. Cell Res. 20, 748–762 [DOI] [PubMed] [Google Scholar]
- 29. Yang Z., Klionsky D. J. (2010) Eaten alive. A history of macroautophagy. Nat. Cell Biol. 12, 814–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Knodler L. A., Celli J. (2011) Eating the strangers within. Host control of intracellular bacteria via xenophagy. Cell Microbiol. 13, 1319–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Korolchuk V. I., Rubinsztein D. C. (2011) Regulation of autophagy by lysosomal positioning. Autophagy 7, 927–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang Y., Weiss L. M., Orlofsky A. (2009) Host cell autophagy is induced by Toxoplasma gondii and contributes to parasite growth. J. Biol. Chem. 284, 1694–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gupta A., Kaul A., Tsolaki A. G., Kishore U., Bhakta S. (2012) Immunobiology 217, 363–374 [DOI] [PubMed] [Google Scholar]
- 34. Dietrich J., Doherty T. M. (2009) Interaction of Mycobacterium tuberculosis with the host. Consequences for vaccine development. APMIS 117, 440–457 [DOI] [PubMed] [Google Scholar]
- 35. Salh B., Wagey R., Marotta A., Tao J. S., Pelech S. (1998) Activation of phosphatidylinositol 3-kinase, protein kinase B, and p70 S6 kinases in lipopolysaccharide-stimulated Raw 264.7 cells. Differential effects of rapamycin, Ly294002, and wortmannin on nitric oxide production. J. Immunol. 161, 6947–6954 [PubMed] [Google Scholar]
- 36. Ortalo-Magné A., Lemassu A., Lanéelle M. A., Bardou F., Silve G., Gounon P., Marchal G., Daffé M. (1996) Identification of the surface-exposed lipids on the cell envelopes of Mycobacterium tuberculosis and other mycobacterial species. J. Bacteriol. 178, 456–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.