Background: Mycobacterium tuberculosis harbors a highly expanded number of toxin-antitoxin (TA) systems.
Results: The M. tuberculosis VapC-mt4 toxin blocks translation and arrests growth through RNA binding at a short recognition sequence.
Conclusion: M. tuberculosis VapC toxins have a function distinct from other characterized TA toxins.
Significance: TA systems may contribute to the slow growth and dormancy characteristic of M. tuberculosis during latent tuberculosis.
Keywords: Bacterial Toxins, Protein Synthesis, Ribonuclease, RNA, Virulence Factors, Antitoxin, Latency, Translation, Tuberculosis
Abstract
The Mycobacterium tuberculosis genome harbors an unusually large number of toxin-antitoxin (TA) modules. Curiously, over half of these are VapBC (virulence-associated protein) family members. Nonetheless, the cellular target, precise mode of action, and physiological role of the VapC toxins in this important pathogen remain unclear. To better understand the function of this toxin family, we studied the features and biochemical properties of a prototype M. tuberculosis VapBC TA system, vapBC-mt4 (Rv0596c-Rv0595c). VapC-mt4 expression resulted in growth arrest, a hallmark of all TA toxins, in Escherichia coli, Mycobacterium smegmatis, and M. tuberculosis. Its expression led to translation inhibition accompanied by a gradual decrease in the steady-state levels of several mRNAs. VapC-mt4 exhibited sequence-specific endoribonuclease activity on mRNA templates at ACGC and AC(A/U)GC sequences. However, the cleavage activity of VapC-mt4 was comparatively weak relative to the TA toxin MazF-mt1 (Rv2801c). Unlike other TA toxins, translation inhibition and growth arrest preceded mRNA cleavage, suggesting that the RNA binding property of VapC-mt4, not RNA cleavage, initiates toxicity. In support of this hypothesis, expression of VapC-mt4 led to an increase in the recovery of total RNA with time in contrast to TA toxins that inhibit translation via direct mRNA cleavage. Additionally, VapC-mt4 exhibited stable, sequence-specific RNA binding in an electrophoretic mobility shift assay. Finally, VapC-mt4 inhibited protein synthesis in a cell-free system without cleaving the corresponding mRNA. Therefore, the activity of VapC-mt4 is mechanistically distinct from other TA toxins because it appears to primarily inhibit translation through selective, stable binding to RNA.
Introduction
Mycobacterium tuberculosis has the unique ability to persist for long periods of time in its host as a latent infection (1). This latent state in which the bacteria are thought to be dormant with markedly altered physiology is pivotal to the survival of the bacteria in the stressful environments it encounters. Importantly, M. tuberculosis cells in the latent state are generally refractory to antibiotics, most of which target processes occurring in actively replicating bacteria (2). Despite ongoing efforts to understand the latent state, the molecular switches that enable M. tuberculosis to slow or stop replication and enter a latent state remain unknown.
Toxin-antitoxin (TA)4 systems (also referred to as addiction or suicide modules) have the potential to regulate M. tuberculosis replication and typically comprise an autoregulated operon encoding a labile antitoxin and a stable toxin. All characterized chromosomal TA toxins have been found to inhibit cell growth by targeted inhibition of an essential cellular process, such as protein synthesis or DNA replication. The activity of the toxin is regulated by its cognate antitoxin, which enables finely tuned control of TA module toxicity during relatively short periods of stress. However, TA toxin-mediated growth arrest is only reversible up to a point of no return after which prolonged activation of the toxin results in cell death (3–6).
Gerdes and co-workers (4) initially reported that the genome of M. tuberculosis harbors a remarkably high number of TA systems (more recently estimated at 88 putative TA systems (7)). They also uncovered a correlation between the number of chromosomal TA systems and growth rate with TA loci being most abundant in organisms with slow growth rates (4). These observations suggest a possible role for TA systems in growth rate regulation. Despite these striking observations, the physiological roles of TA systems are not well understood. In Escherichia coli, activation of toxin-antitoxin systems is triggered by various stresses (8–11) and induces a state of cell growth arrest (5, 6) with striking similarities to the slowly or non-replicating state thought to occur in M. tuberculosis during the latent infection (12). As in E. coli, TA toxins in M. tuberculosis can be activated by selected stresses, including hypoxic conditions and macrophage infection (7). Overexpression of selected individual M. tuberculosis toxins in E. coli, Mycobacterium smegmatis, and M. tuberculosis also results in growth arrest (7, 13, 14). Consistent with their bacteriostatic effects, activation of E. coli TA toxins results in the generation of a persistent population of cells refractory to antibiotics (15–18). Likewise, overexpression or deletion of individual M. tuberculosis RelE family toxins resulted in an increase (for overexpression) or decrease (for deletion) in persister recovery in antibiotic-treated M. tuberculosis cells. However, an M. tuberculosis RelE mutant did not exhibit a persistence defect in a mouse model (13) perhaps because only one of the three M. tuberculosis RelE genes was deleted. Because of the large number of TA modules in M. tuberculosis, the contribution of individual toxins might be marginal, and measurable effects may require the concerted action of multiple toxins in vivo.
The chromosomal TA loci encoded in the genome of M. tuberculosis belong to five families, mazEF, relBE, higBA, parDE, and vapBC (4, 7). MazF toxins from many bacteria have been studied, including several from M. tuberculosis (19–21). In all cases, MazF acts as a sequence-specific endoribonuclease that cleaves mRNA independently of the ribosome, thereby inhibiting translation. The E. coli RelE toxin also perturbs translation by interacting with the ribosome to enable RelE cleavage of mRNAs positioned in the ribosomal A site (22–25). The HigB toxin in the Rts1 plasmid from Proteus vulgaris functions in a manner analogous to RelE and inhibits protein synthesis by associating with the ribosome and cleaving mRNA at A-rich sequences (26, 27). The ParE toxin in the RK2 plasmid, on the other hand, perturbs DNA replication by inhibiting the function of DNA gyrase (28). Therefore, although the mechanism of action is known for the other TA system families found in M. tuberculosis, the precise function of the VapC toxins remains unclear.
The vapBC (virulence-associated protein) module is composed of the VapB antitoxin and the VapC toxin. The VapC toxins present in M. tuberculosis are grouped together based on the presence of a PIN (PilT N-terminal) domain containing a conserved quartet of acidic residues and a fifth invariant serine or threonine residue responsible for coordinating a Mg2+ ion(s) in the catalytic center (29). The presence of a PIN domain suggests a putative role for the VapC toxins as Mg2+-dependent ribonucleases (29–31). The PIN domain-containing protein PAE2754 from Pyrobaculum aerophilum cleaves single-stranded DNA flap structures in a Mg2+-dependent fashion, whereas VapC-1 from nontypeable Haemophilus influenzae cleaves single-stranded RNA, and the VapBC-mt5 toxin-antitoxin protein complex (Rv0626-Rv0627) from M. tuberculosis appeared to cleave double-stranded RNA in vitro in the presence of Mg2+ (30, 32, 33). In contrast, no nuclease activity was detected for FitB (the PIN domain toxin from the fast intracellular trafficking locus) from Neisseria gonorrhoeae, VapC from a Shigella flexneri virulence plasmid, or VapC from the Salmonella enterica genome (34, 35). However, recently the Shigella and Salmonella VapC toxins were shown to cleave tRNAfMet at a single site between the anticodon stem and loop (35). These contradictory results suggest that not all VapCs function by cleaving RNA and that the specific mechanism of action may vary among different members of this family of PIN domain proteins.
To better understand the function of the VapC toxin family, we performed detailed biochemical characterization of the VapC toxin from the vapBC-mt4 (Rv0596c-Rv0595c) TA system. Our results demonstrate that VapC-mt4 targets RNA in a manner distinct from other TA toxins. Whereas other TA toxins, such as MazF, YafQ, HigB, and ChpBK, inhibit translation by cleaving mRNA, cleavage of mRNA is not the primary activity responsible for VapC-mt4-mediated translation inhibition. Instead, VapC-mt4 toxicity appears to result from the stable, selective binding of RNA containing an ACGC or AC(A/U)GC consensus sequence.
EXPERIMENTAL PROCEDURES
Strains, Plasmids, and Reagents
The E. coli strains BL21(DE3)pLysE (F− ompT hsdSβ(rβ−, mβ−) dcm gal (DE3) pLysE (CamR)) (Novagen) and BW25113Δ6 (F− lacIq rrnBT14 ΔlacZWJ16 hsdR514 ΔaraBADAH33 ΔrhaBADLD78 ΔmazEF ΔchpBIK ΔrelBE ΔyefM-yoeB ΔdinJ-yafQ) (37) were used for all protein expression and toxicity studies. E. coli K12 Mach1-T1 cells (F− ΔrecA1398 endA1 tonA Φ80(lacZ)ΔM15 ΔlacX74 hsdR(rk−mk+)) (Invitrogen) were used for all cloning experiments. Plasmids used in this study include pINIII (38–40), pBAD18, pBAD33 (41), pMC1s (42), and pET21c and pET28a (Novagen). The 23 VapBC and VapC ORFs were PCR-amplified from M. tuberculosis H37Rv genomic DNA using primers with 5′NdeI-BamHI/EcoRI3′ ends and cloned into the corresponding sites of pET21c to create pET21c-VapBC and pET21c-VapC, respectively. The pET21c-VapC plasmids were digested with 5′XbaI-HindIII3′ and the resulting VapC-containing fragment was cloned into the corresponding sites of pBAD33 to create pBAD33-VapC. Plasmid pET21c-VapC-mt4 was digested with 5′XbaI-EcoRI3′ and the resulting VapC-containing fragment was cloned into pBAD18 to create pBAD18-VapC-mt4. Plasmid pET21c-VapC-mt4 was digested with 5′NdeI-EcoRI3′ and the resulting VapC-containing fragment was cloned into the corresponding sites of pET28a to create pET28a-His6-VapC-mt4. The D9A, E40A, D98A, T114A, and D116A mutations in the VapC-mt4 ORF were created by PCR amplification using pBAD33-VapC-mt4 and cloned into the 5′XbaI-HindIII3′ sites of pBAD33 to create pBAD33-VapC-mt4-D9A, pBAD33-VapC-mt4-E40A, pBAD33-VapC-mt4-D98A, pBAD33-VapC-mt4-T114A, and pBAD33-VapC-mt4-D116A, respectively. In each case, the residue was mutated to the most abundant E. coli codon for alanine (GCG). The VapB-mt4 ORF was PCR-amplified from the pET21c-VapBC-mt4 plasmid using primers with 5′NdeI-BamHI3′ ends and cloned into the corresponding sites of pINIII and pET28a to create pINIII-VapB-mt4 and pET28a-His6-VapB-mt4, respectively. The VapB-mt4 and VapC-mt4 ORFs were PCR-amplified from the pET21c-VapBC-mt4 plasmid with 5′BamHI-EcoRI3′ ends and additional GA nucleotides on the 5′-end to maintain frame and cloned into the corresponding sites of pET21c to create pET21c-T7-VapB-mt4 and pET21c-T7-VapC-mt4, respectively. The VapBC-mt4 and VapC-mt4 ORFs were PCR-amplified from M. tuberculosis H37Rv genomic DNA using primers with 5′ClaI-EcoRI3′ ends and cloned into the corresponding sites of pMC1s to create pMC1s-VapBC and pMC1s-VapC, respectively.
All E. coli liquid cultures were grown in M9 minimal medium supplemented with 0.2% glucose or 0.1% glycerol at 37 °C. All M. smegmatis and M. tuberculosis liquid cultures were grown in 7H9 Middlebrook medium supplemented with 0.05% Tween 80, 0.5% bovine albumin, 0.2% glucose, and 0.085% NaCl (7H9-TW80-ADN). Spent culture supernatant was prepared from an early stationary phase M. tuberculosis H37Rv culture (A600 between 1.1 and 1.2) by centrifugation (2000 × g, 10 min, 4 °C) followed by filtration through a 0.22-μm filter (Millipore). 1 mm isopropyl 1-thio-β-d-galactopyranoside, 0.2% arabinose, 100 ng/ml anhydrotetracycline (M. smegmatis), 200 ng/ml anhydrotetracycline (M. tuberculosis), 50 μg/ml ampicillin, 20 μg/ml kanamycin (mycobacteria), 50 μg/ml kanamycin (E. coli), and 25 μg/ml chloramphenicol were added to culture medium as required. The accuracy of DNA sequences in PCR products used for cloning was confirmed by DNA sequence analysis. Polyclonal antibodies used for Western analysis were produced in rabbits using purified His6-VapB-mt4 (Rv0596c) or His6-VapC-mt5 (Rv0627) as antigens (Pocono Rabbit Farm and Laboratory, Canadensis, PA).
Purification of Recombinant Protein
E. coli BL21(DE3)pLysE was transformed with pET28a-His6-VapB-mt4, pET28a-His6-VapC-mt4, or pET28a-His6-MazF-mt1 plasmids and grown in 1 liter of M9 minimal medium containing 0.2% glucose at 37 °C until an A600 between 0.6 and 0.8 was reached. Isopropyl 1-thio-β-d-galactopyranoside was then added to a final concentration of 1 mm, and the cultures were incubated for an additional 4 h. The cultures were harvested by centrifugation (6,000 rpm, 10 min, 4 °C) in a Sorvall SA-600 rotor, and the cell pellet was resuspended in 25 ml of lysis buffer (50 mm NaH2PO4 (pH 8.0), 500 mm NaCl, 20 mm imidazole, 10 mm β-mercaptoethanol, 1 mm PMSF, 1 mg/ml lysozyme) and incubated for 30 min on ice. Triton X-100 was then added to a final concentration of 2%. Cells were lysed by sonication (Branson Digital Sonifier 250; 30% amplitude, 30 cycles, 10 s on, 30 s off, 4 °C), and the lysate was cleared by centrifugation (15,000 rpm, 10 min, 4 °C). The supernatant was adsorbed onto 1 ml of pre-equilibrated Ni-NTA-agarose resin (Qiagen) in a 50-ml disposable centrifuge tube with mild agitation (1 h, 4 °C), and the resin was collected by centrifugation (1,000 rpm, 5 min, 4 °C). The adsorbed resin was washed twice with 50 ml of Wash Buffer I (50 mm NaH2PO4 (pH 8.0), 500 mm NaCl, 20 mm imidazole, 5 mm β-mercaptoethanol, 0.5% Triton X-100) with mild agitation (30 min, 4 °C) and collected by centrifugation (1,000 rpm, 5 min, 4 °C). The washed resin was resuspended in 50 ml of Wash Buffer I, loaded into a column, washed extensively (i.e. three times with 20 ml of Wash Buffer I, three times with 20 ml of Wash Buffer II (50 mm NaH2PO4 (pH 8.0), 500 mm NaCl, 43 mm imidazole, 5 mm β-mercaptoethanol, 0.45% Triton X-100), and once with 10 ml of Wash Buffer III (50 mm NaH2PO4 (pH 8.0), 500 mm NaCl, 66 mm imidazole, 5 mm β-mercaptoethanol, 0.4% Triton X-100)), and eluted in 5 ml of elution buffer (50 mm NaH2PO4 (pH 8.0), 500 mm NaCl, 250 mm imidazole, 5 mm β-mercaptoethanol). The purified protein was dialyzed against 1 liter of dialysis buffer (10 mm Tris (pH 8.0), 100 mm NaH2PO4, 10 mm β-mercaptoethanol) (≥2 h, three times) and concentrated using a 3,000 NMWL Amicon Ultra-4 Centrifugal Filter (Millipore) as required. Glycerol was added to a final concentration of 5%, and the purified protein was aliquoted and stored at −80 °C.
Protein Interaction
E. coli BL21(DE3)pLysE was transformed with pET28a or pET28a-His6-VapB/C-mt4 and pET21c-T7-VapB/C-mt4 and grown in 500 ml of M9 minimal medium containing 0.2% glucose at 37 °C until an A600 between 0.6 and 0.8 was reached. Isopropyl 1-thio-β-d-galactopyranoside was then added to a final concentration of 1 mm, and the cultures were incubated for an additional 4 h. The cultures were harvested by centrifugation (6,000 rpm, 10 min, 4 °C) in a Sorvall SA-600 rotor, and the cell pellet was resuspended in 25 ml of lysis buffer (50 mm NaH2PO4 (pH 8.0), 500 mm NaCl, 10 mm imidazole, 20 mm β-mercaptoethanol, 1 mm PMSF, 1 mg/ml lysozyme) and incubated for 30 min on ice. Cells were lysed by sonication (Branson Digital Sonifier 250; 30% amplitude, 30 cycles, 10 s on, 30 s off, 4 °C), and the lysate was cleared by centrifugation (15,000 rpm, 10 min, 4 °C). The supernatant was adsorbed onto 500 μl of pre-equilibrated Ni-NTA-agarose resin (Qiagen) and purified as recommended by Qiagen. The samples were separated by 17.5% SDS-PAGE and visualized by Coomassie staining or Western analysis using a 1:300,000 dilution of the VapB-mt4 polyclonal antibody or 1:50,000 dilution of the VapC-mt5 polyclonal antibody.
Assessment of Protein Synthesis in Vivo
E. coli BW25113Δ6 cells containing pBAD33-VapC-mt4 were grown in M9 minimal medium containing 0.1% glycerol and 1 mm of all amino acids except cysteine and methionine at 37 °C until an A600 between 0.2 and 0.3 was reached. The culture was then split into equal portions, and arabinose was added to one portion at a final concentration of 0.2%, whereas an equal volume of water was added to the other as a control. 500-μl aliquots were removed at 0, 10, 20, 30, 60, 120, and 180 min postinduction and incubated with 30 μCi of [35S]methionine at 37 °C. After 1 min of incorporation, the samples were chased with 0.3 mg of cold methionine at 37 °C for 5 min. 50 μl of the culture was applied to a cellulose filter disc (Whatman). The filters were boiled for 30 min in 10% trichloroacetic acid (TCA) and washed three times with 10% TCA and once with acetone. The amount of radioactivity incorporated was determined using a liquid scintillation counter. Cell pellets were collected by centrifugation (3,000 rpm, 1 min, 4 °C) and resuspended in equivalent volumes of Tris-EDTA buffer (pH 8.0) and 2× Laemmli buffer (125 mm Tris (pH 6.8), 20% glycerol, 4% SDS, 0.01% bromphenol blue). Incorporation was visualized by autoradiography following 17.5% SDS-PAGE.
Assessment of DNA and RNA Synthesis in Vivo
E. coli BW25113Δ6 cells containing pBAD18-VapC-mt4 were grown in M9 minimal medium containing 0.1% glycerol at 37 °C until an A600 between 0.2 and 0.3 was reached. Then [methyl-3H]thymidine or [methyl-3H]uridine was added to a final concentration of 1 μCi/ml. The culture was then split into three equal portions, arabinose was added to one portion at a final concentration of 0.2%, chloramphenicol was added to another at a final concentration of 25 μg/ml, and an equivalent volume of water was added to the remaining portion as a control. At 0, 10, 20, 30, 60, 120, and 180 min postinduction, 50 μl of culture was applied to a cellulose filter disc (Whatman). The filters were washed three times with 10% TCA and once with 95% ethanol. The amount of radioactivity incorporated was determined using a liquid scintillation counter.
Analysis of mRNA Levels in E. coli
E. coli BW25113Δ6 cells containing pBAD33-VapC-mt4 were grown in M9 minimal medium containing 0.1% glycerol at 37 °C until an A600 between 0.3 and 0.4 was reached. The culture was then split into two equal portions, and arabinose was added to one portion at a final concentration of 0.2%, whereas an equal volume of water was added to the other as a control. 45 ml of culture was added to 6 ml of ice-cold Stop Solution (95% ethanol, 5% phenol), and the cells were harvested at 0, 20, 40, 60, 80, and 100 min postinduction by centrifugation (3,000 rpm, 15 min, 4 °C). Bacterial pellets were stored at −80 °C until all time points were collected. Bacterial pellets were resuspended in 500 μl of Buffer A (173 mm sodium dodecyl sulfate (SDS), 20 mm sodium acetate (pH 4.0), 10 mm EDTA) by vortexing, mixed with 500 μl of saturated phenol (pH 4.3) by vortexing, and then incubated at 65 °C for 6 min with mixing every 1 min. The organic and aqueous phases were separated by centrifugation (13,200 rpm, 15 min, 4 °C). The aqueous phase was transferred to a fresh tube and re-extracted with an equal volume of chloroform. The organic and aqueous phases were separated by centrifugation (13,200 rpm, 15 min, 4 °C). The aqueous phase was transferred to a fresh tube containing 1 ml of 100% ethanol and incubated at −70 °C overnight. The RNA was pelleted by centrifugation (13,200 rpm, 30 min, 4 °C), washed with 1 ml of 70% ethanol, and incubated at −70 °C for 30 min. The RNA was pelleted by centrifugation (13,200 rpm, 30 min, 4 °C), dried at room temperature for 5 min, and resuspended in 50 μl of diethyl pyrocarbonate-treated water. The radiolabeled DNA fragments used for hybridization to the Northern blots were derived from PCR products for the open reading frames of the E. coli genes ompA (outer membrane porin protein A) and lpp (major outer membrane lipoprotein) and a 600-bp region immediately upstream of tufA (elongation factor Tu) using the Random Primed DNA Labeling kit (Roche Applied Science).
MS2 RNA Cleavage Assays
MS2 RNA cleavage assays were performed with 0.7 pmol of MS2 RNA (Roche Applied Science), 0.5 μl of RNase inhibitor (Roche Applied Science), and 61.8 pmol of His6-VapC-mt4 or His6-MazF-mt1 in a 10-μl reaction containing 10 mm Tris (pH 7.8), 150 mm NaCl, 10 mm MgCl2. 61.8 pmol of His6-VapC-mt4 was preincubated with 123.6 or 247.2 pmol of His6-VapB-mt4 for 15 min at 37 °C prior to starting the reaction. The reactions were performed at 37 °C, and time points were taken at 0, 1, 3, 6, 12, and 18 h. The reactions were stopped by the addition of 10 μl of sequence loading buffer (95% formamide, 20 mm EDTA, 0.05% bromphenol blue, 0.05% xylene cyanol FF). The samples were incubated at 95 °C for 5 min prior to electrophoresis on an 8% polyacrylamide, 7 m urea gel and stained with ethidium bromide.
Synthetic RNA Cleavage Assays
The synthetic RNAs used were: NWO1355 (+ACGC), 5′-UAAGAAGGAAGAUACGCGAUAUGAAUCAAA-3′; NWO1430, 5′-AGGAAGAUACGCGAUAUGAA-3′; NWO1434, 5′-AGGAAGAUACACGAUAUGAA-3; and NWO1435, 5′-AGGAAGAUACGUGAUAUGAA-3. The RNAs were 5′-end-labeled with [γ-32P]ATP using T4 polynucleotide kinase (New England Biolabs). The cleavage reactions were performed with 0.2 pmol of radioactively labeled RNA, 61.8 pmol of His6-VapC-mt4, and/or 247.2 pmol of His6-VapB-mt4 as described above. RNA ladders were prepared by digesting 0.4 pmol of 5′-end-labeled RNA with 0.1 units of RNaseT1 (Sigma) in 20 mm sodium citrate (pH 5), 7 m urea, 1 mm EDTA for 15 min at 25 °C and by partial alkaline hydrolysis in 100 mm NaOH for 2 min at 75 °C. The reactions were stopped by the addition of 10 μl of sequence loading buffer. The samples were incubated at 95 °C for 5 min prior to electrophoresis on a 15% polyacrylamide, 7 m urea gel followed by autoradiography.
RNA Binding Assays
Thirty nucleotide synthetic RNAs were 5′-end-labeled with [γ-32P]ATP using T4 polynucleotide kinase (New England Biolabs). The RNAs used were: NWO1353 (−ACGC), 5′-CGCAUGGCGUUCGUACUUAAAUAUGGAACU-3′; and NWO1355 (+ACGC), 5′-UAAGAAGGAAGAUACGCGAUAUGAAUCAAA-3′. The 15-μl reactions contained 0.2 pmol of radioactively labeled RNA and 15.45, 30.9, 46.35, or 61.8 pmol of His6-VapC-mt4 in 10 mm Tris (pH 8.0), 10 mm KCl, 1 mm EDTA, 7.4% glycerol. Specific and nonspecific binding was shown using a 50-fold concentration (10 pmol) of unlabeled synthetic RNA. The binding reactions were incubated on ice for 20 min. For electrophoretic mobility shift assays (EMSAs), the samples were separated on a 15% native acrylamide (19:1) gel in 1× Tris-Borate-EDTA buffer and run at 110 V at 4 °C until the dye front was ¾ down the gel. The RNA was transferred to nitrocellulose using a Trans-Blot semidry electrophoretic transfer cell (Bio-Rad) at 0.2 A for 30 min in 0.5× Tris-Borate-EDTA and visualized by autoradiography.
In Vivo Primer Extension Analysis
Total RNA was extracted as described above. Primer extension reactions were carried out at 47 °C for 1 h in a 20-μl reaction containing 0.7 pmol of 5′-end-labeled primer, 1 mm dNTPs, 0.5 μl of RNase inhibitor (Roche Applied Science), and 0.5 μl avian myeloblastosis virus reverse transcriptase (New England Biolabs) in 50 mm Tris (pH 8.3), 75 mm potassium acetate, 8 mm magnesium acetate, 10 mm DTT. The primers were 5′-end-labeled with [γ-32P]ATP using T4 polynucleotide kinase (New England Biolabs). Cleavage products in tufA, ompA, and lpp were detected using the following primers: NWO1169, 5′-GGCGTTCCGTACATCATCGTGT-3′ (Fig. 3, A and B); NWO938, 5′-GGTATCAAAGACGTTGTAACTCAGC-3′ (Fig. 3C); and NWO1121, 5′-GTGAAGTGAAAAATGGCGCACATTGTG-3′ (Fig. 3D).
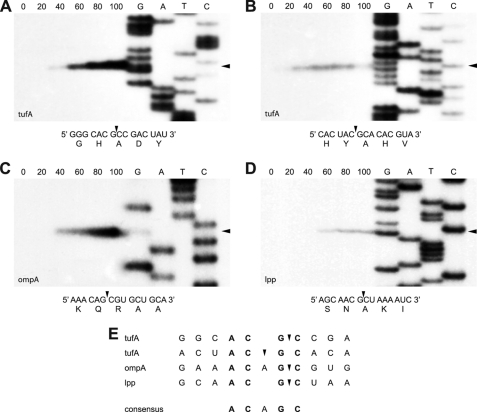
FIGURE 3.
VapC-mt4-mediated sequence-specific RNA cleavage. Primer extension analysis for tufA (A and B), ompA (C), and lpp (D) mRNAs extracted from E. coli BW25113Δ6 cells following induction of VapC-mt4. Numbers indicate the time (min) after VapC-mt4 induction. Cleavage sites/products are indicated by black arrowheads on the right side of gels and in the relevant RNA sequences/corresponding amino acid sequences below each gel. Lanes G, A, T, and C correspond to DNA sequencing ladders prepared using the same primers used in the primer extension reactions. E, alignment of sequences containing VapC-mt4 cleavage sites. The ACGC consensus sequence is in bold.
In Vitro Primer Extension Analysis
The MS2 RNA was incubated as described above with 61.8 pmol of His6-VapC-mt4, 247.2 pmol of His6-VapB-mt4, or 61.8 pmol of His6-VapC-mt4 plus 247.2 pmol of His6-VapB-mt4. Primer extensions were carried out using radioactively labeled primers as described above. The primers used were: NWO1459, 5′-CTCACAACTCGCGACGCAAACC-3′ (Fig. 5A); J1, 5′-CATCAAGTTAGATGGCCGTC-3′ (Fig. 5B); NWO1461, 5′-GACTGGCTCCTACCTGTAGGTAAC-3′ (Fig. 5C); G, 5′-CATTAATCAGGCAACGGCTCT-3′ (Fig. 5D); NWO1462, 5′-CAAGCAACTTCGTAACGGGGTC-3′ (Fig. 5E); E, 5′-CTCTTTATGTATTGATCTTC-3′ (Fig. 5F); NWO1463, 5′-CTTAAGGGACGAATTGCTCACAAAGC-3′ (Fig. 5G); D, 5′-CCCCCGCGCTCTGAGAGCGG-3′ (Figs. 5, H and I); B4, 5′-GTTCTTCGGTGGGACGGACC-3′ (Fig. 5J); and B3, 5′-GTAAACGGGGTGGGTGTGCT-3′ (Fig. 5K).
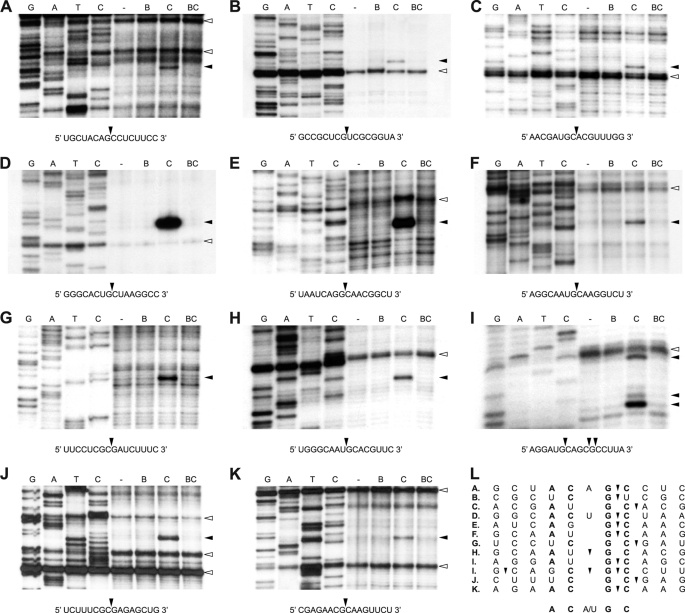
FIGURE 5.
VapC-mt4-mediated sequence-specific MS2 RNA cleavage. A–K, primer extension analysis for MS2 RNA identified 12 VapC-mt4 cleavage sites. Lanes labeled “−” represent control reactions to which no proteins were added. In lanes labeled “B,” purified His6-VapB-mt4 was incubated with the MS2 RNA. In lanes labeled “C,” the addition of His6-VapC-mt4 resulted in the cleavage of the MS2 RNA. In lanes labeled “BC,” preincubation of His6-VapC-mt4 with His6-VapB-mt4 prior to the addition of the MS2 RNA prevented RNA cleavage. Cleavage sites/products are indicated by black arrowheads on the right side of gels and in the relevant RNA sequences below each gel. Apparent secondary structure is indicated by white arrowheads on the right side of gels. Lanes G, A, T, and C correspond to DNA sequencing ladders prepared by reverse transcription using the same primers used in the primer extension reactions. Data shown are representative of two independent experiments. L, alignment of sequences containing VapC-mt4 cleavage sites. The ACGC consensus sequence is in bold.
The DNA sequencing ladders for the in vivo primer extension reactions were prepared with a Sequenase Version 2.0 DNA Sequencing kit (USB Corp.) as recommended by the manufacturer. The DNA sequencing ladders for the in vitro primer extension reactions were also prepared using the Sequenase Version 2.0 DNA Sequencing kit (USB Corp.) with minor modifications. The primer was annealed to the MS2 RNA in a 10-μl reaction containing 1 pmol of primer, 1.4 pmol of MS2 RNA (Roche Applied Science), and 1.4 μl of RNase inhibitor (Roche Applied Science) in 50 mm Tris (pH 8.3), 75 mm potassium acetate, 8 mm magnesium acetate, 10 mm DTT. The DNA sequencing reaction was performed as recommended by the manufacturer using 1 unit of avian myeloblastosis virus reverse transcriptase (New England Biolabs) in place of Sequenase DNA polymerase. The primer extension and sequencing reactions were stopped by the addition of 12 or 6 μl of sequence loading buffer, respectively. The samples were incubated at 95 °C for 5 min prior to electrophoresis on a 6% polyacrylamide, 7 m urea gel followed by autoradiography.
In Vitro Protein Synthesis
Prokaryotic cell-free protein synthesis was carried out using the E. coli T7 S30 Extract System for Circular DNA (Promega). The PinPoint Xa Control vector supplied with the kit was used as the template. The 25-μl reaction mixture contained 154 pmol of His6-VapC-mt4, 616 pmol of His6-VapB-mt4, or 154 pmol of His6-VapC-mt4 plus 616 pmol of His6-VapB-mt4, respectively. The samples were preincubated for 15 min at 37 °C prior to the addition of 0.24 μg of the PinPoint Xa Control vector after which the samples were incubated for an additional 1 h at 37 °C. Samples were separated by 17.5% SDS-PAGE and visualized by autoradiography.
Reverse Transcription-PCR
Total RNA was extracted from the E. coli T7 S30 Extract System with saturated phenol (pH 4.3). The samples were treated with 1 unit of RQ1 RNase-free DNase (Promega) in 40 mm Tris (pH 8.0), 10 mm MgSO4, 1 mm CaCl2 for 30 min at 37 °C. The RQ1 RNase-free DNase was removed by extraction with saturated phenol (pH 4.3) followed by ethanol precipitation. Reverse transcription was carried out on equal volumes of RNA from each sample using the following primer: NWO1443, 5′-TTACGCCCCGCCCTGCCAC-3′. The reverse transcriptase was then heat-inactivated for 5 min at 95 °C, and all samples were treated with DNase-free RNase for 10 min at 37 °C. The samples were then used as templates for PCR using the following primers to amplify the chloramphenicol acetyltransferase (CAT) fusion transcript: NWO1442, 5′-ATGAAACTGAAGGTAACAGTCAACGGC-3′; and NWO1443, 5′-TTACGCCCCGCCCTGCCAC-3′. The PCR reactions were then run on an 1% agarose gel and visualized by staining with EtBr.
Ribosome Profile Analysis
Polysomes were isolated as described previously (43–45). Briefly, E. coli BW25113Δ6 cells containing pBAD18-VapC-mt4 were grown in M9 minimal medium supplemented with 0.1% glycerol at 37 °C until an A600 between 0.3 and 0.4 was reached. The culture was then split into three samples (−VapC, −Clm; −VapC, +Clm; +VapC; −Clm). Arabinose was added to one sample (+VapC, −Clm) at a final concentration of 0.2%, whereas an equal volume of water was added to the other samples (−VapC, −Clm; −VapC; +Clm) as a control. After 60 min, chloramphenicol was added to one sample (−VapC, +Clm) at a final concentration of 0.1 mg/ml, whereas an equal volume of water was added to the other samples (−VapC, −Clm; +VapC, −Clm) as a control. After 3 min, the cells were poured over an equal volume of ice and harvested by centrifugation (14,500 rpm, 5 min, 4 °C) in a Sorvall SA-600 rotor. The cell pellet was resuspended in polysome profile buffer (20 mm Tris (pH 7.5), 100 mm NH4Cl, 10 mm MgCl2, 5 mm DTT). Lysozyme was added to a final concentration of 1 mg/ml, and the cells were frozen in a dry ice/ethanol bath and thawed in a 5 °C water bath three times. 600 μg of total RNA was layered onto a 5–40% (w/v) continuous sucrose gradient in polysome profile buffer and centrifuged (35,000 rpm, 3.5 h, 4 °C) in a Beckman SW41 rotor. Gradients were fractionated and analyzed with continuous monitoring at 254 nm. Fractions corresponding to the polysomes; 70, 50, and 30 S; and tRNAs/RNAs not associated with ribosomes or ribosomal subunits were concentrated by centrifugation (45,000 rpm, 20 h, 4 °C) in a Thermo Fisher Scientific T647.5 rotor and subjected to Western analysis using a 1:50,000 dilution of the VapC-mt5 (Rv0627) polyclonal antibody.
tRNAfMet Cleavage Activity Assay
Purified tRNAfMet from E. coli was purchased from Chemical Block (Moscow, Russia). M. tuberculosis tRNAfMet was synthesized in vivo following the method described by Sisido et al. (46) with minor modifications. Briefly, a synthetic DNA oligonucleotide containing the T7 RNA polymerase promoter and the 5′-end of the M. tuberculosis tRNAfMet gene was annealed to a second oligonucleotide corresponding to the 3′-end of the M. tuberculosis tRNAfMet gene. The annealed oligos were then extended using Taq DNA polymerase to create dsDNA containing the entire tRNAfMet gene preceded by the T7 promoter. The product was then run on a 2% agarose gel to confirm its size and purified using the QIAquick gel extraction kit (Qiagen). The sequence of the product was confirmed by automated DNA sequence analysis. 200 ng of M. tuberculosis tRNAfMet dsDNA was then transcribed in vitro using the RiboMAX Large Scale RNA Production System (Promega) as recommended by the manufacturer. The transcription reaction was then run on a 9% polyacrylamide, 7 m urea gel and visualized by staining with EtBr to confirm the size and purity of the tRNAfMet transcript.
The tRNAfMet transcript was purified from the gel as follows. The RNA band was excised from the gel and incubated for 18 h at 37 °C in elution buffer (1 mm EDTA, 0.5 m ammonium acetate, 10 mm magnesium acetate, 0.1% SDS). The eluate was then collected, and the gel pieces were washed in fresh elution buffer. The elution and wash were combined, and the RNA was collected by ethanol precipitation.
Cleavage of E. coli and M. tuberculosis tRNAfMet by VapC-mt4 was performed in vitro following the assay described by Winther and Gerdes (47). 20 pmol of E. coli tRNAfMet was incubated with 2, 25, or 50 pmol of VapC-mt4, or 2 pmol of M. tuberculosis tRNAfMet was incubated with 0.2, 2.5, or 5 pmol of VapC-mt4 at 37 °C for 15 min in 10 mm HEPES (pH 7.5), 15 mm potassium chloride, 3 mm magnesium chloride, 10% glycerol. The products of these reactions were run on a 9% polyacrylamide, 7 m urea gel and visualized by staining the E. coli tRNAfMet with EtBr and the M. tuberculosis tRNAfMet with SYBR Gold (Invitrogen).
RESULTS
First, we determined whether expression of any of the 23 VapC toxins originally annotated in the genome of M. tuberculosis (4) caused growth arrest in E. coli. This was accomplished by individually expressing each of the 23 VapC toxins using the arabinose-inducible pBAD33 plasmid in an E. coli BW25113Δ6 strain that lacks the loci for the six well characterized E. coli chromosomal TA modules (mazEF, chpBIK, relBE, hipBA, yefM-yoeB, and dinJ-yafQ) (supplemental Fig. S1). Use of the BW25113Δ6 strain in all experiments precluded the activation of these TA toxins as a result of the stress imparted by VapC expression. Expression of four VapC toxins, VapC-mt4 (Rv0595c), VapC-mt11 (Rv1561), VapC-mt19 (Rv2548), and VapC-mt20 (Rv2549c), inhibited cell growth in E. coli. From this group, we elected to focus on the vapBC-mt4 operon as a representative of this class of TA systems.
The vapBC-mt4 module exhibits the features of a typical TA system: the open reading frames overlap or have minimal distance between them (vapB-mt4 and vapC-mt4 overlap by 4 base pairs) (supplemental Fig. S2), both genes encode relatively small proteins (VapB-mt4 is 85 amino acids and 9.7 kDa; VapC-mt4 is 130 amino acids and 14.1 kDa), the proteins have opposing isoelectric points (VapB-mt4 pI is 7.70; VapC-mt4 pI is 4.86), and the proteins form a stable protein complex. This was demonstrated by co-expressing VapB-mt4 with a T7 tag fused to its N terminus and VapC-mt4 with a His6 tag attached to its N terminus. After the resulting cell extract was passed over a Ni-NTA column, we were able to co-purify T7-VapB-mt4 (lacking the His6 tag) with His6-VapC-mt4, demonstrating that VapB-mt4 and VapC-mt4 interact and form a stable protein complex (supplemental Fig. S3A). Complex formation was confirmed by performing the reciprocal experiment using His6-VapB-mt4 and T7-VapC-mt4 (supplemental Fig. S3B).
VapC-mt4 Expression Leads to Growth Arrest in E. coli, M. smegmatis, and M. tuberculosis That Can Be Rescued by Co-expression with Its Cognate Antitoxin VapB-mt4
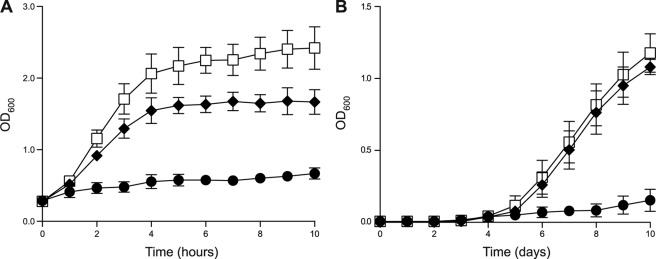
We then showed that the vapBC-mt4 module constitutes a functional TA system as induction of the VapC-mt4 toxin in E. coli resulted in growth arrest that could be rescued by co-expression of the VapB-mt4 antitoxin from an independent plasmid (Fig. 1A). We then demonstrated that the VapBC-mt4 TA module is active in mycobacteria. Expression of VapC-mt4 using the anhydrotetracycline-inducible pMC1s vector in M. smegmatis (a rapidly growing nonpathogenic mycobacterium) or M. tuberculosis inhibited cell growth, whereas co-expression of VapB-mt4 and VapC-mt4 from the same plasmid relieved the growth defect (supplemental Fig. S4 and Fig. 1B). These results demonstrate that vapBC-mt4 constitutes a functional TA system where VapB-mt4 is the cognate antitoxin for VapC-mt4 and validate the use of E. coli for the characterization of VapC-mt4 activity.
FIGURE 1.
Expression of VapC-mt4 leads to growth arrest. A, growth profile for VapB-mt4 and VapC-mt4 uninduced (□), VapB-mt4 and VapC-mt4 induced (♦), or pINIII and VapC-mt4 induced (●) in E. coli BW25113Δ6 containing either pINIII or pINIII-VapB-mt4 and pBAD33-VapC-mt4 grown at 37 °C in M9 minimal medium containing 0.1% glycerol. Both 0.2% arabinose and 1 mm isopropyl 1-thio-β-d-galactopyranoside were added to the induced samples. An empty pINIII vector was used to assess VapC-mt4 toxicity because leaky expression of VapB-mt4 from pINIII-VapB-mt4 abrogated VapC-mt4 toxicity. B, growth profile for pMC1s (□), pMC1s-VapBC-mt4 (♦), or pMC1s-VapC-mt4 (●) in M. tuberculosis H37Rv grown at 37 °C in 7H9-TW80-ADN medium containing 30% spent culture supernatant and 200 ng/ml anhydrotetracycline. Cultures were inoculated with single colonies. Additional anhydrotetracycline was added every 48 h to maintain the anhydrotetracycline concentration between 12.5 and 200 ng/ml. Data points represent the average of three independent experiments; error bars represent the S.D.
VapC-mt4 Inhibits Protein Synthesis
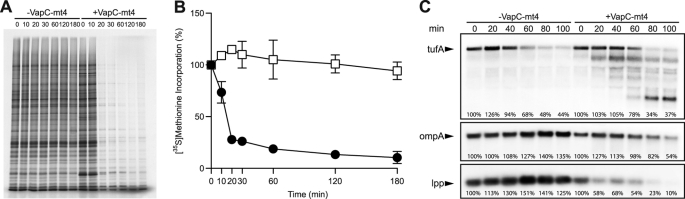
We next sought to identify the cellular process perturbed by VapC-mt4. To determine whether VapC-mt4 expression affected translation, transcription, or DNA replication, we measured the incorporation of radioactive precursors for protein ([35S]methionine), RNA ([3H]uridine), and DNA ([3H]thymidine) synthesis. Expression of VapC-mt4 reduced [35S]methionine incorporation by 76% within 20 min and by as much as 94% after 180 min relative to the control (Fig. 2, A and B). Curiously, VapC-mt4 expression decreased the incorporation of [3H]thymidine after 60 min and increased the incorporation of [3H]uridine after 60 min (supplemental Fig. S5). As VapC-mt4 decreased [35S]methionine incorporation to the greatest extent, we hypothesized that inhibition of protein synthesis was directly responsible for the growth defect. The effects on [3H]thymidine and [3H]uridine incorporation appear to be indirect effects of inhibiting translation because treatment with chloramphenicol (which also arrests growth through translation inhibition) in the absence of toxin expression exhibited the same trends noted for VapC-mt4 expression (supplemental Fig. S5).
FIGURE 2.
Expression of VapC-mt4 leads to translational arrest and RNA cleavage. A, [35S]methionine incorporation into E. coli BW25113Δ6 cells with or without VapC-mt4 expression. Equivalent amounts of cell lysate were subjected to SDS-PAGE followed by autoradiography. Numbers indicate the time (min) after VapC-mt4 induction. Data shown are representative of three independent experiments. B, quantification of [35S]methionine incorporation into E. coli BW25113Δ6 cells with (●) or without (□) VapC-mt4 expression. Data points represent the average of three independent experiments; error bars represent the S.D. Time points correspond to those in A. C, Northern analysis for tufA, ompA, and lpp mRNAs extracted from E. coli BW25113Δ6 cells with or without expression of VapC-mt4. VapC-mt4 expression results in a gradual decrease in the steady-state levels of tufA, ompA, and lpp mRNAs. Numbers above lanes indicate the time (min) after VapC-mt4 induction. Numbers below lanes indicate the band intensity relative to the zero time point.
VapC-mt4 Arrests Translation Elongation
To learn more about the mechanism of VapC-mt4-mediated translation inhibition, we compared the ribosome profiles obtained from cells overexpressing VapC-mt4 (supplemental Fig. S6C) with those from untreated (supplemental Fig. S6A) or chloramphenicol-treated (supplemental Fig. S6B) cells. Expression of VapC-mt4 resulted in a marked increase in the recovery of polysomes, mirroring the polysome stabilization seen for chloramphenicol-treated cells. The accumulation of polysomes, which represent stalled translation elongation complexes, indicates that VapC-mt4 expression inhibits protein synthesis by arresting translation elongation.
To determine whether VapC-mt4 increased the recovery of polysomes through interactions with translated mRNAs or rRNA in ribosomes, we performed Western analysis on the fractions spanning the sucrose gradient. However, we were unable to detect VapC-mt4 in the polysome fraction containing translated mRNA or in the 70, 50, and 30 S fractions containing rRNA. However, we did detect VapC-mt4 in the fraction containing tRNA and RNAs not associated with ribosomes or ribosomal subunits (supplemental Fig. S6C).
VapC-mt4 Does Not Cleave tRNAfMet
In contrast to M. tuberculosis, there is only one VapC toxin in S. flexneri and S. enterica derived from a single plasmid- and chromosomally encoded vapB-vapC TA module, respectively. Each of these VapC toxins has been shown recently to efficiently and specifically cleave initiator tRNA at a single, identical position between the anticodon stem and loop (35). Using comparable experimental conditions, we did not detect any cleavage of E. coli or M. tuberculosis tRNAfMet by VapC-mt4 (supplemental Fig. S7, lanes 2–4). Therefore, unlike the VapC family members in Shigella and Salmonella, VapC-mt4 does not inhibit protein synthesis by cleaving initiator tRNA.
Mutation of Conserved PIN Domain Residues Abolishes VapC-mt4 Toxicity
Sequence and structural similarity suggests that a conserved quartet of acidic residues and a fifth invariant serine or threonine residue compose the active site of PIN domain proteins (supplemental Fig. S8A). To determine whether these residues are required for VapC-mt4 toxicity, we created D9A, E40A, D98A, T114A, and D116A mutations in VapC-mt4 and examined the effect of mutating each of the residues individually. Unlike wild-type VapC-mt4, expression of the mutants did not inhibit growth on plates (supplemental Fig. S8B). Despite the loss of toxicity, the VapC-mt4 mutants retained the ability to form a protein complex with the VapB-mt4 antitoxin, suggesting that the structure of the protein was retained (supplemental Fig. S8C). Together, these results indicate a role for these residues in VapC-mt4 toxicity.
VapC-mt4 Expression Leads to Gradual Decrease in Steady-state Levels of Several mRNAs
A common means by which TA toxins inhibit protein synthesis is by degrading a population of cellular mRNA using their sequence-specific endoribonuclease activity. The presence of a PIN domain, a motif associated with ribonuclease activity, suggested that the VapC-mt4 toxin might also perturb translation by a similar mechanism. We analyzed the effect of VapC-mt4 expression on the stability of several transcripts in vivo. Northern analysis for three E. coli mRNAs, tufA, ompA, and lpp, revealed that VapC-mt4 expression resulted in a gradual decrease in the steady-state levels of each transcript (Fig. 2C). Degradation products were detected below the full-length tufA mRNA, and a marked decrease in the level of the lpp transcript was detected as early as 20 min after VapC-mt4 induction. Although these results are consistent with the function of VapC-mt4 as a ribonuclease, degradation of the three transcripts was not complete even 100 min after VapC-mt4 induction (Fig. 2C).
VapC-mt4 Recognizes mRNA at ACGC and ACAGC Sequences in Vivo
Because VapC-mt4 exhibited ribonuclease activity in vivo, we sought to determine the RNA sequence recognized by VapC-mt4. We isolated total RNA from E. coli BW25113Δ6 cells expressing VapC-mt4 and performed primer extension analysis on the three mRNAs (tufA, ompA, and lpp) subjected to Northern analysis (Fig. 2). We initially used a primer ∼150 nucleotides downstream of the 5′-end of the translation start site for each mRNA and then performed additional primer extension experiments to enable nearly complete coverage of the 1225-nt ompA and 318-nt lpp mRNAs. In total, we detected four VapC-mt4-specific cleavage sites in the mRNAs, revealing a preference for mRNA cleavage at ACGC and ACAGC sequences (Fig. 3E).
VapC-mt4 Binds and Cleaves MS2 RNA in Vitro
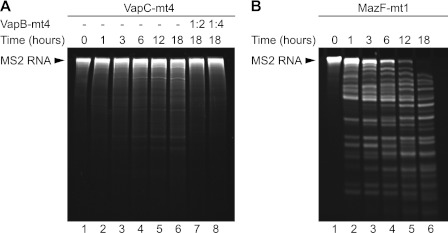
To ensure that the effect on the cellular mRNAs was directly due to VapC-mt4, we tested the RNA binding and cleavage activity of purified recombinant VapC-mt4 using bacteriophage MS2 RNA as the substrate. First, VapC-mt4 was added to a fixed amount of MS2 RNA, and the reaction was monitored over time (Fig. 4A). For comparison, we performed a parallel experiment using MazF-mt1 (Rv2801c), a MazF ortholog from M. tuberculosis that cleaves single-stranded RNA at UAC sequences (Fig. 4B) (14). Although the addition of purified VapC-mt4 resulted in the gradual cleavage of the MS2 RNA over time, there was a marked difference in the ribonuclease activity of VapC-mt4 and MazF-mt1. MazF-mt1 cut the MS2 RNA faster and more extensively than VapC-mt4. This inefficient cleavage of MS2 RNA by VapC-mt4 in vitro was consistent with the slow and incomplete cleavage of cellular transcripts in vivo by VapC-mt4. We also demonstrated that cleavage of the MS2 RNA was specifically due to VapC-mt4 and not a contaminating ribonuclease because preincubation of VapC-mt4 with the purified VapB-mt4 antitoxin prior to the addition of the MS2 RNA prevented RNA cleavage (Fig. 4A, lanes 7 and 8).
FIGURE 4.
VapC-mt4-mediated MS2 RNA cleavage. Equal molar amounts of His6-VapC-mt4 (A) or His6-MazF-mt1 (B) were added to MS2 RNA, and RNA cleavage was monitored over time. Preincubation of His6-VapC-mt4 with His6-VapB-mt4 before the addition of the MS2 RNA prevented RNA cleavage. Data shown are representative of three independent experiments.
VapC-mt4 Specifically Recognizes MS2 RNA at ACGC and AC(A/U)GC Sequences in Vitro
Because of the marked difference in the cleavage activity of VapC-mt4 and MazF-mt1, we wanted to determine whether additional sequences were required for VapC-mt4 RNA binding and cleavage. Toward this end, we performed in vitro primer extension analysis using primers that allowed nearly complete coverage of the 3569-nt MS2 RNA. In doing so, we identified 12 RNA recognition sites in the MS2 RNA that were cleaved by VapC-mt4 (Fig. 5). Importantly, all 12 cleavage products were generated by VapC-mt4 as cleavage was inhibited in the presence of the cognate VapB-mt4 antitoxin.
The combined results of the in vivo (Fig. 3) and in vitro (Fig. 5) experiments revealed that VapC-mt4 specifically recognized RNA at sites containing the ACGC or AC(A/U)GC consensus sequence (Figs. 3E and 5L). Further analysis of the in vitro primer extension results illuminated two notable trends. First, consistent with the mRNA cleavage activity we documented in vivo, all VapC-mt4 cut sites in MS2 mapped to single-stranded regions based on modeling of the MS2 RNA using RNAfold (data not shown). Second, there was strict conservation of the GC sequence at the 3′-end of the VapC-mt4 recognition sequences. In fact, substitution of either the G or C within an internal ACGC cleavage site in a 20-nt RNA substrate precluded VapC-mt4 cleavage (supplemental Fig. S9).
mRNA Cleavage Is Not Required for VapC-mt4-mediated Translation Inhibition
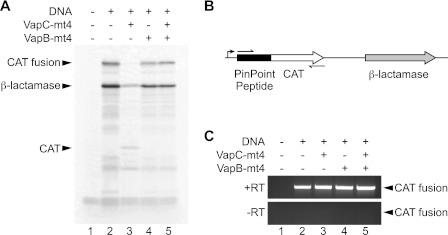
Although VapC-mt4 possessed sequence-specific ribonuclease activity, there was a drastic difference between the cleavage activity of MazF-mt1 and VapC-mt4. These results in combination with the marked time lag from the start of translation inhibition (20 min) to the point where significant (≥50%) mRNA cleavage was observed (80–100 min) raised the possibility that mRNA cleavage was a secondary, downstream effect and was not directly responsible for the translational defect (Fig. 2, A and B). To reconcile these discrepancies, we further examined the effect of VapC-mt4 on protein synthesis in vitro using an S30 extract cell-free transcription/translation system. We tested the effect of adding purified recombinant VapC-mt4 alone or both VapB-mt4 and VapC-mt4 on the transcription/translation of the CAT gene fused to the PinPoint peptide and β-lactamase encoded by the PinPoint Xa vector (Fig. 6A). Purified VapC-mt4 inhibited the synthesis of the PinPoint-CAT fusion protein, and this was reversed by preincubating VapC-mt4 with the VapB-mt4 antitoxin. Surprisingly, the addition of purified VapC-mt4 affected the expression of β-lactamase to a lesser extent and resulted in the appearance of a band corresponding to the full-length CAT protein lacking the N-terminal PinPoint peptide.
FIGURE 6.
mRNA cleavage is not required for VapC-mt4-mediated translation inhibition. A, purified His6-VapC-mt4 inhibits coupled in vitro transcription/translation of the PinPoint-CAT fusion protein (lane 3). His6-VapB-mt4 can rescue the inhibition and reconstitute translation (lane 5). B, schematic representation of the open reading frames encoded in the PinPoint Xa Control vector. The black rectangle denotes the PinPoint peptide. The white and gray arrows correspond to the ORFs encoding the 27.5-kDa CAT protein and the 31.5-kDa β-lactamase, respectively. The 39.5-kDa PinPoint-CAT fusion protein and β-lactamase are the major proteins expressed using the PinPoint Xa Control vector. The bent arrow shows the location of the T7 promoter. Thin arrows correspond to primers used for RT-PCR. C, the RNA was extracted from the coupled in vitro transcription/translation system, and RT-PCR was performed to assess the state of the mRNA encoding the PinPoint-CAT fusion protein. The mRNA encoding the PinPoint-CAT fusion protein was not cleaved by His6-VapC-mt4. Data shown are representative of two independent experiments.
Because VapC-mt4 effectively inhibited expression of the PinPoint-CAT fusion protein, we assessed the state of the corresponding transcript by reverse transcription-PCR. Although purified VapC-mt4 inhibited the synthesis of the PinPoint-CAT fusion protein, the PinPoint-CAT fusion transcript could still be detected in the sample in which translation was inhibited by VapC-mt4 (Fig. 6C, lane 3). Therefore, cleavage of the PinPoint-CAT fusion transcript was not coincident with translation inhibition, suggesting that mRNA cleavage is not required for protein synthesis inhibition by VapC-mt4.
In support of a model in which VapC-mt4 toxicity is not mediated by RNA cleavage, we also observed that total RNA levels increased with VapC-mt4 expression over time (supplemental Figs. S5B and S10A). In contrast, a parallel experiment using the UAC-specific endoribonuclease MazF-mt1 revealed a decrease in total RNA levels following induction (supplemental Fig. S10B). The MazF-mt1 data are also representative of the trend consistently observed upon induction of TA toxins that enlist RNA cleavage as their primary mode of translation inhibition (data not shown).5
VapC-mt4 Preferentially Binds ssRNA Containing ACGC Sequence
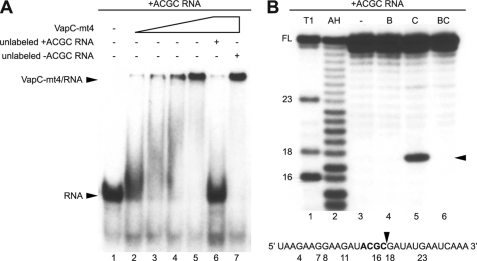
The inhibition of protein synthesis in the absence of mRNA cleavage raised the possibility that VapC-mt4 toxicity is primarily mediated by RNA binding. Subsequent cleavage of the mRNA may be a secondary effect of the prolonged association of VapC-mt4 with the mRNA. EMSAs using purified recombinant VapC-mt4 and a synthetic 30-nt RNA containing an ACGC sequence were performed to assess the RNA binding activity of VapC-mt4. RNA binding experiments were performed in the presence of EDTA, a chelating agent that sequesters Mg2+, to preclude RNA cleavage by VapC-mt4. VapC-mt4 was able to shift the mobility of the RNA containing the ACGC sequence (Fig. 7A, lanes 2–5). The specificity of VapC-mt4 for the RNA containing the ACGC sequence was assessed by competition with a 50-fold molar excess of unlabeled RNA. An unlabeled RNA containing the ACGC sequence (+ACGC) was sufficient to outcompete the labeled RNA containing the ACGC sequence, resulting in a loss of the VapC-mt4·RNA complex (Fig. 7A, lane 6). However, 50-fold excess of unlabeled RNA lacking the ACGC sequence (−ACGC) did not disrupt the VapC-mt4·RNA complex (Fig. 7A, lane 7). Although all visible RNA was present in a VapC-mt4·RNA complex (Fig. 7A, lane 5), less then 2% of the RNA was cleaved by VapC-mt4 in the presence of Mg2+ (Fig. 7B, lane 5) despite using identical amounts of RNA and protein. The inefficient cleavage of the 30-nt RNA by VapC-mt4 in vitro is consistent with the slow and incomplete cleavage of cellular transcripts in vivo by VapC-mt4. This marked difference between the RNA binding versus cleavage activities of VapC-mt4 indicated that a higher proportion of the RNA was stably bound by the toxin than cleaved.
FIGURE 7.
VapC-mt4 selectively binds and cleaves ssRNA containing an ACGC sequence. A, EMSAs were performed in the presence of EDTA, a chelating agent that sequesters Mg2+, to avoid RNA cleavage. His6-VapC-mt4 was able to bind and retard migration of a radioactively labeled synthetic 30-nucleotide RNA containing the ACGC consensus site (lanes 2–5). Sequence specificity was assessed by competition with a 50-fold excess of unlabeled RNA containing (lane 6) or lacking (lane 7) the ACGC consensus site. Data shown are representative of three independent experiments. B, RNA cleavage of the same radioactively labeled synthetic 30-nucleotide RNA containing the ACGC consensus site was performed in the presence of Mg2+, a cation required for VapC-mt4-mediated RNA cleavage. The lane labeled “−” represents a control reaction to which no proteins were added (lane 3). In the lane labeled “B,” purified His6-VapB-mt4 was incubated with the RNA (lane 4). In the lane labeled “C,” the addition of His6-VapC-mt4 resulted in the cleavage of ∼2% of the RNA (lane 5). In the lane labeled “BC,” preincubation of His6-VapC-mt4 with His6-VapB-mt4 prior to the addition of RNA prevented RNA cleavage (lane 6). The cleavage site/product is indicated by a black arrowhead on the right side of the gel and in the relevant RNA sequence below the gel. T1 and AH denote RNA digested with RNase T1 and partially hydrolyzed with alkali, respectively. Numbers indicate the position of G nucleotides. FL indicates the position of full-length RNA. Data shown are representative of two independent experiments.
DISCUSSION
Our initial experiments showing that VapBC-mt4 overexpression caused growth arrest associated with decreased translation in vivo led us to pursue a mechanism of action comparable with previously reported TA toxins, mRNA cleavage. We were able to detect weak ribonuclease activity associated with VapC-mt4 in vivo and identified the consensus recognition sequence, ACGC or AC(A/U)GC, in the RNA products that were cleaved by VapC-mt4. This weak ribonuclease activity was confirmed by in vitro studies comparing the kinetics of RNA cleavage of VapC-mt4 with that of MazF-mt1 (Fig. 4).
An unexpected finding of our work, however, was the large time lag between the point of translation inhibition (20 min postinduction; Fig. 2, A and B) versus when a significant (≥50%) decrease in the steady-state levels of the mRNA transcripts was observed (80–100 min postinduction; Fig. 2C). These findings suggested that mRNA cleavage might not be the primary mechanism of translation inhibition and growth arrest. In this context, we discovered that translation inhibition by VapC-mt4 in a coupled transcription/translation assay was not the consequence of cleaving the RNA transcript (Fig. 6). Using a synthetic 30-nt RNA containing an internal ACGC sequence, we found that although the RNA was readily bound by VapC-mt4 only a small percentage (∼2%) of the RNA was cleaved (Fig. 7). These results support a model of VapC-mt4-mediated translation inhibition that has two novel features: RNA binding at ACGC or AC(A/U)GC sequences and inhibition of translation as the result of RNA binding rather than RNA cleavage.
This model of translational regulation is distinct from the sequence-specific endoribonuclease activity of the extensively characterized MazF family of toxins. In M. tuberculosis, there are at least seven MazF-mt toxins, three of which have been characterized in detail. MazF-mt1 cuts mRNA at U↓AC sequences (14), MazF-mt3 at UU↓CCU and CU↓CCU, and MazF-mt7 at U↓CGCU (48). In each case, the MazF-mt toxins recognize and cleave at a unique recognition sequence. Similarly, if each of the M. tuberculosis VapC toxins possesses a unique RNA recognition profile for RNA binding, the individual VapBC TA systems could regulate translation by selectively targeting distinct subsets of transcripts to yield different physiological consequences.
Although our data clearly indicate that mRNA cleavage is not the primary mechanism by which VapC-mt4 causes translation inhibition and growth arrest, the extent to which the RNA binding model is applicable to this family of TA modules is not known. The recently characterized examples in Shigella and Salmonella exhibit tRNAfMet cleavage activity. However, we demonstrated that VapC-mt4 does not cleave tRNAfMet.
All VapC toxin family members contain a PIN domain. PIN domain-containing proteins were originally suggested to possess nuclease activity based on the properties and structural features of PIN domain proteins in eubacteria, archaebacteria, and eukaryotes. The first PIN domain structure (for the protein PAE2754 from the archaebacterium P. aerophilum) exhibited distant structural homology to several Mg2+-dependent nucleases (30). The four conserved acidic residues in PAE2754 are clustered in a surface pocket facing into a tunnel with a diameter only wide enough to accommodate single-stranded nucleic acids. Additional PIN domain protein structures reported since then have revealed that all PIN domains are structurally similar and thus have some homology to nucleases (34, 36, 49–52). Consistent with our model for VapC-mt4, the x-ray crystal structure of the VapC ortholog, FitB, has a high degree of structural homology to PAE2754 but no detectable nuclease activity (34). Our data suggest that some PIN domain proteins, including members of the VapC-mt family, may function as RNA-binding proteins rather than high efficiency single-stranded RNA endoribonucleases.
Although our data strongly support a novel mechanism of VapC-mt4 translation arrest by RNA binding, a number of questions remain. One is the extent of RNAs targeted by this toxin in vivo. We have demonstrated that VapC-mt4 is able to specifically bind to RNA at sites containing the consensus sequence. It is possible that VapC-mt4 may also interact with other cellular RNAs with the same features to inhibit translation. Although we did not detect any significant loss of cellular tRNAs containing VapC-mt4 recognition sequences by reverse transcription-PCR (data not shown), we cannot exclude the possibility that VapC-mt4 and other VapC-mt toxins act by targeting tRNAs other than tRNAfMet (or rRNA) as well as mRNA. A second question is the mechanism by which binding inhibits translation. Although our Western blot experiments did not identify VapC-mt4 in stabilized polysomes, ribosome stalling resulting from VapC-mt4 binding remains a leading possible mechanism. Answers to these questions will require further investigation of VapC-mt4 and other members of this large TA family.
Supplementary Material
Acknowledgments
We thank Sangita Phadtare for help with the electrophoretic mobility shift assays; Qian Tan and Yoshi Yamaguchi for the construction and use of the BW25113Δ6 strain; Yoshi Yamaguchi for optimizing the protocol used to purify VapB-mt4, VapC-mt4, and MazF-mt1; Ling Zhu for providing the pET28a-His6-MazF-mt1 plasmid (14), Christine Dunham for providing advice and tRNAfMet purified from E. coli cells (ChemBlock); and Krishnamurthy Shankarling for comments on the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant AI072399 from the NIAID R21 (to R. N. H. and N. A. W.) and T32 Training Grant AI07403, Virus-Host Interactions in Eukaryotic Cells, from the NIAID (to J. D. S. and J. W. C. (awarded to G. Brewer)).

This article contains supplemental Figs. S1–S10.
N. A. Woychik and M. Inouye laboratories, unpublished data.
- TA
- toxin-antitoxin
- CAT
- chloramphenicol acetyltransferase
- vap
- virulence associated protein
- PIN
- PilT (pili protein) N-terminal
- fit
- fast intracellular trafficking
- Ni-NTA
- nickel-nitrilotriacetic acid
- nt
- nucleotide.
REFERENCES
- 1. Mwinga A., Bernard Fourie P. (2004) Prospects for new tuberculosis treatment in Africa. Trop. Med. Int. Health 9, 827–832 [DOI] [PubMed] [Google Scholar]
- 2. Russo E. (2005) Scientist 19, 22–23 [Google Scholar]
- 3. Gerdes K., Christensen S. K., Løbner-Olesen A. (2005) Prokaryotic toxin-antitoxin stress response loci. Nat. Rev. Microbiol. 3, 371–382 [DOI] [PubMed] [Google Scholar]
- 4. Pandey D. P., Gerdes K. (2005) Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 33, 966–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pedersen K., Christensen S. K., Gerdes K. (2002) Rapid induction and reversal of a bacteriostatic condition by controlled expression of toxins and antitoxins. Mol. Microbiol. 45, 501–510 [DOI] [PubMed] [Google Scholar]
- 6. Suzuki M., Zhang J., Liu M., Woychik N. A., Inouye M. (2005) Single protein production in living cells facilitated by an mRNA interferase. Mol. Cell 18, 253–261 [DOI] [PubMed] [Google Scholar]
- 7. Ramage H. R., Connolly L. E., Cox J. S. (2009) Comprehensive functional analysis of Mycobacterium tuberculosis toxin-antitoxin systems: implications for pathogenesis, stress responses, and evolution. PLoS Genet. 5, e1000767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Christensen-Dalsgaard M., Jørgensen M. G., Gerdes K. (2010) Three new RelE-homologous mRNA interferases of Escherichia coli differentially induced by environmental stresses. Mol. Microbiol. 75, 333–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hazan R., Sat B., Engelberg-Kulka H. (2004) Escherichia coli mazEF-mediated cell death is triggered by various stressful conditions. J. Bacteriol. 186, 3663–3669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Prysak M. H., Mozdzierz C. J., Cook A. M., Zhu L., Zhang Y., Inouye M., Woychik N. A. (2009) Bacterial toxin YafQ is an endoribonuclease that associates with the ribosome and blocks translation elongation through sequence-specific and frame-dependent mRNA cleavage. Mol. Microbiol. 71, 1071–1087 [DOI] [PubMed] [Google Scholar]
- 11. Singletary L. A., Gibson J. L., Tanner E. J., McKenzie G. J., Lee P. L., Gonzalez C., Rosenberg S. M. (2009) An SOS-regulated type 2 toxin-antitoxin system. J. Bacteriol. 191, 7456–7465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Barry C. E., 3rd, Boshoff H. I., Dartois V., Dick T., Ehrt S., Flynn J., Schnappinger D., Wilkinson R. J., Young D. (2009) The spectrum of latent tuberculosis: rethinking the biology and intervention strategies. Nat. Rev. Microbiol. 7, 845–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Singh R., Barry C. E., 3rd, Boshoff H. I. (2010) The three RelE homologs of Mycobacterium tuberculosis have individual, drug-specific effects on bacterial antibiotic tolerance. J. Bacteriol. 192, 1279–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhu L., Zhang Y., Teh J. S., Zhang J., Connell N., Rubin H., Inouye M. (2006) Characterization of mRNA interferases from Mycobacterium tuberculosis. J. Biol. Chem. 281, 18638–18643 [DOI] [PubMed] [Google Scholar]
- 15. Lewis K. (2005) Persister cells and the riddle of biofilm survival. Biochemistry 70, 267–274 [DOI] [PubMed] [Google Scholar]
- 16. Keren I., Shah D., Spoering A., Kaldalu N., Lewis K. (2004) Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli. J. Bacteriol. 186, 8172–8180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Korch S. B., Henderson T. A., Hill T. M. (2003) Characterization of the hipA7 allele of Escherichia coli and evidence that high persistence is governed by (p)ppGpp synthesis. Mol. Microbiol. 50, 1199–1213 [DOI] [PubMed] [Google Scholar]
- 18. Korch S. B., Hill T. M. (2006) Ectopic overexpression of wild-type and mutant hipA genes in Escherichia coli: effects on macromolecular synthesis and persister formation. J. Bacteriol. 188, 3826–3836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Christensen S. K., Pedersen K., Hansen F. G., Gerdes K. (2003) Toxin-antitoxin loci as stress-response-elements: ChpAK/MazF and ChpBK cleave translated RNAs and are counteracted by tmRNA. J. Mol. Biol. 332, 809–819 [DOI] [PubMed] [Google Scholar]
- 20. Muñoz-Gómez A. J., Santos-Sierra S., Berzal-Herranz A., Lemonnier M., Díaz-Orejas R. (2004) Insights into the specificity of RNA cleavage by the Escherichia coli MazF toxin. FEBS Lett. 567, 316–320 [DOI] [PubMed] [Google Scholar]
- 21. Zhang Y., Zhang J., Hoeflich K. P., Ikura M., Qing G., Inouye M. (2003) MazF cleaves cellular mRNAs specifically at ACA to block protein synthesis in Escherichia coli. Mol. Cell 12, 913–923 [DOI] [PubMed] [Google Scholar]
- 22. Christensen S. K., Gerdes K. (2003) RelE toxins from bacteria and Archaea cleave mRNAs on translating ribosomes, which are rescued by tmRNA. Mol. Microbiol. 48, 1389–1400 [DOI] [PubMed] [Google Scholar]
- 23. Christensen S. K., Mikkelsen M., Pedersen K., Gerdes K. (2001) RelE, a global inhibitor of translation, is activated during nutritional stress. Proc. Natl. Acad. Sci. U.S.A. 98, 14328–14333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pedersen K., Zavialov A. V., Pavlov M. Y., Elf J., Gerdes K., Ehrenberg M. (2003) The bacterial toxin RelE displays codon-specific cleavage of mRNAs in the ribosomal A site. Cell 112, 131–140 [DOI] [PubMed] [Google Scholar]
- 25. Neubauer C., Gao Y. G., Andersen K. R., Dunham C. M., Kelley A. C., Hentschel J., Gerdes K., Ramakrishnan V., Brodersen D. E. (2009) The structural basis for mRNA recognition and cleavage by the ribosome-dependent endonuclease RelE. Cell 139, 1084–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Budde P. P., Davis B. M., Yuan J., Waldor M. K. (2007) Characterization of a higBA toxin-antitoxin locus in Vibrio cholerae. J. Bacteriol. 189, 491–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hurley J. M., Woychik N. A. (2009) Bacterial toxin HigB associates with ribosomes and mediates translation-dependent mRNA cleavage at A-rich sites. J. Biol. Chem. 284, 18605–18613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jiang Y., Pogliano J., Helinski D. R., Konieczny I. (2002) ParE toxin encoded by the broad-host-range plasmid RK2 is an inhibitor of Escherichia coli gyrase. Mol. Microbiol. 44, 971–979 [DOI] [PubMed] [Google Scholar]
- 29. Clissold P. M., Ponting C. P. (2000) PIN domains in nonsense-mediated mRNA decay and RNAi. Curr. Biol. 10, R888–R890 [DOI] [PubMed] [Google Scholar]
- 30. Arcus V. L., Bäckbro K., Roos A., Daniel E. L., Baker E. N. (2004) Distant structural homology leads to the functional characterization of an archaeal PIN domain as an exonuclease. J. Biol. Chem. 279, 16471–16478 [DOI] [PubMed] [Google Scholar]
- 31. Arcus V. L., Rainey P. B., Turner S. J. (2005) The PIN-domain toxin-antitoxin array in mycobacteria. Trends Microbiol. 13, 360–365 [DOI] [PubMed] [Google Scholar]
- 32. Daines D. A., Wu M. H., Yuan S. Y. (2007) VapC-1 of nontypeable Haemophilus influenzae is a ribonuclease. J. Bacteriol. 189, 5041–5048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Miallau L., Faller M., Chiang J., Arbing M., Guo F., Cascio D., Eisenberg D. (2009) Structure and proposed activity of a member of the VapBC family of toxin-antitoxin systems. VapBC-5 from Mycobacterium tuberculosis. J. Biol. Chem. 284, 276–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mattison K., Wilbur J. S., So M., Brennan R. G. (2006) Structure of FitAB from Neisseria gonorrhoeae bound to DNA reveals a tetramer of toxin-antitoxin heterodimers containing pin domains and ribbon-helix-helix motifs. J. Biol. Chem. 281, 37942–37951 [DOI] [PubMed] [Google Scholar]
- 35. Winther K. S., Gerdes K. (2009) Ectopic production of VapCs from enterobacteria inhibits translation and trans-activates YoeB mRNA interferase. Mol. Microbiol. 72, 918–930 [DOI] [PubMed] [Google Scholar]
- 36. Takeshita D., Zenno S., Lee W. C., Saigo K., Tanokura M. (2006) Crystallization and preliminary x-ray analysis of the PIN domain of human EST1A. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 62, 656–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Datsenko K. A., Wanner B. L. (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nakano E. T., Rao M. M., Perucho M., Inouye M. (1987) Expression of the Kirsten ras viral and human proteins in Escherichia coli. J. Virol. 61, 302–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Masui Y., Mizuno T., Inouye M. (1984) Novel high-level expression cloning vehicles: 104-fold amplification of Escherichia coli minor protein. Nat. Biotechnol. 2, 81–85 [Google Scholar]
- 40. Inouye S., Inouye M. (1985) Up-promoter mutations in the lpp gene of Escherichia coli. Nucleic Acids Res. 13, 3101–3110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Guzman L. M., Belin D., Carson M. J., Beckwith J. (1995) Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177, 4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ehrt S., Guo X. V., Hickey C. M., Ryou M., Monteleone M., Riley L. W., Schnappinger D. (2005) Controlling gene expression in mycobacteria with anhydrotetracycline and Tet repressor. Nucleic Acids Res. 33, e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Powers T., Noller H. F. (1990) Dominant lethal mutations in a conserved loop in 16S rRNA. Proc. Natl. Acad. Sci. U.S.A. 87, 1042–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tai P. C., Davis B. D. (1979) Isolation of polysomes free of initiation factors. Methods Enzymol. 59, 362–371 [DOI] [PubMed] [Google Scholar]
- 45. Ron E. Z., Kohler R. E., Davis B. D. (1966) Polysomes extracted from Escherichia coli by freeze-thaw-lysozyme lysis. Science 153, 1119–1120 [DOI] [PubMed] [Google Scholar]
- 46. Sisido M., Ninomiya K., Ohtsuki T., Hohsaka T. (2005) Four-base codon/anticodon strategy and non-enzymatic aminoacylation for protein engineering with non-natural amino acids. Methods 36, 270–278 [DOI] [PubMed] [Google Scholar]
- 47. Winther K. S., Gerdes K. (2011) Enteric virulence associated protein VapC inhibits translation by cleavage of initiator tRNA. Proc. Natl. Acad. Sci. U.S.A. 108, 7403–7407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhu L., Phadtare S., Nariya H., Ouyang M., Husson R. N., Inouye M. (2008) The mRNA interferases, MazF-mt3 and MazF-mt7 from Mycobacterium tuberculosis target unique pentad sequences in single-stranded RNA. Mol. Microbiol. 69, 559–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bunker R. D., McKenzie J. L., Baker E. N., Arcus V. L. (2008) Crystal structure of PAE0151 from Pyrobaculum aerophilum, a PIN-domain (VapC) protein from a toxin-antitoxin operon. Proteins 72, 510–518 [DOI] [PubMed] [Google Scholar]
- 50. Glavan F., Behm-Ansmant I., Izaurralde E., Conti E. (2006) Structures of the PIN domains of SMG6 and SMG5 reveal a nuclease within the mRNA surveillance complex. EMBO J. 25, 5117–5125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Jeyakanthan J., Inagaki E., Kuroishi C., Tahirov T. H. (2005) Structure of PIN-domain protein PH0500 from Pyrococcus horikoshii. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 61, 463–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Levin I., Schwarzenbacher R., Page R., Abdubek P., Ambing E., Biorac T., Brinen L. S., Campbell J., Canaves J. M., Chiu H. J., Dai X., Deacon A. M., DiDonato M., Elsliger M. A., Floyd R., Godzik A., Grittini C., Grzechnik S. K., Hampton E., Jaroszewski L., Karlak C., Klock H. E., Koesema E., Kovarik J. S., Kreusch A., Kuhn P., Lesley S. A., McMullan D., McPhillips T. M., Miller M. D., Morse A., Moy K., Ouyang J., Quijano K., Reyes R., Rezezadeh F., Robb A., Sims E., Spraggon G., Stevens R. C., van den Bedem H., Velasquez J., Vincent J., von Delft F., Wang X., West B., Wolf G., Xu Q., Hodgson K. O., Wooley J., Wilson I. A. (2004) Crystal structure of a PIN domain (AF0591) from Archaeoglobus fulgidus at 1.90 Å resolution. Proteins 56, 404–408 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.